SUMMARY
The ESCRT-II-ESCRT-III interaction coordinates the sorting of ubiquitinated cargo with the budding and scission of intralumenal vesicles into multivesicular bodies. The interacting regions of these complexes were mapped to the second winged-helix domain of human ESCRT-II subunit VPS25 and the first helix of ESCRT-III subunit VPS20. The crystal structure of this complex was determined at 2.0 Å resolution. Residues involved in structural interactions explain the specificity of ESCRT-II for Vps20, and are critical for cargo sorting in vivo. ESCRT-II directly activates ESCRT-III driven vesicle budding and scission in vitro via these structural interactions. VPS20 and ESCRT-II bind membranes with nanomolar affinity, explaining why binding to ESCRT-II is dispensable for the recruitment of Vps20 to membranes. Docking of the ESCRT-II -VPS202 supercomplex reveals a convex membrane-binding surface, suggesting a hypothesis for negative membrane curvature induction in the nascent intralumenal vesicle.
Newly internalized receptors and transporters, lysosomal hydrolases, and other cargo destined for the lysosome arrive through a pathway in which portions of the limiting membrane of endosomes invaginate into the lumen of the endosome (Gruenberg and Stenmark, 2004; Piper and Katzmann, 2007; Russell et al., 2006). Endosomes filled with intralumenal vesicles (ILVs) are referred to as multivesicular bodies (MVBs). The major pathway for the entry of ubiquitinated transmembrane proteins into MVBs is catalyzed by the Endosomal Sorting Complex Required for Transport (ESCRT) machinery (Hurley, 2008; Raiborg and Stenmark, 2009; Saksena et al., 2007; Williams and Urbe, 2007). There are five ESCRT complexes, ESCRT-0, -I, -II, and –III, and the Vps4-Vta1 complex. The system is conserved from yeast to humans (throughout this report, all capitals (non-italicized) “VPS4” denotes human proteins while mixed case “Vps4” denotes either yeast proteins or all orthologs collectively). In human cells it has additional roles in cytokinesis and HIV-1 budding (Carlton and Martin-Serrano, 2009). ESCRT-0, -I, and –II contain ubiquitin-binding domains and are currently thought to cluster ubiquitinated cargo in a membrane patch preparatory to its invagination. ESCRT-III is responsible for the scission of the nascent vesicle into the lumen (Wollert et al., 2009). Vps4-Vta1 functions to disassemble and recycle the ESCRT-III complex following vesicle scission (Babst et al., 1998; Wollert et al., 2009).
The ESCRT system can be conceptually divided into machinery that binds to cargo (ESCRT-0, -I, and –II); machinery that remodels the membrane (ESCRT-III), and recycling machinery (Vps4-Vta1). The pivotal links between these sets of machines are at the ESCRT-II-ESCRT-III and ESCRT-III-Vps4-Vta1 stages. The structural basis for ESCRT-III-Vps4-Vta1 interactions have been characterized (Kieffer et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007 ; Xiao et al., 2008), while the key ESCRT-II-ESCRT-III interaction has not.
ESCRT-II in yeast and humans consists of one copy each of the Vps22 and Vps36 subunits, and two copies of the Vps25 subunit. The subunits are arranged in the shape of the letter “Y” with Vps22 and Vps36 forming one branch of the “Y” and the two copies of Vps25 forming the other branch (Hierro et al., 2004; Im and Hurley, 2008; Teo et al., 2004). The presence of both copies of Vps25 is essential for function in yeast (Hierro et al., 2004). ESCRT-III was first described in yeast, where it consists of the Vps20, Snf7, Vps24, and Vps2 subunits (Babst et al., 2002a). The ESCRT-III subunits assemble from cytosolic monomers into a detergent-insoluble membrane-bound array in the order listed above (Teis et al., 2008). Vps20 is the first subunit and thus is the key initiator of ESCRT-III assembly on endosomes (Teis et al., 2008).
ESCRT-II physically interacts with Vps20 in yeast (Babst et al., 2002b; Bowers et al., 2004) and humans (where it is also known as CHMP6) (Martin-Serrano et al., 2003; von Schwedler et al., 2003; Yorikawa et al., 2005). Vps20 binds to ESCRT-II via the Vps25 subunit of the latter (Teo et al., 2004). Both Vps20 and ESCRT-II interact strongly with membranes. Vps20 is myristoylated at its N-terminus in both yeast and human cells (Babst et al., 2002a; Yorikawa et al., 2005). ESCRT-II binds tightly to membranes due to a phosphoinositide-specific interaction with the GLUE domain in its VPS36 (also known as EAP45) subunit (Slagsvold et al., 2005; Teo et al., 2006), and a basic N-terminal helix in its VPS22 (also known as EAP30) subunit (Im and Hurley, 2008). Progress in understanding how ESCRT-II binds to and activates Vps20 has been hampered because at concentrations used for structural studies, ESCRT-II and Vps20 form an insoluble precipitate (Teo et al., 2004).
In this study, we set out to circumvent this problem and thereby fill in the structural missing link between the ESCRT-II and ESCRT-III complexes. We were able to determine the crystal structure of a complex of the minimal interacting fragments of VPS20 and VPS25. The affinity of the ESCRT-II-VPS20 interaction was characterized in solution. We applied structural insights to design alleles of ESCRT-II and Vps20 selectively disabled for their interaction with one another, and characterized the activation of MVB biogenesis by ESCRT-II in yeast cells. We show that ESCRT-II and Vps20 bind to membranes with nanomolar to tens of nanomolar affinity. The activation of ESCRT-III by ESCRT-II was directly demonstrated using a recently developed assay for ESCRT function in vitro, and shown to depend on the structural interactions. Finally, docking of the VPS20-VPS25 crystal structure and previously solved structures was used to build a structural model of the ESCRT-II-VPS202 supercomplex, revealing a convex curved membrane-binding face with implications for the mechanism of MVB biogenesis.
