Abstract
Sphingosine-1-phosphate lyase (SPL) is a highly conserved enzyme that catalyses the final step of sphingolipid degradation, namely the irreversible cleavage of the carbon chain at position 2-3 of a long chain base phosphate (LCBP), thereby yielding a long-chain aldehyde and phosphoethanolamine. LCBPs are potent signaling molecules involved in cell proliferation, survival, migration, cell-cell interactions and cell stress responses. Therefore, tight regulation of LCBP signaling is required for proper cell function, and perturbations of this system can lead to alterations in biological processes including development, reproduction and physiology. SPL is a key enzyme in regulating the intracellular and circulating levels of LCBPs and is, therefore, gaining attention as a putative target for pharmacological intervention. This review provides an overview of our current understanding of SPL structure and function, mechanisms involved in SPL regulation and the role of SPL in development and disease.
1. Introduction
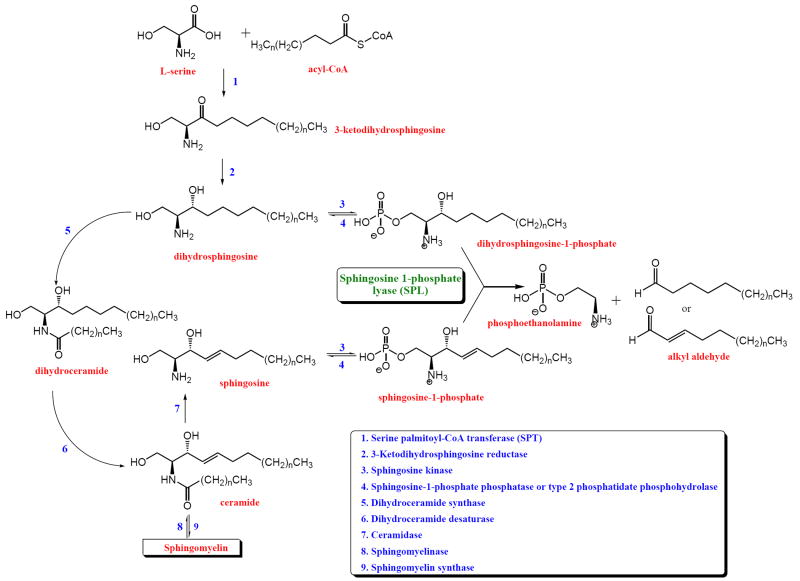
Sphingolipid metabolites such as long chain bases (LCBs), long chain phosphorylated bases (LCBPs), ceramide and ceramide-1-phosphate are bioactive lipids involved in cell fate[1-3]. In general, LCBs and ceramide act as growth inhibitory molecules promoting apoptosis whereas the phosphorylated compounds LCBP and ceramide-1-phosphate stimulate growth and promote proliferation. Maintaining a tight balance between the levels of these metabolites seems to be critical for normal cellular function. The biological effects of sphingosine-1-phosphate (S1P) have been studied extensively. S1P has been shown to promote cell survival, proliferation and migration, ischemic preconditioning, and is essential for angiogenesis and lymphocyte trafficking[4-9]. S1P signaling is likely mediated through two separate mechanisms. Extracellular effects are mediated via a group of G-protein coupled cell surface receptors belonging to the endothelial differentiation gene (EDG) family now known as the S1P receptors[10, 11]. To date, five members of this family have been identified (S1P1 to S1P5) and they show differential expression depending on tissue and cell type. The biological effects of S1P receptor signaling are diverse and involve several pathways including mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase (JNK), extracellular-signal-regulated kinase (ERK), phosphoinositide 3-kinase (PI3K), adenylate cyclase, phospholipase C, phospholipase D and other downstream mediators[12-19]. In contrast to the well-characterized receptor mediated effects of S1P, an intracellular S1P signaling mechanism has been implicated but not proven definitively. Evidence from mammalian systems in which S1P receptor signaling has been eradicated, as well as from non-mammalian systems lacking S1P receptors, indicate a potentially receptor-independent role for S1P in regulating calcium mobilization, heat shock resistance, caspase inhibition, activation of non-receptor tyrosine kinases and activation of the Raf/MAPKK/ERK pathway[12, 20-25]. The sphingolipid metabolic pathway in mammalian systems has been thoroughly studied (Figure 1). LCBPs are formed in a reaction catalyzed by sphingosine kinases[26, 27]. In this reaction, LCBs generated by sphingolipid biosynthesis, sphingolipid recycling and degradation or derived from cellular uptake mechanisms are phosphorylated at the 1-position. Once formed, LCBPs can be metabolized back to LCBs by dephosphorylation in a reaction catalyzed by sphingosine-1-phosphate phosphatases or type 2 phosphatidate phosphohydrolases[22, 23, 28-30]. Alternatively, LCBPs can be irreversible degraded by SPL, which catalyzes the cleavage of the carbon chain at position 2-3, yielding a long-chain aldehyde and phosphoethanolamine[31-33]. This review will summarize our current understanding of SPL structure and function. We discuss current knowledge of how SPL is regulated and the role that SPL plays in biological processes from development to disease.
Figure 1.
Sphingolipid metabolism. LCBs formed from either sphingolipid de novo synthesis (step 1, 2) or from sphingolipid degradation (step 9, 7) can be phosphorylated by sphingosine kinases (step 3). The formed LCBPs can then be dephosphorylated by sphingosine-1-phosphate phosphatase or type 2 phosphatidate phosphohydrolase activity (step 4). Alternatively, LCBPs can be irreversible cleaved by SPL, thereby yielding an alkyl aldehyde and phosphoethanolamine.
2. Biochemical characterization of SPL
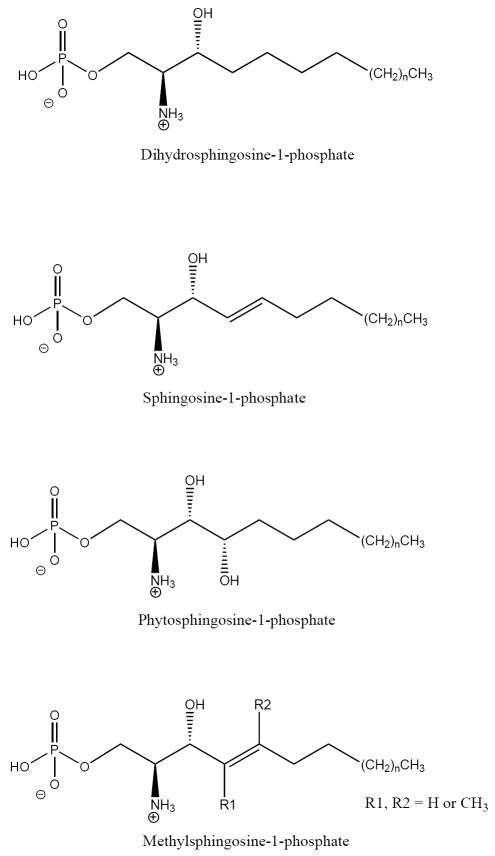
SPL is a pyridoxal-phosphate dependent member of the carbon-carbon lyase subclass of aldehyde-lyases. The substrate is a LCBP with the phosphate group attached at the 1-position. SPL shows specificity towards the stereochemistry of the substrate and only the LCBP containing the naturally occurring D-erythro-isomer of the LCB is used[34]. In contrast, SPL shows very little specificity towards the chain length, degree of unsaturation and substitution of the LCB backbone[35]. In mammalian and yeast cells, the majority of LCBs are compounds containing a 18 carbon backbone. SPL metabolizes the 1-phosphorylated derivatives of the d18:0 saturated LCB dihydrosphingosine (DHS1P), the d18:1 4-trans unsaturated LCB sphingosine (S1P) and the t18:0 4D-hydroxylated LCB phytosphingosine (PHS1P) (Figure 2). In addition, DHS1P substrates with chain lengths from 7 carbons to 20 carbons as well as S1P analogs carrying methyl groups at carbon 4 or carbon 5 are metabolized[35, 36]. SPL interacts with pyridoxal 5’-phosphate as a cofactor. The molecular interactions between SPL, cofactor and substrate have not been established experimentally although the cofactor is believed to form a Schiff base with the free amino group of the LCB[37].
