Abstract
Two transcripts coding for proteins homologous to apyrases were identified by massive sequencing of a Phlebotomus (P.) duboscqi salivary gland cDNA library. The sequence analysis revealed that the amino acids important for enzymatic activity including nucleotidase activity and the binding of calcium and nucleotides were well conserved in these molecules. A recombinant P. duboscqi salivary apyrase was expressed in Escherichia coli and purified. The resulting protein efficiently hydrolyzed ADP and ATP, but not AMP, GDP, CDP or UDP, in a calcium-dependent manner. Further, the recombinant protein inhibited ADP- and collagen-induced platelet aggregation. The results indicated that this salivary protein plays an important role in the blood-feeding process in P. duboscqi. Its unique enzymatic activity makes the salivary apyrase an attractive candidate as a therapeutic agent for the treatment of thrombotic pathologies as well as a reagent for a wide variety of research purposes.
Keywords: Apyrase, Sand fly, Saliva
1. Introduction
Blood-sucking arthropods have evolved potent pharmacologically active components to counteract the hemostatic and inflammatory systems of vertebrate hosts (Ribeiro, 1995; Ribeiro and Francischetti, 2003). When probing the skin for blood, they inject a saliva cocktail containing various physiologically active substances including anticoagulants, vasodilators, and inhibitors of platelet aggregation induced by collagen, adenosine diphosphate (ADP), arachidonic acid, or thrombin (Ribeiro, 1995; Ribeiro and Francischetti, 2003; Valenzuela, 2004, 2005; Champagne, 2005; Andersen et al., 2005). ADP is released by injured cells and dense granules of activated platelets, and is one of the most important physiological mediators for the recruitment and aggregation of platelets and the formation of plugs via the activation of platelet purinergic receptors (Kunapuli, 2002; Benoit and Dogné, 2003; Murugappan et al., 2004). Interestingly, hematophagous arthropods have evolved an ability to inhibit the ADP-mediated activation of host platelets to facilitate feeding on blood (Ribeiro, 1995; Smith and Kirley, 2006). Apyrases are nucleoside triphosphate-diphosphohydrolases present in a variety of organisms, and the salivary apyrases of blood-feeding arthropods hydrolyze ADP and consequently inhibit ADP-induced platelet aggregation when injected into the host (Ribeiro, 1995; Smith and Kirley, 2006). Initially, most apyrases identified were related to the yeast GTPase/CD39 family, which can be grouped into the actin/heat shock 70/sugar kinase superfamily (Smith and Kirley, 1999). In the subsequent studies, another family of apyrases, belonging to the 5′-nucleotidases, was found in the saliva of mosquitoes (Champagne et al., 1995; Mathews et al., 1996). This same family was further identified in arthropods such as Triatomine bugs (Faudry et al., 2004; Ribeiro et al., 2004; Santos et al., 2007; Assumpção et al., 2008). Both families require either Ca2+ or Mg2+ for their actions. Further, a new family of apyrases was identified in the saliva of the bed bug Cimex lectularius, and a recombinant protein derived from the transcript was characterized (Valenzuela et al., 1998). This novel type of apyrase had homology with neither the actin/heat shock 70/sugar kinase superfamily nor the 5′-nucleotidase family of apyrases, and functioned dependent on Ca2+ but not Mg2+, Mn2+ or Zn2+ (Valenzuela et al., 1998). Cimex family apyrases were further identified in Old World sand fly species, Phlebotomus (P.) papatasi, and their activity was characterized (Valenzuela et al., 2001). Active transcriptome-based analyses of salivary glands from New and Old World sand fly species, Lutzomyia (Lu.) longipalpis (Charlab et al., 1999; Valenzuela et al., 2004), P. ariasi (Oliveira et al., 2005), P. argentipes, P. perniciosus (Anderson et al., 2006) and P. duboscqi (Kato et al., 2006), identified transcripts of this family, but the functions of the proteins have not been characterized. Meanwhile, vertebrate homologues of Cimex family apyrases, termed calcium-activated nucleotidases (CANs), were identified (Failer et al., 2002; Smith et al., 2002; Murphy et al., 2003; Smith and Kirley, 2006), and key amino acids for the binding of nucleotides and calcium and nucleotidase activity were studied extensively using mutagenesis (Yang and Kirley, 2004; Dai et al., 2004). Similar to Cimex family apyrases, the activities of CANs were strictly Ca2+-dependent; however, interestingly, the preferred nucleotides were UDP > GDP ≫ ADP (Failer et al., 2002; Smith et al., 2002), whereas Cimex family apyrases from blood-feeding arthropods hydrolyzed ADP most efficiently (Valenzuela et al., 1998, 2001; Smith and Kirley, 2006).
