Abstract
Summary
Carbohydrate arrays, also referred to as glycan arrays, are composed of various oligosaccharides and/or polysaccharides immobilized on a solid support in a spatially-defined arrangement. This technology provides a powerful, high-throughput approach to examining carbohydrate-macromolecule interactions, and glycan arrays have had a significant impact on the field of glycobiology. This review focuses on recent advances in glycan array technology, limitations, and opportunities for improvement. In particular, new methods for the production of natural glycan arrays and chemo-enzymatic approaches are greatly expanding the diversity of structures on arrays. Since multivalent complex formation is general required to achieve tight binding, methods to evaluate and modulate presentation are vital for enhancing the capabilities of this technology.
Introduction
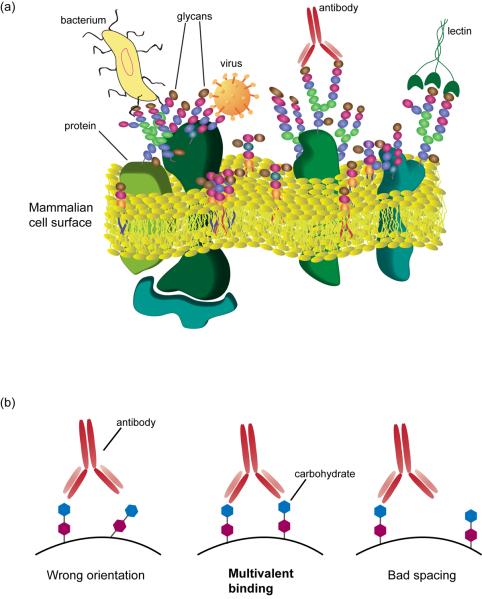
Carbohydrate-macromolecule interactions play a crucial role in basic and applied research (Figure 1a) [1,2]. They mediate a variety of biological processes such as inflammation and development. In addition, many pathogens bind carbohydrates on the surface of host cells as a key step of infection. Furthermore, lectins (carbohydrate binding proteins other than antibodies) and glycan-binding antibodies are used extensively as research tools, diagnostics, and therapeutic agents to detect and target glycans. As a result, there has been significant interest in evaluating the specificity of carbohydrate binding macromolecules, developing ligands that can be used to modulate their activity, and identifying new carbohydrate binding proteins.
Figure 1.
Carbohydrate-macromolecule interactions (a) Interaction of carbohydrate binding proteins and macromolecules with cell surface glycans presented on glycoproteins and glycolipids. (b) Optimal spacing and orientation of carbohydrate ligands is critical to achieve high avidity multivalent binding.
Traditionally, analysis of carbohydrate-macromolecule interactions has been a challenging area of science for several reasons. First, carbohydrates can be very difficult to obtain in large quantities and/or in homogeneous form. With limited access to the ligands, one cannot easily evaluate recognition. Second, traditional methods used to evaluate carbohydrate-protein interactions such as mono- and oligosaccharide inhibition studies, isothermal calorimetry (ITC), surface plasmon resonance (SPR), and enzyme-linked lectin assays (ELLA) can be labor intensive and/or require large quantities of each carbohydrate. Finally, molecular recognition is dependent on carbohydrate presentation. [3,4]. Typically, monovalent interactions between a single carbohydrate ligand and a single binding domain of a protein are very weak. To achieve tight binding, most carbohydrate-binding proteins possess two or more binding sites or assemble into functional units with multiple binding sites to produce high avidity multivalent complexes. Formation of these multivalent complexes, in turn, depends on appropriate spacing and orientation of ligands (Figure 1b). Since the optimal presentation is difficult to predict, it is often necessary to evaluate multiple combinations of structure and presentation.
Array technology overcomes many of these challenges. Carbohydrate arrays, also referred to as glycan arrays, are composed of numerous oligosaccharides and/or polysaccharides immobilized on a solid support in a spatially-defined arrangement. Various forms of “arrays” have been around for many years. For example, glycoconjugates have been immobilized on silica plates [5], synthesized on beads [6], or immobilized in different wells of ELISA plates [7]; however recent advances in high precision robotic arraying and high resolution imaging have transformed the field by permitting substantial miniaturization such that tens of thousands of carbohydrates or other array “features” can be affixed and imaged on a standard size microscope slide. These “microarrays” permit high-throughput analysis of many potential combinations of structure and presentation while using only miniscule amounts of each carbohydrate. Several comprehensive reviews on methods for fabricating glycan arrays have been published [8-11].
Although glycan array technology has become a key tool for glycobiologists, many challenges remain. For example, many screens show no binding, producing results that are deemed inconclusive. In addition, different array platforms can yield significantly different results (for example, compare the results of Gildersleeve [12] with those of the Consortium for Functional Glycomics for monoclonal antibodies F3, 7LE, and T174). Therefore, there is a need to improve this technology and better understand how to interpret results. This review will focus on recent advances and challenges of glycan array technology.
