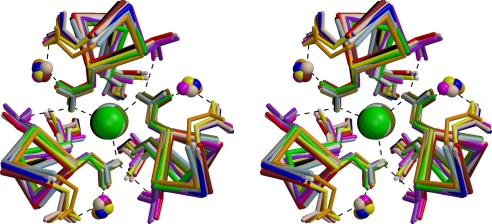Abstract
Most core residues of coiled coils are hydrophobic. Occasional polar residues are thought to lower stability, but impart structural specificity. The coiled coils of trimeric autotransporter adhesins (TAAs) are conspicuous for their large number of polar residues in position d of the core, which often leads to their prediction as natively unstructured regions. The most frequent residue, asparagine (N@d), can occur in runs of up to 19 consecutive heptads, frequently in the motif [I/V]xxNTxx. In the Salmonella TAA, SadA, the core asparagines form rings of interacting residues with the following threonines, grouped around a central anion. This conformation is observed generally in N@d layers from trimeric coiled coils of known structure. Attempts to impose a different register on the motif show that the asparagines orient themselves specifically into the core, even against conflicting information from flanking domains. When engineered into the GCN4 leucine zipper, N@d layers progressively destabilized the structure, but zippers with 3 N@d layers still folded at high concentration. We propose that N@d layers maintain the coiled coils of TAAs in a soluble, export-competent state during autotransport through the outer membrane. More generally, we think that polar motifs that are both periodic and conserved may often reflect special folding requirements, rather than an unstructured state of the mature proteins.
Keywords: ion coordination, protein export, trimeric autotransporter adhesin, polar core residues
Coiled coils are bundles of α-helices that are wound into superhelical structures. They represent one of the most common structural motifs in proteins and show a considerable diversity of forms, the 2 most frequent being parallel homooligomers of 2 or 3 helices (1). As proposed by Crick (2, 3), the hallmark of coiled coils is the distinctive packing of amino acid side chains in the core of the bundle, called knobs into holes, in which a residue from 1 helix (knob) packs into a space surrounded by 4 side chains of the facing helix (hole). This regular meshing requires that side chains occupy periodically equivalent positions along the bundle interface. In undistorted α-helices, with 3.6 residues per turn on average, side-chain positions only repeat after about 18 residues or 5 turns (18/5 = 3.6). Coiled coils reduce this periodicity to 3.5, or 7 residues over 2 turns (the heptad repeat), by compensating for the resulting left-handed drift of side-chain positions across the face of the helices with an equal, left-handed supercoiling of the helices themselves. Coiled-coil helices thus wind around each other to produce a geometrically ideal meshing of side chains at their interface. If the 7 positions of the coiled-coil heptad repeat are labeled a–g, the residues forming the core are in a and d.
Despite their primarily hydrophobic nature, which is the result of the dominant contribution of the hydrophobic effect to protein folding, the residues forming the core also include hydrophilic side chains. As already noted in the first tabulation of residue frequencies in coiled coils (4), polar residues constitute roughly a quarter of the total and are more frequent in 2-stranded than in 3-stranded structures (5), presumably because core residues in 2-stranded coiled coils are less shielded from solvent. We have recently collected all coiled coils of known structure into the CC+ database (6) and observe the same proportion of about 25% polar residues in core positions, with some variation across the different structural classes (O.T. and D.N.W., unpublished work). Burying polar residues is thermodynamically disfavored and weakens the overall stability of the structure, but appears to impart structural specificity. In the best-studied case, replacement of the asparagine buried in core position a of the dimeric GCN4 leucine zipper with valine resulted in a protein with higher stability, but lower structural specificity, as the mutant protein formed mixtures of dimers and trimers (7, 8). Nevertheless, the neutral theory of evolution suggests that most polar core residues are the result of random drift and should not be expected to serve a specific function in their respective proteins. In this view, mutations of hydrophobic to hydrophilic residues in the core would often have no phenotype and therefore be invisible to stabilizing selection, because coiled coils can tolerate a certain proportion of polar core residues without altering their structural properties.