Recently, a transcriptome-based analysis of the salivary gland of the sand fly P. duboscqi resulted in the identification of transcripts with homology with Cimex family apyrases (Kato et al., 2006). In the present study, we prepared a recombinant P. duboscqi apyrase (PduApy) and characterized its activity.
2. Materials and methods
2.1. Salivary gland cDNA library
The P. duboscqi salivary gland cDNA library was prepared as described previously (Kato et al., 2006). Briefly, mRNA was isolated from 55 salivary gland pairs using the Micro-FastTrack mRNA isolation kit (Invitrogen, San Diego, CA, USA). The PCR-based cDNA library was prepared following the instructions for the SMART cDNA library construction kit (BD-Clontech, Palo Alto, CA) with some modifications, and randomly selected clones were sequenced using an Applied Biosystems 3730xl DNA Analyzer (Foster City, CA) (Kato et al., 2006). A bioinformatics analysis was performed with the sequence data as described previously (Kato et al., 2006).
2.2. Phylogenetic analysis
Consensus protein sequences were compared to related sequences from sand flies as well as non-sand fly species obtained from GenBank. Sequences were aligned with CLUSTAL W software (Thompson et al., 1994) and examined using the program MEGA (Molecular Evolutionary Genetics Analysis) version 4.0 (Tamura et al., 2007). Neighbor-joining (NJ) trees were constructed with the distance algorisms available in the MEGA package.
2.3. Production of recombinant P. duboscqi apyrase
A DNA fragment encoding a mature P. duboscqi apyrase (PduApy) was amplified and inserted into the EcoRI site of an N-terminal thioredoxin (Trx)-hexahistidine (His)· tag fusion plasmid vector, pET-32b(+) (Novagen, Drams, Germany). The EcoRI adaptor-ligated primers used for amplification of the mature PduApy-encoding fragment were Eco-PduM39-S (5′-ccgaattccGCTCCAAGTAGTGAAACAAT-3′) and Eco-PduM39-R (5′-gggaattcTCACTCTTTACGTTTCAAAA-3′). Escherichia coli (E. coli) BL21 cells were transformed with the recombinant plasmid and grown in Luria-Bertani (LB) medium containing ampicillin (50 μg/ml). Production of the recombinant PduApy 2 (rPduApy 2) was induced by the addition of isopropyl β-D-thiogalactoside (IPTG) to a final concentration of 1 mM. The BL21 cells suspended in sonication buffer [50 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA and 1 mM DTT] were sonicated, and the insoluble fusion protein was solubilized by a refolding procedure using a urea solution [8 M urea, 50 mM Tris–HCl (pH 8.0), 1 mM EDTA and 1 mM DTT]. The Trx-His-tagged PduApy fusion protein was purified with a His SpinTrap kit (GE Healthcare, Buckinghamshire, UK). Since the purified recombinant protein was insolubilized again in phosphate-buffered saline (PBS), it was finally dialyzed against 100 mM Tris buffer (pH 8.0). Trx-His-tag protein only was expressed and purified to use as a control.
2.4. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis
The samples were treated with 2× sample buffer [125 mM Tris–HCl (pH6.8), 4.5% SDS, 20% glycerol, 0.01% bromophenol blue and 10% 2-mercaptoethanol] and analyzed in a 12.5% polyacrylamide gel. To estimate the molecular weight of the samples, Precision Plus Protein Standards (Bio-Rad Laboratories, Hercules, CA) were used. After electrophoresis, the gel was stained with coomassie brilliant blue.
For the immunoblot analysis, the proteins in the gel were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA). After blocking with 5% skim milk in PBS for 1 h at room temperature, the membrane was incubated with mouse anti-His antibody (GE Healthcare) or P. dubosqi salivary gland homogenate (SGH)-immunized mouse serum overnight at 4 °C. After three washes with PBS containing 0.1% Tween 20 (PBS-T), the membrane was further incubated with alkaline phosphatase (AP)-conjugated goat anti-mouse immunoglobulin (Zymed Laboratories, San Francisco, CA) for 1 h at room temperature. After three washes with PBS-T, the blots were developed by adding a substrate (Alkaline Phosphatase Conjugate Substrate Kit; Bio-Rad Laboratories) and visualized.
