Abstract
The genes that encode for CYP3A4 and CYP3A5 are located in the same region (CYP3A cluster) on chromosome 7. Midazolam (MDZ) is a substrate for both CYP3A4 and CYP3A5. We hypothesize that MDZ disposition in vivo is associated with genotypes of the CYP3A cluster. A meta-analysis of the pharmacokinetic (PK) parameters from 7 clinical trials was performed, in which MDZ was administered both intravenously and orally. DNA samples were available from 116 subjects. There were significant ethnic differences in the allelic frequencies of these 4 common single nucleotide polymorphisms (SNPs) in the CYP3A cluster. Significant linkage disequilibrium was found between CYP3A5*3 and CYP3A4*1A in Caucasians, and between CYP3A5*1 and CYP3A4*1B in African Americans. There were no differences in MDZ disposition in vivo between different genotypes, haplotypes and diplotypes in the CYP3A cluster (P>0.05). No significant differences in MDZ PK parameters were observed between Caucasians and African Americans. Women had higher weight-corrected systemic and oral clearance than men, but dose-adjusted AUC and bioavailability differences were not observed between sexes. The clinical importance of elevated CYP3A activity in women remains to be determined. The rGCs of MDZ PK parameters were between 0.3% and 13.6%. In conclusion, meta-analysis of seven studies suggests that environmental factors explain the majority of CYP3A activity variation. Further studies are necessary to define the functional significance of SNPs in the CYP3A cluster and the effects of CYP3A genotypes on MDZ disposition in vivo.
Keyworks: CYP3A, genetic/environmental variations, midazolam
Introduction
The human CYP3A enzyme subfamily is involved in the oxidative metabolism of a wide range of substrates, including more than 50% of all currently marketed drugs, endogenous steroids and xenobiotic chemicals (1, 2). CYP3A mediated activity exhibits approximately 10-fold difference between individuals, which presents a challenge in predicting drug effect and safety (3). While concomitant drugs and environment factors partly account for the variability, genetic polymorphisms and variation in expression may play a critical role in between-subject variability of CYP3A activity (4, 5).
The CYP3A gene cluster, which is located on chromosome 7q22 and spans ∼220 kb, consists of four genes including CYP3A4, CYP3A5, CYP3A7 and CYP3A43 (6). CYP3A4 and CYP3A5 are the major metabolism enzymes in adults and are expressed primarily in the liver and small intestine. CYP3A7 is expressed in fetal liver and plays an important role in the metabolism of endogenous substrates. CYP3A43 is expressed at very low levels in adult human livers. Its contribution to the elimination of CYP3A substrates is regarded to be negligible (7, 8).
Although a number of single nucleotide polymorphisms (SNPs) at the CYP3A4 locus have been identified, few of them occur frequently enough to contribute to variations in CYP3A activity. No evidence has shown functionally important allelic mutations in the CYP3A4 coding region, the between-subject variability in CYP3A4 activity may be the result of transcriptional regulation (1, 9-11). For instance, the CYP3A4*1B (-392A>G) is a common SNP located in the 5′ promoter region. The role of this polymorphism on enzyme activity in vivo remains controversial (9). Functional SNPs are more commonly observed in the CYP3A5 gene. The CYP3A5*3 (6989A>G) SNP in intron 3 introduces a cryptic splice site that results in a frame shift and truncated protein (12). The CYP3A5*6 (14690G>A) SNP in exon 7 leads to a splicing defect, and the CYP3A5*7 (insertion at 27131_32) SNP in exon 11 results in a premature stop codon (13, 14). It has been suggested that CYP3A5*6 and CYP3A5*7 be considered together in conjunction with CYP3A5*3 in order to predict significantly diminished CYP3A5 expression (4). Previous studies have shown that in Caucasians CYP3A4*1B is in strong linkage disequilibrium with the functional CYP3A5*1. About 80% of Caucasians are homozygous for the CYP3A5*3 and CYP3A4*1A alleles (15, 16).
Midazolam (MDZ), which can be administrated both intravenously and orally, is selectively metabolized by CYP3A4 and CYP3A5 to its primary metabolite, 1′-hydroxymidazolam, and is not a substrate of P-glycoprotein (17, 18). MDZ exhibits most of the desired characteristics to be used as a probe to measure CYP3A activity, although MDZ clearance may be influenced by hepatic blood flow (19-24). Systemic and apparent oral clearances of MDZ are pharmacokinetic (PK) parameters recognized as biomarkers for hepatic and intestinal CYP3A activity (23, 25). Intravenous (IV) administration of MDZ reflects only hepatic CYP3A activity, whereas orally administered MDZ is a measure of intestinal and hepatic CYP3A activities (25-27). Simultaneous IV and oral (PO) MDZ administration has been used to examine the individual contributions of intestinal and hepatic CYP3A to metabolism (25-27).
Although remarkable ethnic differences exist within the CYP3A cluster structure, the genetic component of variability (i.e. between-subject variability) remains uncertain (28-30). Some studies also suggest that there are sex differences in CYP3A activity, but the results are inconsistent (31-33). The objectives of our study are to investigate whether IV and PO MDZ disposition in vivo is associated with genotypes of the CYP3A cluster, ethnicity, sex or age in healthy volunteers and to understand the genetic component of its variability.
