Abstract
Homomeric α7 nicotinic acetylcholine receptors are a well-established, pharmacologically distinct subtype. The more recently identified α9 subunit can also form functional homopentamers as well as α9α10 heteropentamers Current fluorescent probes for α7 nicotinic acetylcholine receptors are derived from α-bungarotoxin. However, α-bungarotoxin also binds to α9* and α1* receptors which are coexpressed with α7 in multiple tissues. We used an analog of α-conotoxin ArIB to develop a highly-selective fluorescent probe for α7 receptors. This fluorescent α-conotoxin, Cy3-ArIB[V11L;V16A], blocked acetylcholine-evoked α7 currents in Xenopus laevis oocytes with an IC50 value of 2.0 nM. Observed rates of blockade were minute-scale with recovery from blockade even slower. Unlike fluorescein isothiocyanate-conjugated α-bungarotoxin, Cy3-ArIB[V11L;V16A] did not block α9α10 or α1β1δε receptors. In competition binding assays, Cy3-ArIB[V11L;V16A] potently displaced [125I]-α-bungarotoxin binding to mouse hippocampal membranes with a Ki value of 21 nM. Application of Cy3-ArIB[V11L;V16A] resulted in specific punctate labelling of KXα7R1 cells but not KXα3β2R4, KXα3β4R2, or KXα4β2R2 cells. This labelling could be abolished by pre-treatment with α-cobratoxin. Thus, Cy3-ArIB[V11L;V16A] is a novel and selective fluorescent probe for α7 receptors.
Keywords: α7 nAChR, α-conotoxin, fluorescent, HEK293 cells
Introduction
Nicotinic acetylcholine receptors (nAChRs) are ligand gated ion channels that are ubiquitously expressed throughout the nervous system but are also found on non-neuronal cells in the brain and periphery. nAChRs are pentameric assemblies of homologous subunits that combine to form ligand-gated ion channels. When activated by agonists, nAChRs open to allow the passage ions across the cell membrane. To date, 17 nAChR subunits have been cloned encoding the α1-α10, β1-β4, δ, ε, and γ subunits; α8 is apparently only expressed in aves. These subunits assemble in various combinations with each other to form heteromeric receptors or, in the case of α7, α8, and α9, homomeric receptors (for review see (Dani & Bertrand 2007)).
For the last several decades α-bungarotoxin (α-BgTx), isolated from the venom of the Taiwanese banded krait Bungarus multicinctus, and α-cobratoxin (α-CbTx), from Naja naja, have been used to pharmacologically identify α7 nAChRs. However, it has recently become apparent that α-BgTx will also block α9 homomers and α9α10 heteromers with nanomolar potency IC50 = 2.1 and 14.0 nM, respectively, (Verbitsky et al. 2000, Sgard et al. 2002) in addition to its well characterized antagonist activity on the α1β1δε and to the avian α8 (Gotti et al. 1997) subtypes. α-CbTx will also block α9α10 (IC50 = 3.8 nM; our unpublished result) and the α1β1δε subtype. Methyllycaconitine (MLA), another widely used α7 antagonist, IC50 = 0.03 nM (Palma et al. 1996), is a plant alkaloid isolated from Delphinum and Consolida species. Unfortunately, MLA also blocks α9 homomers IC50 = 1.1 nM (Verbitsky et al. 2000), and α6* (Klink et al. 2001, Mogg et al. 2002). Thus, none of these ligands can be used to definitively identify α7 nAChRs if α1*, α6*, or α9* nAChR subtypes are also present.
This difficulty in unequivocally identifying α7 nAChRs is particularly problematic in peripheral tissues. For example, multiple nAChR subtypes have been implicated in the modulation of pain and inflammation. The α4β2, α7, and α9α10 subtypes have recently received considerable attention in this regard (Vincler & McIntosh 2007, Damaj et al. 2007, Damaj et al. 1998). The α7 subtype has been determined to be an essential component of the cholinergic anti-inflammatory pathway (Wang et al. 2003), and block of the α9α10 subtype has been demonstrated to be analgesic in animal models of neuropathic pain (Satkunanathan et al. 2005, Vincler et al. 2006). The detection of these two subtypes and the elucidation of their contributions to cellular processes have been complicated by the relative lack of subtype-specific ligands that can discriminate between α7 and α9α10 nAChRs (McIntosh et al. 2005). The ability of a ligand to discriminate between these two subtypes is essential given the likelihood that both may be co-expressed in a variety of cell types including T-lymphocytes (Kawashima & Fujii 2004, Peng et al. 2004) and macrophages (Biallas et al. 2007, Grau et al. 2007), cells especially relevant in pain and inflammatory conditions. In addition, α7, α9, and α10 subunits have been reported to be co-expressed in the dorsal horn of the spinal cord (Genzen & McGehee 2003) and dorsal root ganglia neurons (Genzen et al. 2001, Papadopolou et al. 2004, Rau et al. 2005, Haberberger et al. 2004, Lips et al. 2006, Lips et al. 2002).
We recently described a set of novel α-conotoxins (α-CTx) isolated from the venomous marine snail Conus arenatus (Whiteaker et al. 2007). Directed substitutions in the amino acid sequence of the cloned peptides were made which resulted in ligands that are highly selective for the α7 subtype. One such ligand, ArIB[V11L;V16A], was demonstrated to be >10,000-fold more selective for α7 over α9α10 and α1β1δε. We have also shown that a radioligand version of ArIB[V11L;V16A] can be used in autoradiography and ligand binding assays (Whiteaker et al. 2008). With its selectivity for α7, potency (IC50 = 0.52 nM), and slow off-rate kinetics, we reasoned that ArIB[V11L;V16A] would be an attractive candidate for the development of a fluorescent probe that could be used to definitively identify α7 nAChRs in tissues where there are other receptors that are sensitive to α-BgTx and MLA.
In this study, we describe the development of a fluorescent conjugate of ArIB[V11L;V16A] namely, Cy3-ArIB[V11L;V16A]. We used oocyte electrophysiology, binding, and fluorescence imaging techniques to assess its potency at and selectivity for α7 nAChRs. Cy3-ArIB[V11L;V16A] potently competed with [125I]-α-BgTx for binding sites in mouse hippocampal membranes. In HEK293 cells stably transfected with rat α7 nAChRs (KXα7R1), punctuate labelling of α7 nAChRs was observed. No labelling was observed in HEK293 cells expressing α3β2 (KXα3β2R4), α3β4 (KXα3β4R2), or α4β2 (KXα4β2). Together, these results suggest that Cy3-ArIB[V11L;V16A] is a useful probe for the study of α7 nAChR.
Materials and Methods
Cy3 NHS ester dye and [I25I]-α-BgTx (specific activity 2000 Ci/mmol-1) were purchased from General Electric Healthcare (Piscataway, NJ). Penicillin/streptomycin, minimum essential medium, G418 sulfate [Geneticin], 0.5% Trypsin/EDTA, Hoechst 33342, and calcium and magnesium free phosphate buffered saline (PBS) were from Invitrogen (Carlsbad, CA). Heat inactivated fetal bovine serum (FBS) was from Hyclone (Logan, UT). α-Conotoxin ArIB[V11L;V16A] was synthesized as previously described (Cartier et al. 1996). Fluorescein isothiocyanate-α-bungarotoxin (FITC-α-BgTx), α-CbTx, acetylcholine chloride (ACh), carbachol, poly-L-lysine hydrobromide, acetonitrile (ACN), trifluoroaceitic acid (TFA), atropine sulphate, bovine serum albumin (BSA), phenylmethylsulfonyl fluoride, EGTA, EDTA, leupeptin trifluoroacetate, aprotinin, and pepstatin A were from Sigma Aldrich (St. Louis, MO). HEPES was from Research Organics (Cleveland, OH). Amikacin sulphate, sulfamethoxazole, and trimethoprim were purchased from the University of Utah Health Science Center (Salt Lake City, UT).
