Abstract
Subtilases are serine proteases found in Archae, Bacteria, yeasts, and higher eukaryotes. Plants possess many more of these subtilisin-like endopeptidases than animals, e.g., 56 identified genes in Arabidopsis compared with only 9 in humans, indicating important roles for subtilases in plant biology. We report the first structure of a plant subtilase, SBT3 from tomato, in the active apo form and complexed with a chloromethylketone (cmk) inhibitor. The domain architecture comprises an N-terminal protease domain displaying a 132 aa protease-associated (PA) domain insertion and a C-terminal seven-stranded jelly-roll fibronectin (Fn) III-like domain. We present the first structural evidence for an explicit function of PA domains in proteases revealing a vital role in the homo-dimerization of SBT3 and in enzyme activation. Although Ca2+-binding sites are conserved and critical for stability in other subtilases, SBT3 was found to be Ca2+-free and its thermo stability is Ca2+-independent.
Keywords: calcium, proprotein convertase, protease-associated domain, subtilisin, thermostability
Subtilases constitute the S8 family in clan SB of serine proteases (http://merops.sanger.ac.uk). They are characterized by a catalytic triad of Asp, His, and Ser residues in an arrangement shared with subtilisins from Bacillus species (1). The first eukaryotic subtilase to be identified was kexin. It is involved in the maturation of α-mating factor and killer toxin from their respective precursor proteins in yeast (2). Nine subtilisin-like endopeptidases have since been discovered in mammals, seven of which are related to kexin and also involved in the highly specific processing of precursor proteins. Their substrates include polypeptide hormone precursors, growth factors, receptors, enzymes, and viral surface glycoproteins, which are typically cleaved on the carboxyl side of paired basic residues (3). The remaining two subtilases, PCSK9 and S1P, belong to the proteinase K and pyrolysin subfamilies of subtilases (3). The discovery of mammalian proprotein convertases (PCs), characterized by their exquisite substrate specificity compared with bacterial subtilisins, further stimulated interest in this class of serine proteases.
Plants appear to lack kexin-related PCs but they possess a largely expanded pyrolysin family with 56 genes identified in Arabidopsis thaliana (4). They have been implicated in general protein turnover (5), the regulation of plant development (6), biotic and abiotic stress responses (7, 8), and the processing of precursors of peptide growth factors in plants (9). It therefore seems that the majority of plant subtilases assumed plant-specific functions in the course of evolution. With the physiological roles of plant subtilases beginning to emerge, it will now be interesting to see whether or not the adoption of specific roles in plant physiology is reflected in unique structural or biochemical features that distinguish subtilases in plants from those in other organisms.
To address this question we recently purified and characterized a typical pyrolysin-like subtilase from tomato (Solanum lycopersicum) plants (SlSBT3 or SBT3). SBT3 is an extracellular 79 kDa glycoprotein that exhibits a remarkable level of stability at elevated temperatures and alkaline pH (10). Like most other subtilases, it is synthesized as a prepro-protein and targeted for secretion by an N-terminal signal peptide (10). Distinguishing features of SBT3 compared with mammalian PCs include the C-terminal extension to the catalytic domain and, most notably, a large insertion between the His and Ser residues of the catalytic triad. This so-called protease-associated (PA) domain is also found in most other plant and some of the bacterial subtilases, and in functionally unrelated proteins, including the mammalian transferrin and plant vacuolar-sorting receptors. Suggested functions for the PA domain include regulation of substrate access to the active site, ligand or substrate binding, and protein–protein interactions (11–14). The PA domain of SBT3 was found to be required for protease activity and secretion (10). As part of our efforts to clarify the function of the PA-domain and to identify the factors governing SBT3 activity and stability, the enzyme was crystallized and its structure elucidated. We report here the first structure of a subtilase from plants revealing a role for the PA domain in homo-dimerization and regulation of protease activity. Furthermore, we are unaware of another structure of a subtilase that is Ca2+-free in its native state and independent of Ca2+ with respect to activity and thermo stability.
Results and Discussion
Overall Structure and Domain Architecture.