Figure 2.
LCBPs metabolized by SPL. Dihydrosphingosine-1-phosphate, sphingosine-1-phosphate and phytosphingosine-1-phosphate are the major LCBPs found in mammals and yeast and the naturally occurring D-erythro isomer of these compounds is metabolized by SPL. SPL also metabolizes certain forms of LCBPs containing methyl groups at C4 or C5. SPL shows broad specificity towards the chain length of the LCBP and medium and long chain compounds are used as substrates.
SPL is a membrane-associated enzyme. Therefore, accurate enzyme kinetic parameters are difficult to obtain and depend on the assay conditions used for the analysis. Nevertheless, S1P and DHS1P were found to be degraded at roughly similar rates. The Km values for DHS1P as determined for rat liver microsomes was determined to be from 9 to 16 μM[38]. Similar values were obtained in studies performed on human SPL overexpressed in HEK293 cells. The Km value found for DHS1P in the latter system was 20 μM[39]. The original SPL assay is based on the degradation of a radioactive LCBP substrate, such as [3H]DHS1P. Recently, a new assay based on the degradation of the fluorescent S1P analog omega-(7-nitro-2-1, 3-benzoxadiazol-4-yl)-D-erythro-S1P (Omega-NBD-D-erythro-S1P) was explored. This new assay system was found to be comparable to the original assay, and the Km value found for the fluorescent S1P substrate was 15 μM[39]. As new and more potent fluorescent substrates are developed, the SPL assay will be adapted to increase sensitivity and ability to employ substrates relevant to different species.
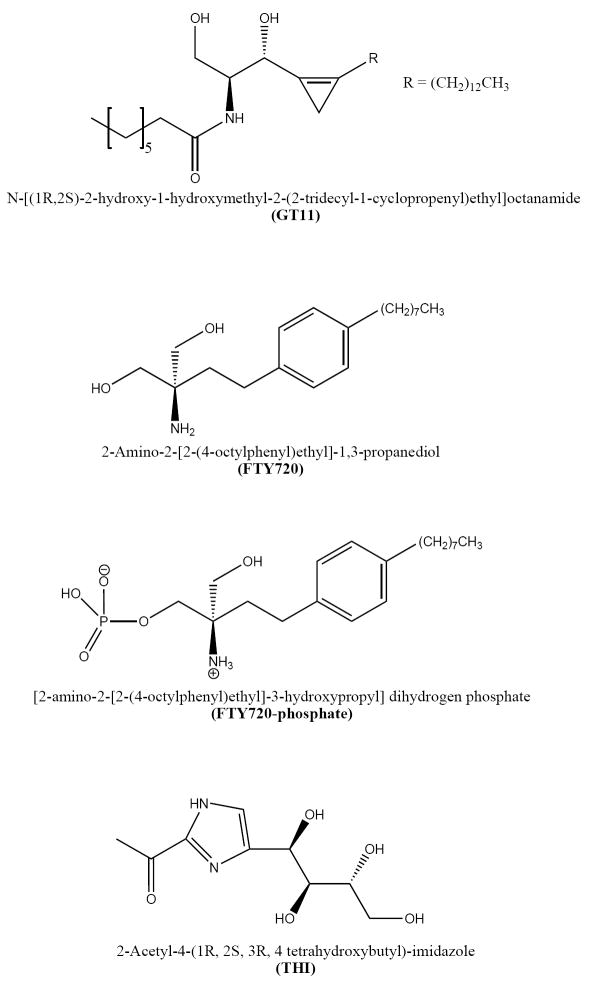
Several SPL inhibitors have been described. The first reported inhibitor of the enzyme was the substrate analog 1-desoxydihydrosphingosine-1-phosphonate[40]. Other substrate analogs found to inhibit the enzyme are the 2D,3L-isomer of DHS1P and 2-vinyldihydrosphingosine-1-phosphate[41, 42]. The Ki values reported for these substrate analogs are the range of 2-10 μM. The ceramide analog and dihydroceramide desaturase inhibitor N-[(1R,2S)-2-hydroxy-1-hydroxymethyl- 2-(2-tridecyl-1-cyclopropenyl) ethyl] octanamide (GT11) also inhibits SPL activity in vivo, although only at concentrations above 5 μM (Figure 3)[43]. GT11 has no effect on SPL activity in vitro suggesting that a structural modification of the inhibitor may be needed. The compound 2-Amino-2-[2-(4-octylphenyl)ethyl]-1,3-propanediol, also knows as Fingolimod (trademark) or FTY720, is a synthetic sphingosine analog (Figure 3). FTY720 is an immunomodulatory drug that targets S1P receptors following its in vivo phosphorylation by sphingosine kinase [44]. In addition, FTY720 was found to be a modest inhibitor of SPL activity both in vivo and in vitro[39, 45]. These results suggest that the non-phosphorylated and possibly the phosphorylated form of FTY720 (FTY720-phosphate) may inhibit SPL activity.
Figure 3.
SPL inhibitors. Several sphingolipid analogs inhibit enzyme activity and lately compounds such as the ceramide analog GT11, the LCB analog FTY720 and the LCBP analog FTY720-phosphate has been shown to inhibit SPL. Moreover, pyridoxal-phosphate analogs including THI, has also been shown to inhibit SPL.
Another group of SPL inhibitors includes pyridoxal-phosphate analogs or compounds that affects the binding of the coenzyme. Deoxypyridoxine acts as a competitive inhibitor of the enzyme and semicarbazide, cyanide, and bisulfite also inhibit the enzyme, which suggest that the coenzyme might be loosely associated. The food colorant 2-acetyl-4-(1R, 2S, 3R, 4-tetrahydroxybutyl)-imidazole (THI), was recently demonstrated to inhibit SPL activity in vivo although the mechanism is not understood [46]. Inhibition of the enzyme by THI was shown to have potent immunomodulatory effects, as discussed below. Finally, SPL activity is inhibited by divalent metal ions such as Ca2+ and Zn2+[38]. Despite the number of small molecules reported to inhibit SPL, none are specific and non-cytotoxic.
In summary, SPL is an intrinsic membrane enzyme that is promiscuous with regard to substrate specificity and is a critical regulator of tissue and circulating S1P levels [47]. Thus, identification of potent and specific small molecule inhibitors of the enzyme remains an important goal.