Conjugation of ArIB[V11L;V16A] with Cy3 NHS ester
To generate the fluorescent conjugate of ArIB[V11L;V16A], the peptide was suspended in a NaHCO3 buffer (final concentration 100 mM), pH 8.5, and the dye in anhydrous DMSO. Two hundred μg of Cy3 NHS ester dye suspended in 14 μl of DMSO was reacted with 50 nanomoles of peptide suspended in 11 μl of NaHCO3 buffer. The reaction was kept protected from light at RT for 1 h with brief vortexing and centrifugation performed at least once during this time period. The reaction was terminated by dilution with 50 μl of DMSO then 150 μl of buffer A (0.1% (vol/vol) TFA in distilled water). The reactants were separated by reverse phase- HPLC (RP-HPLC) using a Vydac C18 column. The buffers used were buffers A and B60 (0.092% (vol/vol) TFA, 60% (vol/vol) ACN). The peptides were eluted using a linear gradient of 20% to 90% B60 over 40 minutes. Quantification of the fluorescent conjugate was calculated using the Absmax for Cy3 at 550 nm (determined using a spectrophotometer), the published molar extinction coefficient (150,000 M-1cm-1), and the Lambert-Beer equation. The masses were verified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) which was performed at the Salk Institute, La Jolla, CA, under the direction of Dr. Jean Rivier.
cRNA preparation and injection
Rat nAChR subunit cDNA clones α2, α3, α4, α7 and β2-β4 were from S. Heinemann (Salk Institute, La Jolla, CA), α9 and α10 were from A.B. Elgoyhen (Universidad de Buenos Aires, Buenos Aires, Argentina), β2 and β3 in the high expressing vector pGEMHE were from C. Luetje (University of Miami, Miami, FL). Construction of the α6/α3 chimera has been previously described (McIntosh et al. 2004) and provided by J. Garret (Cognetix Inc., Salt Lake City, UT). Capped cRNA was made using the mMESSAGE mMACHINE T7 in vitro transcription kit (Ambion, Austin, TX) and purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA). The concentration of cRNA was determined by the absorbance at 260 nm. The mouse muscle subunit cDNA clones in the CMV-based pRBG4 vector were provided by S.M. Sine (Mayo Clinic College of Medicine, Rochester, MN). Fifty nl (at least 5-10 ng) of cRNA or 18.3 nl of cDNA (25 ng/ul of each subunit) were injected into each Xenopus oocyte with a Drummond microdispenser (Drummond Scientific, Broomall, PA) and incubated at 17° C in ND96 (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5 mM HEPES, pH 7.5) containing antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml amikacin sulfate, 160 μg/ml sulfamethoxazole, and 32 μg/ml trimethoprim). Oocytes were injected within 1-2 day of harvesting and recordings were made 3-6 days post-injection.
Electrophysiology
Oocytes were voltage-clamped and exposed to ACh and toxins as previously described (Cartier et al. 1996). Briefly, the oocyte chamber consisting of a cylindrical well (~30 μl in volume) was gravity perfused at a rate of ~2 ml/min with ND96 containing 0.01% (wt/vol) BSA and 1 μM atropine to block endogenous muscarinic receptors. For experiments involving α7 and α9α10, atropine was excluded from the perfusion solution because it has been shown to block these receptor subtypes (Gerzanich et al, 1994). Oocytes were exposed once a minute to a 1 sec pulse of ACh. The ACh concentrations used were 200 μM for α7, 50 μM for α1β1δε, and 100 μM for all other subtypes. Once a stable baseline was achieved, either ND96 alone or ND96 containing varying concentrations of toxins was perfusion-applied during which 1-sec pulses of ACh were applied every minute until a constant level of block by the toxins was achieved. For the baseline response, at least 3 ACh-induced responses were averaged. After a steady-state block had been achieved, the % block by the toxins was determined by taking the final value and dividing by the pre-toxin baseline value to yield a % response. For experiments involving concentrations >1 μM, toxins were bath applied to conserve material. In this case, toxins were applied at the beginning of a 5 minute static bath to ensure equilibrium. For bath applied toxins, the % block was calculated as a % of control response (ND96 + ACh, see above for ACh concentrations used for each subtype) where no toxin was applied. The concentration-response data were fit to the equation Y = 100/(1 + 10^((LogIC50 - Log[Toxin])*Hill Slope)) by nonlinear regression analysis and the observed rate of block data by a one phase exponential decay equation using GraphPad Prism (GraphPad Software, San Diego, CA). Each data point represents the mean ± SEM from at least 3 oocytes. Statistical differences for observed rates of block and recovery were determined by a two-tailed Student's t-test and considered significant if p < 0.05. Kd values were calculated from functional IC50 using the equation IC50/Kd=21/N-1, where N is equal to the number of binding sites and in the case of α7 homomers N=5 (Whiteaker et al. 2007).
Radioligand binding assay
To prepare brain membranes, adult male (60-90 days) C57B6/J mice were sacrificed by cervical dislocation and the brains removed and placed on an ice-cold platform. Tissue was collected from the hippocampus. Hippocampal tissue was chosen because it expresses high concentrations of α7 nAChRs, is relatively large, and is simple to dissect. Individual hippocampal samples were homogenized in ice-cold hypotonic buffer (14.4 mM NaCl, 0.2 mM KCl, 0.2 mM CaCl2, 0.1 mM MgSO4, 2 mM HEPES, pH 7.5) using a glass-Teflon tissue grinder. Particulate fractions were collected by centrifugation at 25,000 x g for 15 min at 4°C (Eppendorf 5417 C centrifuge; Eppendorf North America, New York, NY). The pellets were resuspended in fresh homogenization buffer, incubated on ice for 10 min, and then harvested by centrifugation as described above. Each pellet was washed twice more by resuspension/centrifugation before storage (in pellet form under homogenization buffer) at -70°C until use.