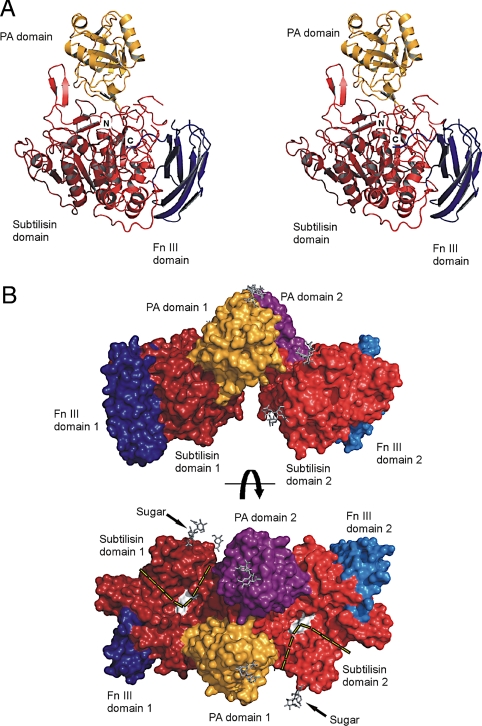
SBT3 was purified from a tomato cell culture as the active enzyme (Thr-113 to Trp-761) with the signal peptide and the prodomain cleaved during zymogen maturation. We determined its structure by MIRAS procedures and refined it at 2.5 Å (crystallographic statistics given in Table S1) with details of the procedure described elsewhere (15). In the final model, density for 639 of 649 residues is well-defined. The polypeptide is folded into three domains, the catalytic (subtilisin) domain (Thr-113-Arg-342 and Leu-473-Tyr-654), an interposed protease-associated (PA) domain (Thr-343-Tyr-472) and a C-terminal Fn III domain (Pro-655-Trp-761). Concerning this domain architecture, the most similar protein is C5α peptidase from Streptococcus pyogenes [PDB entry 3EIF (14)], which also displays a PA domain interposed in the protease domain and shows three C-terminal Fn III-like domains the first of which is similar to the one in SBT3 (Fig. S1). SBT3 crystallizes as dimer with dimerization mediated by the PA domains. The PA domains contact each other directly and bind also to the subtilisin domain of the second monomer. This interaction is established via a β-hairpin (Sβ9-Sβ10; Fig. S1), which is not found in bacterial and mammalian subtilases.
Subtilisin Domain and Active Site.
The core of the subtilisin fold is well conserved between SBT3 and other subtilases from pro and eukaryotes. It consists of a highly twisted β-sheet composed of eight parallel and one anti-parallel β-strands. This β-sheet surrounds two central helices and is flanked by three adjacent and five peripheral helices (Fig. 1A and Fig. S2). Structurally the most similar protein is subtilisin BPN′ from Bacillus amyloliquefaciens (PDB code 1A2Q). 264 Cαs align with an overall rms deviation (RMSD) of 1.9 Å, resulting in a DALI Z score of 35.9. In contrast to subtilisins, which display their active sites in a relatively open and shallow channel on the surface of the molecule (14), the active site of SBT3 resides in a canyon-like crevice with the Ser-538-His-215-Asp-144 active site triad arranged in the center (Fig. 2A). Its accessibility in the homodimer is restricted by the PA-domain, enforcing a 90° kink in the putative substrate binding channel (Fig. 1B).
Fig. 1.
Overall structure of SBT3. (A) Stereoview of the ribbon plot of the SBT3 monomer. The catalytic (subtilisin), protease-associated (PA) and fibronectin (Fn) III-like domains are colored red, orange and blue, respectively. (B) Two perpendicular views of the solid surface representation of the SBT3 dimer with the subtilisin domains in red, the Fn III domains in blue, and the PA domains in orange and magenta. Active site residues are displayed as white surface; N-linked oligosaccharides are displayed as gray sticks. The proposed substrate binding channel is indicated by a dashed line.
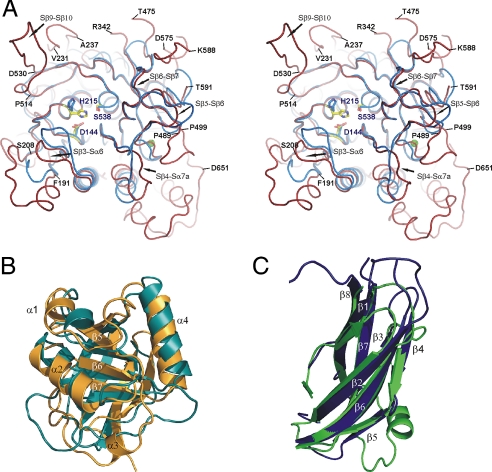
Fig. 2.
Superimpositions of individual SBT3 domains with their nearest structural homolog. (A) Ribbon plot of SBT3′s subtilisin domain (red coil) superimposed with subtilisin BPN′ (blue coil, PDB entry 1A2Q). Residues of the catalytic triad are shown as sticks colored in blue (BPN′) or for SBT3 in yellow (carbon), blue (nitrogen), and red (oxygen). Residue numbering of SBT3 is given as blue labels. Loops bordering the active site are labeled. A calcium ion bound by BPN′ is shown as green sphere. (B) Ribbon plot of SBT3′s PA domain (orange) superimposed with the apical domain of an aminopeptidase from Aneurinibacillus (cyan, PDB entry 2EK8). (C) Ribbon plot of the Fn III domain of SBT3 (blue) superimposed with SSP-19 (sperm-specific protein) from C. elegans (green, PDB entry 1ROW).