Given the underrepresentation of polar residues in the core of coiled coils, we were surprised to find that the coiled coils of trimeric autotransporter adhesins (TAAs) systematically include a large number of polar residues in predicted core positions, often in recurring patterns (9). The most frequent heptad motifs we observed were those with 1 hydrophilic core residue per heptad (always in position d), primarily [I/V]xxNTxx and to a lesser extent LxxTNxx, but there were also several in which both core residues were hydrophilic, such as NxxQDxx, SxxNTxx, QxxHxxx, and QxxDxxx. Coiled-coil segments containing such motifs were often difficult to recognize with software built on canonical scoring matrices and in many cases were predicted as low-complexity, natively unstructured regions.
TAAs are widespread adhesins of proteobacteria and include many well-established pathogenicity factors (10). They follow a basic head–stalk–anchor architecture from N to C terminus, where the head mediates adhesion and autoagglutination, the (typically coiled coil-rich) stalk projects the head from the cell surface, and the anchor provides the export function and attaches the adhesin after export is complete. In some complex adhesins, head and stalk segments may alternate several times before the anchor is reached. The process of autotransport is thought to initiate when 4 β-strands at the C-terminal end of a TAA monomer insert into the outer membrane and trimerize to form a 12-stranded β-barrel, through which the 3 chains exit the periplasm. Once export is complete, the chains fold and the stalk forms a 3-stranded coiled coil that extends into the pore and occludes its opening. Little is known about the export process, how it is energized, and how the chains are maintained in an unfolded but soluble (and thus export-competent) state as they pass through the 1.5 nm-wide pore of the β-barrel. The latter is far from being a simple problem, as many TAAs are thousands of residues long, the largest known to us having 5,399 residues (MS0748 of Mannheimia succiniciproducens).
Here we present the analysis of a coiled-coil segment from the TAA of Salmonella enterica, SadA, a complex adhesin of 1,461 residues with multiple central coiled-coil segments that consist almost entirely of repeating IxxNTxx motifs. We study the occurrence of similar motifs in coiled coils of known structure and also investigate the effects of introducing up to 3 copies of this motif into a coiled-coil model system, the GCN4 leucine zipper. We propose that the polar core residues, which sequester anions to the hydrophobic core, help to maintain the coiled coils in a soluble and unfolded state during export through the outer membrane.
Results and Discussion
Sequence Properties of TAA Coiled Coils.
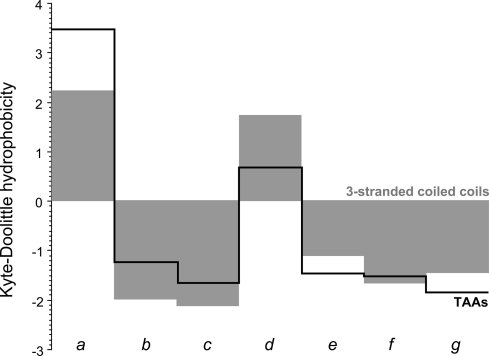
The stalks of TAAs may extend for considerable distances and are rich in coiled-coil segments. The stalk of the Bartonella henselae adhesin BadA, for example, projects the head ≈200 nm away from the cell surface (11) and contains 23 coiled-coil segments along its 2,500 residue length (12). Individual segments vary widely in size, from less than 1 heptad, as in a fragment from the Haemophilus influenzae adhesin Hia (see PDB: 1S7M, 107-VGDLRG-112) to over 1,220 residues in the haemaglutinin-like outer membrane protein of Burkholderia sp. 383 (GenBank accession YP_371201), which is the longest we have observed so far. The latter is built on 68 essentially perfect repeats of the sequence IGSLSTSTSTGLSSANSS and has a skewed residue composition, comprising two-thirds Ser and Thr, which highlights another peculiarity of TAA coiled coils: many have a compositional bias that leads to their erroneous identification as natively unstructured sequences. TAA coiled coils have one further peculiarity in the nature of their core residues. As we observed during the analysis of a representative set of TAA sequences (9), they contain an unusual number of polar residues in position d of the core, but not in position a, which is on average more hydrophobic than that of 3-stranded coiled coils in general (Fig. 1). In our analysis, almost half of residues in d were polar or charged, and of these, fully half were asparagine; we refer to these residues as N@d. Strikingly, they often occur in extended runs, such as for example the 19 consecutive N@d residues in the adhesin BR1846 of Brucella suis 1330 (GenBank accession NP_698826). In 90% of the cases, the asparagines were preceded by Ile or Val in position a and in about 60% of the cases, they were followed by Thr in position e. The dominant heptad motif containing a polar core residue can thus be described as [I/V]xxNTxx. To investigate its structural and biophysical properties, we focused on 1 of 6 central coiled-coil segments from the Salmonella adhesin, SadA, which are largely built of this coiled-coil motif.