Affinity for α7 nAChRs was measured by displacement of [125I]-α-BgTx binding to hippocampal membranes, as described in Whiteaker et al. (2008). Incubations were performed at 22 °C, for 4 h, in 1.2-ml polypropylene tubes arranged in a 96-well format. Hippocampal membranes were incubated with 2 nM [125I]-α-BgTx in a total volume of 30 μl of buffer (144 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgSO4, BSA 0.1% (wt/vol), 20 mM HEPES, pH 7.5) containing protease inhibitors (5 mM EDTA, 5 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml each of aprotinin, leupeptin trifluoroacetate, and pepstatin A), to minimize proteolysis. Displacement of [125I]-α-BgTx binding was assessed in duplicate. Total (no peptide added) and nonspecific binding (defined using 10 μM α-CbTx) was also measured in duplicate. After the initial incubation, 1 ml of isotonic binding buffer (144 mM NaCl, 1.5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 20 mM HEPES, BSA 0.1% (wt/vol), pH 7.2) was added to each tube, and the incubation was continued for a further 5 min at 22°C. This dilution and further incubation step allow some of the nonspecific [125I]-α-BgTx binding to dissociate, but it has no measurable effect on specific binding, increasing the signal-to-noise ratio of the binding assay. Binding reactions were terminated by filtration using a 96-place manifold (Inotech Biosystems, Lansing, MI). Particulate fractions were collected onto single layers of Inotech 0.75 μm retention glass fiber filters that had been soaked in 5% (wt/vol) nonfat skim milk dissolved in isotonic binding buffer. Samples were washed 6 times, and all filtration and washing steps were conducted in a 4°C cold room, using ice-cold binding buffer. Bound radioligand was quantified by gamma counting at ~83 to 85% efficiency, using a Packard Cobra counter (PerkinElmer Life and Analytical Sciences, Waltham, MA). Results for inhibition of [125I]-α-BgTx binding were calculated using a one-site fit: B = B0/(1+I / IC50), where B is ligand bound at inhibitor concentration I, Bo is the binding in the absence of inhibitor, and IC50 is the concentration of inhibitor required to reduce binding to 50% of Bo. Values for Ki (inhibition binding constant) were derived by the Cheng and Prusoff method (Cheng & Prusoff 1973): Ki = IC50/(1 + L/Kd).
Cell transfection and culture
The cells lines KXα3β2R4, KXα3β4R2, KXα4β2R2, and KXα7R1 were established previously by stably transfecting HEK293 cells with rat nAChR subunit genes (Xiao & Kellar 2004, Xiao et al. 1998, Xiao et al. 2009). The cells were routinely grown in a selective growth medium (SGM-7) which consists of minimum essential medium containing 2.2 mg/ml NaHCO3, 10% FBS (vol/vol), 100 U/ml penicillin, 100 μg/ml streptomycin, 0.7 mg/ml Geneticin, and 0.18 mg/ml carbachol, pH 7.4. Cells were grown in 75cm2culture flasks and passaged every 3-4 days upon reaching ~80-90% confluency.
Live cell fluorescence microscopy
Cells were plated on 15 mm glass coverslips (catalog # 64-0703, Warner Instruments, Hamden, CT) treated with 0.1 mg/ml poly-L-lysine and grown in 12 well tissue culture plates. Cells were suspended at RT in SGM-7 at a density of 8 × 104/ml. One ml of the cell suspension was added to each well plus 500 μl of SGM-7 and cultured at 37 °C in 95% air and 5% CO2 for 2-3 days prior to use. For labelling experiments, cells were washed 3 times with RT PBS (pH 7.5) then exposed to a PBS solution containing 2 μM Hoechst 33342 and 100 nM Cy3-ArIB[V11L;V16A] for 45 min at 4°C. To determine non-specific binding, cells were pre-incubated with 2 μM α-CbTx for 30 min at 4°C then with 100 nM Cy3-ArIB[V11L;V16A] in the presence of 2 μM α-CbTx for 45 min at 4°C. Cells were washed 3x with 4°C PBS and immediately examined using a Nikon FN1 microscope equipped with an epiilluminator, a 40x NIR Apo water immersion objective numerical aperture 0.80 and a filter set appropriate for FITC, Hoechst 33342 and Cy3 flurophores. The images were captured at 640x480 resolution with 2×2 binning. The light source was a Lamda LS xenon arc lamp with a 300 watt bulb (Sutter Instruments, Novato, CA). For z-series images, a remote focus adaptor (Sutter Instruments, Novato, CA) was used to capture 1 μm interval optical sections. All images were processed using NIH ImageJ software.
Results
Conjugation of ArIB[V11L;V16A] with Cy3 NHS ester dye
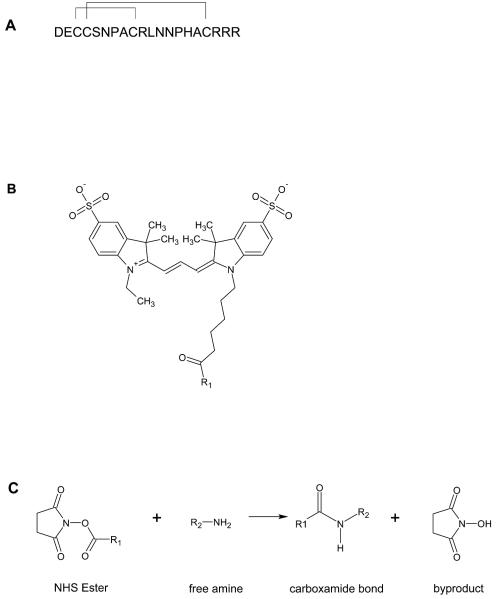
α-CTx ArIB[V11L;V16A] (Fig. 1A) is a synthetic peptide composed of 20 amino acids and 2 disulfide bridges connecting cysteines residues 3 to 8 and 4 to 17. Since there are no lysine residues in the amino acid sequence, the only free amino group available for coupling with Cy3 NHS ester (Fig. 1B) is located at the N-terminus of the peptide. The reaction between the free amino group and the NHS ester proceeds via nucleophilic acyl substitution to form a carboxamide bond between the peptide and the dye (Fig. 1C). The desired product, Cy3-ArIB[V11L;V16A], was separated from the reactants by RP-HPLC (Fig. 2). RP-HPLC analysis revealed that conjugation of the peptide resulted in a shift in retention time for the conjugate compared to the unconjugated peptide indicating increased hydrophobicity. The desired product, Cy3-ArIB[V11L;V16A], eluted at 24.2% B60 and the unconjugated peptide at 17.7% B60. Cy3-ArIB[V11L;V16A] eluted as a single peak indicating a single product. MALDI-TOF MS confirmed the RPHPLC results. The calculated monoisotopic mass for Cy3-ArIB[V11L;V16A] is 2922.17 Da and the observed monoisotopic mass was 2922.6 Da.
Fig. 1.
(A), Amino acid sequence of α-CTx ArIB[V11L;V16A] showing the disulfide connectivity between cysteine residues. (B), chemical structure of Cy3 NHS ester dye. (C), the conjugation reaction between α-CTx ArIB[V11L;V16A] and Cy3 NHS dyes resulting in the formation of a carboxamide bond between the dye and the N-terminus of the peptide.
Fig. 2.
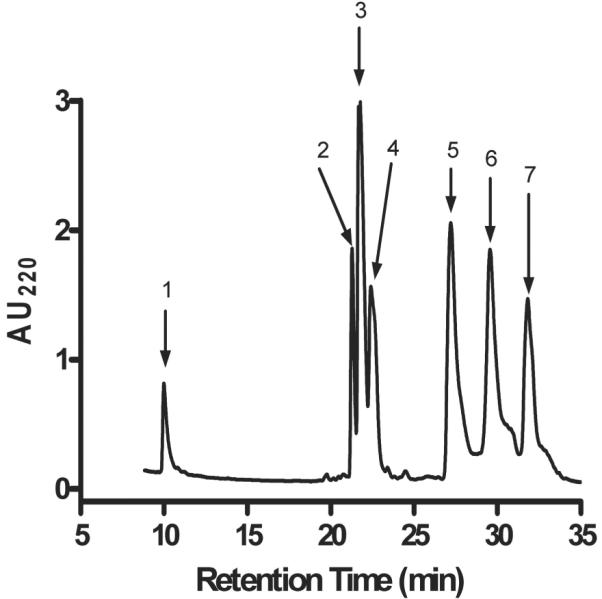
Separation of Cy3-ArIB[V11L;V16A] from the reactants by RP-HPLC. The labelled peaks correspond to unconjugated α-CTx ArIB[V11L;V16A] (1), unconjugated Cy3 NHS ester dye (2), and the fluorescent conjugate Cy3-ArIB[V11L;V16A] (3).