Individual substrate binding pockets in the active site of SBT3 were identified in the structure of peptidyl-chloromethylketone (Ac-Phe-Glu-Lys-Ala-cmk)-inhibited SBT3. The inhibitor is covalently bound to active site residues His-215 and Ser-538 (Fig. S3A) and occupies the active site cleft in a way similar to other eukaryotic subtilases, for example kexin inhibited by Ac-Arg-Glu-Arg-Lys-cmk (16). The S1 pocket of the substrate-binding site is filled only partially by the P1-Ala side chain of the inhibitor and provides space for larger residues like asparagine or glutamine, the latter being in agreement with the substrate preference of SBT3 (10). P2-Lys establishes a salt bridge interaction with the side-chain carboxylate of Asp-213 in pocket S2 (Fig. S3A). The P3-Glu side chain points in the direction of Arg-318 but without contacting it directly. A lysine might be more favorable in this position; in fact oligopeptides with lysine in P2 and P3 were confirmed as preferred substrates of SBT3 in activity assays (10).
Aside from the highly similar core structures, there are numerous loops and insertions that discriminate the catalytic domain of SBT3 from BPN′ and other pro- and eukaryotic subtilases (Fig. 2A). The most significant insertions are the PA-domain (Thr-343-Tyr-472), an additional β-hairpin (Pro-514-Asp-530), and the insertions Val-231-Ala-237 and Phe-191-Ser-208. The latter two are located in the regions of Ca2+ sites 1 and 2, which are conserved in other subtilases but absent from SBT3. Furthermore, a large C-terminal extension of ≈50 aa (Ala-605-Tyr-654) harbors two additional helices (helices Sα12 and Sα13, Figs. S1 and S2) and connects to the C-terminal Fn III-like domain.
PA Domain-Mediated Homodimerization and Activation of SBT3.
The PA domain is present in corresponding positions in the majority of plant subtilases [e.g., 54 of the 56 subtilases in Arabidopsis (4)] and some multidomain bacterial subtilases (14). It is built from three anti-parallel and four parallel β strands forming a seven-stranded β-sheet that is surrounded by three peripheral helices (Figs. 1A and 2B). Structurally the most similar protein domains are found in an aminopeptidase from Aneurinibacillus (PDB entry 2EK8) where 109 Cαs of the apical domain align with an RMSD of 2.7 Å (DALI Z score: 11.4) and the streptococcal C5α peptidase [PDB entry 3EIF (14)] whose PA domain aligns over 112 Cαs with an RMSD of 3.2 Å, resulting in a DALI Z score of 9.7.
PA domains are not restricted to subtilases but have been recruited in several evolutionary independent events to different classes of proteases and functionally unrelated proteins (11). This includes metallopeptidases [glutamate carboxypeptidase II (12), transferrin receptor (13), yeast aminopeptidase Y (17)], aspartic proteases [presenilin homologues (18)], and vacuolar sorting receptors (19, 20). Although the PA domain has been implicated in the interaction of these proteins with their cognate ligands or substrates (11), such a function has clearly been shown only for the PA domains of vacuolar sorting receptors in plants, which interact specifically with targeting signals of their cargo proteins and are required for trafficking to lytic and protein-storage vacuoles, respectively (19, 20). We present here the first structural evidence for the proposed function of the PA domain as a protein–protein interaction module, which is required in SBT3 for homodimerization.
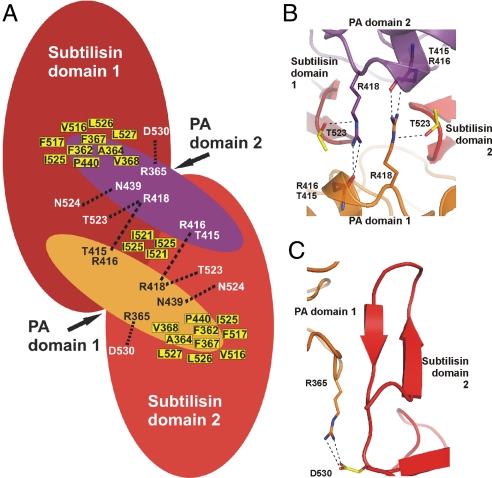
The interface between the two protomers of the SBT3 dimer buries a surface of 2428 Å2. It consists mainly of hydrophobic interactions between the PA domains and the subtilisin domain of the binding partner and, to a lesser extent, also between adjacent subtilisin domains (Figs. 1B and 3A). At the periphery, several hydrophilic interactions are established. Especially eye-catching is the only direct contact between the PA domains, where the terminal η nitrogens of Arg-418 of one PA domain are hydrogen-bonded to the main chain carbonyls of Thr-415 and Arg-416 of the other. This conformation is further stabilized by a contact between the ε nitrogen of Arg-418 and the side chain oxygen of Thr-523 within the same protomer (Fig. 3B).
Fig. 3.
PA domain-mediated homodimerization of SBT3. (A) Schematic representation of the SBT3 homodimer. The subtilisin domains are shown in red, interposed PA domains in orange and magenta, respectively. Residues contributing to the extensive hydrophobic interface are shown as yellow-filled squares. The only direct contact of the subtilisin domains to each other is observed between Ile-521 and Ile-525 of the two β-hairpins. Hydrophilic interactions between the PA and the subtilisin domains are indicated as dotted lines. The PA domains contact each other directly via Arg-418. (B) Detailed view on the contacts between the PA domains. The Arg-418 side chain in each PA domain is hydrogen-bonded to the main chain carbonyls of Thr-415 and Arg-416 of the other. An additional contact is established with Thr-523 in the subtilisin domain of the same protomer. (C) Detailed view on the interaction between Arg-365 from the PA domain and Asp-530 from the subtilisin domain within the same SBT3 molecule.