Fig. 1.
Average hydrophobicity at the 7 positions, a–g, of 3-stranded coiled coils of known structure (gray surface), compared to the average hydrophobicity at the same positions of coiled coils from trimeric autotransporter adhesins (black line).
The Structure of N@d Layers in SadA.
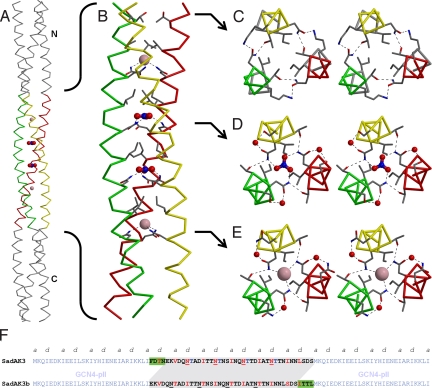
SadA is a complex adhesin with 13 predicted coiled-coil segments (supporting information (SI) Fig. S1). For structural analysis, we expressed residues 479–519, encompassing almost the entire fourth segment and containing 4 consecutive [I/V]xxNTxx motifs. Because we frequently experienced problems with the expression of isolated TAA stalk segments in earlier studies, we fused the fragment in the correct heptad register to the trimeric form of the GCN4 leucine zipper, GCN4-pII, at both the N- and C-terminal ends (Fig. 2F); we named this construct SadAK3 (13). SadAK3 gave a CD spectrum typical for α-helical proteins and unfolded cooperatively upon heating, with a transition midpoint (Tm) at 72 °C (Fig. S3A). Crystallization assays yielded crystals diffracting to 1.85 Å.
Fig. 2.
Structure of SadAK3. (A) The whole crystal structure of SadAK3, including the GCN4-adaptors in gray and the 2 chloride and 2 nitrate anions trapped in the N@d layers. (B) Close-up of the SadA insert. Residues in positions a and d of the N@d layers and the single T@d are shown as sticks. (C–E) Binding details in stereoview upstream the helical bundle, showing I478, T481, N482, and K484 of the single T@d layer (C), I492, N495@d, T496, S498, and a bridging water coordinating nitrate (D) and I506, N509@d, T510, N512, and a bridging water coordinating chloride (E). (F) Sequences of the constructs SadAK3 and SadAK3b. In SadAK3 and SadAK3b residues 479–519 and 483–523 of SadA were fused as an insert between the N- and C-terminal GCN4-pII adaptors. For SadAK3, the fusion was done in register. Relative to SadAK3, the insert of SadAK3b has the first 4 residues deleted and 4 residues added to its end (green boxes). The resulting shift in the sequence is highlighted in gray. For SadAK3 the observed and for SadAK3b the expected core positions (a and d) are highlighted in red. The C-terminal His-tag of SadAK3b (KLHHHHHH) is omitted.
The structure of SadAK3 shows a trimeric coiled coil with a continuous heptad periodicity; the N@d residues are located in the central 4 heptads (Fig. 2). Their amino groups form hydrogen bonds to the hydroxyl groups of threonines in position e of neighboring chains and coordinate an anion in the core of the bundle. Their carbonyl groups interact via a bridging water molecule with the polar head group of the residue in position g of the same chain, i.e., with the carboxyl group of an aspartate in N@d layers 1 and 3, the hydroxyl group of a serine in layer 2, and the amino group of an asparagine in layer 4. Indeed, in revisiting the residue statisitics of N@d heptads in TAAs we find that, cumulatively, Asn, Asp, and Ser are as frequent in position g as Thr is in e.
The N@d asparagines are thus at the center of an elaborate network of polar interactions with highly defined geometry (Fig. 2). The anions are enclosed in a cavity, which is bounded on one side by the asparagines and on the other by the β-branched residues, Ile or Val, in position a. The anions in the first and last N@d layer could be convincingly modeled as chloride (Fig. 2E). The electron density in the middle 2 layers indicated larger ions of triangular, planar shape and could be interpreted best as nitrate (Fig. 2D).