Effect of conjugation of Cy3 NHS ester on ArIB[V11L;V16A] pharmacology
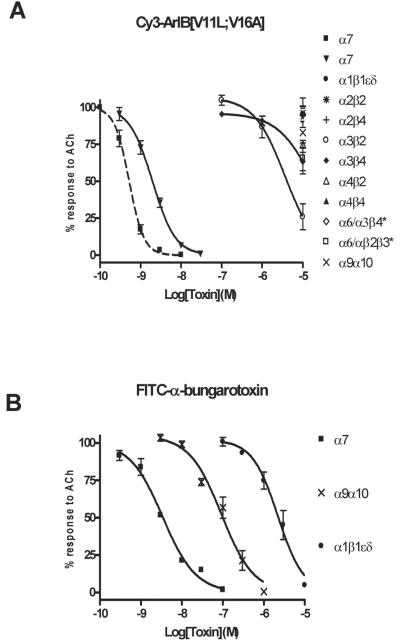
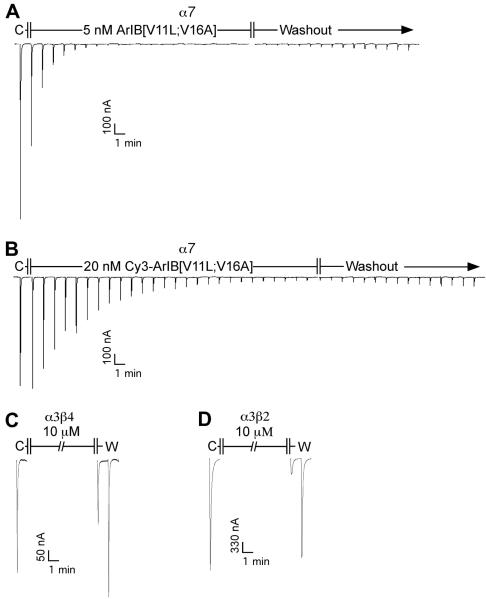
Because substitutions in the amino acid sequence of α-CTx's can dramatically affect the binding affinity and specificity of the peptide for its target receptor, we determined whether conjugation with Cy3 would also affect the pharmacology of the peptide. Conjugation with Cy3 resulted in a right-ward shift in the IC50 value for inhibition of ACh-induced α7 responses in Xenopus laevis oocytes (Fig. 3A) from 0.52 nM to 2.0 nM with corresponding Hill coefficients of 2.4 ± 0.2 (2.0-2.8) and 1.5 ± 0.1 (1.3-1.8), respectively. Assuming there are 5 binding sites per α7 homopentamer, that occupation of only 1 site is required to block function, and all sites are equal with respect to ligand binding affinity (Whiteaker et al. 2007, Palma et al. 1996), this yields a Kd value of 13.2 ± 1.45 nM. In contrast to FITC-α-BgTx (Fig. 3B), Cy3-ArIB[V11L;V16A] (10 μM) did not have any significant activity on rα9α10 or mα1β1δε nAChRs. Next we assessed whether the observed rates of block and unblock were affected by conjugation with Cy3. At the concentrations of toxins used, 5 nM for ArIB[V11L;V16A] and 20 nM for Cy3-ArIB[V11L;V16A], the observed rates of block were similar (Fig. 4) indicating that the observed rate constant (kob) for Cy3-ArIB[V11L;V16A] is substantially smaller than the kob for ArIB[V11L;V16A]. Measurement of current recovery showed no difference (p > 0.05) in the rate of unblock between Cy3-ArIB[V11L;V16A] and the parent peptide indicating similar dissociation rate constants (koff). The ACh response recovered from block by Cy3-ArIB[V11L;V16A] to 5.7 ± 1.8% compared to 5.9 ± 0.7% for ArIB[V11L;V16A] after 15 minutes of wash (Fig. 4A,B). Thus, conjugation of the parent peptide with Cy3 produces an analog with slower on-rate kinetics and with a small loss of potency compared to the parent peptide. However, Cy3-ArIB[V11L;V16A] retains the slow off-rate and remarkable selectivity of the parent peptide for the α7 nAChR subtype. At 100 nM Cy3-ArIB[V11L;V16A], the average block of rα3β4 was 4.6 ± 1.8% (Fig.3A) and for rα3β2 no block was detected (average response was 104.8 ± 3.4%; Fig. 3A). Even at a concentration 5000-fold (10 μM) greater than the IC50 for rα7, significant block was only detected for rα3β4 (37%) (Fig. 4C) and rα3β2 (74%) (Fig. 4D). At more realistic concentrations (5-10x the Kd for α7) little-to-no block of α3* would occur. Furthermore, the off-rate kinetics of Cy3-ArIB[V11L;V16A] on these 3 subtypes are substantially different. Full recovery from block of rα3β2 and rα3β4 by Cy3-ArIB[V11L;V16A] occurred within 60 seconds (Fig. 4C,D).
Fig. 3.
Concentration-response analysis of ACh-induced current block by Cy3-ArIB[V11L;V16A] and FITC-α-BgTx. (A), Cy3-ArIB[V11L;V16A] blocks rα7 with an IC50 of 2.0 (1.7-2.2) nM, nH = 1.5 (1.3-1.8). Conjugation with Cy3 NHS ester shifts the concentration-response curve of ArIB[V11L;V16A] (dashed line) to the right. Note lack of activity for Cy3-ArIB[V11L;V16A] on rα9α10 and mα1β1δε whereas (B), FITC-α-BgTx blocks rα7 with an IC50 of 3.4 (2.3-5.0) nM, nH = 1.1 (0.7-1.4), rα9α10 92.4 (64.3-132.7) nM, nH = 1.1 (0.7-1.5), and mα1β1δε 2.2 (1.7-2.8) μM, nH = 1.4 (1.0-1.8). In graph A, dashed line indicates concentration-response data for ArIB[V11L;V16A] binding to rα7 denoted by ■ rα7, solid lines indicate concentration-response data for Cy3-ArIB[V11L;V16A] binding to ▼ rα7, • mα1β1δε, * rα2β2, + α2β4, ○ α3β2, ◆ α3β4, Δ α4β2, ▲ rα4β4, ◆ α6/α3β4*, □ α6/α3β2β3*, × rα9α10; *, denotes α6/α3 chimera. In graph B, solid lines indicate concentration-response data for FITC-α-BgTx binding to ■ rα7, × rα9α10, and, • mα1β1δε. r, rat; m, mouse; nH, Hill slope; ( ), 95% confidence interval.
Fig. 4.