3. SPL structure and topology
The first report describing the cloning of a SPL gene was published in 1997[31]. In this report, the Saccharomyces cerevisiae DPL1 (DHS1P lyase) gene was identified by its ability to suppress sphingosine-induced growth inhibition. Subsequently, SPL homologs from Mus musculus, Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Dictyostelium discoideum and Leishmania major were identified and confirmed to encode functional SPL enzymes by biochemical assays and functional complementation of yeast dpl1 mutants [32, 33, 48-51]. To date, genomic sequencing has revealed the existence of putative SPL genes in a wide variety of organisms including fungi, plants and mammals. The human SPL gene, Sgpl1, encodes a predicted protein of 568 amino acids with a molecular mass of 63.5 KDa [33]. The amino acid sequence of the murine SPL homolog displays 84% identity and 92% similarity to human SPL. Similarity in primary sequence is also found between SPL homologs from Drosophila melanogaster, Leishmania major, Caenorhabditis elegans, Dictyostelium discoideum and Saccharomyces cerevisiae (Figure 4).
Figure 4.
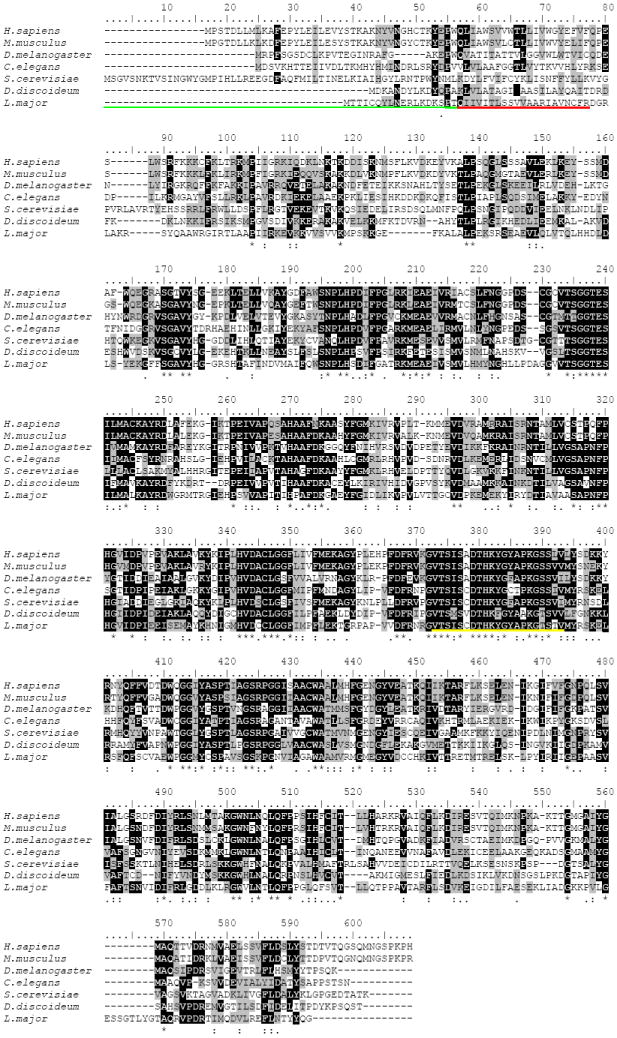
Sequence alignment of SPL. ClustalW alignment of Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, Dictyostelium discoideum and Leishmania major SPL. The underlined regions indicate the ER luminal domain (green), the transmembrane spanning domain (red) and the predicted pyridoxal-phosphate binding domain (yellow). Black shading indicates identical residues. Gray shading indicates biochemically similar residues.
The first description of SPL activity by Stoffel in 1969 indicated that the major activity was found in association with microsomal fractions, whereas a lesser amount was enriched in the inner mitochondrial membrane [35]. Subsequent studies using both immunofluorescence and subcellular fractionation confirmed the primary location of SPL within the endoplasmic reticulum (ER), although the possibility that some SPL may localize to other organelles has not been definitively ruled out [38, 52, 53]. How SPL is specifically localized to the ER has not yet been established, although removal of the first 58 amino acids leads to its expression in the soluble fraction of E. coli [33]. SPL has not been detected in serum, plasma or the extracellular space, and there are no reports of ecto-enzymes or secreted isoforms. Thus, SPL seems to be restricted to the intracellular environment. Its intracellular localization allows SPL to function as an S1P “sink”, generating a gradient between tissue and circulating S1P levels that has physiological relevance, as discussed below.
The topology of the enzyme in the ER membrane has been predicted by structural modeling and biochemical characterization and it belongs to the family of single-pass type III membrane proteins also referred to as type I without a cleavable N-terminal signal sequence [38, 53]. The enzyme contains a single transmembrane segment, which is located close to the N-terminus (see Figure 4). Its N-terminus resides in the ER lumen, whereas the large hydrophilic domain, which is responsible for catalytic activity, is located in the cytoplasmic compartment. A 20 amino acid stretch spanning position 344-364 of the human and mouse SPL is predicted to be involved in binding of the cofactor, since it shows high homology to other pyridoxal-phosphate binding motifs[33]. This region is highly conserved in all SPL homologs and contains two lysine residues (position 382 and 388 in Figure 4) that may be involved in forming an aldimine link with the cofactor. Moreover, site-directed mutagenesis studies of the human SPL have defined two cysteines residues C218 and C317 (position 245 and 346 in Figure 4) that are necessary for proper enzyme activity, likely due to substrate or cofactor binding[33]. The C317 is conserved throughout the species shown in Figure 4. However the C218 is not found in Drosophila melanogaster, Dictyostelium discoideum and Leishmania major. Interestingly, an N-terminal truncated enzyme lacking 58 amino acids including the ER luminal sequence and the membrane spanning domain was found to be active when expressed in bacteria [33].
In summary, SPL structure and ER localization appear to be highly conserved throughout evolution. In the next section, we will explore mammalian SPL tissue distribution as an indicator of the enzyme’s function in vertebrate organisms.
4. Tissue distribution of SPL
SPL is expressed in several tissues. In mice and rats, SPL expression and activity appear to be highest in the small intestine, colon, thymus and liver and lowest in heart and brain, with the exception of certain portions of the cerebellum and the olfactory mucosal epithelium, a neuronal tissue in which the enzyme is highly enriched[54]. SPL expression appears to be highest in tissues marked by rapid cell turnover, consistent with its ability to promote apoptosis (see below). SPL expression is high in the olfactory mucosa, a unique neuronal tissue that is subject to high rates of apoptosis due to inhaled toxicant-induced cell damage and which is unique among adult neuronal tissues in its ability to sustain continuous neurogenesis [55]. The high expression of SPL in intestinal epithelial cells may underlie its role in catabolizing dietary sphingolipids [56]. However, SPL activity may also be required to maintain low S1P levels in the cells at the villus tips, facilitating cell death and turnover in response to oxidant exposure, other cellular stresses and as a mechanism of gut immunity. [57, 58]. (Figure 5). The high expression of SPL in thymus is likely to be necessary for maintaining low tissue S1P levels compared to the surrounding plasma. This S1P concentration gradient between thymus and the plasma enables T cells to exit from thymus into the circulation (see below). However, a role for SPL in regulating lymphocyte development or epithelial functions cannot be ruled out. S1P signaling is essential for vascular maturation during embryogenesis and continues to influence endothelial cell biology in the adult organism. A variety of cells secrete S1P into the blood plasma including platelets, erythrocytes, neutrophils, mast cells, mononuclear cells and endothelial cells as reviewed in [47, 59]. SPL activity in macrophages monocytes and neutrophils, has not been characterized to date [60]. However, SPL activity is absent in platelets and erythrocytes, suggesting that lack of SPL activity or its downregulation may be important for S1P secretion[61]. In fact, SPL expression was found to be downregulated in mouse vascular endothelial cells in response to laminar shear stress, thereby increasing the release of S1P into the plasma[59]. Therefore, SPL may play a dynamic role in the regulation of local S1P levels at the blood/vascular interface.