RESULTS
Structural basis of the ESCRT-II-ESCRT-III interaction
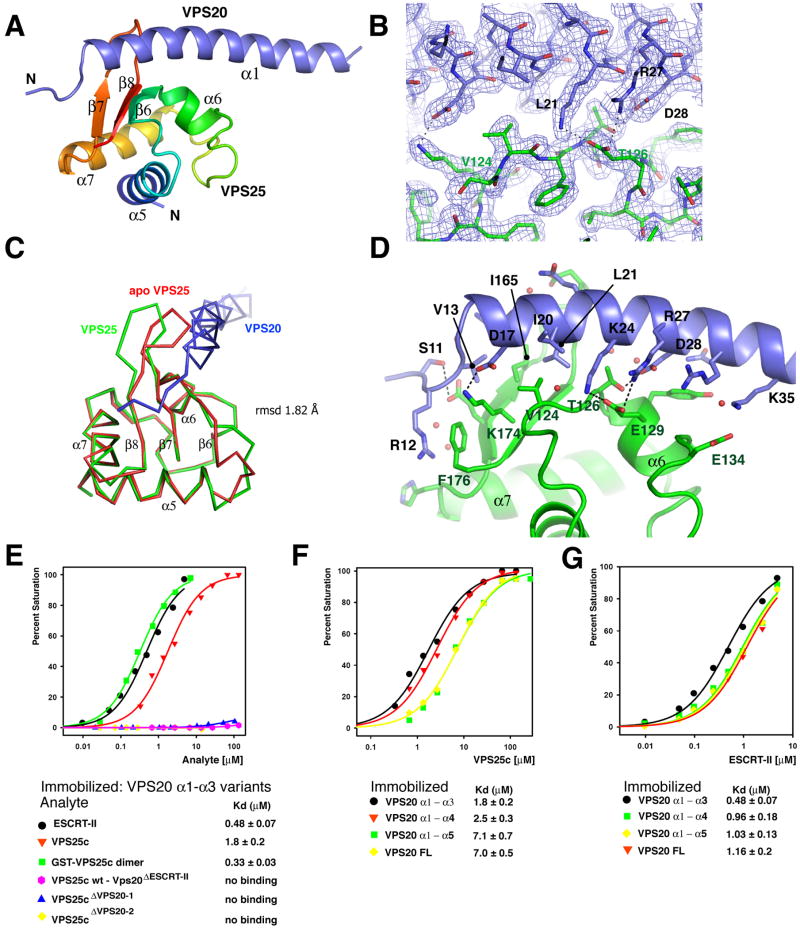
The N-terminal half of VPS20 binds to the C-terminal winged helix (WH2) domain of VPS25 (also known as EAP20) (Langelier et al., 2006), which is referred to hereafter as VPS25c. We screened a series of constructs to precisely map the smallest fragment of VPS20 competent to bind to VPS25. We found that residues 11 – 48 in helix α1 of VPS20 were necessary and sufficient for association with VPS25 (Figure S1). These fragments were co-expressed in order to overcome difficulties with the insolubility of the VPS20 fragment when expressed alone. The VPS25c (residues 102-176) – VPS20 α1 (residue 11-48) complex was crystallized and diffracted to 2.0 Å. The structure was solved by molecular replacement using the structure of VPS25 (Im and Hurley, 2008) (Figure 1A, B; Table 1). The β7 – β8 loop of VPS25 opens up relative to the uncomplexed structure to accommodate the α1 helix of VPS20, with an overall r.m.s.d. between the VPS25c structures of 1.8 Å (Figure 1C).
Figure 1. The VPS20-VPS25 complex.
(A) Overall structure of the complex. The WH2 domain of VPS25 is colored in a blue to red gradient from the N- to C-terminal direction. Secondary structures are numbered as they occur in intact VPS25. (B) Electron density from a final 2FO-FC map contoured at 1.0 σ in the vicinity of VPS25 – VPS20 binding site. The final refined structure was shown in a ball and stick model. (C) Superposition of apo VPS25 and VPS25 – VPS20 complex structures. (D) Interactions between VPS25 and VPS20. VPS25 is colored green and VPS20 blue. The side chains in the interface residues are shown in a ball and stick model. Selected hydrogen bonds and salt bridges are shown in dashed lines. (E) Binding isotherms for ESCRT-II, VPS25c, and GST-VPS25c binding to wild-type VPS20 α1 – α3 (residues 1 – 118) and, where indicated, VPS20ΔESCRT-II α1 – α3. (F) Binding of VPS25c to truncated VPS20 constructs (VPS20 α1 – α4, residue 1 – 145, VPS20 α1 – α5, residue 1 – 167). (G) Binding of full length ESCRT-II to the indicated VPS20 constructs. Binding isotherms from the equilibrium phases were fitted to a 1:1 binding model to determine the dissociation constants and statistical fitting errors shown beneath panels (E-G). All binding experiments were carried out twice with similar results.
Table 1.
Statistics of data collection and crystallographic refinement
| Crystal | Native |
|---|---|
| Constructs | VPS25 (102-176) VPS20 (11-48) |
| Space group, unit cell | P21, a = 58.6 Å, b = 51.0 Å, c = 77.0 Å, β = 90.4 |
| X-ray source | APS 22-ID |
| Wavelength (Å) | 1.0000 |
| Resolution (Å) (last shell) | 2.0 (2.07 – 2.00) |
| No. of unique reflections | 30553 |
| I/σ(I)σ (last shell) | 29.1 (3.0) |
| Rsym a (%) | 5.9 (28.1) |
| Data completeness (%) | 98.4 (87.6) |
| Refinement | |
| R factor b (%) | 23.6 (22.4) |
| Free R factor c (%) | 27.7 (34.8) |
| R.m.s. bond length (Å) | 0.012 |
| R.m.s. bond angle (°) | 1.326 |
| Average B value (Å2) d | 38.8 |
| Number of atoms | protein 3661, water 213 |
The values in parentheses relate to highest resolution shells.
Rsym = Σ h Σ i |Ii(h) -<I>| / Σ h Σ i Ii (h), where I is the observed intensity and <I> is the average intensity of multiple observations of symmetry-related reflections.
R = Σ||Fo|-k|Fc|| /Σ|Fo|, where Fo and Fc are observed and calculated structure factor amplitudes, respectively.
Rfree is calculated for a randomly chosen 5% of reflections; the R factor is calculated for the remaining 95% of reflections used for structure refinement.
Average B value of all atoms in an asymmetric unit.
The region of VPS20 consisting of residues 11 to 35 makes extensive interactions with VPS25c (Figure 1A). The N-terminus of the crystallized portion of VPS20 (residues 11 -14) is a random coil, while residues 15 - 44 form an α-helix (Figure 1A). VPS25c has a hydrophobic surface patch centered on Val-124 on the β-sheet formed by β6, β7 and β8 (Figure 1D). This β-sheet forms the main binding surface for VPS20. The β7- β8 loop of VPS25 wraps partway around the VPS20 α1 helix (Figure 1A). Polar interactions are extensive, with a total of 8 hydrogen bonds (< 3 Å between heteroatoms) and 3 salt bridges. VPS25 Val-124 makes the most extensive hydrophobic interaction with VPS20 (Figure 1D). The side-chain of VPS20 Leu-21 inserts into a hydrophobic pocket formed by Val-124, Ile-165 and the β7 – β8 loop of VPS25. Among key polar interactions, VPS25 Thr-126 makes a hydrogen bond with VPS20 Asp-28, and VPS25 Glu-129 makes salt bridges with Lys-24 and Arg-27 of VPS20. VPS20 binding buries 713 Å2 of VPS25c surface, which amounts to 14.5% of the total VPS25c surface.
ESCRT-II binds to VPS20 with micromolar affinity
Biosensor binding experiments were performed with purified recombinant ESCRT-II, VPS25, and VPS20 proteins. In order to validate the crystallographic interactions, residues observed to interact structurally were mutated. VPS25 mutants V124E and T126K (hereafter referred to as VPS25 ΔVPS20-1 and VPS25 ΔVPS20-2) completely abolished binding to VPS20. The triple mutation in VPS20 (L21R, R27A, D28A; hereafter referred to as VPS20 ΔESCRT-II) also completely abolished the VPS25 - VPS20 interaction (Figure 1E). These data confirm that the VPS25-VPS20 interaction in solution depends on the residues identified in the crystal structure.