Kinetics of block and unblock of ArIB[V11L;V16A] and Cy3-ArIB[V11L;V16A]. (A,B), representative trace recordings of Xenopus laevis oocytes expressing α7 nAChRs. After a stable ACh baseline was achieved, the oocytes were exposed to a concentration of toxin equal to 10x the IC50 for each respective toxin until a steady state level of block was achieved. The toxin solution was then changed to ND96 without toxin and the responses were monitored for recovery. At the concentrations used, the observed rates of block were similar. The responses recovered from block by ArIB[V11L;V16A] and Cy3-ArIB[V11L;V16A] to 5.9 ± 0.7% and 5.6 ± 1.8% respectively, after 15 minutes of wash which were also similar (p > 0.05). (C), 10 μM Cy3-ArIB[V11L;V16A] blocked α3β4 by 36.7 ± 6.2% and α3β2 by 74.1 ± 8.8%. C, ACh control response without toxin; W, washout; ±, SEM from at least 4 individual determinations.
To assess binding affinity of the Cy3 derivative at native α7 nAChRs, we used Cy3-ArIB[V11L;V16A] to compete with [125I]-α-BgTx for binding to α7 nAChRs from mouse hippocampal membranes. [125I]-α-BgTx is a standard radioligand for binding assays involving α7 nAChRs and is considered α7-selective when used on preparations from mammalian brain because other nAChRs that are bound by α-BgTx are not expressed in this tissue. Cy3-ArIB[V11L;V16A] potently displaced [125I]-α-BgTx from mouse hippocampal membranes with a of Ki value of 20.8 ± 1.4 nM and Hill coefficient value of 0.83 ± 0.05 (Fig. 5). Using the equation IC50/Kd=21/N-1 (see methods), this Ki value corresponds to a functional IC50 of 3.0 nM which is in good agreement with the IC50 value (2.0 nM) determined by the preceding Xenopus oocyte experiments. This confirms that addition of Cy3 allows retention of competitive binding at α7 nAChRs, similar to that of the parent compound ArIB[V11L;V16A]. However, conjugation with Cy3 does induce a modest loss of potency compared to ArIB[V11L;V16A]; binding Ki = 10.5 nM (Whiteaker et al. 2008).
Fig. 5.
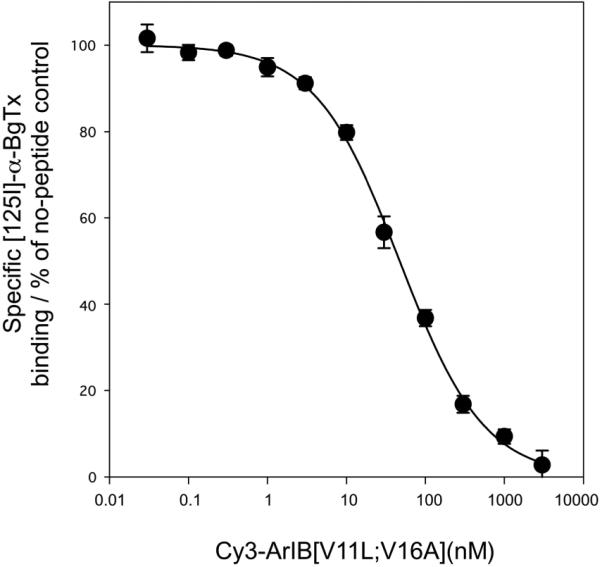
Cy3-ArIB[V11L;V16A] competes with [125I]-α-BgTx for binding sites from mouse hippocampal membranes. A competition binding assay was performed as described in materials and methods and the data were fit using a single-site model. Cy3-ArIB[V11L;V16A] displaced [125I]-α-BgTx with a Ki of 20.8 ± 1.4, nH 0.83 ± 0.05. nH, Hill slope; each point represents the mean ± SEM of 4 individual experimental determinations.
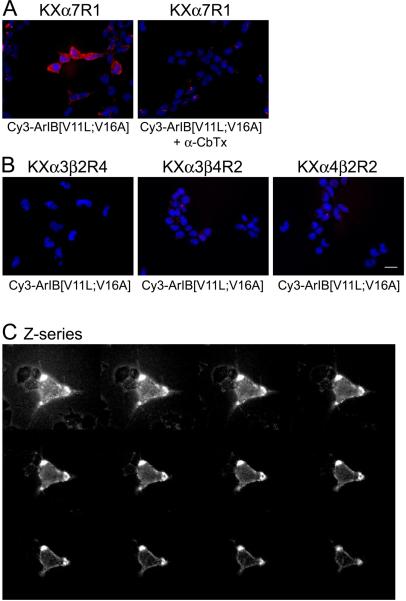
Cy3-ArIB[V11L;V16A] labels α7 nAChRs in KXα7R1 cells
The stably transfected HEK293 cell line KXα7R1 was used to assess the ability of Cy3-ArIB[V11L;V16A] to label α7 nAChRs. Application of 100 nM (Kd = 13.2 nM) Cy3-ArIB[V11L;V16A] to KXα7R1 cells resulted in bright and punctate labelling of the cells. This labelling was prevented by preapplication of the competitive α7 antagonist α-CbTx (Fig. 6A) indicating that Cy3-ArIB[V11L;V16A] labelling of the cells represents specific binding to α7 nAChRs. No labelling was observed in KXα3β2R4, KXα3β4R2, or KXα4β2R2 cells (Fig. 6B) indicating that Cy3-ArIB[V11L;V16A] labelling is specific to the α7 subtype. Using 100 nM (Kd = 23 nM) FITC-α-BgTx under these same conditions, we could not distinguish specific labelling from nonspecific labelling of KXα7R1 cells due to a large amount of apparent nonspecific binding to cellular debris and the coverslip. To determine whether the nonspecific FITC-α-BgTx binding could be reduced, the cells were pre-incubated for 20 min in PBS containing 10% (vol/vol) FBS and 10% (vol/vol) FBS was included in all toxin and wash solutions. Under these conditions, we were still not able to detect specific labelling of KXα7R1 cells although it appeared that nonspecific binding to the coverslip was reduced (see supplemental data).
Fig. 6.
Cy3-ArIB[V11L;V16A] selectively labels cells expressing α7 nAChRs. (A), KXα7R1 cells were labelled with 100 nM Cy3-ArIB[V11L;V16A] (red). The labelling was abolished by pre-incubating the cells with 2 μM α-CbTx. (B), No labelling was observed with 100 nM Cy3-ArIB[V11L;V16A] on KXα3β2R4, KXα3β4R2, or KXα4β2R2 cells. Cells in all images were counter stained with 2 μM Hoechst 33342 (blue) to label the nuclei. Scale bar is 20 μm. (C), 1 μm interval Z-series projection of KXα7R1 cells labelled with 100 nM Cy3-ArIB[V11L;V16A]. All images were taken with a cooled CCD camera at 40x magnification and processed with NIH ImageJ.
Discussion
Multiple subtypes of nAChRs are expressed within the same tissue creating a need for highly specific probes with which to study these subtypes. The 500 species of carnivorous cone snails in aggregate produce thousands of distinct nAChR antagonists that they use to help immobilize their prey. The subtype specificity of these peptides enables their development as novel, specific tools (Olivera et al. 2008). In this study, we utilized the recently reported α-CTx ArIB as a platform to develop a fluorescent ligand for α7 nAChRs. The ArIB[V11L;V16A] analog was chosen due to both its high potency and slow off-rate kinetics. The addition of Cy3 to α-CTx ArIB[V11L;V16A] resulted in a single homogeneous product (Fig. 2) owing to the single conjugation site present at the N-terminus of the synthetic parent peptide. This contrasts to the situation with α-BgTx whose sequence is ~4-fold longer than α-CTx ArIB[V11L;V16A] and that has multiple free amino groups from multiple Lys residues and the N-terminus to which FITC may couple (Basus et al. 1993). HPLC analysis of FTIC-α-BgTx confirms the presence of multiple compounds; this heterogeneity may be compounded by the heterogeneous nature of the parent venom-derived compound (unpublished observations).