Another prominent hydrophilic interaction between Arg-365 and Asp-530 contributes to the association of a characteristic β-hairpin of the SBT3 catalytic domain with the PA domain of the other protomer (Fig. 3C). This β-hairpin is not conserved in bacterial and mammalian subtilases (Fig. S4A) and appears to be of extraordinary importance for homo-dimerization. First, it provides the only direct contact between the two subtilisin domains (Ile-521, Ile-525). Second, it appears to position the PA domain relative to the subtilisin domain. Third, the β-hairpin forms almost the entire interface between the PA domain of one monomer and the subtilisin domain of the other. In the latter interaction, a hydrophobic patch on the surface of the PA domain consisting of the side chains of Phe-362, Ala-364, Phe-367, Val-368, P440 and the main chains of Glu-395 and Lys-420 accommodates the β-hairpin of the neighboring subtilisin domain (Fig. S4B). The binding surface is completed by hydrophilic interactions involving Arg-365, Arg-418 and Asn-439. Both the PA domain and the β-hairpin are thus essential for dimerization of SBT3.
Moreover, the interaction between the PA domain and the β-hairpin appears to be important for the control of SBT3 activity. The β-hairpin is located in vicinity of the active site (Fig. S4C). Its basis is built from potentially flexible loop elements. In the SBT3 dimer, the β-hairpin is forced into a rigid position by contacting both PA-domains and the β-hairpin of the dimerization partner, and movement of the hairpin in the direction of the active site is strongly restricted. In the hypothetical monomer, the mobility of the hairpin and the PA domain is expected to be much higher. It is therefore conceivable that a conformational change of the flap-like β-hairpin may close the active site cavity, interfere with substrate binding, resulting in (partial) inactivation of the protease. PA domain-mediated homo-dimerization may thus be essential for enzyme activation.
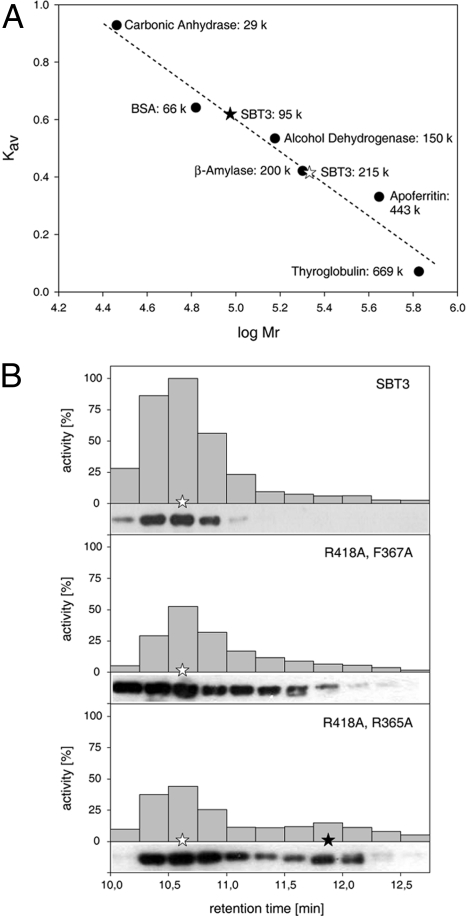
The relevance of critical residues at the dimer interface for homodimerization of SBT3, and the suggested correlation of dimerization status and proteolytic activity, were assessed by site-directed mutagenesis. SBT3 double mutants R418A/F367A and R418A/R365A were generated, transiently expressed in N. benthamiana, and extracted from the leaf apoplast. Compared with the SBT3 wild-type protein, which forms a dimer in solution (Fig. 4A), the R418A/F367A mutant showed a pronounced tailing toward lower molecular weights during size exclusion chromatography indicating a shift in monomer-dimer equilibrium toward the monomeric state (Fig. 4B). This mass shift was more pronounced for the R418A/R365A mutant, for which a second peak was observed at half the mass of the wild-type enzyme. Substitution of Arg-418 in combination with either R365 or F367 with Ala thus reduced the dimerization potential of SBT3, which correlated with a reduced proteolytic activity (Fig. 4 A and B).
Fig. 4.
Quarternary structure and activity of SBT3 and site-directed SBT3 mutants. (A) Analytical gelfiltration. Proteins were separated on a TSK3000 analytical gel filtration column. Wild-type SBT3 (open star) exhibited an apparent molecular mass of 215 k. Tailing toward lower molecular weights and an additional peak at 95 k (filled star) were observed for R418A/F367A and the R418A/R365A double mutants, suggesting a reduced dimerization potential of the mutant enzymes compared with the wild-type. (B) The SBT3 dimer and monomer represent active and inactive states of the enzyme. The activity of SBT3 and its site-directed mutants was assayed in 15-s fractions collected after gelfiltration. The highest activity was observed in fraction 3 for wild-type SBT3 and the activity in all other fractions is expressed in percentage thereof. Data were normalized for slightly different expression levels of the three proteins.