Expanding Structural Diversity on Glycan Arrays
The size and diversity of carbohydrates on a glycan array contribute significantly to the information extracted from an experiment, but acquiring a large collection of glycans in a format suitable for immobilization on a surface is a formidable challenge. Approaches to addressing this issue fall into two general categories: isolation from natural sources and synthesis.
A number of groups have been examining strategies for producing “natural glycan arrays”. The basic approach involves a) isolating mixtures of glycans from natural sources such as cells, tissues, pathogens, milk, or urine, b) derivatizing the glycans with a linker/tag (if necessary) to facilitate purification and allow immobilization on an array surface, and then c) separating the mixture into individual components or sub-fractions. This approach offers a number of advantages: the organism synthesizes the glycans, in principle one can access the entire glycome, and one can focus studies on the glycans found in a particular target cell, tissue, or sample. For this approach to be effective in practice, one needs efficient and reliable methods to derivatize the glycans, powerful separation and purification techniques, and methods to identify and characterize the unknown glycan structures. For glycoproteins and glycolipids, release of the glycan from an attached protein or lipid may also be necessary. In addition, each of these steps must be amenable to very small scale, since many individual glycans are only present in minute amounts.
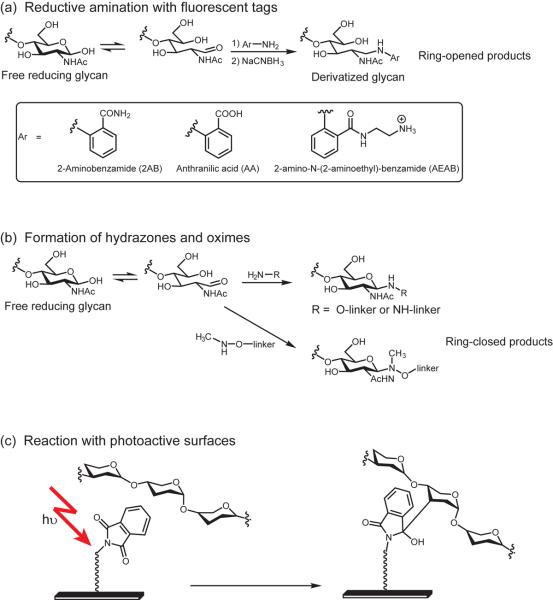
Recent work has focused on identifying optimal tags or linkers that can be attached to the glycans and used to facilitate separation, aid detection, and provide functionality for immobilization on an array surface (Figure 2a). One very useful development was reported by de Boer's group. After release from proteins or lipids, glycans are frequently derivatized with the fluorescent labels 2-aminobenzamide (AB) or 2-aminobenzoid acid (AA) to facilitate separation and detection by HPLC. De Boer's group found that these modified glycans can be covalently attached to epoxide-coated surfaces via reaction with the secondary amine [13]. This work provides a smooth transition between standard analytical techniques for glycans and array fabrication. In related work, the Cummings group has developed a 2-aminobenzamide analog that contains an alkyl amine [14]. Like the AB and AA linkers, it can be coupled to glycans via reductive amination, but the alkyl amine group is more reactive towards surface electrophiles. In either case, the fluorescent tags facilitate determination of the concentration of each separated glycan and provide a label to verify immobilization on the array surface, which can be used for quality control.
Figure 2.
Examples of approaches to expanding structural diversity on glycan arrays. (a) Reductive amination with fluorescent tags facilitate purification and immobilization of isolated natural glycans but results in ring-opening of the reducing end sugar. (b) Formation of hydrazones and oximes with the reducing end sugar enables surface immobilization of glycans via linkers and can afford ring-closed products. (c) Covalent immobilization of underivatized glycans to photoactive surfaces such as phthalimidefunctionalized surfaces.
One disadvantage of reductive amination is that the reducing end sugar ring is destroyed, which can disrupt recognition. To circumvent this problem, a number of groups have designed linkers with aminooxy- [15,16], methylaminooxy- [17], and hydrazide groups [16] (Figure 2b). The reactions of these groups with aldehydes are facile and produce stable products. Unlike reductive amination, the products formed with these linkers/tags can have a significant proportion of the ring closed form.
To circumvent the need for a linker, several groups have investigated direct immobilization of glycans on surfaces to produce natural glycan arrays. Under appropriate conditions, large carbohydrates will non-covalently adhere onto various surfaces without modification. Examples include bacterial polysaccharides [18-21] and plant polysaccharides [22-25]. A second approach involves the development of photo-active surfaces that produce highly reactive intermediates that can covalently attach to glycans (Figure 2c) [26-29]. A third approach involves constructing surfaces with glycan reactive groups such as aminooxy or hydrazide functional groups [30-33].