Materials and Methods
Study design
We reviewed 7 clinical trials conducted from 1998 to 2003 by our research team (Table 1). In each study single-dose MDZ was administrated both IV and PO. In 5 studies, subjects were simultaneously administered a single IV dose (0.05 mg/kg over 30 minutes) of MDZ and an oral dose of 15N-MDZ (3 mg) after an overnight fast. In the additional 2 studies, oral MDZ (4 mg) was administered 24 hrs after the IV dose. All drugs and food known to affect CYP3A activity were prohibited before and for the duration of the studies. For each subject, blood samples for MDZ concentrations were obtained over a period of 12 to 24 hours. The sample sizes varied among studies (Table 1). The MDZ serum concentrations were determined using a previously published method (34, 35). PK parameter estimates were calculated using non-compartmental methods (WinNonLin 4.0; Pharsight, Mountain View, CA). Dose-adjusted IV and PO area under the concentration-time curve (AUC), weight-corrected IV and PO clearance (CL), and bioavailability (F) were used as MDZ PK parameters, since dosages were different in these 7 trials and body size appears to be an important determinant of between-subject variability (36). Blood was also collected for DNA analysis.
Table 1.
Characteristics of the 7 MDZ Clinical Trials
| Study title | Time | # subjects | # DNA samples | Published information |
|---|---|---|---|---|
| Assessment of the contribution of intestinal CYP3A to the first pass metabolism of midazolam in elderly volunteers (643) | 2002 | 16 | 13 | Quinney et al. BJCP 2007; 65(1): 98-109 |
| The effect of age, sex, and rifampin administration on intestinal and hepatic cytochrome P450 3A activity (676) | 2002 | 52 | 24 | Gorski et al. CPT 2003; 74: 275-87 |
| The effect of hormone replacement therapy on CYP3A activity (684) | 1998 | 36 | 17 | Gorski et al. CPT 2000; 68: 412-7 |
| The effect of age and gender on the duration of CYP3A inhibition by clarithromycin in humans (855) | 2001 | 15 | 11 | Not published |
| The effect of clarithromycin administration on intestinal and hepatic CYP3A activity in healthy volunteers: A Pilot Study (887A) | 2002 | 13 | 8 | Not published |
| Effect of multiple doses of Ketoconazole on Cytochrome P450 3A activity (1148c) | 2003 | 24 | 24 | Not published |
| Relationship between CYP3A5 genotypes and effect of verapamil on midazolam pharmacokinetics (1056) | 2002 | 26 | 20 | Jin, Y. et al. CPT 2007; 82: 579-85 |
Study population
These 7 clinical trials were approved by the Institutional Review Board of Indiana University-Purdue University Indianapolis and informed consent was obtained from each of the 176 volunteers who participated. Subjects were healthy as determined by medical history, physical examination, 12-lead electrocardiogram, and laboratory screening. DNA samples were collected from 116 subjects among the cohort (Table 1).
Genotyping Methods
Genomic DNA was extracted from whole blood and isolated using a QIAamp DNA blood Midi Kit (Qiagen, Valencia, CA). DNA concentration was detected by SmartSpec™ Plus spectrophotometry (BIO-RAD, Hercules, CA). All DNA samples were genotyped for CYP3A5*3 (6986A>G), CYP3A5*6 (14690G>A), CYP3A5*7 (insertion at 27131_32) and CYP3A4*1B (-392A>G). The genotyping of CYP3A5*3, *6 and *7 was determined using real time PCR as described previously (14). The CYP3A4*1B allele was detected with a TaqMan™ assay from the CGAP Web site (SNP500Cancer, dsSNP ID: rs2740574) and confirmed by sequencing.
Statistical Considerations
The distributions of demographic data were inspected visually. Genotype frequencies were tested for departure from Hardy–Weinberg Equilibrium using χ2 test with one degree of freedom. Frequencies of the four CYP3A SNPs were highly variable among ethnic groups and therefore population-specific pair wise linkage disequilibrium and haplotype frequencies were estimated using Haploview (Ver 3.1, from http://www.broad.mit.edu/mpg/haploview/). We stratified the genotype data based on ethnic background and determined CYP3A5 and CYP3A4*1B haplotype using PHASE II software (Version 2.1, from http://www.stat.washington.edu/stephens/phase). The overall differences in the PK parameters among subjects with different CYP3A5 and CYP3A4*1B genotypes were tested by F-tests in one-way ANOVA with repeated measures; and (dominance, recessive, gene-dose) effects of these genetic polymorphisms on PK parameters were compared by Student's t-tests. Student's t-test was also used to compare the PK parameters of MDZ with regard to ethnicity, sex and age. PK parameters were log-transformed, and their population between-subject variance, , and within-subject variance, , were estimated with the linear mixed model (PROC MIXED, SAS 9.1 Cary, NC). The genetic component was estimated as (37, 38). Please not that this genetic component estimate is different from the formula proposed by Karlow et al. (37). Because our SAS PROC MIXED and estimates are unbiased for between-subject and within-subject variances, respectively, our genetic component estimate doesn't have the bias correction.
Results
Study Characteristics
PK parameters of MDZ were collected from 7 clinical trials, in which single-dose MDZ was administrated both IV and PO. Results of some these 7 studies have been previously published (Table 1).
Subject Characteristics
A total of 116 subjects in these 7 trials had both MDZ disposition data and CYP3A cluster genotype data and were therefore used in this analysis. Among these 116 subjects, 64 subjects participated in only one trail and 22 subjects participated in 2 to 3 different clinical trials. The characteristics of the total 116 subjects and in groups based on the CYP3A5 haplotype are summarized in Table 2. There were significant differences among the three CYP3A5 haplotype groups with respect to ethnicity, sex and age (P<0.05).
Table 2.