SBT3 Activity and Stability Are Ca2+-Independent.
Metal binding is one of the factors contributing to thermal stability of proteins and is common to many thermozymes (21). In subtilases, the binding of calcium ions is a major factor governing thermal stability. Calcium contributes its binding energy to the overall free energy of folding, stabilizing the native conformation of the enzyme (22).
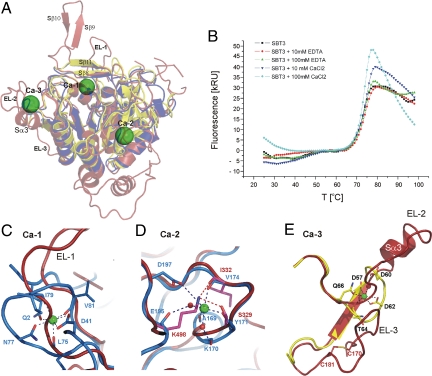
Despite the general calcium dependence of subtilases, we did not detect any calcium in the SBT3 structure. Thermofluor experiments were performed and melting curves recorded to confirm that the stability reported for SBT3 at elevated temperatures and alkaline pH (10) is truly calcium-independent (Fig. 5B). Without EDTA or calcium being added, the median (three independent experiments) melting point of SBT3 was 71.5 °C. The melting points did not differ significantly in presence of 100 mM CaCl2 (71.9 °C) or 100 mM EDTA (71.4 °C; Fig. 5B). Moreover, the activity of SBT3 was also found to be largely unaffected by the addition of CaCl2 or calcium chelators (Fig. S5), and SBT3 autolysis was undetectable over the tested pH range (pH 4.0–9.0) in presence of 5 mM EDTA or 5 mM CaCl2.
Fig. 5.
The structure of SBT3 lacks Ca2+. (A) Superimposed ribbon plots of SBT3 (red), subtilisin BPN′ (dark blue, PDB entry 1A2Q) and thermitase (yellow, PDB entry 1THM). Bound Ca2+ ions are shown as green spheres, the binding sites are labeled Ca-1 to Ca-3. The β-strands Cβ9 and Cβ10 that are part of the hairpin insertion in SBT3 are labeled, and extended loops EL-1 to EL-3. (B) SBT3 melting curves in presence and absence of CaCl2 or EDTA. (C) Ca-1, with residues implicated in Ca2+ coordination in subtilisin BPN′ shown as blue sticks. (D) Ca-2, with Ca2+-coordinating residues of BPN′ shown as blue sticks, and water molecules represented by red spheres. Lys-498 and its interacting residues in SBT3 are shown as magenta sticks. (E) Ca-3, with Ca2+-coordinating residues of thermitase shown as yellow sticks. A stabilizing disulfide bridge of the extended loop 3 of SBT3 is shown as red sticks.
These findings contrast with those for other eukaryotic subtilases and bacterial subtilisins. The activity and stability of kexin in yeast and mammalian PCs depend on calcium (2, 23). In subtilisin BPN′, the rate of thermal inactivation is 1000-fold higher in the absence of calcium, compared with saturating calcium concentrations (22). Similarly, the half-life at 70 °C of the thermophilic subtilisin from Bacillus AK.1 was increased by four orders of magnitude at 5 mM Ca2+ compared with the EDTA-treated enzyme (24).
SBT3 Stability in the Absence of Ca2+-Binding Sites.
Although the general architecture of Ca2+-binding regions is retained in SBT3, the apparent lack of Ca2+-binding sites and Ca2+-independent activity suggest that plants, in the course of evolution, found an alternative way to solve the stability problem in subtilases. In other subtilase structures, there are two highly conserved Ca2+-binding sites (Ca-1, Ca-2) and one less conserved site (Ca-3) (Fig. 5A) (25). Ca-1 is shared between subtilisins (e.g., BPN′, PDB entry 1A2Q), thermitases (e.g., PDB entry 1THM) and the eukaryotic subtilases furin (PDB entry 1P8J) and kexin (PDB entry 1R64). It is situated in a loop at the end of helix C, which harbors the active site His-64 in its first turn (26). This helix is perfectly conserved in SBT3 (helix Sα5), including the active site His-215 (Fig. 2A and Figs. S1 and S6). Deletion of the Ca-1-forming loop in BPN′ resulted in an active enzyme with, however, sharply decreased thermo stability (27). The corresponding loop is much larger in SBT3, comprising 19 (Gly-225-Gly-243) instead of 10 residues (Ala-74-Gly-83 in BPN′). Residues that coordinate Ca2+ in BPN′ are not conserved in SBT3, and the Cα traces differ significantly (Fig. 5C and Fig. S6). The stabilizing effect of Ca2+-binding at the Ca-1 site of BPN′ and other subtilases seems to be achieved in SBT3 by contacts of the extended loop (EL-1) to β-strands Cβ8 and Cβ11 and to the basis of the additional β-hairpin (strands Cβ9 and Cβ10) (Fig. 5A).