Synthesis via chemical, chemo-enzymatic, or enzymatic synthesis provides an alternative to isolating glycans from natural sources. The primary advantages are control of the target structure being synthesized and the ability to produce larger quantities of homogeneous material. Mrksich's group recently published a nice approach wherein chemical and enzymatic syntheses were carried out directly on an array surface [34]. In addition to evaluating glycosyltransferase activity, the approach provides an efficient chemo-enzymatic method for parallel synthesis of large collections of glycans. In another study, Chen's group constructed a diverse sialoside library using a chemoenzymatic approach [35]. The oligosaccharides also possessed a biotin tag which allowed arraying on NeutrAvidin-coated microtiter plates and analysis of binding with lectins. In related work, Blixt et al. produced a series of unnatural sialoside analogs via chemoenzymatic methods [36]. Sialosides are especially difficult to chemically synthesize and many derivatives are degraded in the process of isolation from natural sources. These studies illustrate the power of chemo-enzymatic methods for obtaining difficult to access glycans.
While significant improvements have occurred in the last few years, many areas of diversity such as glycosaminoglycans, glycopeptides, and non-human glycans are not adequately represented on existing arrays. Advances in automated solid phase [37] and fluorous phase [38] carbohydrate synthesis hold tremendous potential to address this issue.
Modulating Carbohydrate Presentation on Glycan Arrays
Presentation is a critical factor for carbohydrate recognition. As mentioned in the introduction, formation of a multivalent complex is often required to obtain a high avidity interaction (see Figure 1) [3,4]. As a result, features of presentation such as spacing and orientation of carbohydrates, linker length and flexibility, and ligand density can have a major impact on recognition. Microarrays are an excellent platform for examining many combinations of carbohydrate structure and presentation. Ideally, a method aimed at modulating presentation should produce variations on the array surface on a scale that is appropriate for biological recognition events and provide information that can readily be translated to other, non-array based applications such as the development of inhibitors or probes. Meeting these criteria is not trivial since many technical parameters can affect presentation.
One key feature of presentation is ligand density. Several strategies have been studied for varying carbohydrate density on an array, but an optimal method has not yet emerged [39-45]. When varying density, several technical factors should be considered. First, the maximum density is generally controlled by the capacity of the surface. Second, when printing material such that the surface is not “saturated”, there is a potential to obtain density gradients within a spot. For example, as a liquid droplet evaporates, one can get higher densities at the center of the spot relative to the border regions. Under other conditions, one can obtain higher densities around the perimeter, sometimes referred to as “ring-like spots”[46]. Third, with certain surface attachment methods, the carbohydrates can move and adopt a range of densities within a spot. While this imparts flexibility to accommodate lectins with varying architectures and presentation requirements, the density is not well defined.
Most arrays are produced by printing monovalent carbohydrates onto a surface. One strategy for modulating density is to vary the concentration of the monovalent carbohydrates in the print solution. With this approach, however, the array surface acts as the multivalent scaffold. Identification of other multivalent scaffolds with similar spacing and presentation of carbohydrates as the array surface can be difficult. An alternative strategy is to first produce multivalent glycoconjugates with varying density, and then print the conjugates onto a solid support to generate an array of different multivalent components. Pieters et al. attached mannose-functionalized dendrimers to an array surface to produce an array of glycodendrimers of valencies ranging from mono- to octavalent [47]. This array chip enabled rapid real time evaluation of multivalency effects on binding of lectins ConA and GNA to mannose. The authors showed that ConA, due to its widely spaced binding sites, exhibited no major multivalency effects, while GNA, which has more closely spaced binding sites, exhibited distinct density-dependent binding. Gildersleeve and coworkers fabricated an array of multivalent glycoconjugates by attaching linker-functionalized mono- and oligosaccharides to BSA to afford neoglycoconjugates [48]. The densities were modulated by varying the number of sugars attached to BSA. The authors evaluated density-dependent binding of lectins, monoclonal antibodies, and polyclonal human serum antibodies and found that, in some cases, different subpopulations of antibodies in humans can recognize different densities of the same antigen. Neoglycoproteins can be used as multivalent inhibitors, immunogens, and in a variety of other assays such as ELISA/ELLA.
Although these studies demonstrate the importance of varying presentation, new developments are critical for the field. First, detailed information on the presentation of glycans on the array surfaces, especially at a molecular level, are crucial for interpreting array results. At present, however, very little characterization has been published and more in depth studies are needed. Second, existing approaches for varying presentation are imprecise; better methods to control and vary features such as spacing and orientation would be invaluable. Third, nature produces an assortment of multivalent binding modes, mechanisms, and scales, but current methods for varying presentation are limited in scope. New strategies for varying presentation are needed to match the diversity found in nature.