In order to probe the role of avidity in the ESCRT-II:VPS20 interaction, the binding to monomeric VPS25c, full length ESCRT-II containing two VPS25 subunits, or an artificially dimerized GST-VPS25c construct was compared (Figure 1E). Full length ESCRT-II showed higher affinities than VPS25c for all VPS20 constructs. Full length ESCRT-II or GST-VPS25c dimer bound more tightly to VPS20 α1 – α3 with Kd values of 0.48 and 0.33 μM respectively (Figure 1E).
In order to determine if C-terminal elements of VPS20 autoinhibited binding, as has been observed for other ESCRT-III interactors, a series of VPS20 constructs of varying length were tested (Figure 1F). VPS25 bound to full length VPS20 with a dissociation constant of 7.0 ± 0.5 μM. VPS25 bound somewhat more tightly to VPS20 constructs lacking the C-terminal α5 helix. VPS25 bound to VPS20 α1 – α3 and VPS20 α1 – α4 with similar affinity, with Kd values of 1.8 ± 0.2 and 2.5 ± 0.3 μM respectively. In the context of the isolated VPS25c domain, the presence of C-terminal sequences after α5 in VPS20 slightly reduce the VPS25 binding affinity. Full length ESCRT-II manifested less difference than VPS25c in its affinities for different VPS20 constructs (Figure 1G), which did not vary by more than a factor of two. This suggests that in the functionally relevant context of the full ESCRT-II complex, autoinhibition by the C-terminus of VPS20 is not playing a major regulatory role.
Determinants for selective recognition of VPS20 by ESCRT-II
All ESCRT-III proteins have a common secondary structure organization with two N-terminal basic α–helices and three to four C-terminal acidic α-helices. The sequences of the α1 helices of Vps20 orthologs are well conserved, particularly in the residues that bind to Vps25 (Figure S2A). These include Leu-21, Arg-27, and Asp-28, which are required for binding in vitro. These residues are identical in animals. In yeast, the only variation is the conservative replacement of Arg-27 by Lys. Key residues on the β sheet of VPS25 involved in VPS20 binding are strictly conserved from yeast to humans (Figure S2B). These residues include Val-124 and Thr-126, essential for binding in vitro. Despite the common overall folding of all ESCRT-III proteins, the ten other known human ESCRT-III proteins have a much lower level of sequence identity in the α1 helix (Figure S2C). VPS25 binding involves 13 residues of VPS20, yet in other ESCRT-III proteins, no more than four of these residues are identically conserved. Leu-21 is conserved only in VPS24. Arg-27 is conservatively replaced by Lys in DID2, but is not conserved in any other ESCRT-III protein. Asp-28 is conservatively replaced by Glu in VPS60 and the N-terminal half of CHMP7, and otherwise not conserved. Thus even among the three residues analyzed mutationally, no other ESCRT-III protein sequence conserves all three positions at once. This sequence analysis indicates that the different species shares a common mode of ESCRT-II -III interaction and explain the selectivity of ESCRT-II for VPS20 over all other ESCRT-III proteins.
ESCRT-II and VPS20 interaction site is required for cargo sorting
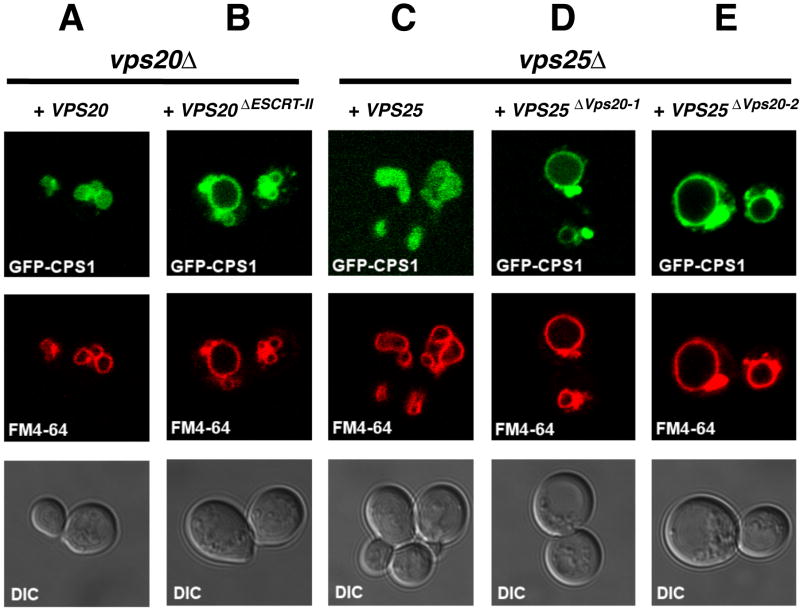
The epitopes used by VPS20 and VPS25 to bind each other are highly conserved, leading us to expect a common function from yeast to animals. Cps1 is a vacuolar hydrolase that is ubiquitinated and sorted to the lumen of the vacuole by the ESCRT pathway. Disruption of the ESCRT pathway can be diagnosed by the mislocalization of Cps1 to the limiting membrane of the vacuole and to the class E compartment. The latter appears by light microscopy as a punctate structure adjoining the vacuole. The localization of the ESCRT substrate Cps1 was assayed in yeast expressing wild type and mutant alleles of VPS25 and VPS20. Deletion of either VPS20 or VPS25 results in the mislocalization of Cps1 (Babst et al., 2002a; Babst et al., 2002b). The wild-type phenotype was completely rescued by expression of a single-copy plasmid bearing wild-type VPS20 or VPS25 (Figure 2A, C), consistent with previous findings (Babst et al., 2002a; Babst et al., 2002b). The triply mutated allele VPS20L18R/K24A/D25A (hereafter referred to as VPS20ΔESCRT-II), in which mutations were engineered in the conserved interaction residues (Figure S2A), was designed to cripple the interaction with ESCRT-II. Expression of VPS20ΔESCRT-II led to the mislocalization of GFP-Cps1 to the class E compartment, and a complete absence of GFP-Cps1 from the vacuolar lumen (Figure 2B); in other words a strong class E phenotype. The mutant alleles VPS25V148E and VPS25T150K (referred to as VPS25ΔVps20-1 and VPS25ΔVps20-2) were designed to cripple the interaction with Vps20, based on the equivalence of yeast Vps25 Val-148 and Thr-150 to Val-124 and Thr-126 in human VPS25 (Figure S2B). Expression of these two alleles resulted in strong class E phenotypes (Figure 2D, E). This indicates that the interaction of ESCRT-II with helix α1 of Vps20 is essential for the cargo-sorting function of ESCRTs.