2.5. Apyrase activity
The apyrase activity of rPduApy 2 was determined by measuring the amount of inorganic phosphate (Pi) released from nucleotides using the method of Fiske and Subbarow (1925) with modifications. Briefly, 30 μl of 3.3 μM rPduApy 2 was mixed with 20 μl of 1 mM ATP, ADP or AMP and 50 μl of 100 mM Tris–HCl buffer containing 2 mM CaCl2 and/or 2 mM MgCl2 in each well of a 96-well microplate, and incubated for 20 min at 37 °C. For color development, 20 μl of acid molybdate solution (1.25% ammonium molybdate in 1.25 M sulfuric acid) and 5 μl of Fiske and Subbarow solution [159 mg/ml Fiske and Subbarow Reducer (Sigma, St. Louis, MO) in distilled water] were added to each well, and the plate was incubated for 10 min at room temperature. The absorbance was read by a MICROPLATE READER Model 450 (BIO-RAD, Anaheim, CA) at a wavelength of 665 nm. The concentrations of Pi were determined from a standard curve prepared using a serially diluted phosphorous standard solution (Wako, Osaka, Japan). All the experiments were performed in triplicate. In some experiments, GDP (Sigma), CDP (Sigma) or UTP (Sigma) was used as a substrate to examine the specificity of the nuclease activity.
2.6. Platelet aggregation assay
Blood was collected from healthy volunteers by veinpuncture in sodium heparin and centrifuged at 70 × g for 15 min to obtain platelet-rich plasma (PRP). Platelet aggregation was measured by using a microplate method as described previously (Mans et al., 2008). Briefly, 30 μl of 13.3 μM Trx-His-tagged rPduApy 2 or Trx-His-tag protein was added to each well of 70 μl of PRP, and platelet aggregation was induced immediately or after incubation at 37 °C for 4 min by the addition of 10 μl of 20 μM ADP (Sigma). The mixture was then incubated for 10 min at 37 °C by shaking on a microplate mixer (Tomy Seiko, Tokyo, Japan), and platelet aggregation was measured by determining the change in light transmission at a wavelength of 630 nm. As a comparative experiment, 10 μl of 0.1 mg/ml of type I collagen (Chrono-log corporation, Havertown, PA) was used as a platelet aggregation inducer.
3. Results
3.1. Sequence analysis
By sequencing a P. dubosqi salivary gland cDNA library, we identified two transcripts (PduM38 and PduM39) coding for proteins homologous to Cimex family apyrases, enzymes that hydrolyze both ADP and ATP to AMP and orthophosphate (Kato et al., 2006). The first transcript (PduM38) containing six cDNA coded for a protein of 336 amino acid residues with a predicted molecular mass of 35.8 kDa in the mature form and a calculated isoelectric point of 7.36 (PduApy 1), and the second transcript (PduM39) containing nine cDNA coded for a protein of 336 amino acid residues with a predicted molecular mass of 35.2 kDa in the mature form and a calculated isoelectric point of 9.03 (PduApy 2) (GenBank accession numbers: DQ834335 and DQ834331). The proteins derived from the two transcripts were identified in a proteomic analysis of P. dubosqi salivary gland homogenate as abundant (Kato et al., 2006), strongly suggesting that they play an important role in the blood-feeding process.
The sequences of PduApy 1 and 2 were aligned with those of Cimex family salivary apyrases from C. lectularius and the sand flies P. papatasi and Lu. longipalpis, and CANs, which are vertebrate homologues of Cimex family apyrases. The level of overall sequence identity was low (Fig. 1); however, conserved regions among species including eight highly conserved motifs (aa 29–38, 85–113, 121–154, 201–211, 217–225, 234–244, 268–289 and 318–322 in PduApy 1) (Failer et al., 2002) were well conserved in PduApy 1 and 2. Notably, four amino acid residues (aa 114, 163, 181 and 300 in human CAN) essential for the nucleotidase activity identified in human CAN (Yang and Kirley, 2004) were strictly conserved in PduApy 1 and 2 (Fig. 2). In addition, four (aa 168, 216, 284 and 345 in human CAN) of the five residues important for binding Ca2+, and six (aa 112, 166, 182, 215, 300 and 394 in human CAN) of thirteen residues important for binding nucleotides in human CAN (Dai et al., 2004), were completely conserved in PduApy 1 and 2 (Fig. 1). Next, a phylogenetic analysis was performed with apyrase-related proteins from different organisms. Interestingly, apyrases from sand fly (Phlebotomus and Lutzomyia) species including P. duboscqi composed a group distinct from vertebrate apyrase-related proteins, salivary apyrases from mosquitoes, Anopheles gambiae, Aedes aegypti and Culex quinquefasciatus, which belong to the 5′-nucleotidase gene family, and unexpectedly, Cimex lectuarius (Fig. 2).
Fig. 1.
Alignment of invertebrate Cimex family salivary apyrases from P. duboscqi (PduApy 1 and 2), P. papatasi, Lu. longipalpis and Cimex lectularias, and their mammalian homologues, calcium-activated nucleotidases (CANs) from mouse, rat and human. Eight highly conserved sequence domains are boxed. The key residues for binding nucleotides and calcium determined in human CAN are represented by pluses (+) and asterisks (*), respectively. Open circles (○) indicate amino acid residues essential for the nucleotidase activity in human CAN. A triangle (△) denotes a glutamic acid, whose substitution with tyrosine increased ADPase activity approximately 5-fold in human CAN. An arrow indicates the first amino acid of the soluble form of human CAN.
