Abstract
Using a high-resolution optical trapping instrument, we directly observed the processive motions of individual Eg5 dimers over a range of external loads and ATP, ADP, and phosphate concentrations. To constrain possible models for dissociation from the microtubule, we measured Eg5 run lengths and also compared the duration of the last step of a processive run to all previous step durations. We found that the application of large longitudinal forces in either hindering or assisting directions could induce Eg5-microtubule dissociation. At a constant moderate force, maintained with a force clamp, the premature binding of ADP strongly promoted microtubule release by Eg5, whereas the addition of ATP or phosphate had little effect on dissociation. These results imply that run length is determined not only by the load, but also by the concentration and type of nucleotides present, and therefore that the biochemical cycles of the two motor domains of the Eg5 dimer are coordinated to promote processive stepping.
Introduction
Eg5 is a homotetrameric kinesin protein (kinesin-5 subclass) that is essential for the formation and maintenance of mitotic and meiotic spindles. Eg5 consists of two pairs of motor domains (heads) situated at the opposite ends of a common stalk. Each head pair can independently bind to a microtubule (MT), so that by moving toward the plus-end, a single Eg5 motor can slide apart two antiparallel MTs that it bridges, a function that is thought to contribute to microtubule flux and chromosome segregation in dividing cells (1,2).
In vitro biophysical assays based on recombinant Eg5 proteins have established many of the fundamental characteristics of Eg5. Optical trapping measurements have shown that human dimeric Eg5 constructs, consisting of a single pair of heads attached to a coiled-coil stalk, move processively, binding to an MT and taking multiple mechanical steps before dissociating (3). However, the run lengths are quite short (∼8 steps) compared to those of other processive cytoskeletal motors. Eg5 dimers can support significant hindering loads (up to ∼6 pN), suffering only a modest reduction in velocity even at the highest loads. There is no indication that dimeric motors stall before MT release. Tetrameric Eg5 motors from Xenopus are also processive, and like the dimers, full length tetramers neither slow significantly nor stall under near-maximum loads (2). However, tetramers were found to dissociate from the MT at much lower forces (∼2 pN), and this observation led to the suggestion that MT release is primarily load-induced. For full-length tetramers, the detailed dependence of velocity and run length upon load is not known.
Solution biochemical experiments have established the basic kinetic mechanism of Eg5 monomers and dimers, and revealed unique features in its kinetic cycle that differ from those of other members of the kinesin superfamily (4–6). Importantly, the initiation of a processive run in dimeric Eg5 is thought to occur differently from the steps that follow (6). Processive stepping evidently begins with both motor domains bound to the MT and free of nucleotide, and requires a slow conformational change before the dimer can enter the normal stepping cycle. Once normal stepping begins, alternating-site catalysis takes place, with the binding of ATP to the forward head triggering the advance of the trailing head (5,6). Although biochemical kinetic measurements are exquisitely sensitive to the first step after formation of the MT-motor “collision complex”, they are less sensitive to any subsequent steps, due to the growing asynchrony in the ensemble of proteins (7). For motors whose initial and subsequent steps are identical, this has not posed a practical challenge, but the unusual kinetic scheme of Eg5 dimers has prompted questions about the biochemical regulation of its motor domains within a processive run, as well as about the factors governing Eg5-MT dissociation.
Optical trapping measurements can supply detailed information about the processive steps within a run, including a direct determination of the cycle time for each mechanical step. In contrast to kinesin-1, the abbreviated run lengths of Eg5 dimers provide an opportunity to score nearly all MT-dissociation events, because these tend to occur within the active zone of the position detector used in optical trapping experiments (which typically subtends <1 μm2 in the specimen plane). Because the step rate of Eg5 is slow compared to that of other kinesin motors (∼12 s−1 at zero force and saturating ATP) (3,6), it becomes possible to discern the stepwise transitions of single Eg5 dimers over a wider range of biochemical and force conditions. Using a high-resolution optical trapping apparatus, we determined the factors governing MT release by Eg5 dimers by directly measuring the behavior of individual molecules as functions of external load, supplied by a force clamp, and the ATP, ADP, and inorganic phosphate (Pi) concentrations. Our data support a model where the premature binding of ADP compromises head-head coordination and thereby decreases processivity. At the highest forces, the probability of load-induced dissociation of both heads from the MT is also increased.