Figure 2. Cargo is mislocalized in yeast expressing VPS20ΔESCRT-II and VPS25ΔVps20 alleles.
(A-E) The uppermost panel of each column shows the sorting of the GFP-Cps1 construct (green) in various strains, as indicated at the top of each column. The middle panels show the limiting membrane of the vacuole as labeled by FM4-64 (red), and the lower panels show the DIC image. Results presented here are characteristic of observations of >100 cells for each strain shown.
ESCRT-II binding is not required for membrane recruitment of Vps20 in yeast
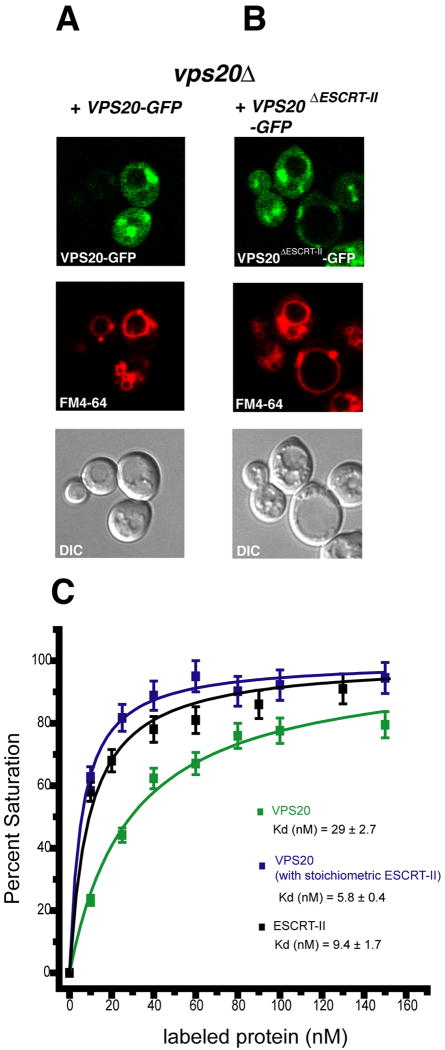
The use of GFP-fusions has been established as an approach to monitor ESCRT-III membrane localization in yeast. These constructs have a dominant-negative phenotype, resulting in the stabilization of their membrane binding on the class E membrane compartment (Figure 3A, B). Vps20-GFP is predominantly localized to the punctate class E structures (Figure 3A), as previously reported (Teis et al., 2008). When visualized with high sensitivity, a diffuse background of cytosolic Vps20-GFP was also observed (Figure 3A). Localization of Vps20ΔESCRT-II-GFP to both punctate structures and cytosol is essentially identical to wild-type (Figure 3B), indicating that abrogating ESCRT-II binding does not affect the localization of Vps20. These findings were confirmed by Western blotting of the cytosol and membrane fractions, indicating that the majority of both wild-type and mutant Vps20-GFP was in the pellet fraction (data not shown).
Figure 3. ESCRT-II is not required for Vps20 membrane localization in yeast.
(A-B) The designated VPS20-GFP alleles were transformed into the indicated yeast strain and their localization imaged by fluorescence microscopy as described in the experimental procedures. (C) Binding of VPS20 and human ESCRT-II to PC:PE:Cholesterol:PI(3)P liposomes monitored by FRET. Error ranges reported in the Kd values were derived from the errors in the non-linear best fit. Error bars in the data points shown in (C) were derived from the propagation of the estimated error in the measurement of ΔImax. All experiments were carried out twice.
VPS20 and ESCRT-II bind strongly to membranes in vitro
In order to probe the basis for the ESCRT-II-independent endosomal localization of Vps20, the membrane binding was assessed in vitro by monitoring FRET between rhodamine-labeled proteins and fluorescein-labeled liposomes. Recombinant unmyristoylated VPS20 was labeled on a unique engineered Cys residue at residue 119, which is outside of the ESCRT-II binding site and the membrane binding basic face. VPS20 bound with Kd = 29 nM to liposomes containing 3 mol % PI(3)P in an endosome-like background of PC, PS, and cholesterol (Figure 3C). Human ESCRT-II labeled on native Cys residues bound to the same lipid mixture with Kd = 9 nM (Figure 3C). The affinity of the ESCRT-II-VPS20 supercomplex for membranes was monitored using labeled VPS20 and unlabeled ESCRT-II. ESCRT-II was preincubated with liposomes at each step in the titration in order to avoid difficulties with the insolubility of the supercomplex. In the context of the supercomplex, VPS20 bound to membranes with Kd = 6 nM (Figure 3C). These results show synergism in the membrane binding of ESCRT-II and VPS20, consistent with the interaction of the proteins with each other. The binding of each protein or complex to liposomes by themselves, however, is very tight, explaining their independent targeting to endosomes in vivo.
ESCRT-II accelerates ESCRT-III-dependent ILV budding in vitro
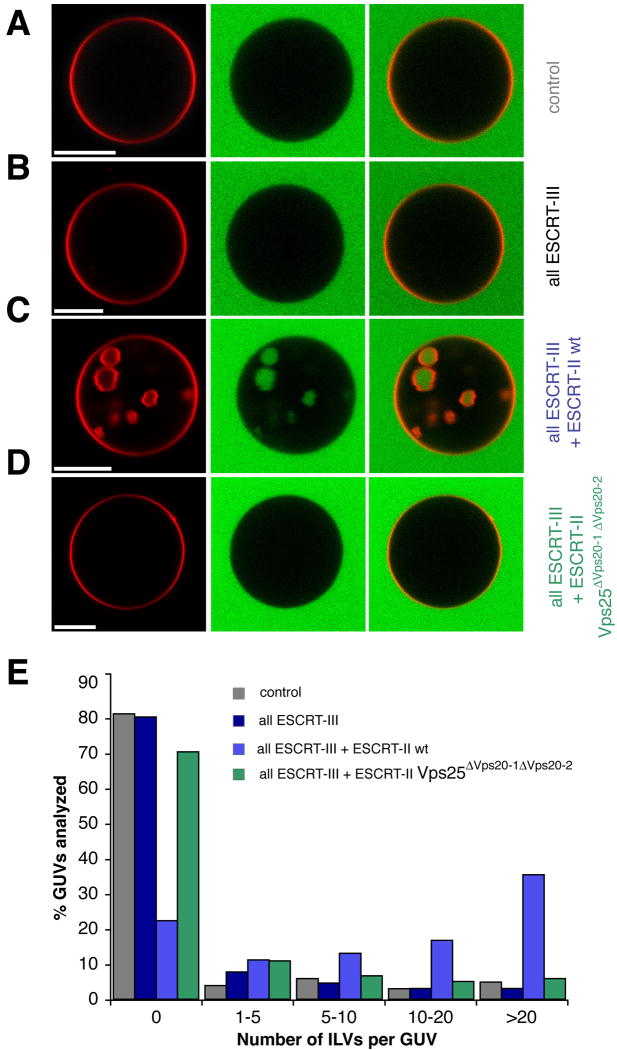
Purified yeast ESCRT-III subunits are capable of supporting budding and scission of ILVs into giant unilamellar vesicles (GUVs) (Wollert et al., 2009), providing that superphysiological protein concentrations and a partially activated C-terminal deletion mutant of Vps20 are used. When full-length versions of all of the ESCRT-III subunits are used at concentrations closer to physiological levels (40:200:40:40 nM Vps20:Snf7:Vps24:Vps2), no ILVs are observed above background (Figure 4A, B). Addition of 40 nM ESCRT-II to this mixture led to the production of high levels of ILVs (Figure 4C), while addition of 40 nM ESCRT-II alone produced no ILVs (not shown). Addition of the doubly mutated ESCRT-II-Vps25ΔVps20-1 ΔVps20-2 led to essentially no ILV production above background (Figure 4D). This demonstrates that ESCRT-II potently and directly activates ESCRT-III for membrane budding and scission in the absence of cargo or other proteins, and that it does so in a manner that depends on the structural interaction between Vps25 and Vps20.