In the heptad preceeding the N@d layers, the positions of the asparagine and threonine are reversed, corresponding to the second most frequent polar core motif of TAA coiled coils. In this layer, the threonines point their side-chain methyl groups toward the core and coordinate the amino groups of asparagines in position e of neighboring chains via their hydroxyl groups, providing a reversed network of interactions relative to the N@d layers, but without the anion (Fig. 2C). Here also, the polar network is extended by the residues in position g.
N@d Layers in Coiled Coils of Known Structure.
Asparagines in position d of coiled-coil structures from bacterial and viral surface proteins have been noted previously (ref. 14 and references therein). We explored their occurrence systematically in coiled coils of known structure using CC+ (6), a relational database of coiled coils of known structure compiled from the Protein Data Bank. We focused specifically on occurrences in parallel, homooligomeric coiled coils of at least 11 residues. The resulting 37 nonredundant examples were filtered to remove coiled coils with noncanonical structure, insufficient resolution, or other problematic aspects, as listed in Table S1. The final subset contained 5 occurrences in dimeric, 16 in trimeric, and 1 in tetrameric coiled coils. Although this set contains too few examples in dimeric and tetrameric coiled coils for a reliable comparative analysis, several observations seem warranted:
The 5 examples from dimeric coiled coils are structurally fairly heterogeneous and do not contain ions in the core (Fig. S2A).
In contrast, the 16 examples from trimeric coiled coils can be superimposed very closely and show the same conformation of asparagine side chains around a central chloride ion as SadAK3. In most cases the polar network is also extended by additional residues, including 6 with Thr in e of the neighboring chain and 7 with Asn or Asp in g of the same chain, all with the same geometry as in SadAK3, including the bridging water molecule (Fig. 3).
The single example of a tetrameric N@d layer (Fig. S2C) also shows a symmetrical arrangement of asparagine side chains around a central ion, but the ion is a cation (Ca2+) and the asparagine head groups have a reversed orientation, with the carbonyl groups pointing inward to coordinate the ion.
Fig. 3.
Superposition of 16 trimeric N@d layers of known structure, including 3 exemplars from this study as listed in Table S1, all coordinating chloride. In 6 cases the polar network is extended by Thr in e of the neighboring chain and in 7 cases by Asn or Asp in g of the same chain, all with the same geometry as in SadAK3, including the bridging water molecule. Dimeric, tetrameric, and 2 further trimeric instances are shown in Fig. S2.
It would thus appear that N@d layers are more common in trimeric than in dimeric coiled coils and that this may be the result of their ability to form highly ordered, symmetrical networks of polar interactions in the former, but not in the latter. The variable nature of the residues extending the polar networks and their absence in some structures suggest that the presence of asparagines in d may be sufficient to bring about ordered N@d layers.
N@d Layers Orient Themselves Specifically into the Core.
Given their unusual nature, do N@d layers form inherently, or do they only occur when a specific heptad register is imposed by flanking domains? To explore this question, we shifted the register of the SadA insert by 4 residues out of frame relative to the GCN4 adaptors. Were the insert to follow the register set by the flanking coiled coils, the isoleucines previously found in position a would shift to position d and the threonines, previously in position e, would shift into the core to occupy position a, yielding the heptad pattern Txx[I/V]xxN. This seemed an attractive alternative, because threonine is the most frequent polar residue in position a of trimeric coiled coils and the fourth most frequent residue overall, after Leu, Ile, and Val (M. Gruber and A.N.L., unpublished work). This construct, SadAK3b, contains residues 483–523 of SadA and differs from SadAK3 by the deletion of 4 residues at the N-terminal end of the insert and the addition of 4 residues at its C-terminal end (Fig. 2F).