Materials and Methods
Assays
The details of the assay have been described (3). Briefly, biotinylated MTs were prepared by polymerizing a 1:10 mixture of biotinylated and unmodified bovine brain tubulin (Cytoskeleton, Denver, CO) at 37°C for 1 h. MTs were taxol-stabilized, then pelleted via centrifugation and resuspended in fresh buffer to remove free tubulin. To prepare beads for coupling to motor proteins, 0.44-μm-diameter streptavidin-coated polystyrene beads (Spherotech, Libertyville, IL) were incubated with biotin-conjugated anti-5X-His antibodies (Qiagen, Valencia, CA) for 1 h at room temperature. Unbound antibodies were removed by centrifugation and resuspension in fresh buffer. Antibody-coated beads were incubated with Eg5-513-5His (a generous gift of S.P. Gilbert, Rensselaer Polytechnic Institute, Troy, NY; called “Eg5” herein) in assay buffer (80 mM PIPES at pH 6.9, 1 mM EGTA, 4 mM MgCl2, 200 mM KCl, 50 mM K-Acetate, 2 mM DTT, 7 μM taxol, 200 nM ATP, and excess free biotin) at 4°C for 3–9 h. Eg5 concentrations were sufficiently dilute that less than one motor, on average, could bind to a bead and support motion along the MT (3). A glucose oxidase-based oxygen-scavenging system, along with the desired amount of ATP, ADP, and inorganic phosphate (Pi) (as KH2PO4), were added immediately before each measurement. During data collection, the lowest concentration of ATP used was 8 μM. For collection of ADP- and Pi-dependent data, at least 100 μM ATP was present in the sample buffer. Control measurements performed with an additional 400 mM KCl added to the assay buffer verified that velocity and run length did not vary appreciably with higher ionic strength (data not shown).
Flow cells were assembled using glass slides, double-stick tape, and plasma-cleaned coverglasses. To reduce the nonspecific binding of Eg5, the surfaces were incubated sequentially with biotinylated poly-L-lysine-graft-polyethlylene glycol copolymers (Surface SolutionS, Zurich, Switzerland), streptavidin (Prozyme, San Leandro, CA), and biotinylated MTs. Unbound reagents were washed away with buffer after each step. Immediately before measurement, the slide was incubated with assay buffer for several minutes and the Eg5-coated beads were introduced to the sample chamber. The slide was viewed immediately, and data were collected for up to 1 h.
Instrumentation and data collection
Details of the optical trapping instrument, calibration, and data collection have been described (3,8). Briefly, a computer-controlled, steerable optical trap was used to apply constant longitudinal hindering or assisting loads to individual Eg5-coated beads using a feedback-based force-clamp arrangement (9). Automated positioning and calibration procedures were performed on each bead before data collection to reduce measurement error. The photodetector response was calibrated by raster-scanning each bead over the detection region (10). Trap stiffness was determined by measuring both the mean-squared displacement and the corner frequency of the Lorentzian power spectrum for every bead (11). Bead motion was measured using light scattered from an independent, low-power single-mode diode laser onto a position sensitive detector (PSD); bead and trap positions were sampled at 2 kHz and Bessel-filtered at 1 kHz.
For measurements of unloaded bead motion, weakly trapped Eg5-coated beads were placed near an MT. Images of bead position were then recorded at video rates using Nomarski differential interference contrast microscopy and analyzed using a custom-written centroid-tracking algorithm. Once a bead had moved >∼35 nm from the center of the optical trap, the trapping beam was shuttered and the bead position recorded. Beads were automatically recaptured after detachment and returned to their starting positions to facilitate measurements of multiple runs.
Data analysis
The average velocity, v, was determined by dividing the total distance traveled (run length) by the total time for each run, and weighting this individual measurement by the run length. To determine the randomness for each run, the variance [x(t + Δt) − (x(t) + 〈v〉Δt)]2 was calculated as a function of lag time, Δt, where x is the bead position and the angle brackets indicate an average over all runs at that condition. A line fit was performed to the variance over the first 20-nm 〈v〉−1 time interval, excluding the first 3.5 ms to account for the viscous relaxation time of the bead (12). The randomness, r, was calculated by dividing the slope of the variance versus Δt by d〈v〉, where d is the molecular step size of 8.1 nm. Average randomness values were weighted by the run length. Errors were calculated by averaging the standard deviation weighted by the run length and dividing by the square root of the number of events (standard error).
Average run lengths were calculated from the arithmetic mean of the run-length distribution at each condition. For very small displacements, it is difficult to distinguish the directional motion of Eg5 from the free diffusion of a trapped particle, particularly at low trap stiffness. Therefore, we scored only those runs where the length exceeded a spatial cutoff, ξ, of 25 nm (∼3 steps). To account for missing events below ξ, we applied a correction factor that assumed an exponential distribution of run lengths: L = Lapp − ξ, where L is the corrected mean run length and Lapp is the mean for runs above ξ (3,13,14). Due to the modest run length of Eg5 dimers, this correction ranged from ∼18% of the uncorrected length for the longest runs to ∼50% for the shortest. At low forces (|F| < 2 pN), the thermally driven motion of the bead was significantly larger than the molecular step size, precluding the identification of individual steps during the observation periods. For larger forces, clear stepwise transitions could be observed. For those records that displayed a series of at least four molecular steps before dissociation, we identified the location of stepwise transitions and recorded the dwell times between each step. For each condition, we calculated the average duration of the last dwell before dissociation and the average dwell time of all other steps in a processive run using the arithmetic means of the respective distributions (see Fig. S1 in the Supporting Material). Standard errors were determined from the standard deviation divided by the square root of the number of events.