Figure 5.
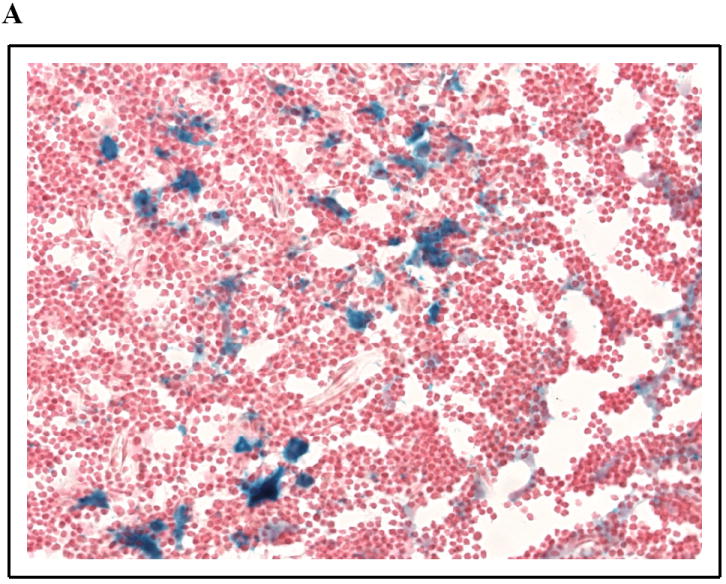
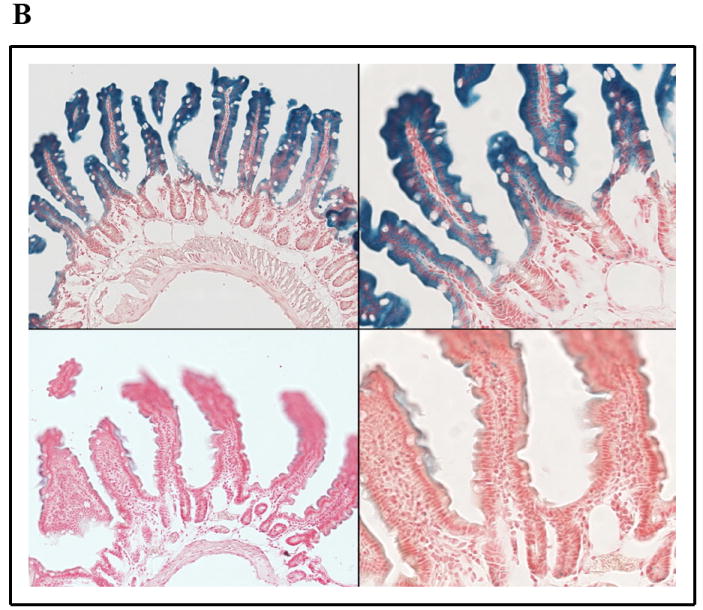
SPL expression in murine thymic epithelium and intestinal mucosa. β-galactosidase staining was performed as described on glutaraldehyde-fixed, frozen sections of SPL reporter mouse tissues [108]. SPL expression can be appreciated in the cytoplasm of the epithelial cells but not lymphocytes of the thymus (a) and in the differentiated enterocytes in the villus tips of the jejunum (b).
In summary, SPL’s tissue distribution likely reflects its role in digestion of dietary sphingolipids, regulation of cell turnover, and circulating S1P levels.
5. SPL regulation
Studies in Drosophila melanogaster and Caenorhabiditis elegans revealed that SPL expression is largely restricted to the developing and adult gut [48, 49, 62]. Moreover, during murine development, SPL mRNA expression is temporarily regulated and is involved in early endodermal differentiation [53, 63]. These observations led to the identification of a conserved mechanism of transcriptional regulation of SPL by GATA transcription factors operational in nematodes and human cells [64]. Additionally, SPL was identified as an immediate early gene and transcriptional target of platelet-derived growth factor (PDGF)[65]. Generation of an SPL knockout mouse revealed phenotypes consistent with PDGF signaling abnormalities, including defects of cell migration, vascular developmental abnormalities and skeletal defects. Analysis of the 5’ flanking region of the SPL gene suggests that other mechanisms of transcriptional control may contribute to the regulation of SPL expression. An investigation of the promoter region of murine Sgpl1 identified cis-elements such as early growth response factor, zinc-binding protein factor and GC-box factor motifs [66]. Sgpl1 was also found along with other pro-apoptotic genes to be a downstream target of mSin3A, a member of a co-repressor complex involved in embryonic development and in regulating cellular functions such as cell cycle progression, proliferation, DNA repair, cell survival and p53 activation [67]. In this regard, it was found that DNA damage induced by etoposide treatment increased the expression of a human SPL reporter construct [60].
In addition to transcriptional regulation, post-translational regulation of human SPL has also been suggested by the finding of SPL in a screen for nitrosylated proteins [68]. SPL is predicted to be nitrosylated on tyrosine residues Y356 and Y366 [68]. By altering the charge of the tyrosine residues, nitrosylation may interfere with enzyme-substrate binding. Multiple protein kinase specific phosphorylation sites are also predicted by protein sequence analysis, but these have not been verified experimentally [69].
Thus, SPL activity is likely to be controlled at various levels, including transcriptional, epigenetic and post-translational mechanisms of regulation.
6. SPL in development
Observations from experiments performed on simple organisms have revealed essential roles for SPL in development. In the slime mold Dictyostelium discoideum, mutations of the SPL gene sglA affects multiple developmental stages in the unique life cycle of this organism [50]. The observed phenotypes are attributed to defects in actin cytoskeletal organization, resulting in impaired pseudopodia formation, cell migration and chemotaxis. In the protozoan parasite Leishmania major, SPL mutants were recently found to be defective in stationary phase differentiation and virulence, raising the possibility that SPL may serve as a useful target for antibiotic therapy for leishmanial diseases [51]. In the fruit fly Drosophila melanogaster, null mutants of the SPL homolog Sply were flightless, and further analysis revealed a severe myopathy affecting the thoracic flight muscles (Figure 6). In addition, Sply null mutants reproduced poorly and demonstrated supernumerary spermathecae, degenerative ovaries and severely reduced testes. A reproductive defect was also found in the nematode Caenorhabditis elegans, following knockdown of SPL expression using RNA interference. The reproductive defects found included withering of the reproductive tract, impaired egg laying and asynchronous egg development [49].
Figure 6.
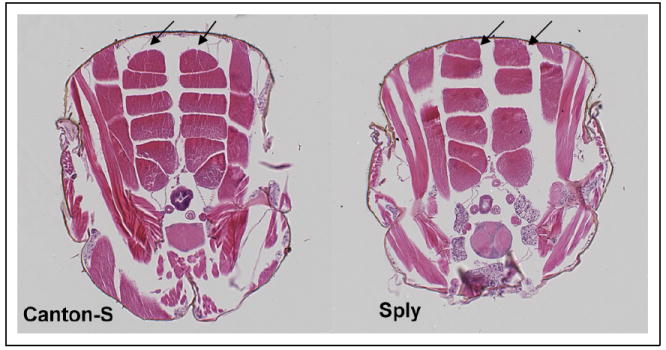
Dorsal longitudinal flight muscle (DLM) abnormalities in the Drosophila SPL null mutant Sply. Whereas the hemithoraces of the Canton-S wild-type fly invariably contains six DLM fibers of equal proportion, Sply homozygotes exhibit a general pattern of missing fibers, asymmetric development and compensatory hypertrophy of remaining fibers. Shown here is a Sply homozygote with only four DLM fibers in the right hemithorax. The upper DLM fiber in each row is marked with an arrow.