Recent applications of carbohydrate microarray technology
The most common application of glycan array technology is the analysis of binding specificity of lectins and antibodies. Arrays are used to determine if a protein recognizes carbohydrates, identify natural ligands, and develop probes/inhibitors to modulate the activity of lectins. To date, hundreds of proteins have been screened and glycan array profiling has become routine. Binding has primarily been detected using fluorescently labeled samples or secondary reagents; however, other methods of detection, such as mass spectroscopy [34] and surface plasmon resonance [49,50], have also been used. When interpreting binding profiles, the assay conditions and array platform should be considered. For example, at high concentrations a lectin may bind many different glycan structures while at lower concentrations it may only bind a small subset of carbohydrates. In addition, the method of carbohydrate presentation can significantly affect binding.
One of the best resources for glycan array screening is the Consortium for Functional Glycomics (CFG). The CFG has developed one of the largest glycan arrays in the world and has provided routine screening for many investigators. Some recent examples include the screening of galectin 8 [51], human ficolin [52], and Candida glabrata adhesion [53]. Some other labs have developed and utilized glycan array technology for lectin and antibody screening. A nice example comes from the Feizi laboratory. Feizi and coworkers used a carbohydrate microarray to reveal the structure of the preferred ligand for a novel protein, malectin [54]. Although it was determined from NMR studies that the protein recognizes glucose oligomers, the exact structure of the ligand was not immediately apparent. By profiling the protein in a glycan array, the authors were able to determine that malectin bound a diglucosylated high mannose-N-glycan structure with exquisite selectivity. In another example, Gildersleeve and coworkers used a carbohydrate microarray to evaluate the specificities of a set of lectins and antibodies used as reagents to detect the tumor-associated Tn antigen [12,55,56]. Many of these reagents were found to cross-react with other glycans. Based on this information, Gildersleeve and coworkers were able to re-interpret previous studies and carry out additional studies on the expression of the Tn antigen in prostate tumors. They found that the Tn antigen is only expressed in a subset of prostate tumors, suggesting that pre-selection of this subset could improve clinical outcomes with Tn vaccines.
One of the growing applications of glycan array technology is serum antibody profiling. Human serum contains a wide variety of carbohydrate-binding antibodies, and the populations of these antibodies change as a result of disease, exposure to pathogens, and vaccination. Several recent examples illustrate the utility of carbohydrate antigen arrays for high-throughput profiling of serum antibodies.
Glycan arrays have been developed for the detection of pathogen specific antibodies for the serodiagnosis of infectious agents. Seeberger and coworkers reported preparation of a microarray comprised of synthetic P. falciparum glycosylphosphatidylinositol (GPI) glycans [57]. The microarray was used to compare anti-GPI IgG levels in donors from malaria-endemic areas with non-exposed individuals and to determine the effect of exposure to the malaria parasite in previously non-exposed individuals. Results revealed distinct differences in GPI antigens recognized as a result of malaria exposure. Other recent examples include profiling of mellidosis patients and animals vaccinated or infected with anthrax or tularemia-causing bacteria [20,21], salmonellosis patients [58], and Schistosoma mansoni infected individuals [50].
Microarrays have also seen many applications in cancer research. One recent report highlights the utility and sensitivity of a glycan microarray to measure antibody levels to a tumor-associated carbohydrate antigen, Globo H, and related structures [59]. In this study, the authors found that breast cancer patients had significantly higher levels of anti-Globo-H antibodies in their blood when compared with the normal individuals, which suggests the potential for development of a diagnostic based on the Globo H antigen on a microarray platform.
Glycan arrays have also been used to profile antibody responses in xenotransplants. Although it is established that anti-α-Gal antibodies are responsible for the majority of hyperacute rejection of pig-human organ xenotransplants, it is believed that non-α-Gal antibodies are involved in the rejection of porcine islet xenografts. Using a glycan array of 200 different structures, Blixt et al. investigated antibody responses to porcine cells in transplant patients [60]. The authors identified antibody responses post-transplant to novel carbohydrate determinants.
These preliminary studies show great promise for glycan microarray technology. To reach its full potential, however, more information regarding experimental and biological variability is needed. Microarray data are notoriously variable. At present, only minimal technical validation of glycan microarray reproducibility and reliability has been published, and these have focused on homogeneous, purified lectins which may perform better than complex, heterogeneous serum. For larger clinical studies, analysis of samples over multiple batches of slides and over longer periods of time will be necessary. To account for gradual changes in performance of scanners and reagents, it will also be necessary to have robust normalization methods. In addition to experimental issues, more information on the normal biological variation between individuals and variation over time within individuals will be useful.