Results
Dissociation kinetics
To probe Eg5-MT dissociation, we measured several physical parameters that supply information about the underlying biochemical reactions: the average cycle time for all steps that led to another forward step, 〈τ〉, the average cycle time for all steps that terminated stepping by dissociation, 〈τlast〉, and the run length, L (Fig. 1). Because our optical trapping assay is sensitive to displacement, 〈τ〉 and 〈τlast〉 measure the timing of a mechanical cycle that both starts and ends with a force-dependent transition. If 〈τlast〉 is approximately equal to 〈τ〉, then the biochemical pathway leading to MT dissociation is overall similar to the pathway that leads to forward stepping. This implies that the branch point between these alternative outcomes occurs late in the pathway, for example, just before translocation. However, if 〈τlast〉/〈τ〉 << 1, then dissociation occurs well before completion of the stepping cycle, so the branch point likely occurs at some earlier transition. In this situation, L is directly related to the branching ratio between stepping and dissociation: for example, a motor with an average run length of 100 steps has an ∼1% likelihood of dissociation per step. By measuring how forces and nucleotides affect 〈τ〉, 〈τlast〉, and L, it is possible to identify those biochemical transitions where dissociation is most likely to take place.
Figure 1.
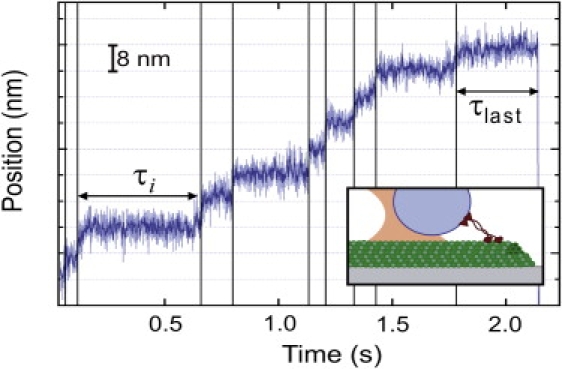
A representative data record showing the processive stepping of a single Eg5-513-His dimer versus time. The median-filtered bead position (25-point window, dark trace) is superimposed on the unfiltered position (light gray trace), and displays clear stepwise transitions (vertical lines) and the final dissociation event. Horizontal gridlines are spaced at 8 nm to indicate the approximate molecular step size. The dwell times for steps (except the last) in all records, τi, were recorded and averaged as 〈τ〉; the dwell times for the final steps were separately recorded and averaged as 〈τlast〉. (Inset) Cartoon representation of the experimental geometry (not to scale). An Eg5-513-His homodimer is attached to a bead held in an optical trap as it walks along the MT under force-clamped conditions.
To assess the role of ATP binding in dissociation, we determined L and the ratio 〈τlast〉/〈τ〉 as functions of ATP concentration (Fig. 2 A). We found that under hindering loads, L was independent of [ATP] and 〈τlast〉/〈τ〉 was comparable to unity for ATP concentrations ranging from 16 μM (equal to the apparent Michaelis constant for ATP binding, KM (3)) to saturation at 2 mM. By contrast, at F = 0, L decreased by nearly a factor of 3 at limiting ATP levels (<50 μM) compared to its value at saturating ATP levels. These data suggest that, just as for kinesin-1 (15), the ATP waiting state is prone to dissociation, and that processivity can be enhanced by the application of moderate external loads.
Figure 2.