Like SadAK3, SadAK3b gave a CD spectrum typical for α-helical proteins, but unfolded in 2 steps upon thermal denaturation, with transition midpoints at Tm1 = 60 °C and Tm2 = 90 °C (Fig. S3A). Its crystal structure, obtained at a resolution of 2.65 Å, shows that both the GCN4 adaptors and the SadA insert retained their original register, leading to the formation of heptad discontinuities at the junctions between them. These discontinuities belong to 2 different types, which have been described previously and are well understood: the deletion of 4 residues is called a stammer and the insertion of 4 residues a stutter. Because 4 residues are slightly more than 1 helical turn, their deletion causes a decrease in the local periodicity and thus a further increase in the local degree of supercoiling, whereas their insertion has the exactly opposite effect. The extent to which they perturb the local periodicity depends on the degree to which their effect is delocalized along the sequence. Thus, if delocalized over about 10 residues, the stammer decreases the local periodicity from 3.5 residues per turn to about 3.3, whereas the stutter increases it to about 3.7 (14 residues − 4 = 10, over 3 helical turns: 10/3 = 3.33; and 7 residues + 4 = 11, over 3 helical turns: 11/3 = 3.67).
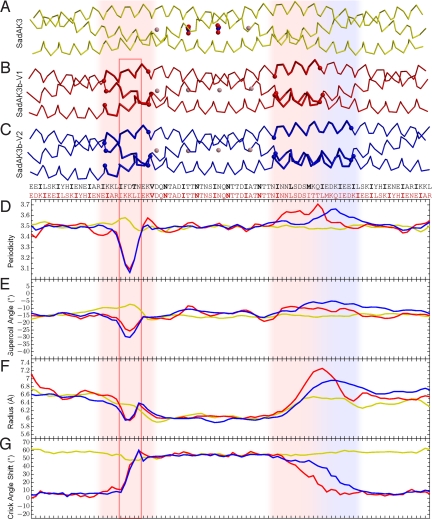
In SadAK3b, the junction between the N-terminal adaptor and the insert contains a stammer, and the junction between the insert and the C-terminal adaptor a stutter. A detailed analysis of the coiled-coil parameters shows that the stammer is highly focused to the junction (Fig. 4). Most of the distortion is confined to the last 3 residues of the N-terminal adaptor, leading to a local drop in periodicity below 3.2 residues per turn. The resulting steepening of the crossing angle is clearly visible in the structure (Fig. 4B). The discontinuity is however not fully compensated for by an increase in supercoiling. Instead, part of the distortion is absorbed by an overwinding of the α-helices themselves, leading to the formation of a short 310-helix (Fig. S3D). This is, to our knowledge, the first structure of a stammer in an oligomeric coiled coil.
Fig. 4.
The structures of SadAK3, SadAK3b-V1 (V1), and SadAK3b-V2 (V2) are depicted together with their sequences and plots of their coiled-coil parameters. Structures, sequences, and plots are in scale to each other and aligned on their inserts. The colors in the plots correspond to the colors of the structures; the sequences for SadAK3 and SadAK3b are black and red, respectively. The discontinuities in the SadAK3b structures are highlighted by vertical shades throughout the figure, colored light red for V1, and additionally light blue for V2. The red box indicates the 310 helix within the stammer (see also Fig. S3D). In the plots for periodicity, supercoil angle, and coiled-coil radius, the SadAK3b structures locally deviate from the SadAK3 structure, whereas the deviations at the stammer are more pronounced and localized than at the stutter. This is in line with the plot of the Crick angle shift, showing the deviation of the observed from the expected Crick angle for heptad repeat. These curves were calculated under the assumption that all structures have a continuous heptad repeat. Therefore the Crick angles for V1 and V2 start to depart from the expected value at the beginning of the stammer and reach their maximum deviation at its end. For the length of the N@d layers, which are again in heptad repeat, this deviation stays constant, until the stutter mediates the transition back to the heptad register of the C-terminal GCN4 adaptor. Because the alignment is based on the inserts, the Crick angle deviation for SadAK3 is constantly at the maximum deviation of the SadAK3b structures.
In contrast, the stutter is less sharply localized. The distortion of coiled-coil parameters occurs rather evenly over about the last 11 residues of the insert, providing a smooth transition and an only minor shallowing of the crossing angle. Surprisingly, we found evidence for heterogeneity in the accommodation of the stutter. While attempting to obtain a higher resolution data set for the SadAK3b structure, we found a second crystal species under the same crystallization condition as for the already examined species (SadAK3b-V1). The structure of this other species (SadAK3b-V2), determined at 2.6 Å, suggested an even more gradual accommodation of the stutter over about 18 residues (Fig. 4C). In contrast, the region of the stammer was identical in both species. It seems possible that the stutter is dynamic in solution, “floating” between the 2 observed conformations.