As in simple metazoans, SPL function appears to be critical for mammalian development. Sgpl1 expression was observed throughout development in mouse embryos [53, 70]. Homozygous Sgpl1 knockout mice do not survive beyond 3-4 weeks after birth, and they demonstrate significant growth failure and anemia. Several congenital defects were reported, including vascular abnormalities, skeletal defects, thoracic malformations of sternum, ribs and vertebrae, and renal abnormalities. Embryonic fibroblasts from Sgpl1 knockout mice demonstrated defects of migration in vitro. These vascular and cell migration defects are reminiscent of pathological changes observed in PDGF receptor and S1P1 receptor knockout mice [71]. Together with the identification of Sgpl1 as a downstream target of PDGF signaling, these observations suggest that SPL may play a role in the regulation of mammalian angiogenesis and other developmental processes.
The etiology of SPL knockout model phenotypes appears to vary in individual species. A marked increase in the level of sphingolipid metabolites including LCBPs, LCBs and ceramides were observed in Sply mutant flies [48]. Introduction of a second mutation inhibiting sphingolipid synthesis normalized lipid levels and attenuated many of the mutant phenotypes, strongly suggesting that tight control of sphingolipid intermediates is required for normal growth and development. In contrast to Drosophila melanogaster, wherein sphingolipid intermediates were found to be responsible for the mutant phenotypes, in Leishmania major a lack of phosphoethanolamine production through the SPL reaction seem to be accountable for the defects of mutants as ethanolamine supplementation completely reversed the viability and differentiation defects resulting in stationary phase survival[51]. SPL may also mediate biological effects through the generation of its products in Drosophila melanogaster, where phosphoethanolamine formed in the SPL reaction was shown to be important for the formation of phosphatidylethanolamine required for the processing of sterol regulatory element binding protein (SREBP), a key regulator of lipid biosynthetic gene transcription [72]. Although it is not clear how SPL products affect mammalian cells, it is interesting to note that in one study performed in F9 murine embryonal carcinoma cells, SPL was found to promote proliferation through an S1P-independent mechanism likely to involve its products [73]. In fact, ethanolamine is associated with increased phosphatidylethanolamine synthesis and proliferation in hepatocytes, is mitogenic in insulin-treated fibroblasts and is increased in cancer cells and in response to carcinogens [74, 75]. Phosphoethanolamine can be converted to CDP-ethanolamine by ethanolamine phosphate cytidydyltransferase, a step necessary for subsequent incorporation into the important membrane glycerophospholipid phosphatidylethanolamine [76]. In addition, phosphoethanolamine methyltransferase can transfer a methyl group from S-adenosyl methionine to phosphoethanolamine, yielding N-methylethanolamine phosphate. Deletion of the latter enzyme in plants has been shown to induce sterility and salt sensitivity [77]. Alternatively, phosphoethanolamine can be metabolized to acetaldehyde, NH3, phosphate and water by another pyridoxal 5’-phosphate-dependent enzyme, ethanolamine-phosphate phospho-lyase. The fate and importance of the aldehyde formed in the SPL reaction is not well understood. The product is unstable and converts to the corresponding alcohol and fatty acid, which could then re-enter the lipid pool. Due to the reactivity of the aldehyde, it is also feasible that it may react with other cellular components, including DNA and proteins.
Thus, SPL expression seems pivotal for development, as its activity is needed for proper regulation of bioactive sphingolipid metabolites. Moreover, at least in simple organisms, SPL activity provides cells with phosphoethanolamine that is needed for phosphatidylethanolamine biosynthesis and regulation of genes involved in lipid metabolism.
7. SPL in cell growth and apoptosis
Sphingolipid metabolites such as LCBPs and ceramide are bioactive molecules with opposing effects on cell fate. Cells appear to maintain a dynamic balance between these molecules [21, 23]. SPL has the ability to influence cell fate by removing pro-proliferative LCBPs from the sphingolipid pool and thereby shifting the balance. The potential role of SPL in regulating cell fate and stress responses has been investigated in several systems. For example, Saccharomyces cerevisiae dpl1 null mutants are resistant to heat stress and nutrient deprivation[25, 78]. An insertional mutagenesis screen in Dictyostelium discoideum identified mutations in the SPL gene as the etiology of enhanced resistance to the anticancer drug cisplatin [79]. Conversely, overexpression of SPL at different levels enhanced the sensitivity of Dictyostelium discoideum cells to the drug [80]. An enhanced sensitivity to chemotherapy drugs including cisplatin was also found in mammalian cells overexpressing SPL. In these cells increased SPL activity resulted in a decrease in the ratio of S1P to ceramide and thereby a shift in the balance between the opposing MAPKs ERK and p38, favoring p38 activation [81]. Cisplatin was recently shown to cause a translocation of dihydroceramide synthase 1 from ER to Golgi, a process that may contribute to mediating cell death by cisplatin; sphingosine kinase 1 blocks both translocation and Cisplatin-induced cell death [82]. SPL may also influence this process, although this possibility was not directly tested. SPL was also found to sensitize mammalian cells to serum deprivation, DNA damage and other stressful stimuli by the ability to promote apoptosis through the action of p53 and p38 tumor suppressor signaling pathways [52, 60, 81]. A lack or reduction in SPL activity can also lead to cell death as evidenced by increased apoptosis found in the reproductive organs of the Drosophila melanogaster Sply null mutant fly[83]. The sphingolipid metabolites responsible for this effect are unknown, although these tissues show a profound accumulation of LCBs including Δ4,6-sphingadienes, which were found to promote apoptosis in Drosophila melanogaster cell lines[84]. A similar accumulation of LCBs was also found in the Drosophila melanogaster cell line Schneider-2 (S2) following RNA interference against Sply[84].
Thus, SPL can affect cell growth decisions by modulating the level of LCBPs and thereby shifting the balance of pro-survival and cytotoxic sphingolipids.
8. SPL in disease
The importance of SPL in regulating S1P levels, levels of other sphingolipid intermediates, and cell fate is likely to contribute to tissue homeostasis and, alternatively, its dysregulation could contribute to the pathophysiology of disease. S1P signaling plays an important role in the immune system by affecting cell survival, migration and cytokine secretion. The immunomodulatory drug FTY720 is rapidly phosphorylated by sphingosine kinases [44, 85, 86]. The phosphorylated compound acts as an agonist for S1P receptors on thymocytes and lymphocytes although chronic treatment with FTY720 leads to “functional antagonism” presumably due to the disappearance of the receptor from the cell surface [87-89]. By reducing cell surface bound S1P receptors it is believed that FTY720 treatment prevents the exit of these cells from thymus and secondary lymphoid organs. However since FTY720 has both agonist and antagonist properties, the means by which it inhibits cell egress from these sites remains controversial. FTY720 has also been shown to inhibit SPL activity, which raises the possibility that SPL inhibition may contribute to the immunological effects of the drug [45]. Inhibition of SPL by THI also prevents T cell egress from thymus and secondary lymphoid organs [46]. This effect was explained by a reduction in the S1P gradient between thymus and the circulation. Like the S1P receptor analogs, SPL inhibitors may, thus, define yet another class of immunosuppressant drugs that may be useful in the treatment of autoimmune disease and prevention of allograft rejection.