Figure 4. ESCRT-II enhances ESCRT-III driven vesicle scission.
GUVs (red) were incubated with buffer (A), ESCRT-III (B), ESCRT-II and ESCRT-III (C), or ESCRT-II Vps25 ΔVps20-1 ΔVps20-2 and ESCRT-III (D). ILV formation was observed in the presence of GFP (green) as a marker for the bulk phase. ESCRT-II increases the ILV formation in GUVs dramatically (C). (E) Summary of the experiment (n = 100). Each color corresponds to a single experiment. In the control, only buffer was added to GUVs (grey). Scale bars, 10 μm. ESCRT-III: Vps20, Snf7, Vps24, and Vps2.
The ESCRT-II-VPS202 complex has a convex membrane-binding surface
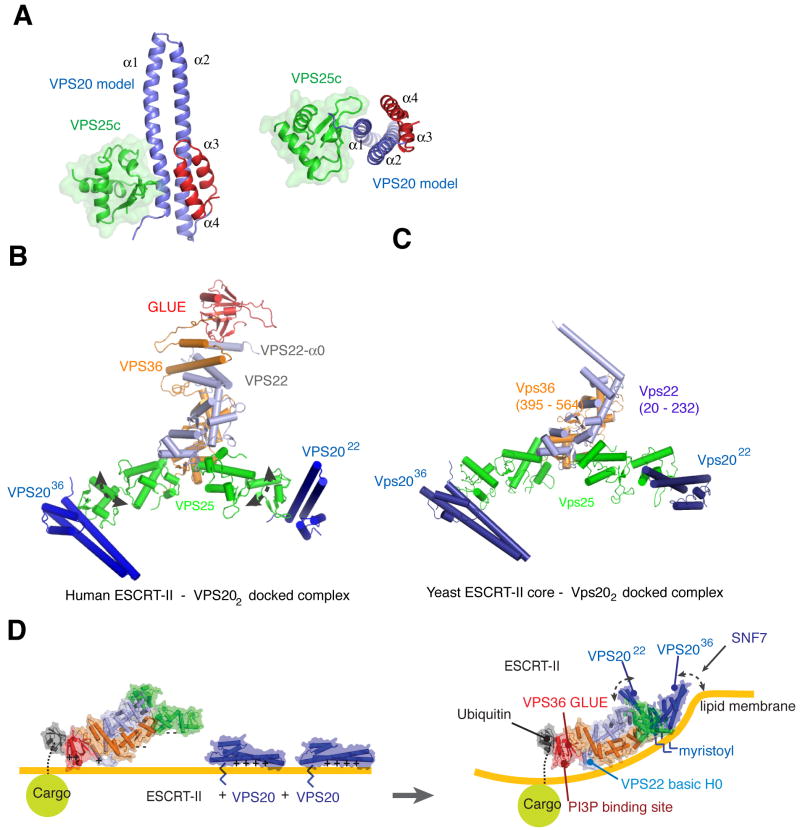
To gain insight into the interaction of ESCRT-II and full length VPS20, we modeled a structure of ESCRT-II and VPS20 complex based on the crystal structures of human ESCRT-II (Im and Hurley, 2008) and VPS24 (Muziol et al., 2006). The conservation of the structures allowed us to build a VPS20 α1 - α4 model docked onto VPS25 (Figure 5A). The C-terminal regions were omitted owing to disorder in the α4 - α5 loop in the crystallized VPS24 protein (Muziol et al., 2006). The modeling shows that VPS25 binds to the opposite side of the VPS20 monomer from helix α3 and α4, and it does not make direct contact with α2 - α4 of VPS20.
Figure 5. The complete ESCRT-II-VPS202 supercomplex and its docking to a concave membrane.
(A) α2 – α4 of VPS20 were modeled based on the structure of VPS24. The non-crystallographic VPS20 α1 dimer contact was used for superposition with VPS24. (B) Model of human ESCRT-II - VPS20 complex. The complex model was made by superimposing the VPS25 – VPS20 complex from (A) onto the VPS25 subunits of ESCRT-II. The curved dashed arrows indicate flexibility about the VPS25 WH1-WH2 linker. (C) Yeast ESCRT-II – Vps20 complex model. Vps20 was docked onto the yeast ESCRT-II by superimposing the Vps25 – Vps20 complex. (D) Membrane docking of the human ESCRT-II-VPS202 complex and proposed mechanism for promoting negative curvature. The curved portion of the membrane is drawn consistent with a radius of curvature of 28 nm, the mean size of ILVs in mammalian cells (Murk et al., 2003).
The model was extended to include full length human ESCRT-II (Im and Hurley, 2008) by superimposing two copies of the VPS25c -VPS20 complex on the VPS25 subunits of ESCRT-II (Figure 5B). Two VPS20 molecules bind to the tips of VPS25 subunits of the ESCRT-II complex. The base of four helices α1 - α4 of VPS20 sits close to the VPS25 and the α1 - α2 tips of VPS20 point outward from the core of the ESCRT-II complex. One copy is bound to the VPS25 that is in turn bound to VPS36, and it will be referred to as VPS2036. The second molecule is bound to the copy of VPS25 that is in turn bound to VPS22, and we refer to it as VPS2022. VPS2022 is nearly perpendicular to the plane of the Y. The N-terminal myristoylation site of VPS20 is eight amino acids upstream from the N-terminus of the VPS20 model, which would allow the myristoyl groups of both VPS20 molecules in the complex to touch the lipid membrane. The ESCRT-II - VPS202 super-complex has multiple points for membrane binding such as ubiquitinated cargo, the GLUE domain, VPS22-α0, and two N-terminal myristoyl groups of VPS20. The two VPS20 molecules are separated by 110 Å between the α1 - α2 tips of VPS20 molecules. A similar orientation of Vps20 molecules was observed in a model of yeast ESCRT-II – Vps202 complex. However, Vps2022 is rotated 40 degrees towards the inside of the Y, reducing the tip-to-tip distance to 90 Å in the yeast complex (Figure 5C).