Ca-2 is a low affinity Ca2+-binding site that is highly conserved in subtilisins, thermitases, proteinase K (e.g., PDB entry 2PRK), and eukaryotic PCSK9 (PDB entry 2QTW). It is established in BPN′ by residues from a loop just before β-strand 6 (Fig. 5D and Fig. S6). Both loop and β-strand are nicely conserved in SBT3. The structural stabilization evoked in other subtilases by binding of a positively charged ion at this site is mimicked by the positively charged side chain of a lysine (Lys-498). This situation is reminiscent of pancreatic kallikreins that lack the stabilizing Ca2+-binding site located in a loop between β-strands 4 and 5 of the related trypsins. Rather than by cation binding, this part of the protein is stabilized in kallikreins by an arginine (Arg-70) substituting for glutamate [Glu-70 in trypsin (28)].
Ca-3 is localized in a loop between the second β-strand and the second α-helix (e.g., in thermitase, PDB entry 1THM) (Fig. 5E). Again, these two secondary structure elements are conserved in SBT3 but the connecting loop (EL-2) is considerably larger, introducing an additional short helix (Sα3) (Fig. 5E). Another extended loop in close proximity (EL-3) harbors an internal disulfide bridge (Cys-170-Cys-181) that may contribute to the stability of this region in the absence of a Ca2+ binding site (Fig. 5E).
In addition to the substitution of the Ca2+ binding sites, further stabilization in SBT3 seems to be afforded by the C-terminal Fn III domain, which consists of a sandwich built by a three-strand and a four-strand anti-parallel β-sheet that is curved to assume a β-barrel-like shape (Fig. 2C). The closest structural neighbor is a single-domain sperm-specific protein (SSP-19) from C. elegans [PDB entry 1ROW (29)], which aligns over 82 Cαs with an RMSD of 1.8 Å (DALI Z score: 9.2). Structurally related Fn III domains are also found in numerous other proteases including C5α peptidase [PDB entry 3EIF (14)], human factor XIII [PDB entry 1GGT (30)], alkaline serine protease KP-43 from Bacillus sp. [PDB entry 1WMD (31)], and mammalian PCs, where it was proposed to stabilize the highly negatively charged active site (23, 32).
In SBT3, the interface of the Fn III domain and the subtilisin domain is largely hydrophobic. The Fn III domain may thus stabilize the protein by shielding hydrophobic surface patches from solvent. Interdomain contacts are established between Fβ1 and parts of the large Pβ9 - Sβ7 loop, between Fβ2 and the Sα11–Sα12 loop, and by interactions of Fβ8 with Sα9 and Sα10. Furthermore, the five C-terminal amino acids fold back onto the subtilisin domain, and the side chain of the ultimate residue, Trp-761, inserts into a hydrophobic pocket in the vicinity of the substrate binding channel (Fig. S7). This interaction appears to stabilize the loop system near the active site. Further stabilization is achieved by a “hydrophobic clamp” formed by Trp-761, Val-760, Ile-758, and Ile-157, which braces the Fn III and subtilase domains (Fig. S7). Consistent with the proposed stabilizing function, the Fn III domain and the “hydrophobic clamp” were shown to be required for SBT3 activity. SBT3 deletion mutants lacking the entire Fn III domain or as little as 5 aa from its C terminus were impaired in autocatalytic processing activity and accumulated intracellularly as unprocessed zymogens (10). Dimerization, which has not been observed for any other subtilase, is likely to be another factor contributing to SBT3 stability.
Subtilisins as Detergent Additives.
The instability of subtilisins under chelating (water-softening) conditions is a major impediment to their industrial use as additives in laundry and dishwashing detergents. Following the introduction of subtilisin Carlsberg from Bacillus licheniformis as the first detergent additive in the 1960s, considerable efforts have been undertaken to improve the performance of subtilisins under chelating conditions. Gallagher and coworkers deleted the entire loop harboring the high-affinity Ca-1 site of subtilisin BPN′ (27). The folding stability of the resulting calcium-independent mutant was subsequently enhanced by directed evolution (33). Thermal inactivation of this subtilisin variant in 10 mM EDTA was 1,000 times slower compared with the wild-type enzyme (33). Directed evolution was also successfully used to develop a thermostable variant of subtilisin E. Eight amino acid substitutions were found to be sufficient to convert the mesophilic enzyme into a functional equivalent of thermitase (34).
There are limitations to this approach, however, because the incremental improvement of a given protein by the successive accumulation of single mutations will inevitably result in solutions that are close to the starting structure, and optimal solutions may not be accessible. The identification of SBT3, which evolved as a calcium-free but stable subtilase in plants, may open new avenues for the generation of calcium-independent subtilisins. As evident from the SBT3 structure, calcium-binding is not the only approach nature has taken to solve the stability problem in subtilases. The SBT3 structure may thus serve as a starting point to guide further efforts in the improvement of subtilisins for industrial applications.
Materials and Methods
Purification and Crystallization of SBT3.