Subject Characteristics of the Cohort Population and base on the CYP3A5 Genotype
| All subjects | CYP3A5*1/*1 | CYP3A5*1/*0 | CYP3A5*0/*0 | P value | |
|---|---|---|---|---|---|
| N | 116 | 10 | 24 | 82 | |
| Ethnicity | |||||
| Caucasian | 88 | 1 | 11 | 76 | 0.000 |
| African American | 26 | 7 | 13 | 6 | |
| Asian | 2 | 2 | 0 | 0 | |
| Gender | |||||
| Male | 55 | 7 | 14 | 34 | 0.039 |
| Female | 61 | 3 | 10 | 48 | |
| Age (year) † | 48.3±21.8 | 40.3±19.1 | 64.5±11.2 | 52.6±22.2 | 0.003 |
| ≤60 | 62 | 10 | 17 | 35 | 0.000 |
| >60 | 54 | 0 | 7 | 45 | |
| Weight (kg) | 76.2±16.0 | 73.7±17.0 | 82.3±15.7 | 74.8±15.7 | 0.106 |
There were two subjects whose age records were missing.
Genotype Determination
In the total 116 subjects, the allele frequencies of CYP3A5*1, *3, *6 and *7 were 19.0% 78.4%, 1.7% and 0.9% respectively. The most common genotype was CYP3A5*3*3. There was only 1 CYP3A5*6*6 and no CYP3A5*7*7 homozygotes in this cohort. The allele frequency of CYP3A4*1B was 20.7% in this population. The distribution of each allele satisfied the Hardy–Weinberg equilibrium (Table 3). We combined *3, *6, and *7 variant alleles into one CYP3A5 non-expresser allele group referred to as the *0 allele, because none of them produce any functional CYP3A5 protein (14). Ten subjects (8.6%) were classified as *1*1 homozygous expressers who were *1*1 for all 3 SNPs. Twenty four subjects (20.7%) were classified as *1*0 heterozygous expressers who carried only one of either *3, *6 or *7. Eighty two subjects (70.7%) were classified as non-expressers who were either homozygous for *3*3, *6*6 or *7*7 or carried either one *3 and *6, one *3 and *7, or one *6 and *7.
Table 3.
Genotype Frequency of the Cohort (N=116)
| Frequency (%) | CYP3A5*3 | CYP3A5*6 | CYP3A5*7 | CYP3A4*1B |
|---|---|---|---|---|
| Heterozygous | 25 (21.6%) | 2 (1.7%) | 2 (1.7%) | 18 (15.5%) |
| Homozygous Mutation | 78 (67.2%) | 1 (0.9%) | 0 (0%) | 15 (12.9%) |
| P value of Hardy–Weinberg Equilibrium | 0.997 | 0.871 | 0.500 | 0.966 |
Diplotype is the most likely pair of genotypes in each individual. We combined CYP3A5 genotype and CYP3A4*1B genotype to determine the diplotype of the CYP3A cluster and coded them into 9 groups. The diplotype distribution of the CYP3A cluster was significantly different in Caucasians and in African Americans (Table 4). In Caucasians, neither CYP3A5*6 nor CYP3A5*7 was found. 81.8% of subjects homozygous for CYP3A5*3 were also homozygous for CYP3A4*1A. Significant linkage disequilibrium was found between CYP3A5*3 and CYP3A4*1A in Caucasians (D′=0.716). The diplotypes in African Americans were more decentralized than in Caucasians, because the frequency of CYP3A4*1B allele was much higher in African Americans (78.8%) than in Caucasians (4.0%). A linkage disequilibrium was found between CYP3A5*1 and CYP3A4*1B in African Americans (D′=0.581).
Table 4.
Diplotype frequency of combined CYP3A5 genotypes with CYP3A4*1B genotypes in Caucasians and African Americans
| Diplotype Code | CYP3A5 genotype | CYP3A4*1B genotype | Caucasian (n=88) | African American (n=26) | ||
|---|---|---|---|---|---|---|
| Number | Frequency (%) | Number | Frequency (%) | |||
| 1 | *1*1 | *1B*1B | 0 | 0 | 6 | 23.1 |
| 2 | *1*1 | *1A*1B | 1 | 1.1 | 1 | 3.9 |
| 3 | *1*1 | *1A*1A | 0 | 0 | 0 | 0 |
| 4 | *1*0 | *1B*1B | 0 | 0 | 6 | 23.1 |
| 5 | *1*0 | *1A*1B | 3 | 3.4 | 7 | 26.9 |
| 6 | *1*0 | *1A*1A | 8 | 9.1 | 0 | 0 |
| 7 | *0*0 | *1B*1B | 0 | 0 | 3 | 11.5 |
| 8 | *0*0 | *1A*1B | 3 | 3.4 | 3 | 11.5 |
| 9 | *0*0 | *1A*1A | 73 | 83.0 | 0 | 0 |
2 of 116 subjects were Asians whose diplotypes were both code 3.
We combined *3, *6, and *7 variant alleles into one CYP3A5 non-expresser allele group referred to as the *0 allele.
The Association Between CYP3A Genotype and MDZ PK
Since CYP3A5 exhibits overlap in its cDNA sequence identity and substrate specificities with CYP3A4, genotyping for CYP3A5/3A4 haplotype or diplotype is necessary to understand the variations in the metabolism and clinical toxicity of drugs (39, 40). With regard to CYP3A5*3 and CYP3A4*1B polymorphisms, there were no significant differences in PK parameters of MDZ, including dose-adjusted AUC and weight-corrected CL following IV or PO administration, or bioavailability (Table 5). There were no significant differences in MDZ disposition among 9 CYP3A diplotype groups (P>0.05, data not shown). The groups of diplotype code 1 (homozygous for CYP3A5*1/*1 and CYP3A4*1B/*1B) and the group of code 9 (homozygous for CYP3A5*0/*0 and CYP3A4*1A/*1A) may be considered to show the most discrepancy between diplotypes, because both CYP3A5*1 and CYP3A4*1B may increase CYP3A activity. However, there were no significant differences in IV or PO MDZ PK parameters between these two diplotype groups.