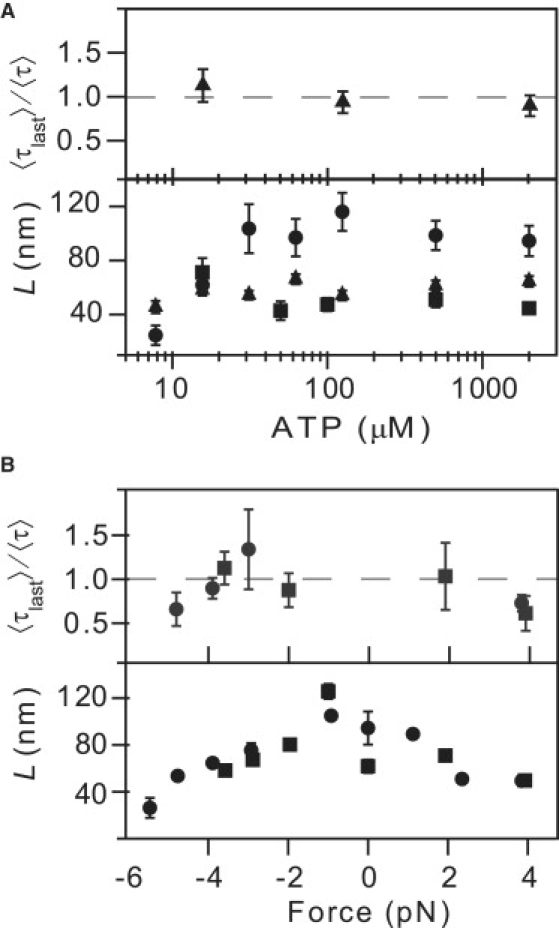
Eg5 processivity as a function of ATP concentration and force. (A) Run length, L (lower), as a function of [ATP] for F = 0 pN (circles; N = 31–71 runs at each condition,), F = −3.0 ± 0.1 pN (triangles; N = 20–207), and F = −3.8 ± 0.1 pN (squares; N = 44–110). Run length was nearly independent of [ATP] for all forces. The ratio of the average dwell time for the last and preceding steps (upper) remains near unity over three orders of magnitude in ATP concentration (F = −3.8 pN). (B) Run length L (lower) as a function of force for [ATP] = 2000 μM (circles; N = 11–277) and [ATP] = 16 μM (squares; N = 72–97). L decreases weakly with the magnitude of applied force, for both assisting and hindering loads. The ratio of the average dwell time for the last and preceding steps (upper) remains near unity for |F| < ∼4 pN, but decreases for forces beyond this value.
To determine the role of force, we measured L and 〈τlast〉/〈τ〉 as functions of both hindering (F < 0) and assisting (F > 0) longitudinal loads (Fig. 2 B). We found that the run length displayed a gentle dependence on force under both saturating ATP conditions and when [ATP] ≈ KM, with L decreasing from a peak value of ∼12 steps at zero load to ∼4 steps under a hindering load of 5 pN under saturating ATP conditions. We found that 〈τlast〉/〈τ〉 was constant and near unity for moderate forces, where individual step transitions can be observed, suggesting that the applied load did not destabilize the final step, and therefore that force-induced unbinding is not the primary mode of detachment. When loads beyond 4 pN were applied in either hindering or assisting directions, 〈τlast〉/〈τ〉 decreased slightly, indicating that for sufficiently large forces, load-induced unbinding occurred. For somewhat lower forces, around −3 pN, 〈τlast〉/〈τ〉 was slightly larger than unity. This may, in principle, indicate entry into an off-pathway state. However, the increase was smaller than what is typically described as a stall condition for kinesin-1 (15).
To determine the roles of nucleotide binding and release of hydrolysis products in dissociation, we measured L and 〈τlast〉/〈τ〉 as functions of [Pi] and [ADP]. The force was kept constant at −3 pN, a load regime where force-induced unbinding is unlikely. As a control, we confirmed that the ATP dependence of the velocity and randomness at F = −3 pN were consistent with previously published data (see Fig. S2). Like other kinesin-related motors, Eg5 releases Pi before ADP, so we assume that phosphate binds to the active site of Eg5·ADP to generate Eg5·ADP·Pi, a posthydrolysis state (16–18). We found that both 〈τlast〉/〈τ〉 and L were independent of phosphate concentration (Fig. 3 A), implying that phosphate binding to Eg5·ADP either did not occur, or did not promote the dissociation of Eg5 from the MT.
Figure 3.