There is evidence that S1P plays a role in inflammation. Sphingosine kinase 1 was shown to be activated downstream of the inflammatory mediator TNFα in monocytes and to contribute to subsequent intracellular signaling, degranulation, cytokine production, and activation of NFκB in these cells [90]. Both S1P and sphingosine kinase 1 also were shown to be necessary for the ability of TNFα to induce expression of COX-2 and PGE2 production [91]. Cells present in inflammatory infiltrates express SPL [60], which raises the possibility that SPL may play a role in the inflammatory process. Interestingly, the expression of human Sgpl1 was found to be increased in skin of patients suffering from atopic dermatitis, a chronic relapsing inflammatory skin disease often associated with autoimmune diseases [92].
The role of sphingolipids in regulating cell fate and stress response raises the possibility that genes of sphingolipid metabolism may be altered in cancer and/or targets for cancer therapeutics. This notion is consistent with the recent finding that administration of a monoclonal S1P antibody attenuated human xenograft progression and angiogenesis in vivo [93], as well as the observed effects of FTY720 on inhibition of tumor angiogenesis and S1P receptor downregulation [94, 95]. Numerous studies have reported sphingosine kinase 1 to be upregulated in human cancers and in some cases enzyme activity and elevated LCBP levels have been demonstrated [96-102]. Conversely, SPL expression and activity were found to be downregulated during intestinal tumorigenesis in the ApcMin/+ mouse model and in human colon cancer specimens compared to adjacent uninvolved tissues (Figure 7) [60]. These results suggest that SPL may function in tumor suppressor/cancer surveillance pathways, and that loss of SPL expression or activity could potentially contribute to tumorigenesis. This is consistent with the finding that Sgpl1 is among a set of genes significantly downregulated in metastatic tumor tissues compared to primary tumors from the same patients [103]. However, it should be noted that SPL upregulation has also been identified as a feature of some malignant tissues. For example, in ovarian cancer, where lysophospholipids including S1P and LPA are elevated in malignant ascites and tissues and stimulate tumor cell proliferation, SPL was found to be upregulated [104-106]. If SPL downregulation in intestinal adenomas is a reversible phenomenon, it might be exploited as an adjuvant approach to enhance the therapeutic response of tumor cells to DNA damaging agents. Conversely, SPL may also be a target for pharmacological intervention, to transiently raise S1P levels as a means of affording protection of reproductive function in patients receiving cytotoxic therapy and radiation [107].
Figure 7.
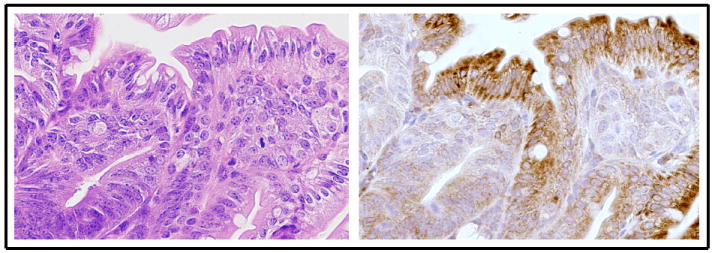
SPL downregulation in intestinal tumorigenesis. Immunohistochemistry with a murine SPL-specific antibody used as described previously shows robust SPL expression in normal differentiated epithelial cells of the villus of the small intestine (brown staining) in the ApcMin/+ mouse model of colon cancer, whereas undifferentiated adenomatous areas in the submucosa do not express SPL [60]. (Left) hematoxylin & eosin; (Right) immunohistochemistry.
9. Summary and future perspectives
It is now well established that LCBPs such as S1P are potent signaling molecules involved in controlling cell fate. Tight regulation of these molecules is essential for proper cell functioning, and SPL may be an important gatekeeper in this process. Evidence suggests that SPL is dynamically regulated through transcriptional and post-translational mechanisms, enabling it to respond to changes in lipid metabolism, nutrient availability and stressful conditions. In simple organisms, reduced or absent SPL activity caused by gene mutations or RNA interference results in severe developmental consequences. Loss of SPL expression in mammals is equally deleterious, leading to early death. Changes in SPL expression and activity are associated with intestinal tumor progression, and inhibition of SPL activity has profound effects on the immune system and lymphocyte trafficking. Whether the role of SPL in these processes can be exploited for medical benefit remains to be determined. The development of conditional SPL knockout mouse models and the identification of small molecule inhibitors of SPL should help to establish the role of SPL in physiology and disease.
Although the first SPL was cloned more than 10 years ago, our knowledge of enzyme structure and regulation remains somewhat limited. Mechanisms responsible for SPL transcriptional regulation are beginning to emerge. Nitrosylation and other modifications such as phosphorylation and glycosylation have not been confirmed experimentally but are likely, based on analysis of the polypeptide. The topology of the enzyme has been analyzed, and its orientation in the membrane and active site location have been determined, but other functions and aspects of polypeptide organization may yet be uncovered. A more in depth analysis of SPL structure and regulation is needed in order to better understand this important enzyme. Such work will aid in the discovery and design of new SPL inhibitors as well as new ways to reactivate the enzyme, potentially for therapeutic purposes.
Acknowledgments
Dr. Saba is supported by National Institutes of Health grants 2R01CA77528, 1R01GM66954 and CA129438. We thank Alexander D. Borowsky and Lisa Dillard-Telm for histology and photography of murine intestinal and thymic tissues. We thank Greg L. Harris and Deron R. Herr for histology and photography of Drosophila flight muscles. We apologize to the many colleagues whose work could not be mentioned directly in this review due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannun YA, Luberto C, Argraves KM. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry. 2001;40:4893–4903. doi: 10.1021/bi002836k. [DOI] [PubMed] [Google Scholar]
- 2.Gómez-Muñoz A, Kong J, Parhar K, Wang S, Gangoiti P, González M, Eivemark S, Salh B, Duronio V, Steinbrecher U. Ceramide-1-phosphate promotes cell survival through activation of the phosphatidylinositol 3-kinase/protein kinase B pathway. FEBS Lett. 2005;579:3744–3750. doi: 10.1016/j.febslet.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 3.Wijesinghe D, Lamour N, Gomez-Munoz A, Chalfant C. Ceramide kinase and ceramide-1-phosphate. Methods Enzymol. 2007;434:265–292. doi: 10.1016/S0076-6879(07)34015-9. [DOI] [PubMed] [Google Scholar]
- 4.Gardell S, Dubin A, Chun J. Emerging medicinal roles for lysophospholipid signaling. Trends Mol Med. 2006;12:65–75. doi: 10.1016/j.molmed.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Karliner J. Mechanisms of cardioprotection by lysophospholipids. J Cell Biochem. 2004;92:1095–1103. doi: 10.1002/jcb.20129. [DOI] [PubMed] [Google Scholar]