Analysis of the functional effects of Cy3-ArIB[V11L;V16A] indicates that it retains the high potency and selectivity of ArIB[V11L;V16A] for α7 nAChRs. However, because of the very slow off-rate kinetics, we were unable to determine precise koff values for Cy3-ArIB[V11L;V16A]. Functional studies often introduce variables that can make accurate measurements of peptide kinetics difficult. In this case, full recovery from block by both the parent peptide and the Cy3 conjugate takes hours. This exceeds the practical limitations of oocyte recording time. Furthermore, current run-up or current run-down in oocytes can also hinder accurate measurements of recovery rates. For these reasons, we chose to provide values for percent recovery after 15 minutes of toxin wash-out rather than koff values. Since we cannot accurately measure koff, determining kon is also difficult. The values provided allow only an estimate of peptide kinetics but nevertheless provides useful information for functional and labelling experiments. Despite the difficulties in accurately measuring kinetics, estimates of peptide affinities determined by concentration-response studies in oocytes are in good agreement with the affinities estimated by radioligand binding assays. Of note, the slow reversibility of Cy3-ArIB[V11L;V16A] allows long wash times which reduces non-specific binding in labelling experiments. The combination of properties possessed by Cy3-ArIB[V11L;V16A] make it well-suited for use in fluorescent labelling applications and it should be especially useful in native tissues where multiple nAChR subtypes are expressed.
Next we used stably transfected mammalian cell lines that express rα7, rα3β2, rα3β4, and rα4β2 to assess the fluorescent labelling properties of Cy3-ArIB[V11L;V16A]. Cy3-ArIB[V11L;V16A] labelled rα7 nAChRs in KXα7R1 cells and the labelling could be prevented by preincubation with the competitive antagonist α-CbTx. The labelling had the appearance of puncta located on the membrane of the cells. The majority of the observed labelling is likely to be associated with receptors expressed on the cell surface as the binding was performed at 4°C to prevent active internalization of the ligand-bound receptor by endocytotic mechanisms (Wilkins et al. 2008). Furthermore, when 1 μm interval z-series images or 3D volume renditions of a z-series stack were examined, the labelling appeared to be associated only with the membrane (Fig 6C). In contrast to these results, we did not observe specific labelling in KXα7R1 exposed to FITC-α-BgTx. This indicates that under the conditions used in this study, Cy3-ArIB[V11L;V16A] has superior specific labelling properties. We did not detect any significant Cy3-ArIB[V11L;V16A] labelling in HEK293 cells expressing rα3β2, rα3β4, or rα4β2.
Recent recognition that α7 nAChRs are co-expressed with α9α10 and α1* nAChRs has made the creation of an α7-selective fluorescent ligand a priority. The detection of α7 nAChRs using standard pharmacological techniques has been complicated due to the relative lack of highly selective ligands for the α7 nAChRs. This is particularly problematic in tissues that express multiple nAChR subtypes that are sensitive to α-BgTx and MLA. These receptor subtypes include α1*, α6*, α7, and α9*. Although α9* is not expressed in mammalian brain, it is co-expressed with α7 and α1* in several peripheral tissues. It is well established that DRG neurons express α7 nAChRs (Genzen et al. 2001, Genzen & McGehee 2003, Rau et al. 2005, Haberberger et al. 2004) and there is increasing evidence to support DRG expression of α9* (Lips et al. 2006, Rau et al. 2005, Haberberger et al. 2004). In Rau et al. (2005), the authors suggest the presence of a nAChR subtype with a distinct pharmacological profile but were unable to definitively identify this receptor as α9α10 because of the lack of selective antagonists of α9α10 nAChRs.
The α9α10 nAChR has been most extensively characterized in the auditory system where it functions as a ligand-gated ion channel (Zorrilla de San Martin et al. 2007, Gomez-Casati et al. 2009, Taranda et al. 2009, Vetter et al. 2007, Ellison et al. 2006, Elgoyhen et al. 1994). The α1β1δγ (fetal) and α1β1δε (adult) nAChR subtypes are generally thought of as being expressed exclusively at the neuromuscular junction and are sensitive to α-BgTx. However, a recent report suggests the presence of the fetal subtype in the vestibular and cochlear systems of the mouse inner ear (Scheffer et al. 2007). Immune cells, including lymphocytes and macrophages, also express the α7 subtype which has been extensively implicated in the inflammatory roles of these cell types (for review see (de Jonge & Ulloa 2007). Macrophages and lymphocytes also express α9α10 receptors (Peng et al. 2004, Grau et al. 2007, Biallas et al. 2007, Kawashima & Fujii 2004) but as is with the case of DRG, the functional role of this subtype expressed by these cells is not known. In addition, α7 and α9α10 nAChRs have been reported in human sperm (Kumar & Meizel 2005) and keratinocytes (Chernyavsky et al. 2007, Arredondo et al. 2002). The α10 subunit has been suggested to require the α9 subunit for functional expression (Sgard et al. 2002). Interestingly, there are several sites where α7 and α10 have been detected but not α9. In the rat, these sites include the heart (Dvorakova et al. 2005), thoracic DRG neurons (Haberberger et al. 2004) and sympathetic ganglia (Lips et al. 2006) but the functional significance of α10 expression in the absence of α9 is not known.
The overlapping pharmacological profiles and expression patterns of α1*, α7, α9*, and α10* nAChRs presents a difficult problem when studying these receptor subtypes. Other techniques for identifying α7 receptors present their own unique caveats. For example, the specificity of commercially available α7 antibodies has been questioned following two recent reports of antibody labelling in α7-knockout mice (Moser et al. 2007, Herber et al. 2004). In the present study we show that Cy3-ArIB[V11L;V16A] binds α7 nAChRs but fails to bind to cells that lack this subtype. Thus, Cy3-ArIB[V11L;V16A] represents a valuable new addition to the collection of available nAChR probes.
Supplementary Material
Acknowledgement
Work supported by MH53631 and GM48677 to J.M.M, DA12242 to P.W., and NIH DA13199 to Y.X.We thank Dr. Doju Yoshikami for thoughtful discussion of the data and assistance with the microscopy experiments.