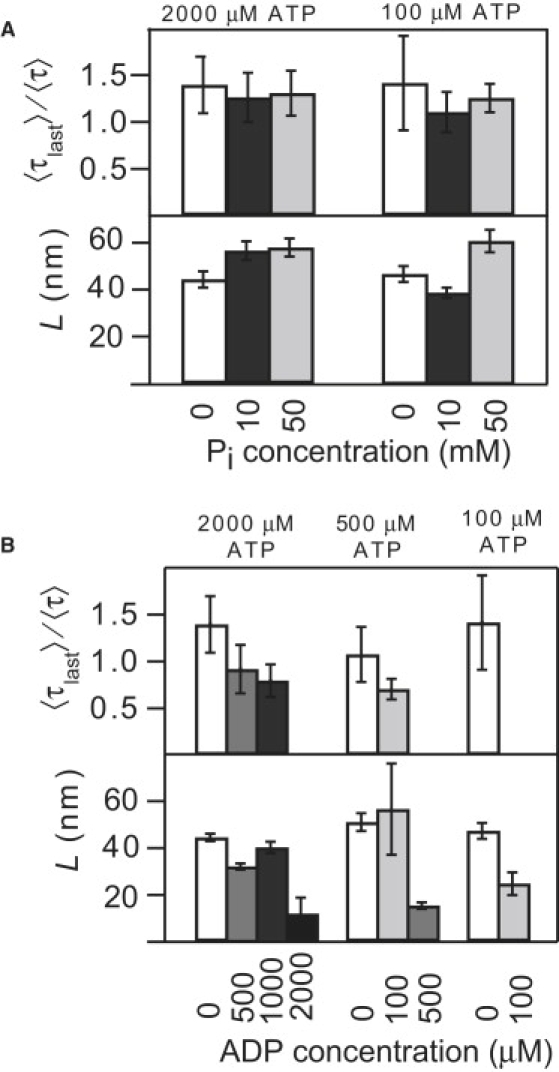
Eg5 processivity as a function of ADP and Pi concentrations. (A) Run length L (lower) and the dwell time ratio for the last and preceding steps (upper) does not show a clear trend with Pi concentration for [ATP] = 100 μM and [ATP] = 2000 μM (N = 96–212). (B) Run length L (lower) and the ratio of the average dwell time for the last and preceding steps (upper) both decrease with ADP concentration (N = 10–207). The processivity is also ATP-dependent: L decreased by ∼50% when [ADP] = [ATP].
By contrast, both 〈τlast〉/〈τ〉 and L decreased with increasing ADP concentration (Fig. 3 B). The magnitude of the decrease depended upon the ATP concentration, as expected, since ADP and ATP compete for the same nucleotide binding site. Under all ATP conditions, the run length dropped by nearly a factor of 2 when [ADP] ≈ [ATP]. At high [ADP]/[ATP] ratios, few long runs were observed, and it became difficult to collect sufficient data to determine 〈τlast〉 and 〈τ〉 accurately. Over the range of accessible ADP concentrations, 〈τlast〉/〈τ〉 decreased monotonically as [ADP] increased, consistent with a pathway in which ADP binding to Eg5 produces a dissociation-prone state.
Enzyme inhibition by phosphate and ADP
To probe the effect of phosphate on stepping, we measured Eg5 velocity as a function of ATP in the presence of either 0 or 10 mM Pi (Fig. 4). The data were consistent with a competitive inhibition mechanism obeying v = vmax[ATP]/([ATP] + KM(1 + [Pi]/KIPi)), where vmax is the maximum velocity and KIPi is a phosphate-dependent inhibition constant. From curve fits, we found that vmax = 68 ± 1 nm/s, KM = 14 ± 3 μM, and KIPi = 6 ± 2 mM (mean ± SE). The randomness, r, did not display strong dependence on [Pi], and r < 1 for all [Pi], suggesting that phosphate release is not rate-limiting.
Figure 4.
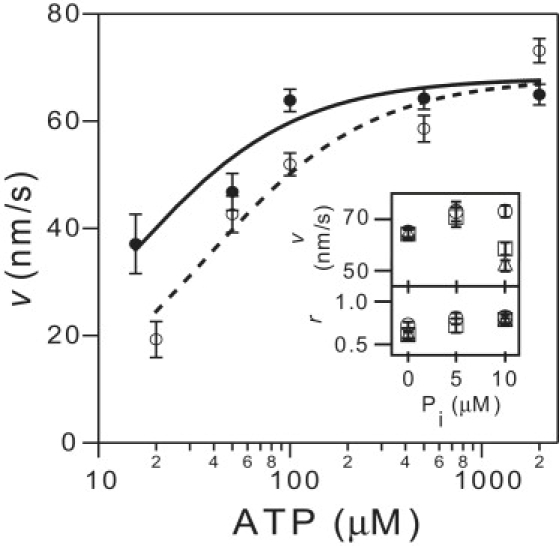
Eg5-513-His velocity, v, versus ATP concentration with (open circles; N = 20–209) and without 10 mM Pi (solid circles; N = 7–200) for F = −3.0 ± 0.1 pN. Solid and dashed lines are fits to a competitive inhibition model, with constant KI = 6 ± 2 mM. (Inset) Velocity and randomness for 2000 μM (circles; N = 60–212), 500 μM (squares; N = 44–115), and 100 μM (triangles; N = 91–150) versus phosphate concentration.
The high probability of dissociation from the MT at high [ADP]/[ATP] ratios made it difficult to collect data for precise velocity and randomness determinations over a wide range of buffer conditions. For low ADP concentrations, the addition of ADP led to a moderate decrease in velocity (Fig. 5). When the data were fit to the competitive inhibition model, we found KIADP to be ∼200 ± 100 μM (mean ± SE); r did not display strong dependence on [ADP], and r was always less than unity, suggesting that ADP release is never rate-limiting (see Fig. S3).