Fig. 2.
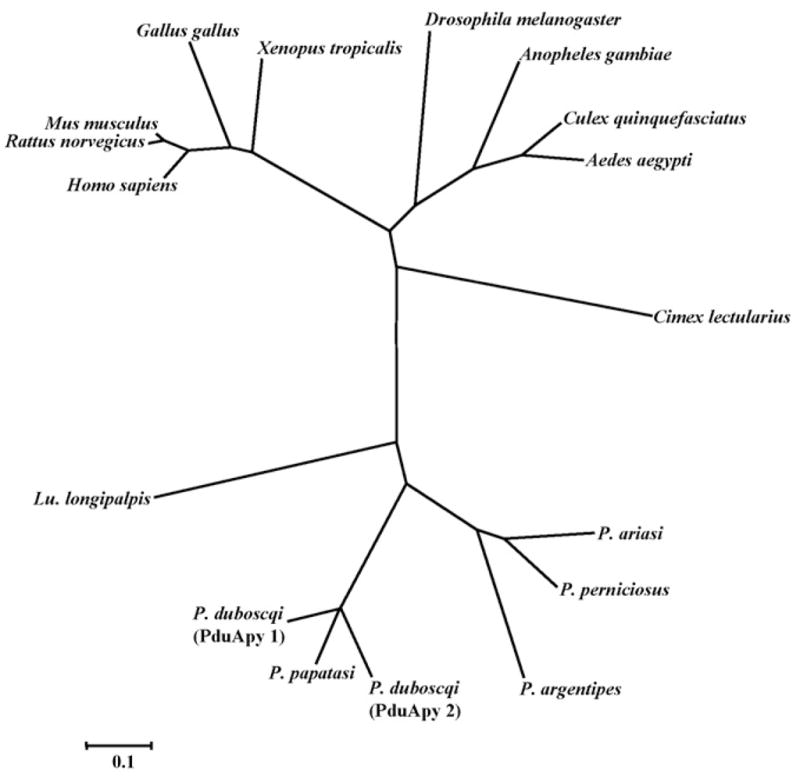
A phylogenetic tree of apyrase-related proteins. P. duboscqi salivary apyrase-like protein sequences (PduApy 1 and 2), along with the related sequences, were aligned and a phylogenetic analysis was performed. The GenBank accession numbers of the proteins are as follows: P. papatasi, AF261768; P. argentipes, DQ136150; P. ariasi, AY845193; P. perniciosus, DQ192491; Lu. longipalpis, AF131933; Cimex lectularias, AF085499; Anopheles gambiae, NM_144734; Aedes aegypti, XM_001652093; Culex quinquefasciatus, XM_001849046; Drosophila melanogaster, AM294107; Homo sapiens, NM_138793; Mus musculus, NM_029502; Rattus norvegicus, NM_144734; Gallus gallus, NM_001031581; Xenopus tropicalis, NM_203609.
3.2. Production and purification of the recombinant P. duboscqi apyrase
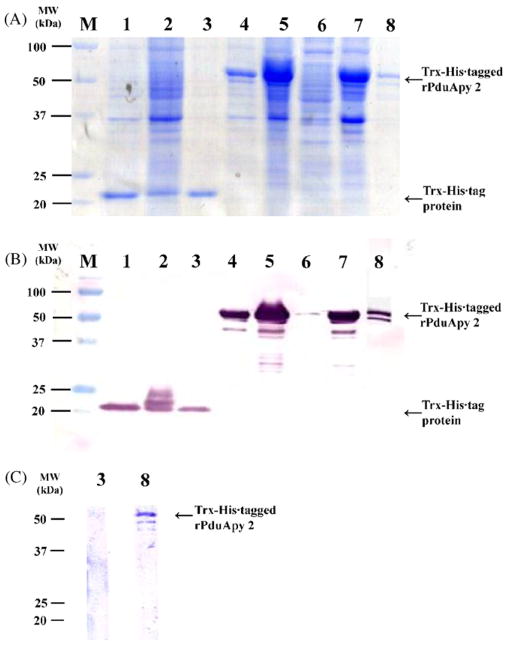
To test whether a PduApy cDNA clone codes for an enzymatically active apyrase, the recombinant PduApy 2 was expressed in E. coli as a Trx-His-tagged fusion protein and purified through a refolding procedure. As a control, Trx-His-tag protein only was also produced from an empty vector and purified. The recombinant PduApy 2 (rPduApy 2) fusion protein had a molecular mass of approximately 50 kDa based on polyacrylamide gel electrophoresis (Fig. 3A) and reacted to the anti-His antibody in an immunoblot analysis (Fig. 3B). The antigenicity of rPduApy 2 was assessed by immunoblotting with P. dubosqi SGH-immunized mouse serum. The immune serum reacted with rPduApy 2 but not the Trx-His-tag protein, indicating that the recombinant protein retained the antigenicity of a P. duboscqi salivary protein (Fig. 3C).