Abbreviations
- ACh
acetylcholine
- nAChR
nicotinic acetylcholine receptor
- α-BgTx
α-bungarotoxin
- α-CbTx
α-cobratoxin
- FITC-α-BgTx
fluorescein isothiocyanate-α-bungarotoxin
- [125I]-α-BgTx
monoiodinated α-bungarotoxin
- α-CTx
α-conotoxin
- MLA
methyllycaconitine
- FBS
fetal bovine serum
- BSA
bovine serum albumin
- DMSO
dimethylsulfoxide
- RT
room temperature
- PBS
phosphate buffered saline
- RP-HPLC
reverse phase-HPLC
- MALDI-TOF MS
matrix assisted laser desorption ionization-time of flight mass spectrometry
- CCD
charged coupled device
- DRG
dorsal root ganglia
- HEK293
human embryonic kidney 293
Footnotes
denotes the possible presence of additional nicotinic acetylcholine receptor subunits
References
- Arredondo J, Nguyen VT, Chernyavsky AI, Bercovich D, Orr-Urtreger A, Kummer W, Lips K, Vetter DE, Grando SA. Central role of alpha7 nicotinic receptor in differentiation of the stratified squamous epithelium. J Cell Biol. 2002;159:325–336. doi: 10.1083/jcb.200206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basus VJ, Song G, Hawrot E. NMR solution structure of an alpha-bungarotoxin/nicotinic receptor peptide complex. Biochemistry. 1993;32:12290–12298. doi: 10.1021/bi00097a004. [DOI] [PubMed] [Google Scholar]
- Biallas S, Wilker S, Lips KS, Kummer W, Grando SA, Padberg W, Grau V. Immunohistochemical detection of nicotinic acetylcholine receptor subunits alpha9 and alpha10 in rat lung isografts and allografts. Life Sci. 2007;80:2286–2289. doi: 10.1016/j.lfs.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem. 1996;271:7522–7528. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Chernyavsky AI, Arredondo J, Vetter DE, Grando SA. Central role of alpha9 acetylcholine receptor in coordinating keratinocyte adhesion and motility at the initiation of epithelialization. Exp Cell Res. 2007;313:3542–3555. doi: 10.1016/j.yexcr.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaj MI, Fei-Yin M, Dukat M, Glassco W, Glennon RA, Martin BR. Antinociceptive responses to nicotinic acetylcholine receptor ligands after systemic and intrathecal administration in mice. J Pharmacol Exp Ther. 1998;284:1058–1065. [PubMed] [Google Scholar]
- Damaj MI, Fonck C, Marks MJ, Deshpande P, Labarca C, Lester HA, Collins AC, Martin BR. Genetic approaches identify differential roles for alpha4beta2* nicotinic receptors in acute models of antinociception in mice. J Pharmacol Exp Ther. 2007;321:1161–1169. doi: 10.1124/jpet.106.112649. [DOI] [PubMed] [Google Scholar]
- Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu Rev Pharmacol Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- de Jonge WJ, Ulloa L. The alpha7 nicotinic acetylcholine receptor as a pharmacological target for inflammation. Br J Pharmacol. 2007;151:915–929. doi: 10.1038/sj.bjp.0707264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorakova M, Lips KS, Bruggmann D, Slavikova J, Kuncova J, Kummer W. Developmental changes in the expression of nicotinic acetylcholine receptor alpha-subunits in the rat heart. Cell Tissue Res. 2005;319:201–209. doi: 10.1007/s00441-004-1008-1. [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S. Alpha 9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell. 1994;79:705–715. doi: 10.1016/0092-8674(94)90555-x. [DOI] [PubMed] [Google Scholar]
- Ellison M, Haberlandt C, Gomez-Casati ME, Watkins M, Elgoyhen AB, McIntosh JM, Olivera BM. Alpha-RgIA: a novel conotoxin that specifically and potently blocks the alpha9alpha10 nAChR. Biochemistry. 2006;45:1511–1517. doi: 10.1021/bi0520129. [DOI] [PubMed] [Google Scholar]
- Genzen JR, McGehee DS. Short- and long-term enhancement of excitatory transmission in the spinal cord dorsal horn by nicotinic acetylcholine receptors. Proc Natl Acad Sci U S A. 2003;100:6807–6812. doi: 10.1073/pnas.1131709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genzen JR, Van Cleve W, McGehee DS. Dorsal root ganglion neurons express multiple nicotinic acetylcholine receptor subtypes. J Neurophysiol. 2001;86:1773–1782. doi: 10.1152/jn.2001.86.4.1773. [DOI] [PubMed] [Google Scholar]
- Gerzanich V, Anand R, Lindstrom J. Homomers of alpha 8 and alpha 7 subunits of nicotinic receptors exhibit similar channel but contrasting binding site properties. Mol Pharmacol. 1994;45:212–220. [PubMed] [Google Scholar]
- Gomez-Casati ME, Wedemeyer C, Taranda J, Lipovsek M, Dalamon V, Elgoyhen AB, Katz E. Electrical Properties and Functional Expression of Ionic Channels in Cochlear Inner Hair Cells of Mice Lacking the alpha10 Nicotinic Cholinergic Receptor Subunit. J Assoc Res Otolaryngol. 2009 doi: 10.1007/s10162-009-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Maggi R, Longhi R, Hanke W, Klinke N, Clementi F. Alpha7 and alpha8 nicotinic receptor subtypes immunopurified from chick retina have different immunological, pharmacological and functional properties. Eur J Neurosci. 1997;9:1201–1211. doi: 10.1111/j.1460-9568.1997.tb01475.x. [DOI] [PubMed] [Google Scholar]
- Grau V, Wilker S, Hartmann P, Lips KS, Grando SA, Padberg W, Fehrenbach H, Kummer W. Administration of keratinocyte growth factor (KGF) modulates the pulmonary expression of nicotinic acetylcholine receptor subunits alpha7, alpha9 and alpha10. Life Sci. 2007;80:2290–2293. doi: 10.1016/j.lfs.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Haberberger RV, Bernardini N, Kress M, Hartmann P, Lips KS, Kummer W. Nicotinic acetylcholine receptor subtypes in nociceptive dorsal root ganglion neurons of the adult rat. Auton Neurosci. 2004;113:32–42. doi: 10.1016/j.autneu.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Herber DL, Severance EG, Cuevas J, Morgan D, Gordon MN. Biochemical and histochemical evidence of nonspecific binding of alpha7nAChR antibodies to mouse brain tissue. J Histochem Cytochem. 2004;52:1367–1376. doi: 10.1177/002215540405201013. [DOI] [PubMed] [Google Scholar]
- Kawashima K, Fujii T. Expression of non-neuronal acetylcholine in lymphocytes and its contribution to the regulation of immune function. Front Biosci. 2004;9:2063–2085. doi: 10.2741/1390. [DOI] [PubMed] [Google Scholar]
- Klink R, de Kerchove d'Exaerde A, Zoli M, Changeux JP. Molecular and physiological diversity of nicotinic acetylcholine receptors in the midbrain dopaminergic nuclei. J Neurosci. 2001;21:1452–1463. doi: 10.1523/JNEUROSCI.21-05-01452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Meizel S. Nicotinic acetylcholine receptor subunits and associated proteins in human sperm. J Biol Chem. 2005;280:25928–25935. doi: 10.1074/jbc.M502435200. [DOI] [PubMed] [Google Scholar]
- Lips KS, Konig P, Schatzle K, Pfeil U, Krasteva G, Spies M, Haberberger RV, Grando SA, Kummer W. Coexpression and spatial association of nicotinic acetylcholine receptor subunits alpha7 and alpha10 in rat sympathetic neurons. J Mol Neurosci. 2006;30:15–16. doi: 10.1385/JMN:30:1:15. [DOI] [PubMed] [Google Scholar]