Figure 5.
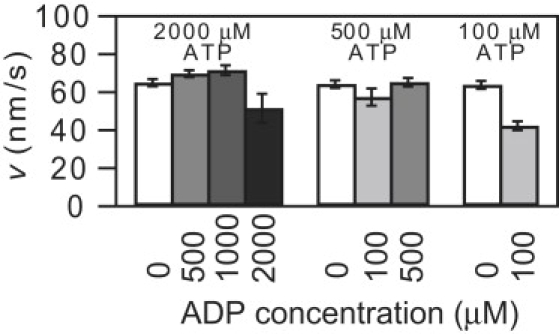
Eg5-513-His velocity, v, versus ADP concentration (0–2 mM; N = 10–207) measured at three different ATP concentrations at F = −3.0 ± 0.1 pN.
Discussion
We propose a model for processive stepping by Eg5 where a dissociation event, taken to be off-pathway, competes with on-pathway advancement (Fig. 6). Under moderate forces (|F| ≤ 4 pN), 〈τlast〉/〈τ〉 was approximately equal to unity under all ATP concentrations studied, and at F = 0, L decreased by a factor of ∼3 under limiting ATP concentrations compared to its value at saturating ATP. Taken together, these data suggest that at low loads and in the absence of additional ADP or Pi, MT dissociation occurs late in the mechanochemical stepping cycle, but likely before ATP binding (we favor state S3). The molecular configuration of Eg5 in the ATP waiting state is presently unknown, but a nucleotide-free, one-head-bound state would seem particularly vulnerable compared to two-heads-bound states, where any imposed load is shared between the heads, and an (unlikely) double-dissociation event would be required for release. Recent evidence suggests that the ATP waiting state for conventional kinesin (kinesin-1) is precisely such a one-head-bound state (19–22). At larger forces (|F| ≥ 4 pN), L and 〈τlast〉/〈τ〉 decreased. Interestingly, MT detachment was insensitive to the direction of applied load, consistent with a purely mechanical disruption of the MT-motor bond. Load-induced unbinding could therefore occur from any biochemical state; however, one-head-bound states (S3, or possibly S0 and S1) would again seem to be particularly susceptible.
Figure 6.
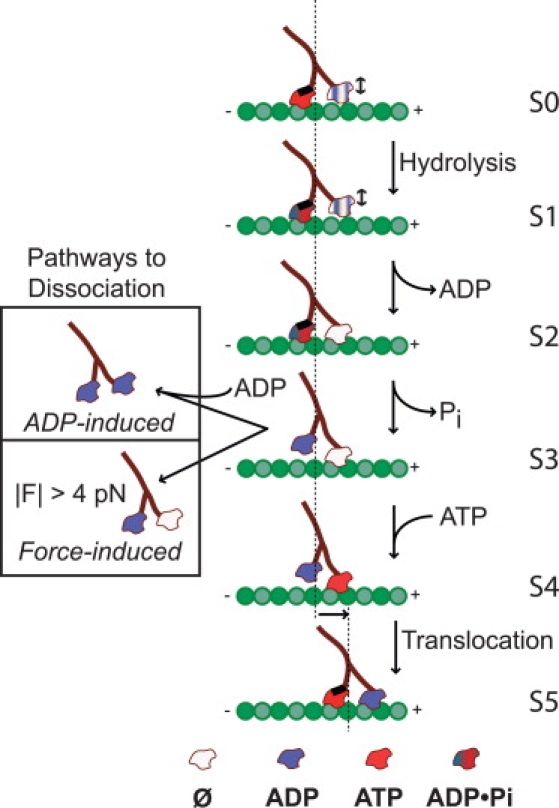
A five-state kinetic scheme (S0–S5) for Eg5 stepping during a processive run, in which Eg5-microtubule dissociation occurs from a one-head-bound state before ATP binding (S3). The model is consistent with single-molecule and solution biochemical data for human Eg5-513 dimers (6). State S0 occurs immediately after the rapid translocation step, the starting point for the cycle, which is easily identified in single-molecule records. The trailing head binds ATP and the advancing head binds ADP, until it forms a tight connection to the forward binding site on the MT. In states S0 and S1, there is a rapid equilibrium at the leading head between the ADP-bound and no-nucleotide states, indicated by the blue-and-white coloring; arrows indicate the resultant transition between weak and strong MT binding. The neck linker of the trailing head is docked (heavy black line). Hydrolysis by the trailing head in S1 is followed by complete ADP release from the leading head in S2. Then, phosphate is released from the trailing head to form the ATP waiting state (S3), in which one head is bound to the MT while its partner is free. Either the premature binding of ADP to the MT-bound head or the application of large hindering or assisting loads can promote MT dissociation from this state. In this model, dissociation competes with ATP binding to the MT-bound head (S4). Successful ATP binding leads to a subsequent forward translocation, restarting the mechanochemical cycle. The new starting state (S5) is biochemically and structurally identical to the original (S0), but the centroid of the motor has now advanced by ∼8 nm along the MT lattice.