Membrane docking of the ESCRT-II-VPS202 supercomplex was initially guided by the docking of the phospholipid binding site of the VPS36 GLUE domain and the basic helix α0 of VPS22, as described (Im and Hurley, 2008). The positioning of the GLUE domain and VPS22- α0 in the composite structure of full-length human ESCRT-II is approximate, but appeared reasonable in that it placed the myristoylation sites of both VPS20 molecules on the same side of the membrane, and allowed the VPS2036 molecule to present its basic membrane-binding face on the same side of the overall supercomplex (Figure 5D). The membrane binding face is non-planar, however. The WH2 domain of VPS25 is attached to the rest of the ESCRT-II core complex through a short linker that has some flexibility as judged by the difference in orientation between the domains in human and yeast, and the high B-factors for this domain in both human and yeast structures. Even allowing for linker movements, it was not possible to devise any orientation of the GLUE domain, VPS22-α0, and either VPS20 that occupied a common plane. However, a concave membrane surface can be readily modeled such that the basic faces of GLUE domain, VPS22-α0, the myristoylation sites of both VPS20 molecules, and the basic face of VPS2036 all contact the membrane without any movement of the VPS25 linker (Figure 5D). It appears that the basic face of VPS2022 can be modeled onto this curved membrane provided the WH2 of VPS2522 is rotated about the WH1-WH2 connecting loop, although Figure 5D shows the geometry without domain rotations or any other adjustments.
DISCUSSION
The ESCRT-II-Vps20 interaction is central to MVB biogenesis, and the structure of the minimal complex explains the specificity of ESCRT-II for Vps20. The simplest model for ESCRT-II activation of ESCRT-III would invoke recruitment of Vps20 to membranes by ESCRT-II. However, we found that Vps20ΔESCRT-II had normal localization. This unexpected result was explained by the finding that both Vps20 and ESCRT-II have nanomolar affinities for endosome-like synthetic liposomes. The normal localization of Vps20ΔESCRT-II stands in sharp contrast to the strong block in cargo sorting observed when the VPS20ΔESCRT-II allele was expressed. ESCRT-II coordinates ESCRT-III scission to cargo locations, which could account for the functional requirement for the direct ESCRT-II-Vps20 interaction. Using a GUV-based assay, the direct activation of ESCRT-III by ESCRT-II was visualized. The GUV assay does not contain cargo, so while the coordination of cargo to ILV biogenesis is undoubtedly important, these results show that there is also a fundamental, cargo-independent aspect to ESCRT-III activation.
One of the central questions in MVB biogenesis is how the limiting membrane of the endosome is bent into a negatively curved ILV (Barelli and Antonny, 2009). ESCRT-III subunits overexpressed in cells coat the inside of plasma membrane evaginations, promoting negative curvature (Hanson et al., 2008). Recombinant ESCRT-III subunits mixed with liposomes (Lata et al., 2008; Saksena et al., 2009) have similar effects. These results lead to a conundrum. The coating of the interior of the evaginations or invagination leaves no room for transmembrane cargo, and following scission, would also lead to uptake of the ESCRT-III proteins into ILVs and thus to their degradation. This is at odds with the lack of reports of ESCRT-III localization in ILVs, and with the observation that ESCRT-III is recycled from endosomes by Vps4 (Babst et al., 1998). The source of negative curvature in MVB biogenesis has therefore remained obscure. The ESCRT-II - VPS202 super-complex has a convex curvature that is complementary to that of a nascent ILV, and this shape is only formed upon the assembly of the supercomplex (Figure 5D). This leads us to speculate that the ESCRT-II - VPS202 super-complex could play a key role in scaffolding negative membrane curvature by imposing its shape on the membrane. An analogous mechanism is well-established for positive curvature stabilization by the crescent-shaped BAR domains (McMahon and Gallop, 2005).
The ESCRT system is complex, and nothing in the above is inconsistent with the concept that ESCRT-II causes an activating conformational change in Vps20 (Saksena et al., 2009). We find robust ILV formation with full-length Vps20, while the use of a partially truncated and activated Vps20 construct must be added at relatively high levels to support budding in vitro without ESCRT-II (Wollert et al., 2009). The curvature of the large in vitro ILVs that can be visualized by light microscopy is lower than in the physiological setting and does not match the curvature of the ESCRT-II-VPS20 supercomplex. The allosteric activation mechanism offers the most appealing explanation for the potent activation of ESCRT-III by ESCRT-II in the GUV system. Similarly, we do not discount that other factors such as ESCRT-I or ALIX could also contribute to regulating ILV size (Kostelansky et al., 2007) or promoting negative curvature (Kim et al., 2005).
ESCRT-II has a special role in MVB biogenesis, in that overexpression of ESCRT-II rescues the loss of ESCRT-I, but not the converse (Babst et al., 2002b). In contrast, ESCRT-I is critical, but ESCRT-II dispensable, for HIV-1 budding (Langelier et al., 2006) and cytokinesis (Morita et al., 2007). In HIV-1 budding, self-assembly of Gag drives membrane curvature. The membrane neck between daughter cells is also established independent of the ESCRTs. In contrast to MVB biogenesis, HIV-1 budding and cytokinesis use other mechanisms to establish membrane curvature, explaining why these processes do not require ESCRT-II. The proposed model (Movie S1) rationalizes the dramatic differences in the ESCRT-II requirement in these three ESCRT-dependent pathways.
In summary, we have elucidated the determinants for the ESCRT-II-Vps20 interaction, which has multiple and vital roles in MVB biogenesis. First, the interaction coordinates cargo binding to ESCRT-II with the initiation of ESCRT-III assembly. Second, the interaction promotes an activating conformational change in Vps20 (Saksena et al., 2009). Finally, the composite structure of the ESCRT-II-VPS20 supercomplex suggests that the interaction could directly promote the negative membrane curvature required for ILV biogenesis. The structural analysis explains how this crucial interaction is conserved from yeast to humans. The conserved specificity of ESCRT-II for Vps20, as compared to all other ESCRT-III subunits highlights the central role for this interacting pair in MVB biogenesis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The codon optimized synthetic gene for VPS25c (residues 102-176) was subcloned into the pGST1 vector (Sheffield et al., 1999). VPS25c was tagged with an N-terminal glutathione S-transferase (GST) followed by a tobacco etch virus (TEV) protease cleavage site. DNA coding for GST-VPS25c subcloned into the first cassette of the polycistronic pST39 vector (Tan, 2001). Various VPS20 constructs with an N-terminal hexahistidine tag followed by a TEV cleavage site were subcloned into the second cassette of pST39. The resulting bicistronic plasmid was transformed into E. coli strain BL21(DE3) Star and expressed overnight at 30 °C. Cells were resuspended in buffer (2X PBS plus 20 mM imidazole) and lysed by sonication. The resulting VPS25c-VPS20 complexes were isolated using sequential Ni2+ and glutathione affinity chromatography. The eluate was concentrated and the GST and histidine tags were removed by cleavage with TEV protease. The VPS25c-VPS20 complexes were further purified by Superdex S200 size exclusion chromatography. The fractions containing the complex were concentrated to 10 mg/ml in buffer 10 mM Tris-HCl (pH 8.0), 100 mM NaCl for crystallization.