Conclusions
Over the last few years, glycan arrays have become powerful tools for analysis of carbohydrate-macromolecule interactions. This technology has been used to evaluate the specificities of hundreds of lectins, antibodies, cells, and viruses. In addition, it is now being applied to clinical research for antigen detection, vaccine development, and biomarker discovery. Nevertheless, a number of challenges remain. First, existing arrays contain only a small fraction of the carbohydrate diversity found in nature. Moreover, these glycans are presented in a limited number of ways on array surfaces. To fully exploit the potential of arrays, it will be necessary to increase the quantity and diversity of carbohydrate structures as well as develop more methods for presenting them. Second, array screening provides large quantities of data and the ability to extract useful information from this data is vital. Better methods to mine the data and use array information to guide additional biological studies will be necessary. Currently, different laboratories disseminate glycan array information using different formats and standards. Efforts to standardize information and results could facilitate meta-analyses. Third, glycan array data provide extensive information on relationships between structure and recognition by glycan-binding proteins, but the design and development of probes and inhibitors using these data are not straightforward. Better methods to translate array data would be beneficial. Finally, glycan array data is not always easily interpretable, particularly to non-specialists, so making array information more accessible will increase the value of this technology to those less familiar with glycobiology.
Acknowledgements
O.O. and J.C.G. are supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Annotations
*13 (de Boer). The authors developed a natural glycan array by releasing glycans from glycoproteins and glycolipids and coupling to a fluorescent linker that facilitated purification and immobilization of glycans to an epoxide surface. Using a single array chip and SPR analysis, they evaluated and compared binding of human serum antibodies in 21 samples.
*14 (Song). The authors fabricated a natural glycan array by coupling glycans to a bifunctional fluorescent linker having an aryl and an alkylamine. Glycans were coupled to the arylamine by reductive amination and the derivatived glycans were attached to the surface via the alkylamine. The array was used to evaluate binding specificity of two galectins.
**34. (Ban). The authors demonstrate on-chip chemical and enzymatic synthesis of oligosaccharides on gold-based self-assembled monolayers and the use of mass spectrometry to directly monitor reactions and evaluate glycosyltransferase activity on the surface.
**35. (Chokhawala). The authors demonstrate chemoenzymatic synthesis of a library of biotinylated sialosides in microtiter plates using a one-pot three-enzyme protocol. After immobilization on NeutrAvidin-coated plates, fluorescently labeled lectins recognizing unreacted acceptor or product were used to determine the efficiency of the reactions. The array was used to evaluate specificities of sialic acid-binding proteins.
*36. (Blixt). The authors synthesize a library of 9-acyl-substitude sialoside analogs by chemoenzymatic synthesis and fabricate a glycan array, which was used to screen for high affinity ligands of sialic acid binding proteins.
**47. (Branderhorst) The fabrication of a flow-through chip consisting of glycodendrimers of different valencies and real time analysis of multivalency effects for two lectins are reported. The authors were able to derive kinetic and thermodynamic data from, and conduct inhibition studies on the arrayw.
**48. (Oyelaran) The fabrication of a carbohydrate microarray consisting of BSA neoglycoproteins having different densities of carbohydrate ligands is reported. The authors used the array to evaluate density dependent binding of lectins, monoclonal antibodies, and serum antibodies and showed that this approach produces variations in ligand presentation in a biologically relevant range.
*54. (Schallus) The authors define the binding specificity of the novel lectin, malectin, by using nuclear magnetic resonance and glycan array analysis. The glycan array revealed that the lectin bound to a diglucose N-glycan probe with remarkable selectivity.
*55. (Li) The authors use antibodies to measure expression of the tumor associated carbohydrate antigen Tn in prostate tumors and then use a carbohydrate microarray to evaluate the binding profiles of the antibodies. The study revealed that only a small subset of prostate tumors express the appropriate Tn antigen.
*59. (Wang) A glycan array of cancer-associated carbohydrate antigen Globo H and its analogs was fabricated and used to evaluate the binding specificity of monoclonal antibodies as well as differences in serum antibody levels between cancer patients and healthy donors. The authors found higher levels of antibodies to Globo H in cancer patients compared to the normal donors.