Fig. 3.
SDS-PAGE and immunoblot analysis of rPduApy 2 expressed in E. coli. E. coli was transformed with the Trx-His (lanes 1–3) or Trx-His-tagged rPduApy 2 (lanes 4–8)-expressing plasmid vector, and expression of the recombinant protein was induced by IPTG. The whole cell lysate with (lanes 2 and 5) or without (lanes 1 and 4) IPTG treatment, soluble (lane 6) and insoluble (lane 7) fractions after sonication, and purified protein (lanes 3 and 8) were subjected to SDS-PAGE (A). Immunoblot analyses were performed with anti-His antibody (B) or P. dubosqi salivary gland homogenate-immunized mouse serum (C). Lane M, protein molecular weight marker.
3.3. Activity of recombinant P. duboscqi apyrase
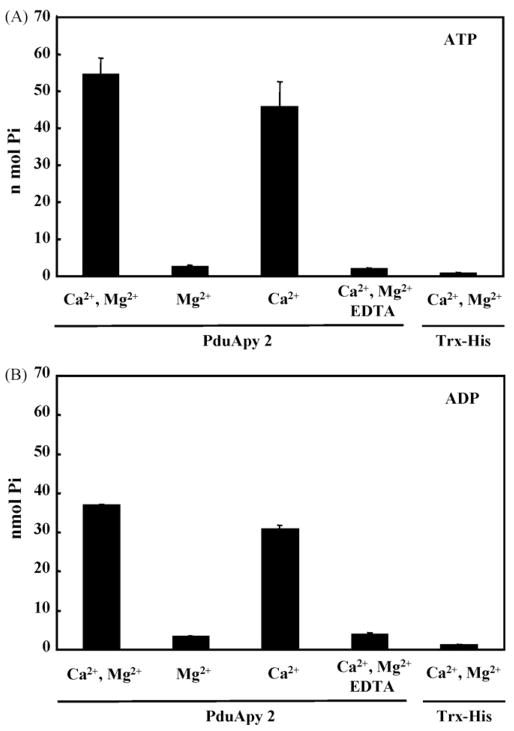
Purified rPduApy 2 was assessed for nucleotide-hydrolyzing activity. As shown in Fig. 4, rPduApy 2, but not Trx-His-tag protein, displayed ATPase and ADPase activities, which were strictly dependent on Ca2+ but not Mg2+. No AMP-hydrolyzing activity was detected (data not shown). These Ca2+-dependent nucleotidase activities were completely inhibited by a chelating agent, EDTA (Fig. 4). The ATPase and ADPase activities were tested using Tris buffer at a pH of 7.2–9.0, and similar levels of activity were detected under these conditions (data not shown). The preference of rPduApy 2 was assessed using ADP, GDP, CDP or UDP as a substrate. rPduApy 2 hydrolyzed ADP but not GDP, CDP or UDP (data not shown).
Fig. 4.
Apyrase activity of the recombinant P. dubosqi apyrase-like protein PduApy 2. Trx-His-tagged rPduApy 2 (PduApy 2) or Trx-His-tag protein only (Trx-His) was incubated with ATP (A) or ADP (B) in the presence of CaCl2 (Ca2+) and/or MgCl2 (Mg2+), and the amount of inorganic phosphate release was measured. EDTA was used to chelate the effect of metals. The results are expressed as the mean ± SD of triplicate measurements.
Since ADP is one of the most important physiological agonists for platelet aggregation (Benoit and Dogné, 2003; Murugappan et al., 2004), the effect of rPduApy 2 on ADP-induced platelet aggregation was assessed. ADP-induced platelet aggregation was inhibited by about 40% in the presence of rPduApy 2 (Fig. 5A) and by approximately 90% when the platelets were preincubated with rPduApy 2 (Fig. 5B), indicating that in P. duboscqi, PduApy acts as an inhibitor of platelet aggregation during the blood-feeding process. In addition, collagen-induced aggregation was also inhibited by about 80% when the platelets were preincubated with rPduApy 2 (data not shown).
Fig. 5.