For the biosensor binding assay, various VPS20 constructs were subcloned into a vector providing an N-terminal His6-tagged maltose binding protein (MBP) followed by a TEV cleavage site. The MBP–VPS20 constructs were expressed as described above and purified by Ni-NTA affinity chromatography. The samples were further purified by Hi-trapQ anion-exchange or size exclusion chromatography. The purified MBP-VPS20 proteins were diluted in 1 × PBS buffer for biosensor binding assay. Full length human ESCRT-II complex was prepared as previously described (Im and Hurley, 2008).
Yeast ESCRT-III subunits Vps20, Snf7, Vps24 and Vps2 for GUV experiments were expressed and purified as described (Wollert et al., 2009). Briefly, N-terminal His6-MBP fusion proteins were expressed at 30°C for 3 hours after induction with 0.5 mM IPTG at an optical density (OD 600 nm) of 0.8. The proteins were affinity purified using Ni-NTA resin (Qiagen). Elution fractions were cleaved using TEV protease and further purified by size exclusion chromatography (SEC; Superdex 200 column). Purified proteins were immediately flash frozen in liquid nitrogen and stored at -80°C until use. Full-length yeast ESCRT-II was purified from E. coli with an N-terminal His6 fusion tagged to Vps22 using Ni-NTA affinity purification and SEC as described (Hierro et al., 2004).
Crystallization and crystallographic analysis
Crystals of VPS25c – VPS20 complex were grown by vapor-diffusion methods at 25 °C over a reservoir of 100 mM Tris HCl (pH 8.0), 35% PEG 4000 for one week. Crystals were cryoprotected in Paratone and flash frozen using liquid nitrogen. Native data were collected to 2.0 Å resolution from a single frozen crystal with an MAR CCD detector at beamline 22-ID, APS. All data were processed and scaled using HKL2000 (HKL Research). The structure of the complex was determined by molecular replacement with the program PHASER (McCoy et al., 2007) using the VPS25c portion of the human ESCRT-II structure (Im and Hurley, 2008) as a starting model. The initial model of VPS20 was built manually into the density-modified map using the programs O (Jones et al., 1991) and Coot (Emsley and Cowtan, 2004). The refinement was carried out using CNS (Brunger et al., 1998) and Refmac (CCP4, 1994) with TLS (Murshudov et al., 1999) and anisotropic B factor refinement. The final model consisted of four copies of the VPS25c:VPS20-α1 complex, with residues 102-176 from VPS25, residues 10-44 from VPS20. Residues 45-48 of VPS20 were present in the construct but not visualized in the structure. There are 99.5% of the residues in the most favored and additional allowed regions of the Ramachandran plot. All structural figures were prepared using the program PyMOL (W. Delano, http://pymol.sourceforge.net/).
Modeling
The asymmetric unit of the VPS25 - VPS20 crystal contains four copies of the complex. VPS20 α1 forms antiparallel coiled-coil dimers in the crystal (Figure S3A). The second copy of helix α1 occupies the position corresponding to that of α2 of VPS20 in full length VPS20. The hydrophobic residues of VPS20 α1 are engaged in packing contacts between the helices that are equivalent to the interactions between α1 and α2 in a full-length ESCRT-III monomer (Figure S3B). The α1 dimer of VPS20 superimposes with the structure of VPS24 α1 - α2 with an rmsd of 0.95 Å (Figure S3C). Consistent with the structural similarity, a structure-based sequence alignment of the coiled-coil residues of VPS20 and VPS24 showed high similarity in the interface residues (Figure S3D). This finding indicates that the isolated VPS20 α1 used for crystallization has a very similar conformation to the α1 of full length VPS20, and that VPS25 binding cause little conformational change in VPS20 α1 helix. Other aspects of modeling are described in the results.
Biosensor binding experiments
Binding of the VPS25c and ESCRT-II to VPS20 was analyzed using a Biacore T100 instrument at 25 °C using a CM5 sensor chip. Approximately 9,000 response units (RU) of anti-MBP monoclonal antibody (Abcam) was immobilized on two flow cells using amine-coupling chemistry. The MBP-VPS20 constructs (α1 - α3 residues 1-118, α1 - α3 mutant L21R/R28A/D29A (ΔESCRT-II), α1 - α4 residues 1-145, α1 - α5 residues 1-167 and full length) were captured to densities of 2500 – 3000 RU. Binding studies were performed in duplicate by passing the wild-type or mutant VPS25c and full length ESCRT-II over the captured MBP-VPS20 proteins at a flow rate of 10 μl min−1 in a 1 × PBS buffer. The surface was regenerated with an injection of 10 mM Glycine-HCl at pH 2.0 at a flow rate of 10 μl min−1 for 30 s. The data were fit with the following equation:
where [VPS25] is the protein concentration of the flowing analyte, Kd is the dissociation constant, Rmax is the maximal response, and ‘offset’ is the background signal. The data was processed using BiaEvaluation (Biacore) software and the figures were made using Sigmaplot (GraphPad Software).
Plasmid construction and yeast strains
DNA coding for the complete expression cassettes for VPS20 and VPS25 were amplified from yeast genomic DNA and cloned into YCplac111 (Gietz and Sugino, 1988). Point mutations in YCplac111-VPS20 and YCplac111-VPS25 were introduced by Quickchange mutagenesis (Stratagene). DNA coding for green fluorescent protein (GFP) was fused to the 3’ end of the coding region of the VPS20 using PCR based mutagenesis. Plasmids encoding the VPS20 or VPS25 gene, and the pGO45 (pRS426 GFP-CPS1) vector (Odorizzi et al., 1998) were transformed to wild-type and mutant strains. The following yeast strains were used: BY4742 vps25Δ∷KanR and BY4742 vps20Δ∷KanR (Open Biosystems).