- Lips KS, Pfeil U, Kummer W. Coexpression of alpha 9 and alpha 10 nicotinic acetylcholine receptors in rat dorsal root ganglion neurons. Neuroscience. 2002;115:1–5. doi: 10.1016/s0306-4522(02)00274-9. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Azam L, Staheli S, Dowell C, Lindstrom JM, Kuryatov A, Garrett JE, Marks MJ, Whiteaker P. Analogs of alpha-conotoxin MII are selective for alpha6-containing nicotinic acetylcholine receptors. Mol Pharmacol. 2004;65:944–952. doi: 10.1124/mol.65.4.944. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Plazas PV, Watkins M, Gomez-Casati ME, Olivera BM, Elgoyhen AB. A novel alpha-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat alpha9alpha10 and alpha7 nicotinic cholinergic receptors. J Biol Chem. 2005;280:30107–30112. doi: 10.1074/jbc.M504102200. [DOI] [PubMed] [Google Scholar]
- Mogg AJ, Whiteaker P, McIntosh JM, Marks M, Collins AC, Wonnacott S. Methyllycaconitine is a potent antagonist of alpha-conotoxin-MII-sensitive presynaptic nicotinic acetylcholine receptors in rat striatum. J Pharmacol Exp Ther. 2002;302:197–204. doi: 10.1124/jpet.302.1.197. [DOI] [PubMed] [Google Scholar]
- Moser N, Mechawar N, Jones I, et al. Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures. J Neurochem. 2007;102:479–492. doi: 10.1111/j.1471-4159.2007.04498.x. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Quik M, Vincler M, McIntosh JM. Subtype-selective conopeptides targeted to nicotinic receptors: Concerted discovery and biomedical applications. Channels (Austin) 2008;2 doi: 10.4161/chan.2.2.6276. [DOI] [PubMed] [Google Scholar]
- Palma E, Bertrand S, Binzoni T, Bertrand D. Neuronal nicotinic alpha 7 receptor expressed in Xenopus oocytes presents five putative binding sites for methyllycaconitine. J Physiol. 1996;491(Pt 1):151–161. doi: 10.1113/jphysiol.1996.sp021203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopolou S, Hartmann P, Lips KS, Kummer W, Haberberger RV. Nicotinic receptor mediated stimulation of NO-generation in neurons of rat thoracic dorsal root ganglia. Neurosci Lett. 2004;361:32–35. doi: 10.1016/j.neulet.2003.08.064. [DOI] [PubMed] [Google Scholar]
- Peng H, Ferris RL, Matthews T, Hiel H, Lopez-Albaitero A, Lustig LR. Characterization of the human nicotinic acetylcholine receptor subunit alpha (alpha) 9 (CHRNA9) and alpha (alpha) 10 (CHRNA10) in lymphocytes. Life Sci. 2004;76:263–280. doi: 10.1016/j.lfs.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Rau KK, Johnson RD, Cooper BY. Nicotinic AChR in subclassified capsaicin-sensitive and -insensitive nociceptors of the rat DRG. J Neurophysiol. 2005;93:1358–1371. doi: 10.1152/jn.00591.2004. [DOI] [PubMed] [Google Scholar]
- Satkunanathan N, Livett B, Gayler K, Sandall D, Down J, Khalil Z. Alpha-conotoxin Vc1.1 alleviates neuropathic pain and accelerates functional recovery of injured neurones. Brain Res. 2005;1059:149–158. doi: 10.1016/j.brainres.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Scheffer D, Sage C, Plazas PV, et al. The alpha1 subunit of nicotinic acetylcholine receptors in the inner ear: transcriptional regulation by ATOH1 and co-expression with the gamma subunit in hair cells. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2007.04980.x. [DOI] [PubMed] [Google Scholar]
- Sgard F, Charpantier E, Bertrand S, Walker N, Caput D, Graham D, Bertrand D, Besnard F. A novel human nicotinic receptor subunit, alpha10, that confers functionality to the alpha9-subunit. Mol Pharmacol. 2002;61:150–159. doi: 10.1124/mol.61.1.150. [DOI] [PubMed] [Google Scholar]
- Taranda J, Maison SF, Ballestero JA, et al. A point mutation in the hair cell nicotinic cholinergic receptor prolongs cochlear inhibition and enhances noise protection. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitsky M, Rothlin CV, Katz E, Elgoyhen AB. Mixed nicotinic-muscarinic properties of the alpha9 nicotinic cholinergic receptor. Neuropharmacology. 2000;39:2515–2524. doi: 10.1016/s0028-3908(00)00124-6. [DOI] [PubMed] [Google Scholar]
- Vetter DE, Katz E, Maison SF, Taranda J, Turcan S, Ballestero J, Liberman MC, Elgoyhen AB, Boulter J. The alpha10 nicotinic acetylcholine receptor subunit is required for normal synaptic function and integrity of the olivocochlear system. Proc Natl Acad Sci U S A. 2007;104:20594–20599. doi: 10.1073/pnas.0708545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincler M, McIntosh JM. Targeting the alpha9alpha10 nicotinic acetylcholine receptor to treat severe pain. Expert Opin Ther Targets. 2007;11:891–897. doi: 10.1517/14728222.11.7.891. [DOI] [PubMed] [Google Scholar]
- Vincler M, Wittenauer S, Parker R, Ellison M, Olivera BM, McIntosh JM. Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc Natl Acad Sci U S A. 2006;103:17880–17884. doi: 10.1073/pnas.0608715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yu M, Ochani M, et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- Whiteaker P, Christensen S, Yoshikami D, Dowell C, Watkins M, Gulyas J, Rivier J, Olivera BM, McIntosh JM. Discovery, synthesis, and structure activity of a highly selective alpha7 nicotinic acetylcholine receptor antagonist. Biochemistry. 2007;46:6628–6638. doi: 10.1021/bi7004202. [DOI] [PubMed] [Google Scholar]
- Whiteaker P, Marks MJ, Christensen S, Dowell C, Collins AC, McIntosh JM. Synthesis and characterization of 125I-alpha-conotoxin ArIB[V11L;V16A], a selective alpha7 nicotinic acetylcholine receptor antagonist. J Pharmacol Exp Ther. 2008;325:910–919. doi: 10.1124/jpet.108.136895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins ME, Li X, Smart TG. Tracking cell surface GABAB receptors using an alpha-bungarotoxin tag. J Biol Chem. 2008;283:34745–34752. doi: 10.1074/jbc.M803197200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Abdrakhmanova GR, Baydyuk M, Hernandez S, Kellar KJ. Rat neuronal nicotinic acetylcholine receptors containing alpha7 subunit: pharmacological properties of ligand binding and function. Acta Pharmacol Sin. 2009 doi: 10.1038/aps.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Kellar KJ. The comparative pharmacology and up-regulation of rat neuronal nicotinic receptor subtype binding sites stably expressed in transfected mammalian cells. J Pharmacol Exp Ther. 2004;310:98–107. doi: 10.1124/jpet.104.066787. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Meyer EL, Thompson JM, Surin A, Wroblewski J, Kellar KJ. Rat alpha3/beta4 subtype of neuronal nicotinic acetylcholine receptor stably expressed in a transfected cell line: pharmacology of ligand binding and function. Mol Pharmacol. 1998;54:322–333. doi: 10.1124/mol.54.2.322. [DOI] [PubMed] [Google Scholar]
- Zorrilla de San Martin J, Ballestero J, Katz E, Elgoyhen AB, Fuchs PA. Ryanodine is a positive modulator of acetylcholine receptor gating in cochlear hair cells. J Assoc Res Otolaryngol. 2007;8:474–483. doi: 10.1007/s10162-007-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.