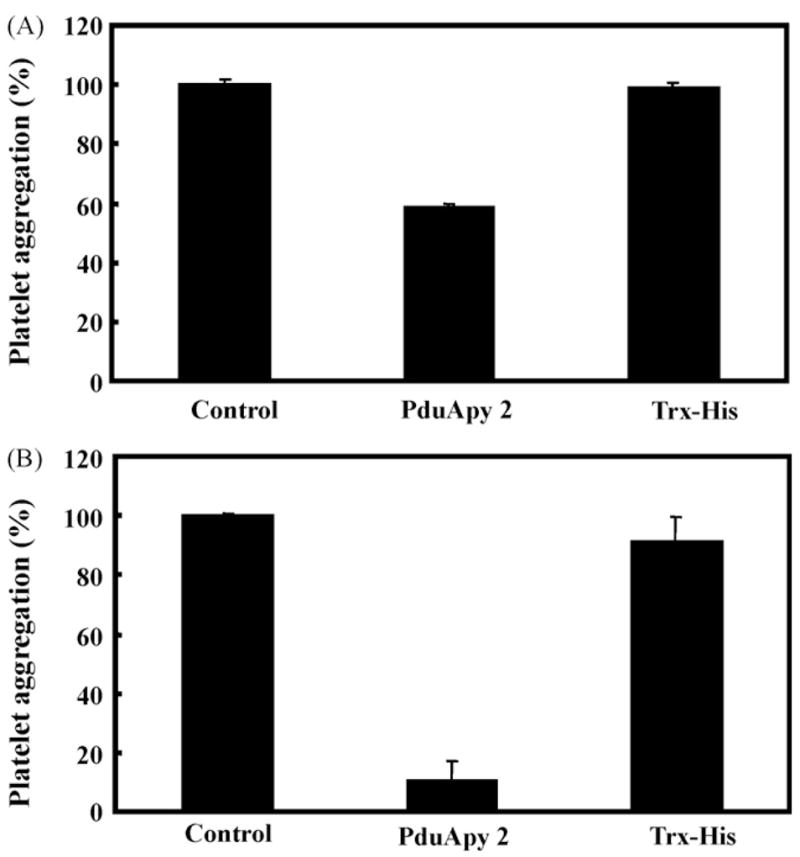
Inhibition of ADP-induced platelet aggregation by rPduApy 2. Tris buffer (Control), Trx-His-tagged rPduApy 2 (rPduApy 2) or Trx-His-tag protein (Trx-His) was added to each well of platelet-rich plasma, and platelet aggregation was induced immediately (A) or after incubation at 37 °C for 4 min (B) by addition of ADP. Platelet aggregation was measured by determining the change in light transmission at the wavelength of 630 nm.
4. Discussion
We recently sequenced a large number of transcripts from the salivary gland of P. duboscqi and identified two transcripts (PduM38 and 39) coding for a protein homologous to Cimex family apyrases (Kato et al., 2006). In the present study, a recombinant protein was produced from the transcript PduM39, and its enzymatic activity was characterized. The protein, rPduApy 2, efficiently hydrolyzed ADP and ATP dependent on Ca2+, but not on AMP, GDP, CDP or UDP. Further, rPduApy 2 inhibited ADP- and collagen-induced platelet aggregation, indicating that this salivary protein plays an important role in the blood-feeding process.
Vertebrate homologues of Cimex family apyrases, termed CANs, have been identified, and interestingly, shown to hydrolyze UDP and GDP rather than ADP (Failer et al., 2002; Smith et al., 2002; Murphy et al., 2003; Smith and Kirley, 2006). Extensive mutagenesis and structural analyses of human CAN have identified those amino acids important for the binding of nucleotides and calcium and for nucleotidase activity (Yang and Kirley, 2004; Dai et al., 2004). Four residues (aa 114, 163, 181 and 300 in human CAN) essential for the nucleotidase activity in human CAN (Yang and Kirley, 2004) were strictly conserved in PduApy 1 and 2 (Fig. 1). In human CAN, 13 amino acid residues were identified as key to the binding of nucleotides (Dai et al., 2004; Smith and Kirley, 2006). Of these, six (aa 112, 166, 182, 215, 300 and 394 in human CAN) were completely conserved in all the species analyzed (Fig. 1). Although the other seven residues (aa 228, 229, 231, 233, 234, 282 and 307 in human CAN) were not completely conserved among species, most of them were well conserved among insect salivary apyrases and among vertebrate CANs (Fig. 1). The difference may reflect the specificity of these enzymes, that is, the nucleotides preferred by vertebrate CANs are UDP > GDP ≫ ADP (Failer et al., 2002; Smith et al., 2002) while insect salivary apyrases efficiently hydrolyze ADP (Ribeiro, 1995). In human soluble CAN, the substitution of one glutamic acid (aa 160 in human CAN) with tyrosine resulted in a marked increase in nucleotidase activities: about a 2-fold increase for GDPase and 5-fold increase for ADPase (Yang and Kirley, 2004). The corresponding amino acid (aa 83 in PduApy 1) was tyrosine in sand fly species, P. duboscqi and P. papatasi, and in C. lectuarius. Since salivary apyrases of these species showed strong ADPase activities, tyrosine at this position was considered to be associated with hydrolyzing activity specific for ADP.