The 2-step thermal denaturation of SadAK3b suggests that the heptad discontinuities have uncoupled the unfolding of the insert from that of the adaptors. We therefore treated the protein with proteinase K both at room temperature and at the first transition point Tm1 = 60 °C. Whereas SadAK3b was largely proteinase K resistant at room temperature, we observed the formation of 2 main proteolytic fragments of 4 kDa and 5 kDa at 60 °C (Fig. S3B). Their analysis by mass spectrometry identified the 4-kDa fragment as the N-terminal adaptor plus the first 6 residues of the insert and the 5-kDa fragment as the last 16 residues of the insert plus the C-terminal adaptor (Fig. S3C). For comparison, the in-register construct SadAK3 was fully protease resistant at 60 °C. We conclude that the first thermal transition point corresponds to the unfolding of the 4 central N@d layers, which now do not benefit from a continuous register with the more stable GCN4-pII adaptors.
N@d Layers in the GCN4 Leucine Zipper.
To gain a better biophysical understanding of N@d layers in coiled coils, we studied their effect in a well-understood model system, the GCN4-p1-N16V leucine zipper. In this GCN4 variant, the core asparagine in position a of the third heptad is substituted with valine, resulting in an increased stability but decreased structural specificity, as this protein forms mixtures of dimers and trimers (15). Into this background, we introduced the N@d motifs VxxNxxx, VxxNTxx, IxxNxxx, and IxxNTxx in 1 (M1), 2 (M2), or 3 (M3) consecutive heptads (Fig. 5). The folding of all M1 peptides was strongly concentration dependent, with the optimum reached at 300 μM (Fig. S4B), pH 9–10 (Fig. S4 C and E), and high ionic strength (Fig. S4F). At concentrations below 200 μM, the absorption spectra indicated the presence of substantial amounts of unfolded material. Under optimum conditions, the melting temperatures of the M1 peptides were around 30–35 °C (see also ref. 16). The presence of Ile or Val in position a of the motif did not affect the results, and the only observable effect of Thr in e was a stronger focusing of the pH optimum to the range around 9–10. We were surprised by this, as Thr is strongly enriched at this position in N@d heptads and therefore under positive selection. The interchain contacts it forms suggested to us a stabilizing effect, but, in light of these results, a role in structural specificity seems more likely.
Fig. 5.
Sequence of synthetic peptides used in this work. The sequence of GCN4-p1 N16V was modified by mutating specific residues (highlighted in gray) to create V/IxxNxx and V/IxxNTxx motifs.
In contrast to the M1 peptides, GCN4-p1-N16V reached its folding optimum at 100 μM (Tm > 100 °C) and showed no dependence on pH and ionic strength. Its helical content at these conditions was clearly higher than that of the M1 peptides. Thus, the insertion of even a single N@d layer strongly reduced stability. None of the M2 and M3 peptides, which contained more than 1 N@d layer, were folded in solution under any condition we explored. Nevertheless, all peptides we attempted to crystallize yielded crystals diffracting to high resolution. We therefore conclude that even the M3 peptides fold at high concentrations as they prevailed in the crystallization trials, which were more than a factor of 10 higher than the maximum concentration we used in CD spectroscopy.
As representatives of the minimal N@d motif we solved the structures of peptides M1-VxxNxxx and M2-VxxNxxx. As anticipated, their N@d layers coordinate chloride ions. Whereas the anion binding site in M1-VxxNxxx was fully occupied, the equivalent site in M2-VxxNxxx was only partly occupied and, in the unoccupied state, 1 of the asparagine side chains rotated into the core to form hydrogen bonds with the other 2 asparagines (Fig. S5). The second binding site in M2-VxxNx was fully occupied. The partial breakdown of the binding site may be indicative of a role of the flanking threonine in structural specificity, because this residue is missing in these constructs. However, in a crystal obtained under different conditions, both sites were fully occupied and we also did not find any case of binding-site breakdown in our survey of N@d layers in coiled coils of known structure.