Table 5.
PK parameters of MDZ in different genotype groups (Mean ± SE)
| PK parameters | Genotype | P value | ||
|---|---|---|---|---|
| CYP3A5 | ||||
| *1/*1 | *1/*0 | *0/*0 | ||
| N | 10 | 24 | 82 | |
| AUCiv (μg/L·h) | 147.9±19.7 | 142.0±9.2 | 158.2±6.6 | 0.566 |
| CLiv (L/h·kg) | 0.43±0.05 | 0.39±0.03 | 0.40±0.02 | 0.343 |
| AUCpo (μg/L·h) | 47.3±14.7 | 34.3±3.3 | 45.2±2.9 | 0.404 |
| CLpo (L/h·kg) | 1.87±0.32 | 1.87±0.27 | 1.90±0.18 | 0.996 |
| F (%) | 27.8±3.2 | 24.1±1.7 | 29.7±1.7 | 0.188 |
| CYP3A5*3 | ||||
| *1/*1 | *1/*3 | *3/*3 | ||
| N | 13 | 25 | 78 | |
| AUCiv (μg/L·h) | 139.7±15.9 | 150.8±9.0 | 157.4±6.9 | 0.451 |
| CLiv (L/h·kg) | 0.43±0.05 | 0.36±0.03 | 0.40±0.02 | 0.796 |
| AUCpo (μg/L·h) | 43.4±11.4 | 36.7±3.4 | 45.1±3.0 | 0.193 |
| CLpo (L/h·kg) | 1.90±0.39 | 1.99±0.28 | 1.86±0.17 | 0.934 |
| F | 28.0±4.2 | 24.0±1.7 | 29.6±1.6 | 0.210 |
| CYP3A*1B | ||||
| *1B/*1B | *1A/*1B | *1A/*1A | ||
| N | 15 | 18 | 83 | |
| AUCiv (μg/L·h) | 154.1±15.5 | 145.7±9.2 | 155.7±6.7 | 0.799 |
| CLiv (L/h·kg) | 0.39±0.04 | 0.41±0.03 | 0.40±0.02 | 0.889 |
| AUCpo (μg/L·h) | 41.0±10.0 | 38.6±3.6 | 44.4±2.9 | 0.672 |
| CLpo (L/h·kg) | 1.98±0.33 | 1.93±0.30 | 1.86±0.17 | 0.955 |
| F | 23.8±2.6 | 26.8±2.0 | 29.4±1.6 | 0.290 |
The Association Between Population Traits and MDZ PK
Population estimates of the PK parameters of MDZ are summarized in Table 6. Women exhibited a 19.4% higher IV weight-corrected CL (P=0.016) and 38.2% higher PO weight-corrected CL (P=0.026) than men (Fig. 1). There were no statistically significant differences for IV and PO dose-adjusted AUC or bioavailability between women and men (P>0.05). No statistically significant differences for IV and PO MDZ disposition were observed between Caucasians and African Americans or between young and elderly subjects (29.5±1.1 year vs. 70.8±0.6 year). In the 62 young subjects who were 19 to 55 years old, there were no significant differences in IV or PO MDZ PK parameters relative to the CYP3A cluster genotype (data not shown).
Table 6.
PK parameters of MDZ in different population groups (Mean ± SE)
| PK parameters | Population | P value | ||
|---|---|---|---|---|
| Sex | ||||
| Male | Female | |||
| N=116 | 55 | 61 | ||
| AUCiv (μg/L·h) | 150.0±7.2 | 157.5±7.8 | 0.486 | |
| CLiv (L/h·kg) | 0.36±0.02 | 0.43±0.02 | 0.016 | |
| AUCpo (μg/L·h) | 42.7±3.3 | 43.4±3.7 | 0.893 | |
| CLpo (L/h·kg) | 1.57±0.14 | 2.17±0.23 | 0.026 | |
| F (%) | 28.8±1.7 | 27.8±1.8 | 0.687 | |
| Race | ||||
| Caucasians | Africa Americans | |||
| N=114† | 88 | 26 | ||
| AUCiv (μg/L·h) | 155.7±6.4 | 148.0±10.2 | 0.552 | |
| CLiv (L/h·kg) | 0.39±0.02 | 0.41±0.03 | 0.530 | |
| AUCpo (μg/L·h) | 44.0±2.7 | 39.7±6.1 | 0.485 | |
| CLpo (L/h·kg) | 1.84±0.17 | 2.07±0.27 | 0.506 | |
| F | 29.1±1.5 | 25.2±1.9 | 0.191 | |
| Age | ||||
| <65 | ≥65 | |||
| N=114‡ | 62 | 52 | ||
| AUCiv (μg/L·h) | 155.3±7.2 | 151.3±8.1 | 0.711 | |
| CLiv (L/h·kg) | 0.40±0.02 | 0.40±0.02 | 0.828 | |
| AUCpo (μg/L·h) | 44.3±3.6 | 42.3±3.6 | 0.696 | |
| CLpo (L/h·kg) | 1.86±0.18 | 1.91±0.23 | 0.870 | |
| F | 27.3±1.2 | 30.0±2.3 | 0.296 | |
2 of 116 subjects were Asians.
2 of 116 subjects had no record of age.
Figure 1.