Fluorescence microscopy
Yeast strains expressing the appropriate alleles were harvested at an A660 of 0.4-0.6, and labeled with FM4-64 for vacuolar membrane staining (Vida and Emr, 1995). Uptake of FM4-64 by live cells was performed at 30 °C for 1 hr, after which cells were resuspended in selection media and incubated for 30 min at 30°C. Visualization of cells was performed on an LSM510 fluorescence microscope (Carl Zeiss MicroImaging) equipped with fluorescein isothiocyanate (FITC) and rhodamine filters and captured with a digital camera. GUVs were visualized in a 200 μl observation chamber (Lab-Tek chambered #1.0 Borosilicate) was coated using 1 mg ml-1 BSA and rinsed with buffer (50 mM Tris HCl pH 7.4, 300 mM NaCl). 100 μl of GUVs were mixed with this buffer and incubated for 5 min. ESCRT-II was added to the GUV buffer mixture to yield a final concentration of 40 nM. The solution was stirred to accelerate protein distribution and binding. After 5 min of incubation at room temperature, Vps20 (final concentration of 40 nM), Snf7 (200 nM), Vps24 (40 nM), Vps2 (40 nM), and Vps4 (40 nM) were added in that order, separated by 5 min incubation intervals. GFP was added before Snf7 and after Vps20 addition, yielding a final concentration of 1.65 μM. One buffer control was performed for each experiment. Stirring and incubation times were the same for the control and protein additions. Images were taken in multi-tracking mode on a Zeiss LSM510 confocal laser-scanning microscope with a 63 × Plan Apochromat 1.4 NA objective and a 488/543 dichroic mirror at a resolution of 512 × 512 pixel. GFP was excited using the 488 nm line and the rhodamine was excited with a 543 nm HeNe laser. GFP emission was collected with a 505-530 nm bandpass filter. Rhodamine emission was collected with a 560 nm longpass filter. The pinholes for each channel were set for an 2 μm optical slice. Laser power was 9 μW and 24μW for the 543 nm and 488 nm channels, respectively. Images were analyzed using the LSM Examiner software. Randomly chosen field of views were evaluated to reveal the number of ILV per GUV. Reactions were performed as described above and all GUVs in the particular field of view were scanned in the z direction. All ILVs per GUV were counted and for each experimental condition 100 GUVs were analyzed. Results were summarized in histograms.
Cell fractionation and Western blotting
Yeast cells grown to OD660 = 0.9 were harvested and resuspended in 50mM Tris-HCl pH 7.4 with the addition of yeast protease inhibitor cocktail (Sigma). The cells were disrupted by glass beads with a Beadbeater. The resulting cell lysate was centrifuged at 500 × g for 5min to remove unlysed cells and cell walls. The resulting extracts were separated by centrifugation at 200,000 × g for 20 min into soluble and membrane-bound pellet fractions. The samples were analyzed for the presence of Vps20-GFP by Western blotting using an antibody specific for the GFP-tag. Quantification of the resulting blot was performed by the LabWorks 4.6 program (UVP).
Membrane binding in vitro
The engineered single-Cys mutant C109S/S119C of VPS20, and wild-type ESCRT-II, were incubated with tetramethylrhodamine-5-iodoacetamide dihydoiodide (5-TMRIA, Molecular Probes) overnight at 4° C at a 20-fold molar ratio of dye to VPS20 and 2.5-fold molar ratio of dye to ESCRT-II. Unreacted dye was removed by gel filtration. This procedure yielded VPS20 labeled at ~100% and ESCRT-II at ~140%. Fluorescence intensity measurements were performed using a Fluorolog-3 (HoribaJobinYvon) fluorescence spectrometer at room temperature with 0.1 mg/ml liposomes consisting of POPC (61 mol%), POPS (10 mol%), cholesterol (25 mol%), PI(3)P (3 mol%) and fluorescein-PE (1 mol%). Increasing amounts of rhodamine labeled protein were titrated into the cuvette. In the case of VPS20 binding in the presence of ESCRT-II, ESCRT-II was titrated first into the cuvette and was allowed to equilibrate for one minute, then a stoichiometric amount of VPS20 was titrated into the cuvette. At each concentration, the intensity of fluorescence was recorded (excitation at 460 nm and emission at 520 nm). Percent saturation was calculated as ΔI / ΔImax where ΔI=I-Io (I represents the measured intensity of fluorescence, Io represents the intensity of fluorescence prior addition of any protein, ΔI represents the change of the fluorescence intensity, ΔImax the change of fluorescence intensity at saturation). Percent saturation was plotted against the protein concentration and fitted by Langmuir isotherm to obtain the Kd values.
Preparation of giant unilamellar vesicles (GUVs)
GUVs were prepared as described (Wollert et al., 2009). Briefly, lipid mixtures containing POPC (62 mol%), POPS (10 mol%), cholesterol (25 mol%), PI(3)P (3 mol%) and lissamine-rhodamine-PE (0.1 mol%) were applied to indium-tin oxide covered glass slides and placed into a custom-made Teflon chamber. An electric AC-field (1V, 10 Hz) was applied for 4 h at 60°C (Angelova and Dimitrov, 1986). The following lipids were purchased from Avanti Polar Lipids: 1-palmitoyl-2-oleoyl-sn- glycero-3-phospho-L-serine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, cholesterol, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl). Phosphatidylinositol 3-phosphate (diC16) was purchased from Echelon.
Supplementary Material
Acknowledgments
We thank Boris Baibakov for assistance with microscopy, Jurrien Dean and Jennifer Lippincott-Schwartz for use of their microscopes, Will Prinz for providing yeast strains and discussions, Adriana Rojas for help with crystallographic data collection, Ethan Tyler for producing Movie S1, and the SER-CAT staff for user support at the Advanced Photon Source (APS). Use of the APS was supported by the U. S. DOE, Basic Energy Sciences, Office of Science, under Contract No.W-31-109-Eng-38. This research was supported by NIH intramural support, NIDDK and IATAP. T. W. is an EMBO long term fellow.
Footnotes
COORDINATES Crystallographic coordinates have been deposited in the Protein Data Bank with the accession code 3HTU.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angelova MI, Dimitrov DS. Liposome electroformation. Faraday Discuss Chem Soc. 1986;81:303–311. [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelli H, Antonny B. Cell biology: Detached membrane bending. Nature. 2009;458:159–160. doi: 10.1038/458159a. [DOI] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect A. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, Hurley JH. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004;431:221–225. doi: 10.1038/nature02914. [DOI] [PubMed] [Google Scholar]
- Hurley JH. ESCRT Complexes and the Biogenesis of Multivesicular Bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron- Density Maps and the Location of Errors in These Models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kieffer C, Skalicky J, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky MS, Schluter C, Tam YYC, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger H, Weissenhorn W. Helical Structures of ESCRT-III are Disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi S, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk JLAN, Humbel BM, Ziese U, Griffith JM, Posthuma G, Slot JW, Koster AJ, Verkleij AJ, Geuze HJ, Kleijmeer MM. Endosomal compartmentalization in three dimensions: Implications for membrane fusion. Proc Natl Acad Sci USA. 2003;100:13332–13337. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr Sect D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Devel Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Russell MRG, Nickerson DP, Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr Opin Cell Biol. 2006;18:422–428. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional Reconstitution of ESCRT-III Assembly and Disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Express Purific. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiano D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M, Skalicky J, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Tan S. A Modular Polycistronic Expression System for Overexpressing Protein Complexes in Escherichia coli. Protein Express Purific. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered Assembly of the ESCRT-III Complex on Endosomes Is Required to Sequester Cargo during MVB Formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: Crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Teo HL, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A New Vital Stain for Visualizing Vacuolar Membrane Dynamics and Endocytosis in Yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xia H, Zhou J, Azmi I, Davies BA, Katzmann DJ, Xu Z. Structural basis of Vta1 function in the multi-vesicular body sorting pathway. Dev Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.