- 6.Milstien S, Spiegel S. Targeting sphingosine-1-phosphate: a novel avenue for cancer therapeutics. Cancer Cell. 2006:148–150. doi: 10.1016/j.ccr.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 7.McVerry B, Garcia J. In vitro and in vivo modulation of vascular barrier integrity by sphingosine 1-phosphate: mechanistic insights. Cell Signal. 2005;17:131–139. doi: 10.1016/j.cellsig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Oskouian B, Saba JD. Death and taxis: what non-mammalian models tell us about sphingosine-1-phosphate. Semin Cell Dev Biol. 2004;15:529–540. doi: 10.1016/j.semcdb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Oskouian B, Saba J. Sphingosine-1-phosphate metabolism and intestinal tumorigenesis: lipid signaling strikes again. Cell Cycle. 2007;6:522–527. doi: 10.4161/cc.6.5.3903. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez T, Hla T. Structural and functional characteristics of S1P receptors. J Cell Biochem. 2004;92:913–922. doi: 10.1002/jcb.20127. [DOI] [PubMed] [Google Scholar]
- 11.Rosen H. Chemical approaches to the lysophospholipid receptors. Prostaglandins Other Lipid Mediat. 2005;77:179–184. doi: 10.1016/j.prostaglandins.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Pyne S, Pyne NJ. Sphingosine 1-phosphate signalling in mammalian cells. Biochem J. 2000;349:385–402. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyne S, Pyne N. Sphingosine 1-phosphate signalling via the endothelial differentiation gene family of G-protein-coupled receptors. Pharmacol Ther. 2000;88:115–131. doi: 10.1016/s0163-7258(00)00084-x. [DOI] [PubMed] [Google Scholar]
- 14.Spiegel S, Milstien S. Sphingosine-1-phosphate: signaling inside and out. FEBS Lett. 2000;476:55–57. doi: 10.1016/s0014-5793(00)01670-7. [DOI] [PubMed] [Google Scholar]
- 15.Hla T. Sphingosine 1-phosphate receptors. Prostaglandins Other Lipid Mediat. 2001;64:135–142. doi: 10.1016/s0090-6980(01)00109-5. [DOI] [PubMed] [Google Scholar]
- 16.Spiegel S, Milstien S. Functions of a new family of sphingosine-1-phosphate receptors. Biochim Biophys Acta. 2000;1484:107–116. doi: 10.1016/s1388-1981(00)00010-x. [DOI] [PubMed] [Google Scholar]
- 17.Hla T. Signaling and biological actions of sphingosine-1-phosphate. Pharmacol Res. 2003;47:401–407. doi: 10.1016/s1043-6618(03)00046-x. [DOI] [PubMed] [Google Scholar]
- 18.Kono M, Mi Y, Liu Y, Sasaki T, Allende ML, Wu YP, Yamashita T, Proia RL. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J Biol Chem. 2004;279:29367–29373. doi: 10.1074/jbc.M403937200. [DOI] [PubMed] [Google Scholar]
- 19.Cyster J. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 20.Spiegel S, Cuvillier O, Edsall L, Kohama T, Menzeleev R, Olah Z, Olivera A, Pirianov G, Thomas D, Tu Z, Brocklyn JV, Wang F. Sphingosine-1-phosphate in cell growth and cell death. Ann NY Acad Sci. 1998;845:11–18. doi: 10.1111/j.1749-6632.1998.tb09658.x. [DOI] [PubMed] [Google Scholar]
- 21.Cuvillier O, Pirianov G, Kleuser B, Vanek P, Coso OA, et al. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 22.Mandala SM, Thornton R, Galve-Roperh I, Poulton S, Peterson C, Olivera A, Bergstrom J, Kurtz MB, Spiegel S. Molecular cloning and characterization of a lipid phosphohydrolase that degrades sphingosine-1-phosphate and induces cell death. Proc Natl Acad Sci U S A. 2000;97:7859–7864. doi: 10.1073/pnas.120146897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci U S A. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birchwood CJ, Saba JD, Dickson RC, Cunningham KW. Calcium influx and signaling in yeast stimulated by intracellular sphingosine -1-phosphate accumulation. J Biol Chem. 2001;276:11712–11718. doi: 10.1074/jbc.M010221200. [DOI] [PubMed] [Google Scholar]
- 25.Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohama T, Olivera A, Edsall L, Nagiec M, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Sugiura M, Nava VE, Edsall LC, Kono K, Poulton S, Milstien S, Kohama T, Spiegel S. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem. 2000;275:19513–19520. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 28.Mao C, Wadleigh M, Jenkins GM, Hannun YA, Obeid LM. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 29.Qie L, Nagiec MM, Baltisberger JA, Lester RL, Dickson RC. Identification of a Saccharomyces gene, LCB3, necessary for incorporation of exogenous long chain bases into sphingolipids. J Biol Chem. 1997;272:16110–16117. doi: 10.1074/jbc.272.26.16110. [DOI] [PubMed] [Google Scholar]
- 30.Le Stunff H, Peterson C, Liu H, Milstien S, S S. Sphingosine-1-phosphate and lipid phosphohydrolases. Biochim Biophys Acta. 2002;1582:8–17. doi: 10.1016/s1388-1981(02)00132-4. [DOI] [PubMed] [Google Scholar]
- 31.Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Saba J. Identification of the first mammalian sphingosine phosphate lyase gene and its functional expression in yeast. Biochem Biophys Res Commun. 1998;242:502–507. doi: 10.1006/bbrc.1997.7993. [DOI] [PubMed] [Google Scholar]
- 33.Van Veldhoven PP, Gijsbers S, Mannaerts GP, Vermeesch JR, Brys V. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22. Biochimica et Biophysica Acta. 2000;1487:128–134. doi: 10.1016/s1388-1981(00)00079-2. [DOI] [PubMed] [Google Scholar]
- 34.Stoffel W, Bister K. Stereospecificities in the metabolic reactions of the four isomeric sphinganines (dihydrosphingosines) in rat liver. Hoppe-Seyler’s Z Physiol Chem. 1973;354:169–181. doi: 10.1515/bchm2.1973.354.1.169. [DOI] [PubMed] [Google Scholar]
- 35.Stoffel W, LeKim D, Sticht G. Distribution and properties of dihydrosphingosine-1-phosphate aldolase (sphinganine-1-phosphate alkanal-lyase) Hoppe Seylers Z Physiol Chem. 1969;350:1233–1241. doi: 10.1515/bchm2.1969.350.2.1233. [DOI] [PubMed] [Google Scholar]
- 36.van Echten-Deckert G, Zschoche A, Bar T, Schmidt R, Raths A, Heinemann T, Sandhoff K. cis-4-methylsphingosine decreases sphingolipid biosynthesis by specifically interfering with serine palmitoyltransferase activity in primary cultured neurons. J Biol Chem. 1997;272:15825–15833. doi: 10.1074/jbc.272.25.15825. [DOI] [PubMed] [Google Scholar]
- 37.Van Veldhoven PP. Sphingosine-1-phosphate lyase. In: Merrill AH Jr, Hannun YA, editors. Sphingolipid Metabolism and Cell Signaling Part A. Vol. 311. Academic Press; New York: 2000. pp. 244–254. [Google Scholar]
- 38.Van Veldhoven PP, Mannaerts GP. Subcellular localization and membrane topology of sphingosine-1-phosphate lyase in rat liver. J Biol Chem. 1991;266:12502–12507. [PubMed] [Google Scholar]