To further study the mechanochemistry of Eg5-MT dissociation, we measured the effect of Pi and ADP concentrations on MT detachment. Increasing the phosphate concentration had no effect on L or 〈τlast〉/〈τ〉, suggesting that Pi release occurs before the branch point to the dissociation pathway. The ADP·Pi-dependent rates were all relatively fast, and did not contribute significantly to the overall dwell time between steps, as indicated by a millimolar-range inhibition constant and the insensitivity of the randomness parameter to [Pi]. Our data would predict that under physiological phosphate conditions (approximately the millimolar range), the overall velocity of Eg5 would slow slightly, due to inhibition of ATP binding, but there would be a negligible effect on dissociation. Although a high-resolution structure of the posthydrolysis Eg5·ADP·Pi state is not yet available, there is a significant amount of evidence to suggest that the ADP·Pi state is strongly bound to the MT in the case of kinesin-1 (19,23,24). Eg5 exhibits binding trends that are entirely consistent with those of kinesin-1 in the presence of ADP, no nucleotide, and AMP-PNP (a nonhydrolyzable ATP analog) (25). We therefore assume here that the ADP·Pi state is strongly bound for Eg5 as well.
The addition of ADP decreased both L and 〈τlast〉/〈τ〉, indicating that an ADP-dependent process induces MT dissociation, in agreement with fluorescence data showing that ADP-bound Eg5 monomers associate only weakly with MTs (26). Phosphate release is rapid for Eg5 (estimated to be >100 s−1 (6)), and therefore, state S2 is short-lived. Our data may be explained either by the failure of ADP to release from the leading head before phosphate release from the trailing head in state S2 (resulting in both heads carrying ADP and detaching from the MT) or by the rebinding of ADP to the empty bound head in state S3 (or both).
Precise coordination of the mechanochemical cycles of each head of a processive dimer, such as kinesin-1 or myosin-V, ensures that at least one head remains tightly bound to its filament at all times and can thereby traverse a substantial distance while sustaining load. In kinesin-1, ADP release by the leading head rapidly follows ATP binding by the trailing head, leading to a two-heads-bound state. Intramolecular strain in the neck linker region, a short polypeptide chain that connects the head to the stalk, is thought to block premature ATP binding to the leading head until hydrolysis and phosphate release (which is rate-limiting) occur at the trailing head (14). This coordination is essential to processive forward stepping, since it prevents the desynchronization of the biochemical cycles of the two motor domains.
The very limited processivity of Eg5 dimers raises questions about the extent to which its two heads coordinate. Kinetic data suggest that Eg5 is much more susceptible to entry into a detached state than kinesin-1: ATP binding at the rear head only partially stimulates ADP release at the forward head, ATP hydrolysis is rate-limiting, and phosphate release is very rapid (6). Despite these kinetic differences, our data indicate that head-head coordination is nevertheless present in Eg5 dimers, and that this coordination can be compromised by changing the type and concentration of nucleotides present in solution. Moreover, we found that at limiting ATP concentrations, the application of moderate hindering loads rescues processivity, and that run length becomes insensitive to [ATP] for F < 0. We postulate that in this regime, the application of load increases intramolecular strain and enhances head-head coordination within the dimer.
There are clear differences in the stepping behaviors of human-derived Eg5 dimers and Xenopus-derived Eg5 tetramers, and important questions remain regarding the role of oligomerization in determining the nanoscale mechanical properties of Eg5 (27,28). It is striking that the maximum force that tetramers appear to support is <30% of that of the dimeric constructs (2). Further experiments will be required to determine whether these differences arise solely from variations between the two species, or whether tetramers are somehow regulated to dissociate at lower forces than isolated dimers withstand, as well as to define better the role of processivity in the activity of Eg5 in vivo.
Summary
This study demonstrates the utility of single-molecule optical trapping measurements in studies of the mechanochemistry of molecular motors. We directly observed stepping and eventual MT dissociation of individual Eg5 dimers over a range of ATP, ADP, and phosphate concentrations and forces. Based on these data, we found that the premature binding of ADP is the main determinant of Eg5-MT dissociation (and therefore the loss of processivity) under low to moderate loads. The application of large loads could also induce Eg5-MT dissociation, but this mechanical disruption was insensitive to the direction of applied force. The data therefore suggest that external load is not the sole governor of processivity, but that the biochemical cycles of the two heads of the Eg5 dimer are coordinated. The addition of excess ADP leads to a loss of this coordination and therefore to a decrease in processivity.