rPduApy 2 expressed in the insoluble fraction of E. coli was purified through refolding using a urea solution. The protein retained marked levels of nucleotidase activity. One important characteristic of this family is their stability compared to other nucleotidases. For example, P. papatasi apyrase was successfully renatured after SDS-PAGE (Valenzuela et al., 2001), and C. lectuarius apyrase was resistant to denaturation by 0.1% trifluoroacetic acid and acetonitrile treatment used for the purification process (Valenzuela et al., 1998). In addition, human CAN was reported to be stable for over 24 h at 37 °C, and for more than one week at room temperature (Yang and Kirley, 2004). These observations suggest that the activity of apyrases is long-lasting in the host after injection by blood-sucking arthropods. In this study, preincubation of platelets with rPdyApy 2 inhibited of platelet aggregation more efficiently (Fig. 5). At present, the underlying mechanism is not known; however, it may be possible that prior binding of apyrase with Ca2+ enhances the enzymatic activity. Another possibility is that apyrase may have some direct effect on platelets. We observed that rPduApy 2 markedly affected the collagen-induced as well as ADP-induced aggregation of platelets. A similar result has been reported for the salivary apyrase of C. lectularius (Valenzuela et al., 1998). These results indicated that ADP plays an essential role in the collagen-induced aggregation process. Interestingly, the apyrase from Ornithodoros savignyi disaggregated platelets that had aggregated due to ADP (Mans et al., 2000). Together with the broad pH range of the enzymatic activity shown in the present study, these findings indicated that apyrases have marked potential for therapeutic use against thrombosis-associated disorders as well as for research purposes.
In conclusion, the P. duboscqi salivary apyrase, a Cimex family apyrase, was produced by E. coli, and an active recombinant protein was purified and examined for ADPase and ATPase activities. Because of its unique activity and stability, this protein is considered to play an important role in the blood-feeding process. Salivary apyrases derived from hematophagous arthropods would be potential anti-thrombotic proteins for the treatment of thrombotic pathologies in addition to reagents for research. In a recent study, it was reported that increased membrane nucleotidase activity of Leishmania or addition of adenosine at the moment of infection with the parasite resulted in an increase in lesion size and parasitism as well as a delay in lesion healing (de Almeida Marques-da-Silva et al., 2008). Therefore, apyrase in sand fly saliva co-injected with the parasite through sand fly bite may have similar effect on the disease progression.
References
- Andersen JF, Gudderra NP, Francischetti IM, Ribeiro JM. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Archives of Insect Biochemistry and Physiology. 2005;58:97–105. doi: 10.1002/arch.20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Oliveira F, Kamhawi S, Mans BJ, Reynoso D, Seitz AE, Lawyer P, Garfield M, Pham M, Valenzuela JG. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics. 2006;7:52. doi: 10.1186/1471-2164-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção TC, Francischetti IM, Andersen JF, Schwarz A, Santana JM, Ribeiro JM. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas’ disease. Insect Biochemistry and Molecular Biology. 2008;38:213–232. doi: 10.1016/j.ibmb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit P, Dogné JM. Platelet ADP receptors and their antagonists. Mini Reviews in Medicinal Chemistry. 2003;3:145–148. doi: 10.2174/1389557033405412. [DOI] [PubMed] [Google Scholar]
- Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. The Proceedings of the National Academy of Sciences USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DE. Antihemostatic molecules from saliva of blood-feeding arthropods. Pathophysiology of Haemostasis and Thrombosis. 2005;34:221–227. doi: 10.1159/000092428. [DOI] [PubMed] [Google Scholar]
- Charlab R, Valenzuela JG, Rowton ED, Ribeiro JM. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. The Proceedings of the National Academy of Sciences USA. 1999;96:15155–15160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Liu J, Deng Y, Smith TM, Lu M. Structure and protein design of a human platelet function inhibitor. Cell. 2004;116:649–659. doi: 10.1016/s0092-8674(04)00172-2. [DOI] [PubMed] [Google Scholar]
- de Almeida Marques-da-Silva E, de Oliveira JC, Figueiredo AB, de Souza Lima Júnior D, Carneiro CM, Rangel Fietto JL, Crocco Afonso LC. Extracellular nucleotide metabolism in Leishmania: influence of adenosine in the establishment of infection. Microbes and Infection. 2008;10:850–857. doi: 10.1016/j.micinf.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Failer BU, Braun N, Zimmermann H. Cloning, expression, and functional characterization of a Ca2+-dependent endoplasmic reticulum nucleoside diphosphatase. Journal of Biological Chemistry. 2002;277:36978–36986. doi: 10.1074/jbc.M201656200. [DOI] [PubMed] [Google Scholar]