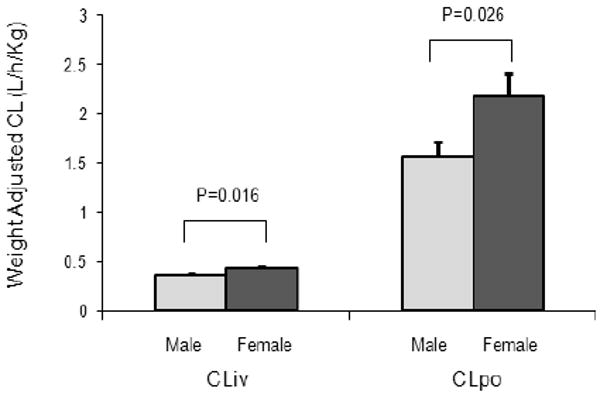
Women (n=61, dark grey bar) have significantly higher IV and oral weight-corrected CL than men (n=55, light gray bar).
Population Between-/Within-subject Variances and Genetic Component of Variability of MDZ PK Parameters
Table 7 displays the between- and within-subject variance for the MDZ PK parameters. The IV variances appear smaller than their PO counter parts. Interestedly, between-subject variance, , were relatively much smaller than within-subject variance, . The rGC s were between 0.3% and 13.6%.
Table 7.
Genetic Component of the MDZ PK Parameter Variances
| PK parameters | Variance Estimates | Genetic Component | ||||
|---|---|---|---|---|---|---|
| Log(AUCiv) | 0.0004 (0.0035) | 0.1262 (0.0166) | 0.3% | |||
| Log(AUCpo) | 0.0521 (0.0646) | 0.3483 (0.0746) | 13.0% | |||
| Log(CLiv) | 0.0181 (0.0267) | 0.1211 (0.0291) | 13.0% | |||
| Log(CLpo) | 0.0460 (0.0742) | 0.3742 (0.0845) | 10.9% | |||
| Log(F) | 0.0431 (0.0413) | 0.2621 (0.0508) | 13.6% | |||
and are population between-subject and within-subject variances, respectively. In log-scale of PK parameters, and can be interpreted as coefficients of variances of PK parameters in the raw scale.
Discussion
Although some previous studies have demonstrated that CYP3A5*1 alleles are associated with greater intestinal CYP3A activity and that affecting CYP3A4 transcription is relevant for between-subject differences in CYP3A4 enzyme expression, the primary in vivo mechanism and contribution of non-genetic factors to CYP3A variability is not clear (4, 11, 30, 41, 42).
Our study found that the allelic frequencies of these 4 SNPs in the CYP3A cluster are highly different between ethnic groups. The most common variant CYP3A5*3 had a frequency of 92.0% in Caucasians and 36.5% in African Americans, consistent with literature reports of 85-95% in Caucasians, 60-73% in Asians, and 27-50% in African Americans (6, 13, 14). The allelic frequencies of CYP3A4*1B in African Americans and in Caucasians in our study were 78.8% and 4.0%, respectively. This is also consistent with the CYP3A4*1B allele frequency being much higher in African Americans (54.6%) than in Hispanic Americans (9.3%) and Caucasians (3.6%) in other studies (5, 11). The haplotypes and diplotypes in African Americans were significantly different from Caucasians. Linkage disequilibrium was observed between CYP3A5*3 and CYP3A4*1A alleles in Caucasians, but between the CYP3A5*1 and CYP3A4*1B alleles in African Americans.
Although the allelic frequencies of the CYP3A genes were highly different among ethnic groups, no significant difference in MDZ disposition in vivo between Caucasians and African Americans was observed (P>0.05). Genetic polymorphisms in the CYP3A cluster were not associated with MDZ disposition. No significant association was observed between genotypes, haplotypes, or diplotypes in the CYP3A locus and MDZ PK parameters (P>0.05). These results were consistent with previous studies of smaller size (29, 43, 36, 44). Using MDZ as an in vivo probe in 57 healthy European- and African-American subjects, Floyd et al. reported that the variability of hepatic and intestinal CYP3A activity was modest (29). In addition, the common polymorphisms of CYP3A4*1B, CYP3A5*3, CYP3A5*6 and CYP3A5*7 did not appear to have important functional significance. Similarly, no significant genotype-phenotype or haplotype-phenotype associations for any of the CYP3A4*1B, CYP3A5*3, and CYP3A5*6 SNPs or haplotypes were found in 27 healthy volunteers receiving oral MDZ (43). Tateishi et al. found no statistically significant or clinically important inter-ethnic differences in CYP3A activity evident between 20 Caucasians and 22 Japanese, using MDZ as an in vivo probe (36). Despite affecting metabolism of MDZ in human liver microsomes, CYP3A5 genotype had no effect on the systemic or apparent oral clearances of MDZ or alfentanil among 99 healthy volunteers (44). Clearances were not different between African Americans (n=25) and Caucasians (n=68) or among CYP3A5 genotype groups within African Americans (44). These results indicate that the CYP3A genetic variants identified so far have only a limited impact on MDZ metabolism in vivo.