*60. (Blixt) The authors used a glycan array to further define the specificities of anti-carbohydrate antibodies produced in response to porcine fetal islet-like cell cluster transplantation in 7 patients. They found elevated levels of antibodies to novel non-α-Gal antigens in some patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- 2.Dwek RA. Glycobiology: Toward understanding the function of sugars. Chem. Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 3.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Lundquist JJ, Toone EJ. The cluster glycoside effect. Chem. Rev. 2002;102:555. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 5.Tang PW, Gooi HC, Hardy M, Lee YC, Feizi T. Novel approach to the study of the antigenicities and receptor functions of carbohydrate chains of glycoproteins. Biochem. Biophys. Res. Comm. 1985;132:474–480. doi: 10.1016/0006-291x(85)91158-1. [DOI] [PubMed] [Google Scholar]
- 6.Liang R, Yan L, Loebach J, Ge M, Uozumi Y, Sekanina K, Horan N, Gildersleeve J, Thompson C, Smith A, et al. Parallel synthesis and screening of a solid phase carbohydrate library. Science. 1996;274:1520–1522. doi: 10.1126/science.274.5292.1520. [DOI] [PubMed] [Google Scholar]
- 7.Roy R. Syntheses and some applications of chemically defined multivalent glycoconjugates. Curr. Opin. Struct. Biol. 1996;6:692–702. doi: 10.1016/s0959-440x(96)80037-6. [DOI] [PubMed] [Google Scholar]
- 8.Culf AS, Cuperlovic-Culf M, Ouellette RJ. Carbohydrate microarrays: Survey of fabrication techniques. Omics. 2006;10:289–310. doi: 10.1089/omi.2006.10.289. [DOI] [PubMed] [Google Scholar]
- 9.Park S, Lee M-R, Shin I. Carbohydrate microarrays as powerful tools in studies of carbohydrate-mediated biological processes. Chem. Comm. 2008:4389–4399. doi: 10.1039/b806699j. [DOI] [PubMed] [Google Scholar]
- 10.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat. Chem. Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 11.Liang P-H, Wu C-Y, Greenberg WA, Wong C-H. Glycan arrays: biological and medical applications. Curr. Opin. Chem. Biol. 2008;12:86. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 13.De Boer AR, Hokke CH, Deelder AM, Wuhrer M. General microarray technique for immobilization and screening of natural glycans. Anal. Chem. 2007;79:8107. doi: 10.1021/ac071187g. [DOI] [PubMed] [Google Scholar]
- 14.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel Fluorescent Glycan Microarray Strategy Reveals Ligands for Galectins. Chem. Biol. 2009;16:36. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Feizi T, Campanero-Rhodes MA, Childs RA, Zhang Y, Mulloy B, Evans PG, Osborn HMI, Otto D, Crocker PR, et al. Neoglycolipid Probes Prepared via Oxime Ligation for Microarray Analysis of Oligosaccharide-Protein Interactions. Chem. Biol. 2007;14:847. doi: 10.1016/j.chembiol.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa JI, Shinohara Y, Kuramoto H, Miura Y, Shimaoka H, Kurogochi M, Nakano M, Nishimura SI. Comprehensive approach to structural and functional glycomics based on chemoselective glycoblotting and sequential tag conversion. Anal. Chem. 2008;80:1094–1101. doi: 10.1021/ac702124d. [DOI] [PubMed] [Google Scholar]
- 17.Bohorov O, Andersson-Sand H, Hoffmann J, Blixt O. Arraying glycomics: a novel bifunctional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology. 2006;16:21C–27C. doi: 10.1093/glycob/cwl044. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate Microarrays for the Recognition of Cross-Reactive Molecular Markers of Microbes and Host Cells. Nat. Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 19.Parthasarathy N, DeShazer D, England M, Waag DM. Polysaccharide microarray technology for the detection of Burkholderia pseudomallei and Burkholderia mallei antibodies. Diagnostic Microbiology and Infectious Disease. 2006;56:329. doi: 10.1016/j.diagmicrobio.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parthasarathy N, DeShazer D, Peacock SJ, Wuthiekanun V, England MJ, Norris SL, Waag DM. Application of polysaccharide microarray technology for the serodiagnosis of Burkholderia pseudomallei infection (melioidosis) in humans. J. Carbohydr. Chem. 2008;27:32–40. [Google Scholar]