Acknowledgments
We thank P. Fordyce, S. Gilbert, N. Guydosh, and J. Shaevitz for helpful discussions.
The authors acknowledge support from a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (M.T.V.), and from a grant from the National Institutes of Health R01-GM51453 (S.M.B.).
Supporting Material
References
- 1.Kapitein L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 2.Korneev M., Lakämper S., Schmidt C. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur. Biophys. J. 2007;36:675–681. doi: 10.1007/s00249-007-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valentine M.T., Fordyce P.M., Krzysiak T.C., Gilbert S.P., Block S.M. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cochran J.C., Krzysiak T.C., Gilbert S.P. Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry. 2006;45:12334–12344. doi: 10.1021/bi0608562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krzysiak T.C., Gilbert S.P. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J. Biol. Chem. 2006;281:39444–39454. doi: 10.1074/jbc.M608056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krzysiak T.C., Grabe M., Gilbert S.P. Getting in sync with dimeric Eg5: initiation and regulation of the processive run. J. Biol. Chem. 2008;283:2078–2087. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert S.P., Mackey A.T. Kinetics: a tool to study molecular motors. Methods. 2000;22:337–354. doi: 10.1006/meth.2000.1086. [DOI] [PubMed] [Google Scholar]
- 8.Lang M.J., Fordyce P.M., Engh A.M., Neuman K.C., Block S.M. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Block S.M., Asbury C.L., Shaevitz J.W., Lang M.J. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc. Natl. Acad. Sci. USA. 2003;100:2351–2356. doi: 10.1073/pnas.0436709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang M.J., Asbury C.L., Shaevitz J.W., Block S.M. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuman K.C., Block S.M. Optical trapping. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnitzer M.J., Block S.M. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 13.Colquhoun D., Hawkes A.G. The principles of stochastic interpretation of ion-channel mechanisms. In: Sakmann B., Neher E., editors. Single Channel Recording. Plenum; New York: 1995. pp. 397–482. [Google Scholar]
- 14.Rosenfeld S.S., Fordyce P.M., Jefferson G.M., King P.H., Block S.M. Stepping and stretching: how kinesin uses internal strain to walk processively. J. Biol. Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter N., Cross R. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 16.Cochran J.C., Gilbert S.P. ATPase mechanism of Eg5 in the absence of microtubules: insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry. 2005;44:16633–16648. doi: 10.1021/bi051724w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackney D.D. Kinesin ATPase: rate-limiting ADP release. Proc. Natl. Acad. Sci. USA. 1988;85:6314–6318. doi: 10.1073/pnas.85.17.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu T., Sablin E., Vale R.D., Fletterick R., Pechatnikova E. Expression, purification, ATPase properties, and microtubule-binding properties of the ncd motor domain. Biochemistry. 2002;34:13259–13266. doi: 10.1021/bi00040a042. [DOI] [PubMed] [Google Scholar]
- 19.Asenjo A.B., Krohn N., Sosa H. Configuration of the two kinesin motor domains during ATP hydrolysis. Nat. Struct. Biol. 2003;10:836–842. doi: 10.1038/nsb984. [DOI] [PubMed] [Google Scholar]
- 20.Asenjo A.B., Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc. Natl. Acad. Sci. USA. 2009;106:5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori T., Vale R.D., Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 22.Guydosh N.R., Block S.M. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009 doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klumpp L.M., Hoenger A., Gilbert S.P. Kinesin's second step. Proc. Natl. Acad. Sci. USA. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sosa H., Peterman E.J.G., Moerner W.E., Goldstein L.S.B. ADP-induced rocking of the kinesin motor domain revealed by single-molecule fluorescence polarization microscopy. Nat. Struct. Mol. Biol. 2001;8:540–544. doi: 10.1038/88611. [DOI] [PubMed] [Google Scholar]
- 25.Krzysiak T.C., Wendt T., Sproul L.R., Tittmann P., Gross H., Gilbert S.P., Hoenger A. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–2273. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenfeld S.S., Xing J., Jefferson G.M., King P.H. Docking and rolling, a model of how the mitotic motor Eg5 works. J. Biol. Chem. 2005;280:35684–35695. doi: 10.1074/jbc.M506561200. [DOI] [PubMed] [Google Scholar]
- 27.Kapitein L.C., Kwok B.H., Weinger J.S., Schmidt C.F., Kapoor T.M. Microtubule cross-linking triggers the directional motility of kinesin-5. J. Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok B.H., Kapitein L.C., Kim J.H., Peterman E.J.G., Schmidt C.F. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat. Chem. Biol. 2006;2:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