As a representative of the most abundant N@d motif we solved the structure of the peptide M3-IxxNTxx. We initially obtained the structures of M3-IxxNTxx-chloride and M3-IxxNTxx-bromide from different crystallization conditions. Subsequently, we exchanged the chloride ions with iodide and with nitrate by soaking the crystals, and obtained the structures for M3-IxxNTxx-iodide and M3-IxxNTxx-nitrate (we were however unable to substitute chloride with fluoride, sulfide, or carbonate). The halogen ions fully occupied their binding sites in the respective structures, with coordination distances that increased with the radius of the ion. Nitrate displaced the chloride fully only in the central N@d layer. In all structures, the amino groups of the core asparagines formed hydrogen bonds to the hydroxyl groups of threonines in position e of neighboring chains, with the same geometry as already described for SadAK3. Fig. S6 illustrates this astonishing combination of great specificity in binding geometry with considerable flexibility in the size of the ion that is bound.
Conclusions
We have undertaken a detailed study of asparagine in position d of coiled coils (N@d). In dimeric coiled coils, N@d residues lead to structural heterogeneity, but in trimeric coiled coils, they form a highly ordered network of polar interactions with an anion at its center. N@d residues may thus prove useful for specifying the oligomeric state of designed coiled coils. Moreover, because of their ability to specifically bind bromide and iodide with high occupancy, they may provide a convenient tool for structure determination via anomalous dispersion (SAD/MAD). In fact we solved the M3-IxxNT-bromide structure via this technique. We therefore plan to incorporate N@d layers into the GCN4 adaptors of our expression system (13) for experimental phasing.
Despite their specific geometry, N@d layers have a destabilizing effect on coiled coils, which we attribute to the fact that they sequester ions away from solvent. Their introduction into consecutive heptads of a thermostable structure is sufficient to abolish folding under physiological conditions, yet the resulting constructs are highly soluble and will fold at elevated concentrations (for example in crystallization assays). Placing them into the context of folded domains, such as the GCN4-pII leucine zipper, provides an efficient mechanism to raise their local concentration to the point where stable folding can occur. In such a context they will adopt their structure with great specificity, even against conflicting information from the flanking domains. Considering the requirement of trimeric autotransporters to keep their passenger domains in an unfolded but soluble state, while 3 chains of hundreds to thousands of residues traverse the outer membrane through a pore of about 15 Å diameter, the properties we observed for N@d layers readily suggest a functional role in maintaining the export-competent state. Once their export is complete, the folding of these coiled-coil segments can then be triggered by the folding of an adjacent domain.
The observation that coiled coils of trimeric autotransporters are frequently predicted to be natively unstructured by bioinformatic disorder predictors, even though they are folded in the mature proteins, points to a weakness of these programs in capturing the structural properties of proteins with special folding requirements. In cases where proteins show periodically recurrent sequence motifs, and more particularly where these motifs are also conserved in homologs, evolutionary considerations indicate that the motifs are under positive selection. We think that in most cases this conservation reflects an underlying propensity to form a repeating, folded structure, a few examples to the contrary notwithstanding (such as the FG repeats of nucleoporins). Sequences predicted to be natively unstructured, but having a detectable underlying sequence periodicity, are encountered throughout prokaryotes and eukaryotes. We submit that the extent to which they are actually unstructured in the mature proteins remains to be established.
Materials and Methods
The SadAK3 construct contains residues 479–519 of SadA (GenBank accession NP_462591), fused at both the N and C termini in the correct heptad register to GCN4 leucine zippers in the expression vector pIBA-GCN4tri as described in ref. 13. SadAK3b is a similar construct containing residues 483–523, out of register by 4 residues to the leucine zippers, the heptad register at the junctions being ..fga fga.. and ..bcd abc… Peptides were synthesized by the core facility of the Max Planck Institute of Biochemistry or purchased from EMC Microcollections.