Native SBT3 was expressed in a transgenic tomato cell suspension culture and purified to homogeneity from culture supernatants (15). SBT3 crystals were grown at 4 °C by the hanging drop vapor diffusion method (15). Ac-Phe-Glu-Lys-Ala-cmk was synthesized by a combination of solid and solution phase chemistry according to published procedures (35). For complexation with SBT3, native crystals were soaked in reservoir solution supplemented with 10 mM inhibitor, transferred to 10 mM inhibitor in cryoprotective solution, and flash-cooled in liquid nitrogen.
Structure Determination.
Crystallization, data collection, and heavy atom search have been described (15). The initial phases as determined with SOLVE and RESOLVE (36) were barely good enough to trace the backbone of the protein but could be used to extract noncrystallographic symmetry (NCS). The NCS data were fed to RESOLVE, which resulted in electron density maps that allowed tracing of the protein. The model was iteratively built and refined with COOT (37) and REFMAC (38).
Site-Directed Mutagenesis and Transient Expression in N. benthamiana.
The R365A, F367A, and R418A single mutants were generated by site-directed mutagenesis of the SBT3 cDNA using complementary oligonucleotides comprising the desired mutations as primers in PCR. Reaction products were digested with DpnI (Fermentas) and transformed into E. coli XL1 blue (Stratagene). A single XhoI restriction site was used to recombine the mutations in position 418 and positions 357/367 to generate the R418A/R365A and R418A/F367A double mutants. The mutated cDNAs were cloned into the vector pART27, transformed into Agrobacterium tumefaciens, and infiltrated into Nicotiana benthamiana leaves as described in ref. 10. Five days after agroinfiltration, apoplastic washes were prepared to recover transiently expressed proteins and reduced to 1/20 volume by ultrafiltration (10).
Size-Exclusion Chromatography and Assay of SBT3 Activity.
Analytical gelfiltration of SBT3 wild-type and mutant proteins was performed on a TSK-GEL G3000SWXL column (Tosoh Bioscience) in 50 mM Tris·HCl pH 7.4, 200 mM NaCl at a flow rate of 0.8 mL/min. Standard proteins for column calibration were from Sigma–Aldrich. Collected fractions (15 s, 200 μL) were analyzed on Western blots for the presence of the SBT3 protein (10). A fluorigenic peptide substrate (Abz-SKRDPPKKQTD(NO2)Y, 20 μM; JPT Peptide Technologies) was used to assay SBT3 activity in a total volume of 200 μL of 50 mM Tris·HCl pH 7.5.
Thermofluor Analysis.
Samples were set up in a 96-well thin-wall PCR plate (Applied Biosystems) in a total volume of 25 μL per well. SBT3 was used in a final concentration of 1 μM in 20 mM phosphate buffer pH 7.5, 150 mM NaCl and CaCl2 or EDTA as indicated. 1 μL of a 25× Sypro Orange solution (Invitrogen) was added and the protein was heated from 25 °C to 95 °C in 1 °C steps of 1 min each in a 7500 Fast RT-PCR system (Applied Biosystems). The VIC filter was used to monitor the fluorescence increase resulting from binding of Sypro orange to hydrophobic regions of the unfolding protein.
Supplementary Material
Acknowledgments.
We thank Brigitte Rösingh and Erika Wenzelburger for excellent technical assistance; the staff of the Swiss Light Source (SLS), Villigen, Switzerland for technical support; and Ilme Schlichting, Wulf Blankenfeldt and Eckhard Hofmann for data collection. This work was supported by German Research Foundation Grant SCHA591/2 (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3IS6 (apo SBT3) and 3I74 (cmk-inhibited enzyme)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0907587106/DCSupplemental.