Since between-subject variations in CYP3A expression and activity are due to a combination of genetic and non-genetic factors such as hormone, health status, and environmental stimuli (4), these non-genetic factors may play a greater role in CYP3A variability. We examined this using a modified statistical estimate proposed by Kalow et al. (37, 38) to calculate the rGC. The lack of evidence of CYP3A genetic effect is partially reflected by the low genetic component, rGC, of the MDZ PK parameters. Estimates of rGC among the 22 subjects who participated in more than one trial ranged from 0.3% to 13.6%. Ozdemir et al. used a similar repeated drug administration method to evaluate the genetic component of variability in CYP3A activity. They calculated the rGC for hepatic CYP3A4 activity through MDZ plasma clearance as 0.96 (95% Cl = 0.92-0.98) (38). However, multiple MDZ doses for this study were obtained within 3 months (45), while multiple data from our subjects were obtained within a time-span of up to 3 years. In addition, the majority of subjects enrolled in multiple studies in our dataset were elderly individuals, compared with the study by Ozdemir et al. where subjects were much younger. In both studies, all subjects were nonsmokers, did not use concomitant medications and were monitored for their diet to avoid exposure to food effects. Hence, the environmental exposure was minimized, while the genetic component was maximized. In our meta-analysis from multiple studies, we found that the rGCs of CYP3A activity by MDZ clearance and AUC ranged from 0.3% to 13.6%. In other words, between-subject variances are much smaller than within-subject variances. This observation hints that environmental factors are more dominant and important than inherit genetic factors in CYP3A activity, if more extensive environmental effects are considered. However, given that CYP3A genetic effect is only a part of rGC, it is difficult to tease CYP3A genetic effect out with only a small or moderate sample size.
Studies regarding sex differences in CYP3A activity have produced conflicting results. Many CYP3A substrates show higher clearance in women than in men. Wolbold et al. found a 2-fold higher CYP3A4 messenger RNA (mRNA) transcripts in human liver in female compared with male samples (33). Zhu et al. reported that CYP3A activity was higher in women than in men, and CYP3A activity differed across the phases of the menstrual cycle in Chinese subjects (46). Chen et al. reviewed data from 13 previous studies and found women exhibited 11% higher mean weight-corrected total MDZ clearance and 28% higher oral clearance compared with men. Although significantly greater hepatic and intestinal CYP3A activities were shown in women, only a minor sex difference in AUC was observed (31, 32). On the other hand, studies by He et al. and Greenblatt et al. showed no sex differences in CYP3A activity (43, 47). In our present study, women had higher weight-corrected systemic and oral clearance than men, indicating that CYP3A activity may be great in women. However, there were no significant differences of systemic and oral clearance between women and men, because women had much lower body weight than men. In addition, dose-adjusted AUC and bioavailability differences were not observed between sexes. Therefore the clinical importance of elevated CYP3A activity in women remains to be determined.
Greenblatt et al. evaluated the age effect on CYP3A activity with triazolam. AUC increased and clearance declined with increased age in men, while age had no significant effect on AUC or clearance in women. Although the findings were consistent with reduced clearance in elderly men, individual variability was large and was not explained by identifiable demographic or environmental factors (32). We did not find any age effect on MDZ PK parameters in our study.
Conclusions
Significant ethnic differences were found in CYP3A allelic frequencies within the CYP3A cluster. The haplotypes and diplotypes in African Americans were significantly different from Caucasians. No significant difference was observed in MDZ disposition in vivo between genotypes, haplotypes and diplotypes in the CYP3A locus (P>0.05), nor between Caucasians and African Americans. Women had higher weight-corrected systemic and oral clearance than men, but dose-adjusted AUC and bioavailability differences were not detected between sexes. The clinical importance of elevated CYP3A activity in women remains to be determined. The rGCs of MDZ PK parameters were between 0.3% and 13.6%, indicating that the environmental factors explain a much larger variation in CYP3A activity than genetics. However, the effects of CYP3A genotype on drug disposition in vivo continue to warrant further investigation.
Acknowledgments
Drs. Lang Li, Sara K. Quinney, Seongho Kim, and Stephen Hall's researches are supported by NIH grant GM74217 (LL).
References
- 1.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annual Review of Pharmacology & Toxicology. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 3.Chen M, Nafziger AN, Bertino JS., Jr Drug-metabolizing enzyme inhibition by ketoconazole does not reduce interindividual variability of CYP3A activity as measured by oral midazolam. Drug metabolism and disposition: the biological fate of chemicals. 2006;34:2079–82. doi: 10.1124/dmd.106.011742. [DOI] [PubMed] [Google Scholar]
- 4.Wojnowski L, Kamdem LK. Clinical implications of CYP3A polymorphisms. Expert Opinion On Drug Metabolism & Toxicology. 2006;2:171–82. doi: 10.1517/17425255.2.2.171. [DOI] [PubMed] [Google Scholar]