Crystallization conditions for all crystals used in the diffraction experiments are listed in Table S2. Crystals of M3-IxxNT-chloride were soaked in their mother liquor supplemented with either Nal or KNO3 to substitute chloride by iodide or nitrate. The structures of SadAK3, SadAK3b-V1, SadAK3b-V2, M1-VxxNx, and M2-VxxNx were solved by molecular replacement using the GCN4-structure 1GCM as a search model, the structure of M3-IxxNT-bromide was solved via single-wavelength anomalous dispersion with data collected slightly above the Br K edge. M3-IxxNT-chloride, M3-IxxNT-iodide, and M3-IxxNT-nitrate were later solved on the basis of the M3-IxxNT-bromide coordinates. Data collection and refinement statistics are summarized in Table S3. Local coiled-coil and α-helical parameters were determined using TWISTER (17). Further methodological details can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank Ines Wanke for technical assistance with sample preparation and crystallization and Dirk Linke for valuable discussions. We are grateful for the continuous support from the staff of beamline PXII/Swiss Light Source. This work was supported by institutional funds from the Max Planck Society and by the German Science Foundation (FOR449/LU1165 and SFB766/B4) and the CC+ database by a Biotechnology and Biological Sciences Research Council Grant (BB/D003016/1) (to D.N.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2WPQ (SadAK3), 2WPR (SadAK3b-V1), 2WPS (SadAK3b-V2), 2WPY (M1-VxxNx), 2WPZ (M2-VxxNx), 2WQ0 (M3-IxxNT-chloride), 2WQ1 (M3-IxxNT-bromide), 2WQ2 (M3-IxxNT-iodide), and 2WQ3 (M3-IxxNT-nitrate)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0907256106/DCSupplemental.
References
- 1.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 2.Crick FH. Is alpha-keratin a coiled coil? Nature. 1952;170(4334):882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- 3.Crick FHC. The Fourier transform of a coiled-coil. Acta Crystallographica. 1953;6(8–9):685–689. [Google Scholar]
- 4.Parry DA. Coiled-coils in alpha-helix-containing proteins: Analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep. 1982;2(12):1017–1024. doi: 10.1007/BF01122170. [DOI] [PubMed] [Google Scholar]
- 5.Conway JF, Parry DA. Three-stranded alpha-fibrous proteins: The heptad repeat and its implications for structure. Int J Biol Macromol. 1991;13(1):14–16. doi: 10.1016/0141-8130(91)90004-e. [DOI] [PubMed] [Google Scholar]
- 6.Testa OD, Moutevelis E, Woolfson DN. CC+: A relational database of coiled-coil structures. Nucleic Acids Res. 2009;37(database issue):D315–D322. doi: 10.1093/nar/gkn675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez L, Jr, Woolfson DN, Alber T. Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat Struct Biol. 1996;3(12):1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 8.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 9.Szczesny P, Lupas A. Domain annotation of trimeric autotransporter adhesins–daTAA. Bioinformatics. 2008;24(10):1251–1256. doi: 10.1093/bioinformatics/btn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006;14(6):264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Szczesny P, et al. Structure of the head of the Bartonella adhesin BadA. PLoS Pathog. 2008;4(8):e1000119. doi: 10.1371/journal.ppat.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riess T, et al. Bartonella adhesin a mediates a proangiogenic host cell response. J Exp Med. 2004;200(10):1267–1278. doi: 10.1084/jem.20040500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez Alvarez B, et al. A new expression system for protein crystallization using trimeric coiled-coil adaptors. Protein Eng Des Sel. 2008;21(1):11–18. doi: 10.1093/protein/gzm071. [DOI] [PubMed] [Google Scholar]
- 14.Guardado-Calvo P, Fox GC, Llamas-Saiz AL, van Raaij MJ. Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre. J Gen Virol. 2009;90(3):672–677. doi: 10.1099/vir.0.008276-0. [DOI] [PubMed] [Google Scholar]
- 15.Knappenberger JA, Smith JE, Thorpe SH, Zitzewitz JA, Matthews CR. A buried polar residue in the hydrophobic interface of the coiled-coil peptide, GCN4–p1, plays a thermodynamic, not a kinetic role in folding. J Mol Biol. 2002;321(1):1–6. doi: 10.1016/s0022-2836(02)00592-2. [DOI] [PubMed] [Google Scholar]
- 16.Akey DL, Malashkevich VN, Kim PS. Buried polar residues in coiled-coil interfaces. Biochemistry. 2001;40(21):6352–6360. doi: 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- 17.Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J Struct Biol. 2002;137(1–2):54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.