References
- 1.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 2.Fuller RS, Brake A, Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci USA. 1989;86:1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidah NG, Khatib AM, Prat A. The proprotein convertases and their implication in sterol and/or lipid metabolism. Biol Chem. 2006;387:871–877. doi: 10.1515/BC.2006.110. [DOI] [PubMed] [Google Scholar]
- 4.Rautengarten C, et al. Inferring hypotheses on functional relationships of genes: Analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol. 2005;1:e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Book AJ, Yang PZ, Scalf M, Smith LM, Vierstra RD. Tripeptidyl peptidase II. An oligomeric protease complex from Arabidopsis. Plant Physiol. 2005;138:1046–1057. doi: 10.1104/pp.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- 7.Vera P, Conejero V. Pathogenesis-related proteins of tomato. P-69 as an alkaline endoproteinase. Plant Physiol. 1988;87:58–63. doi: 10.1104/pp.87.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J-X, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19:4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava R, Liu JX, Howell SH. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 2008;56:219–227. doi: 10.1111/j.1365-313X.2008.03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cedzich A, et al. The protease-associated (PA) domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3) J Biol Chem. 2009;284:14068–14078. doi: 10.1074/jbc.M900370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahon P, Bateman A. The PA domain: A protease-associated domain. Protein Sci. 2000;9:1930–1934. doi: 10.1110/ps.9.10.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis MI, Bennett MJ, Thomas LM, Bjorkman PJ. Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase. Proc Natl Acad Sci USA. 2005;102:5981–5986. doi: 10.1073/pnas.0502101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence CM, et al. Crystal structure of the ectodomain of human transferrin receptor. Science. 1999;286:779–782. doi: 10.1126/science.286.5440.779. [DOI] [PubMed] [Google Scholar]
- 14.Kagawa TF, et al. Model for substrate interactions in C5α peptidase from Streptococcus pyogenes: A 1.9 Å crystal structure of the active form of ScpA. J Mol Biol. 2009;386:754–772. doi: 10.1016/j.jmb.2008.12.074. [DOI] [PubMed] [Google Scholar]
- 15.Rose R, et al. Purification, crystallization and preliminary X-ray diffraction analysis of a plant subtilase. Acta Crystallogr F. 2009;65:522–525. doi: 10.1107/S1744309109013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheatley JL, Holyoak T. Differential P1 arginine and lysine recognition in the prototypical proprotein convertase Kex2. Proc Natl Acad Sci USA. 2007;104:6626–6631. doi: 10.1073/pnas.0701983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishizawa M, Yasuhara T, Nakai T, Fujiki Y, Ohashi A. Molecular cloning of the aminopeptidase Y gene of Saccharomyces cerevisiae. Sequence analysis and gene disruption of a new aminopeptidase. J Biol Chem. 1994;269:13651–13655. [PubMed] [Google Scholar]
- 18.Krawitz P, et al. Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J Biol Chem. 2005;280:39515–39523. doi: 10.1074/jbc.M501645200. [DOI] [PubMed] [Google Scholar]
- 19.Cao X, Rogers SW, Butler J, Beevers L, Rogers JC. Structural requirements for ligand binding by a probable plant vacuolar sorting receptor. Plant Cell. 2000;12:493–506. doi: 10.1105/tpc.12.4.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park M, Lee D, Lee GJ, Hwang I. AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J Cell Biol. 2005;170:757–767. doi: 10.1083/jcb.200504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li WF, Zhou XX, Lu P. Structural features of thermozymes. Biotechnol Adv. 2005;23:271–281. doi: 10.1016/j.biotechadv.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Alexander PA, Ruan B, Bryan PN. Cation-dependent stability of subtilisin. Biochemistry. 2001;40:10634–10639. doi: 10.1021/bi010797m. [DOI] [PubMed] [Google Scholar]
- 23.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: Explanation of their specificity. J Mol Biol. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 24.Toogood HS, Smith CA, Baker EN, Daniel RM. Purification and characterization of Ak. 1 protease, a thermostable subtilisin with a disulphide bond in the substrate-binding cleft. Biochem J 350 Pt. 2000;1:321–328. [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CA, Toogood HS, Baker HM, Daniel RM, Baker EN. Calcium-mediated thermostability in the subtilisin superfamily: The crystal structure of Bacillus Ak. 1 protease at 1.8 A resolution. J Mol Biol. 1999;294:1027–1040. doi: 10.1006/jmbi.1999.3291. [DOI] [PubMed] [Google Scholar]
- 26.Pantoliano MW, et al. Large increases in general stability for subtilisin BPN′ through incremental changes in the free energy of unfolding. Biochemistry. 1989;28:7205–7213. doi: 10.1021/bi00444a012. [DOI] [PubMed] [Google Scholar]
- 27.Gallagher T, Bryan P, Gilliland GL. Calcium-independent subtilisin by design. Proteins. 1993;16:205–213. doi: 10.1002/prot.340160207. [DOI] [PubMed] [Google Scholar]
- 28.Tschesche H, Mair G, Godec G. The primary structure of porcine glandular kallikreins. Adv Exp Med Biol. 1979;120A:245–260. doi: 10.1007/978-1-4757-0926-1_25. [DOI] [PubMed] [Google Scholar]
- 29.Schormann N, Symersky J, Luo M. Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans. Acta Crystallogr D. 2004;60:1840–1845. doi: 10.1107/S0907444904017846. [DOI] [PubMed] [Google Scholar]
- 30.Yee VC, et al. Three-dimensional structure of a transglutaminase: Human blood coagulation factor XIII. Proc Natl Acad Sci USA. 1994;91:7296–7300. doi: 10.1073/pnas.91.15.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonaka T, et al. The crystal structure of an oxidatively stable subtilisin-like alkaline serine protease, KP-43, with a C-terminal beta-barrel domain. J Biol Chem. 2004;279:47344–47351. doi: 10.1074/jbc.M409089200. [DOI] [PubMed] [Google Scholar]
- 32.Henrich S, et al. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10:520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 33.Strausberg SL, et al. Directed evolution of a subtilisin with calcium-independent stability. Nat Biotech. 1995;13:669–673. doi: 10.1038/nbt0795-669. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H, Arnold FH. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 1999;12:47–53. doi: 10.1093/protein/12.1.47. [DOI] [PubMed] [Google Scholar]
- 35.Hauske P, et al. Selectivity profiling of DegP substrates and inhibitors. Bioorg Med Chem. 2009;17:2920–2924. doi: 10.1016/j.bmc.2009.01.073. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.