- 21.Parthasarathy N, Saksena R, Kovác P, DeShazer D, Peacock SJ, Wuthiekanun V, Heine HS, Friedlander AM, Cote CK, Welkos SL, et al. Application of carbohydrate microarray technology for the detection of Burkholderia pseudomallei, Bacillus anthracis and Francisella tularensis antibodies. Carbohydr. Res. 2008;343:2783. doi: 10.1016/j.carres.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Marcus SE, Verhertbruggen Y, Herve C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WG, Knox JP. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8 doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moller I, Marcus SE, Haeger A, Verhertbruggen Y, Verhoef R, Schols H, Ulvskov P, Mikkelsen JD, Knox JP, Willats W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2008;25:37. doi: 10.1007/s10719-007-9059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moller I, Sørensen I, Bernal AJ, Blaukopf C, Lee K, Øbro J, Pettolino F, Roberts A, Mikkelsen JD, Knox JP, et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 2007;50:1118. doi: 10.1111/j.1365-313X.2007.03114.x. [DOI] [PubMed] [Google Scholar]
- 25.Øbro J, Sørensen I, Moller I, Skjøt M, Mikkelsen JD, Willats WGT. High-throughput microarray analysis of pectic polymers by enzymatic epitope deletion. Carbohydr. Polym. 2007;70:77. [Google Scholar]
- 26.Wang DN, Carroll GT, Turro NJ, Koberstein JT, Kovac P, Saksena R, Adamo R, Herzenberg LA, Herzenberg LA, Steinman L. Photogenerated glycan arrays identify immunogenic sugar moieties of Bacillus anthracis exosporium. Proteomics. 2007;7:180–184. doi: 10.1002/pmic.200600478. [DOI] [PubMed] [Google Scholar]
- 27.Carrol GT, Wang D, Turro NJ, Koberstein JT. Photochemical Micropatterning of Carbohydrates on a Surface. Langmuir. 2006;22:2899–2905. doi: 10.1021/la0531042. [DOI] [PubMed] [Google Scholar]
- 28.Pei Z, Yu H, Theurer M, Walden A, Nilsson P, Yan M, Ramstrom O. Photogenerated Carbohydrate Microarrays. Chembiochem. 2007;8:266–268. doi: 10.1002/cbic.200600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycoprofiling with Micro-Arrays of Glycoconjugates and Lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Zhou J. Oligosaccharide microarrays fabricated on aminooxyacetyl functionalized glass surface for characterization of carbohydrate-protein interaction. Biosens. Bioelectron. 2006;21:1451–1458. doi: 10.1016/j.bios.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Vila-Perello M, Gallego RG, Andreu D. A simple approach to well-defined sugar-coated surfaces for interaction studies. Chembiochem. 2005;6:1831–1838. doi: 10.1002/cbic.200500125. [DOI] [PubMed] [Google Scholar]
- 32.Park S, Lee MR, Shin I. Construction of carbohydrate microarrays by using one-step, direct immobilizations of diverse unmodified glycans on solid surfaces. Bioconjugate Chem. 2009;20:155–162. doi: 10.1021/bc800442z. [DOI] [PubMed] [Google Scholar]
- 33.Lee M, Shin I. Facile preparation of carbohydrate microarrays by site-specific, covalent immobilization of unmodified carbohydrates on hydrazide-coated glass slides. Org. Lett. 2005;7:4269–4272. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 34.Ban L, Mrksich M. On-chip synthesis and label-free assays of oligosaccharide arrays. Angew. Chem. Int. Ed. 2008;47:3396. doi: 10.1002/anie.200704998. [DOI] [PubMed] [Google Scholar]
- 35.Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial Chemoenzymatic Synthesis and High-Throughput Screening of Sialosides. ACS Chem. Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blixt O, Han S, Liao L, Zeng Y, Hoffmann J, Futakawa S, Paulson JC. Sialoside analogue arrays for rapid identification of high affinity siglec ligands. J. Am. Chem. Soc. 2008;130:6680. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plante OJ, Palmacci ER, Seeberger PH. Automated solid-phase synthesis of oligosaccharides. Science. 2001;291:1523–1527. doi: 10.1126/science.1057324. [DOI] [PubMed] [Google Scholar]
- 38.Jaipuri FA, Pohl NL. Toward solution-phase automated iterative synthesis: Fluorous-tag assisted solution-phase synthesis of linear and branched mannose oligomers. Org. Biomol. Chem. 2008;6:2686–2691. doi: 10.1039/b803451f. [DOI] [PubMed] [Google Scholar]
- 39.Smith EA, Thomas WD, Kiessling LL, Corn RM. Surface Plasmon Resonance Imaging Studies of Protein-Carbohydrate Interactions. J. Am. Chem. Soc. 2003;125:6140–6148. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 40.Houseman BT, Mrksich M. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem. Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 41.Ngundi MM, Taitt CR, McMurry SA, Kahne D, Ligler FS. Detection of bacterial toxins with monosaccharide arrays. Biosens. Bioelectron. 2006;21:1195–1201. doi: 10.1016/j.bios.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chevolot Y, Bouillon C, Vidal S, Morvan F, Meyer A, Cloarec JP, Jochum A, Praly JP, Vasseur JJ, Souteyrand E. DNA-based carbohydrate biochips: A platform for surface glyco-engineering. Angew. Chem. Int. Ed. 2007;46:2398–2402. doi: 10.1002/anie.200604955. [DOI] [PubMed] [Google Scholar]