- Faudry E, Lozzi SP, Santana JM, D’Souza-Ault M, Kieffer S, Felix CR, Ricart CA, Sousa MV, Vernet T, Teixeira AR. Triatoma infestans apyrases belong to the 5′-nucleotidase family. Journal of Biological Chemistry. 2004;279:19607–19613. doi: 10.1074/jbc.M401681200. [DOI] [PubMed] [Google Scholar]
- Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. Journal of Biological Chemistry. 1925;66:375–400. [Google Scholar]
- Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, Sangare CS, Samake S, Sissoko I, Garfield M, Sigutova L, Volf P, Doumbia S, Valenzuela JG. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya) BMC Genomics. 2006;7:226. doi: 10.1186/1471-2164-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunapuli SP. P2 receptors and platelet activation. Scientific World Journal. 2002;2:424–433. doi: 10.1100/tsw.2002.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Coetzee J, Louw AI, Gaspar AR, Neitz AW. Disaggregation of aggregated platelets by apyrase from the tick, Ornithodoros savignyi (Acari: Argasidae) Experimental and Applied Acarology. 2000;24:271–282. doi: 10.1023/a:1006440714276. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Andersen JF, Schwan TG, Ribeiro JM. Characterization of anti-hemostatic factors in the argasid, Argas monolakensis: implications for the evolution of blood-feeding in the soft tick family. Insect Biochemistry and Molecular Biology. 2008;38:22–41. doi: 10.1016/j.ibmb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews GV, Sidjanski S, Vanderberg JP. Inhibition of mosquito salivary gland apyrase activity by antibodies produced in mice immunized by bites of Anopheles stephensi mosquitoes. American Journal of Tropical Medicine and Hygiene. 1996;55:417–423. doi: 10.4269/ajtmh.1996.55.417. [DOI] [PubMed] [Google Scholar]
- Murphy DM, Ivanenkov VV, Kirley TL. Bacterial expression and characterization of a novel, soluble, calcium-binding, and calcium-activated human nucleotidase. Biochemistry. 2003;42:2412–2421. doi: 10.1021/bi026763b. [DOI] [PubMed] [Google Scholar]
- Murugappan S, Shankar H, Kunapuli SP. Platelet receptors for adenine nucleotides and thromboxane A2. Seminars in Thrombosis and Hemostasis. 2004;30:411–418. doi: 10.1055/s-2004-833476. [DOI] [PubMed] [Google Scholar]
- Oliveira F, Kamhawi S, Seitz AE, Pham VM, Guigal PM, Fischer L, Ward J, Valenzuela JG. From transcriptome to immunome: identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2005;24:374–390. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infectious Agents. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annual Review of Entomology. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochemistry and Molecular Biology. 2004;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Santos A, Ribeiro JM, Lehane MJ, Gontijo NF, Veloso AB, Sant’ Anna MR, Nascimento Araujo R, Grisard EC, Pereira MH. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae) Insect Biochemistry and Molecular Biology. 2007;37:702–712. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TM, Kirley TL. The calcium activated nucleotidases: a diverse family of soluble and membrane associated nucleotide hydrolyzing enzymes. Purinergic Signalling. 2006;2:327–333. doi: 10.1007/s11302-005-5300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TM, Hicks-Berger CA, Kim S, Kirley TL. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Archives of Biochemistry and Biophysics. 2002;406:105–115. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- Smith TM, Kirley TL. Site-directed mutagenesis of a human brain ectoapyrase: evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry. 1999;38:321–328. doi: 10.1021/bi9820457. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG. Exploring tick saliva: from biochemistry to ‘sialomes’ and functional genomics. Parasitology. 2004;129:S83–94. doi: 10.1017/s0031182004005189. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG. Blood-Feeding Arthropod Salivary Glands and Saliva. In: Marquardt WC, Black WC, Freier JE, Hagedorn HH, Hemingway J, Higgs S, James AA, Kondratieff B, Moore CG, editors. Biology of Disease Vectors. 2. Elsevier; San Diego: 2005. pp. 377–386. [Google Scholar]
- Valenzuela JG, Charlab R, Galperin MY, Ribeiro JM. Purification, cloning, and expression of an apyrase from the bed bug Cimex lectularius. A new type of nucleotide-binding enzyme. Journal of Biological Chemistry. 1998;273:30583–30590. doi: 10.1074/jbc.273.46.30583. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. Journal of Experimental Biology. 2001;204:229–237. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. Journal of Experimental Biology. 2004;207:3717–3729. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- Yang M, Kirley TL. Site-directed mutagenesis of human soluble calcium-activated nucleotidase 1 (hSCAN-1): identification of residues essential for enzyme activity and the Ca2+-induced conformational change. Biochemistry. 2004;43:9185–9194. doi: 10.1021/bi049565o. [DOI] [PubMed] [Google Scholar]