- 5.Ball SE, Scatina J, Kao J, Ferron GM, Fruncillo R, Mayer P, et al. Population distribution and effects on drug metabolism of a genetic variant in the 5′ promoter region of CYP3A4. Clinical Pharmacology & Therapeutics. 1999;66:288–94. doi: 10.1016/S0009-9236(99)70037-8. [DOI] [PubMed] [Google Scholar]
- 6.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. American journal of human genetics. 2004;75:1059–69. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burk O, Tegude H, Koch I, Hustert E, Wolbold R, Glaeser H, et al. Molecular mechanisms of polymorphic CYP3A7 expression in adult human liver and intestine. Journal of Biological Chemistry. 2002;277:24280–8. doi: 10.1074/jbc.M202345200. [DOI] [PubMed] [Google Scholar]
- 8.Thompson EE, Kuttab-Boulos H, Yang L, Roe BA, Di Rienzo A. Sequence diversity and haplotype structure at the human CYP3A cluster. Pharmacogenomics Journal. 2006;6:105–14. doi: 10.1038/sj.tpj.6500347. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Martin E, Martinez C, Pizarro RM, Garcia-Gamito FJ, Gullsten H, Raunio H, et al. CYP3A4 variant alleles in white individuals with low CYP3A4 enzyme activity. Clinical Pharmacology & Therapeutics. 2002;71:196–204. doi: 10.1067/mcp.2002.121371. [DOI] [PubMed] [Google Scholar]
- 10.Wandel C, Witte JS, Hall JM, Stein CM, Wood AJ, Wilkinson GR. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5′-promoter region polymorphism. Clinical Pharmacology & Therapeutics. 2000;68:82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M. Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochemical & Biophysical Research Communications. 2005;338:299–305. doi: 10.1016/j.bbrc.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature Genetics. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 13.Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–9. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Wang YH, Miao J, Li L, Kovacs RJ, Marunde R, et al. Cytochrome P450 3A5 genotype is associated with verapamil response in healthy subjects. Clinical Pharmacology & Therapeutics. 2007;82:579–85. doi: 10.1038/sj.clpt.6100208. [DOI] [PubMed] [Google Scholar]
- 15.Dally H, Edler L, Jager B, Schmezer P, Spiegelhalder B, Dienemann H, et al. The CYP3A4*1B allele increases risk for small cell lung cancer: effect of gender and smoking dose. Pharmacogenetics. 2003;13:607–18. doi: 10.1097/00008571-200310000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Wojnowski L, Hustert E, Klein K, Goldammer M, Haberl M, Kirchheiner J, et al. Re: modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. Journal of the National Cancer Institute. 2002;94:630–1. 1–2. doi: 10.1093/jnci/94.8.630. author reply. [DOI] [PubMed] [Google Scholar]
- 17.Kim RB. Drugs as P-glycoprotein substrates, inhibitors, and inducers. Drug Metabolism Reviews. 2002;34:47–54. doi: 10.1081/dmr-120001389. [DOI] [PubMed] [Google Scholar]
- 18.Kim RB, Wandel C, Leake B, Cvetkovic M, Fromm MF, Dempsey PJ, et al. Interrelationship between substrates and inhibitors of human CYP3A and P-glycoprotein. Pharmaceutical research. 1999;16:408–14. doi: 10.1023/a:1018877803319. [DOI] [PubMed] [Google Scholar]
- 19.Masica AL, Mayo G, Wilkinson GR. In vivo comparisons of constitutive cytochrome P450 3A activity assessed by alprazolam, triazolam, and midazolam. Clinical Pharmacology & Therapeutics. 2004;76:341–9. doi: 10.1016/j.clpt.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Krupka E, Venisse N, Lafay C, Gendre D, Diquet B, Bouquet S, et al. Probe of CYP3A by a single-point blood measurement after oral administration of midazolam in healthy elderly volunteers. European journal of clinical pharmacology. 2006;62:653–9. doi: 10.1007/s00228-006-0159-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee JI, Chaves-Gnecco D, Amico JA, Kroboth PD, Wilson JW, Frye RF. Application of semisimultaneous midazolam administration for hepatic and intestinal cytochrome P450 3A phenotyping. Clinical Pharmacology & Therapeutics. 2002;72:718–28. doi: 10.1067/mcp.2002.129068. [DOI] [PubMed] [Google Scholar]
- 22.Lown KS, Kolars JC, Thummel KE, Barnett JL, Kunze KL, Wrighton SA, et al. Interpatient heterogeneity in expression of CYP3A4 and CYP3A5 in small bowel. Lack of prediction by the erythromycin breath test. Drug Metabolism & Disposition. 1994;22:947–55. erratum appears in Drug Metab Dispos 1995 Mar;23(3):followi. [PubMed] [Google Scholar]
- 23.Liu YT, Hao HP, Liu CX, Wang GJ, Xie HG. Drugs as CYP3A probes, inducers, and inhibitors. Drug Metab Rev. 2007;39:699–721. doi: 10.1080/03602530701690374. [DOI] [PubMed] [Google Scholar]
- 24.Lee LS, Bertino JS, Jr, Nafziger AN. Limited sampling models for oral midazolam: midazolam plasma concentrations, not the ratio of 1-hydroxymidazolam to midazolam plasma concentrations, accurately predicts AUC as a biomarker of CYP3A activity. Journal of clinical pharmacology. 2006;46:229–34. doi: 10.1177/0091270005283466. [DOI] [PubMed] [Google Scholar]
- 25.Streetman DS, B JS, Jr, N A. Phenotyping of drug-metabolizing enzymes in adults: a review of in-vivo cytochrome P450 phenotyping probes. Pharmacogenetics. 2000;10:187–216. doi: 10.1097/00008571-200004000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Thummel KE, O'Shea D, Paine MF, Shen DD, Kunze KL, Perkins JD, et al. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clinical pharmacology and therapeutics. 1996;59:491–502. doi: 10.1016/S0009-9236(96)90177-0. [DOI] [PubMed] [Google Scholar]