- 43.Liang PH, Wang SK, Wong CH. Quantitative Analysis of Carbohydrate-Protein Interactions Using Glycan Microarrays: Determination of Surface and Solution Dissociation Constants. J. Am. Chem. Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- 44.Mercey E, Sadir R, Maillart E, Roget A, Baleux F, Lortat-Jacob H, Livache T. Polypyrrole oligosaccharide array and surface plasmon resonance imaging for the measurement of glycosaminoglycan binding interactions. Anal. Chem. 2008;80:3476. doi: 10.1021/ac800226k. [DOI] [PubMed] [Google Scholar]
- 45.Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Quantifiable fluorescent glycan microarrays. Glycoconj. J. 2008;25:15. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 46.Deng Y, Zhu XY, Kienlen T, Guo A. Transport at the air/water interface is the reason for rings in protein microarrays. J. Am. Chem. Soc. 2006;128:2768–2769. doi: 10.1021/ja057669w. [DOI] [PubMed] [Google Scholar]
- 47.Branderhorst HM, Ruijtenbeek R, Liskamp RMJ, Pieters RJ. Multivalent carbohydrate recognition on a glycodendrimer-functionalized flow-through chip. ChemBioChem. 2008;9:1836–1844. doi: 10.1002/cbic.200800195. [DOI] [PubMed] [Google Scholar]
- 48.Oyelaran OO, Li Q, Farnsworth DF, Gildersleeve JC. Microarrays with Varying Carbohydrate Density Reveal Distinct Subpopulations of Serum Antibodies. J. Proteome Res. 2009;8 doi: 10.1021/pr9002245. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karamanska R, Clarke J, Blixt O, MacRae JI, Zhang JQ, Crocker PR, Laurent N, Wright A, Flitsch SL, Russell DA, et al. Surface plasmon resonance imaging for real-time, label-free analysis of protein interactions with carbohydrate microarrays. Glycoconjugate J. 2008;25:69. doi: 10.1007/s10719-007-9047-y. [DOI] [PubMed] [Google Scholar]
- 50.De Boer AR, Hokke CH, Deelder AM, Wuhrer M. Serum antibody screening by surface plasmon resonance using a natural glycan microarray. Glycoconj. J. 2008;25:75. doi: 10.1007/s10719-007-9100-x. [DOI] [PubMed] [Google Scholar]
- 51.Diskin S, Cao Z, Leffler H, Panjwani N. The role of integrin glycosylation in galectin-8-mediated trabecular meshwork cell adhesion and spreading. Glycobiology. 2009;19:29–37. doi: 10.1093/glycob/cwn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krarup A, Mitchell DA, Sim RB. Recognition of acetylated oligosaccharides by human L-ficolin. Imm. Lett. 2008;118:152. doi: 10.1016/j.imlet.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zupancic ML, Frieman M, Smith D, Alvarez RA, Cummings RD, Cormack BP. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 2008;68:547–559. doi: 10.1111/j.1365-2958.2008.06184.x. [DOI] [PubMed] [Google Scholar]
- 54.Schallus T, Jaeckh C, Fehér K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T, et al. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell. 2008;19:3404–3414. doi: 10.1091/mbc.E08-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q, Anver M, Butcher D, Gildersleeve JC. Resolving Conflicting Data on Expression of the Tn Antigen and Implications for Clinical Trials with Cancer Vaccines. Mol. Cancer. Res. 2009;8:971–979. doi: 10.1158/1535-7163.MCT-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manimala J, Li Z, Jain A, VedBrat S, Gildersleeve JC. Carbohydrate array analysis of anti-Tn antibodies and lectins reveals unexpected specificities: Implications for diagnostic and vaccine development. ChemBioChem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 57.Kamena F, Tamborrini M, Liu X, Kwon YU, Thompson F, Pluschke G, Seeberger PH. Synthetic GPI array to study antitoxic malaria response. Nat. Chem. Biol. 2008;4:238–240. doi: 10.1038/nchembio.75. [DOI] [PubMed] [Google Scholar]
- 58.Blixt O, Hoffmann J, Svenson S, Norberg T. Pathogen specific carbohydrate antigen microarrays: A chip for detection of Salmonella O-antigen specific antibodies. Glycoconj. J. 2008;25:27–36. doi: 10.1007/s10719-007-9045-0. [DOI] [PubMed] [Google Scholar]
- 59.Wang C-C, Huang Y-L, Ren C-T, Lin C-W, Hung J-T, Yu J-C, Yu AL, Wu C-Y, Wong CH. Glycan microarray of Globo H and related structures for quantitative analysis of breast cancer. Proc. Natl. Acad. Sci. 2008;105:11661–11666. doi: 10.1073/pnas.0804923105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blixt O, Kumagai-Braesch M, Tibell A, Groth CG, Holgersson J. Anticarbohydrate antibody repertoires in patients transplanted with fetal pig islets revealed by glycan arrays. Am. J. Transplant. 2009;9:83. [Google Scholar]