- 27.Gorski JC, Jones DR, Haehner-Daniels BD, Hamman MA, O'Mara EM, Jr, Hall SD. The contribution of intestinal and hepatic CYP3A to the interaction between midazolam and clarithromycin. Clinical pharmacology and therapeutics. 1998;64:133–43. doi: 10.1016/S0009-9236(98)90146-1. [DOI] [PubMed] [Google Scholar]
- 28.Eap CB, Buclin T, Hustert E, Bleiber G, Golay KP, Aubert AC, et al. Pharmacokinetics of midazolam in CYP3A4- and CYP3A5-genotyped subjects. European journal of clinical pharmacology. 2004;60:231–6. doi: 10.1007/s00228-004-0767-7. [DOI] [PubMed] [Google Scholar]
- 29.Floyd MD, Gervasini G, Masica AL, Mayo G, George AL, Jr, Bhat K, et al. Genotype-phenotype associations for common CYP3A4 and CYP3A5 variants in the basal and induced metabolism of midazolam in European- and African-American men and women. Pharmacogenetics. 2003;13:595–606. doi: 10.1097/00008571-200310000-00003. see comment. [DOI] [PubMed] [Google Scholar]
- 30.Shih PS, Huang JD. Pharmacokinetics of midazolam and 1′-hydroxymidazolam in Chinese with different CYP3A5 genotypes. Drug Metabolism & Disposition. 2002;30:1491–6. doi: 10.1124/dmd.30.12.1491. [DOI] [PubMed] [Google Scholar]
- 31.Chen M, Ma L, Drusano GL, Bertino JS, Jr, Nafziger AN. Sex differences in CYP3A activity using intravenous and oral midazolam. Clinical pharmacology and therapeutics. 2006;80:531–8. doi: 10.1016/j.clpt.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Greenblatt DJ, Harmatz JS, von Moltke LL, Wright CE, Shader RI. Age and gender effects on the pharmacokinetics and pharmacodynamics of triazolam, a cytochrome P450 3A substrate. Clinical Pharmacology & Therapeutics. 2004;76:467–79. doi: 10.1016/j.clpt.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Wolbold R, Klein K, Burk O, Nussler AK, Neuhaus P, Eichelbaum M, et al. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38:978–88. doi: 10.1053/jhep.2003.50393. [DOI] [PubMed] [Google Scholar]
- 34.Gorski JC, Wang Z, Haehner-Daniels BD, Wrighton SA, Hall SD. The effect of hormone replacement therapy on CYP3A activity. Clinical Pharmacology & Therapeutics. 2000;68:412–7. doi: 10.1067/mcp.2000.110560. [DOI] [PubMed] [Google Scholar]
- 35.Carrillo JA, Ramos SI, Agundez JA, Martinez C, Benitez J. Analysis of midazolam and metabolites in plasma by high-performance liquid chromatography: probe of CYP3A. Therapeutic drug monitoring. 1998;20:319–24. doi: 10.1097/00007691-199806000-00013. [DOI] [PubMed] [Google Scholar]
- 36.Tateishi T, Watanabe M, Nakura H, Asoh M, Shirai H, Mizorogi Y, et al. CYP3A activity in European American and Japanese men using midazolam as an in vivo probe. Clinical pharmacology and therapeutics. 2001;69:333–9. doi: 10.1067/mcp.2001.115447. [DOI] [PubMed] [Google Scholar]
- 37.Kalow W, O V, Tang BK, Tothfalusi L, Endrenyi L. The science of pharmacological variability: An essay. Clinical Pharmacology & Therapeutics. 1999;66:445–7. doi: 10.1016/S0009-9236(99)70006-8. [DOI] [PubMed] [Google Scholar]
- 38.Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, Endrenyi L, et al. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10:373–88. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 39.Xie HG, Wood AJJ, Kim RB, Stein CM, Wilkinson GR. Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics. 2004;5:243–72. doi: 10.1517/phgs.5.3.243.29833. [DOI] [PubMed] [Google Scholar]
- 40.Lee SJ, Goldstein JA. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics. 2005;6:357–71. doi: 10.1517/14622416.6.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Mirghani RA, Sayi J, Aklillu E, Allqvist A, Jande M, Wennerholm A, et al. CYP3A5 genotype has significant effect on quinine 3-hydroxylation in Tanzanians, who have lower total CYP3A activity than a Swedish population. Pharmacogenet Genomics. 2006;16:637–45. doi: 10.1097/01.fpc.0000230411.89973.1b. [DOI] [PubMed] [Google Scholar]
- 42.Pinto AG, Wang YH, Chalasani N, Skaar T, Kolwankar D, Gorski JC, et al. Inhibition of human intestinal wall metabolism by macrolide antibiotics: effect of clarithromycin on cytochrome P450 3A4/5 activity and expression. Clinical Pharmacology & Therapeutics. 2005;77:178–88. doi: 10.1016/j.clpt.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 43.He P, Court MH, Greenblatt DJ, Von Moltke LL. Genotype-phenotype associations of cytochrome P450 3A4 and 3A5 polymorphism with midazolam clearance in vivo. Clinical Pharmacology & Therapeutics. 2005;77:373–87. doi: 10.1016/j.clpt.2004.11.112. see comment. [DOI] [PubMed] [Google Scholar]
- 44.Kharasch ED, Walker A, Isoherranen N, Hoffer C, Sheffels P, Thummel K, et al. Influence of CYP3A5 genotype on the pharmacokinetics and pharmacodynamics of the cytochrome P4503A probes alfentanil and midazolam. Clinical pharmacology and therapeutics. 2007;82:410–26. doi: 10.1038/sj.clpt.6100237. [DOI] [PubMed] [Google Scholar]
- 45.Kashuba AD, Bertino JS, Jr, Rocci ML, Jr, Kulawy RW, Beck DJ, Nafziger AN. Quantification of 3-month intraindividual variability and the influence of sex and menstrual cycle phase on CYP3A activity as measured by phenotyping with intravenous midazolam. Clin Pharmacol Ther. 1998;64:269–77. doi: 10.1016/S0009-9236(98)90175-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhu B, Liu ZQ, Chen GL, Chen XP, Ou-Yang DS, Wang LS, et al. The distribution and gender difference of CYP3A activity in Chinese subjects. British journal of clinical pharmacology. 2003;55:264–9. doi: 10.1046/j.1365-2125.2003.01728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenblatt DJ, Abernethy DR, Locniskar A, Harmatz JS, Limjuco RA, Shader RI. Effect of age, gender, and obesity on midazolam kinetics. Anesthesiology. 1984;61:27–35. [PubMed] [Google Scholar]