- 39.Bandhuvula P, Fyrst H, Saba J. A rapid fluorescent assay for sphingosine-1-phosphate lyase enzyme activity. J Lipid Res. 2007;48:2769–2778. doi: 10.1194/jlr.D700010-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Stoffel W, Grol M. Chemistry and biochemistry of 1-desoxysphinganine 1-phosphonate (dihydrosphingosine-1-phosphonate) Chem Phys Lipids. 1974;13:372–388. doi: 10.1016/0009-3084(74)90011-5. [DOI] [PubMed] [Google Scholar]
- 41.Boumendjel A, Miller S. Synthesis of sphingosine-1-phosphate and dihydrosphingosine-1-phosphate. J Lipid Res. 1994;35:2305–2311. [PubMed] [Google Scholar]
- 42.Stoffel W, Bauer E, Stahl J. The metabolism of sphingosine bases in Tetrahymena pyriformis. Sphingosine kinase and sphingosine-1-phosphate lyase. Hoppe Seylers Z Physiol Chem. 1974;355:61–74. doi: 10.1515/bchm2.1974.355.1.61. [DOI] [PubMed] [Google Scholar]
- 43.Triola G, Fabrias G, Dragusin M, Niederhausen L, Broere R, Llebaria A, van Echten-Deckert G. Specificity of the dihydroceramide desaturase inhibitor N-[(1R,2S)-2-hydroxy-1-hydroxymethyl-2-(2-tridecyl-1-cyclopropenyl)ethyl]o ctanamide (GT11) in primary cultured cerebellar neurons. Mol Pharmacol. 2004;66:1671–1678. doi: 10.1124/mol.104.003681. [DOI] [PubMed] [Google Scholar]
- 44.Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, Bruns C, Prieschl E, Baumruker T, Hiestand P, Foster CA, Zollinger M, Lynch KR. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J Biol Chem. 2002;277:21453–21457. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 45.Bandhuvula P, Tam Y, Oskouian B, Saba J. The immune modulator FTY720 inhibits sphingosine-1-phosphate lyase activity. J Biol Chem. 2005;280:33697–33700. doi: 10.1074/jbc.C500294200. [DOI] [PubMed] [Google Scholar]
- 46.Schwab S, Pereira J, Matloubian M, Xu Y, Huang Y, Cyster J. Lymphocyte sequestration through S1P lyase inhibition an disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 47.Venkataraman K, Lee Y, Michaud J, Thangada S, Ai Y, Borikovsky H, Parikh N, Habrukowich C, Hla T. Vascular endothelium as a contributor of plasma sphingosine-1-phosphate. Circ Res. 2008 doi: 10.1161/CIRCRESAHA.107.165845. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herr DR, Fyrst H, Phan V, Heinecke K, Georges R, Harris GL, Saba JD. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development. 2003;130:2443–2453. doi: 10.1242/dev.00456. [DOI] [PubMed] [Google Scholar]
- 49.Mendel J, Heinecke K, Fyrst H, Saba JD. Sphingosine phosphate lyase expression is essential for normal development in Caenorhabditis elegans. J Biol Chem. 2003;278:22341–22349. doi: 10.1074/jbc.M302857200. [DOI] [PubMed] [Google Scholar]
- 50.Li G, Foote C, Alexander S, Alexander H. Sphingosine-1-phosphate lyase has a central role in the development of Dictyostelium discoideum. Development. 2001;128:3473–3483. doi: 10.1242/dev.128.18.3473. [DOI] [PubMed] [Google Scholar]
- 51.Zhang K, Pompey J, Hsu F, Turk J, Bandhuvula P, Saba J, Beverley S. Redirection of sphingolipid metabolism towards de novo synthesis of ethanolamine in Leishmania. EMBO J. 2007;26 doi: 10.1038/sj.emboj.7601565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiss U, Oskouian B, Zhou J, Gupta V, Sooriyakumaran P, Kelly S, Wang E, Merrill AH, Jr, Saba JD. Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J Biol Chem. 2004;279:1281–1290. doi: 10.1074/jbc.M309646200. [DOI] [PubMed] [Google Scholar]
- 53.Ikeda M, Kihara A, Igarashi Y. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5’-phosphate binding domain exposed to the cytosol. Biochem Biophys Res Commun. 2004;325:338–343. doi: 10.1016/j.bbrc.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 54.Genter MB, Van Veldhoven PP, Jegga AG, Sakthivel B, Kong S, Stanley K, Witte DP, Ebert CL, Aronow BJ. Microarray-based discovery of highly expressed olfactory mucosal genes: potential roles in the various functions of the olfactory system. Physiol Genomics. 2003;16:67–81. doi: 10.1152/physiolgenomics.00117.2003. [DOI] [PubMed] [Google Scholar]
- 55.Genter MB. Molecular biology of the nasal airways: how do we assess cellular and molecular responses in the nose? Toxicol Pathol. 2006;34:274–280. doi: 10.1080/01926230600713491. [DOI] [PubMed] [Google Scholar]
- 56.Vesper H, Schmelz E, Nikolova-Karakashian M, Dillehay D, Lynch D, M A., Jr Sphingolipids in food and the emerging importance of sphingolipids to nutrition. J Nutr. 1999;129:1239–1250. doi: 10.1093/jn/129.7.1239. [DOI] [PubMed] [Google Scholar]
- 57.Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- 58.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 59.Sanchez T, Skoura A, Wu M, Casserly B, Harrington E, Hla T. Induction of vascular permeability by the sphingosine-1-phosphate receptor-2 (S1P2R) and its downstream effectors ROCK and PTEN. Arterioscler Thromb Vasc Biol. 2007;27:1312–1318. doi: 10.1161/ATVBAHA.107.143735. [DOI] [PubMed] [Google Scholar]
- 60.Oskouian B, Sooriyakumaran P, Borowsky A, Crans A, DIllard-Telm L, Tam Y, Bandhuvula P, Saba J. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is downregulated in colon cancer. Proc Natl Acad Sci USA. 2006;103:17384–17389. doi: 10.1073/pnas.0600050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito K, Anada Y, Tani M, Ikeda M, Sano T, Kihara A, Igarashi Y. Lack of sphingosine 1-phosphate-degrading enzymes in erythrocytes. Biochem Biophys Res Commun. 2007;357:212–217. doi: 10.1016/j.bbrc.2007.03.123. [DOI] [PubMed] [Google Scholar]
- 62.Renault AD, Starz-Gaiano M, Lehmann R. Metabolism of sphingosine 1-phosphate and lysophosphatidic acid: a genome wide analysis of gene expression in Drosophila. Mech Dev. 2002;119(Suppl 1):S293–301. doi: 10.1016/s0925-4773(03)00131-x. [DOI] [PubMed] [Google Scholar]
- 63.Kihara A, Ikeda M, Kariya Y, Lee E, Lee Y, Igarashi Y. Sphingosine-1-phosphate lyase is involved in the differentiation of F9 embryonal carcinoma cells to primitive endoderm. J Biol Chem. 2003;278:14578–14585. doi: 10.1074/jbc.M211416200. [DOI] [PubMed] [Google Scholar]
- 64.Oskouian B, Mendel J, Shocron E, Lee MA, Jr, Fyrst H, Saba JD. Regulation of sphingosine-1-phosphate lyase gene expression by members of the GATA family of transcription factors. J Biol Chem. 2005;280:18403–18410. doi: 10.1074/jbc.M410928200. [DOI] [PubMed] [Google Scholar]
- 65.Alvarez R, Kantarjian H, Cortes J. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin Proc. 2006;81:1241–1257. doi: 10.4065/81.9.1241. [DOI] [PubMed] [Google Scholar]
- 66.Hutton J, Jegga A, Kong S, Gupta A, Ebert C, Williams S, Katz J, Aronow B. Microarray and comparative genomic-based identification of genes and gene regulatory regions of the mouse immune system. BMC Genomics. 2004;5:82. doi: 10.1186/1471-2164-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal Biochem. 2006;354:279–289. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 69.Huang H, Lee T, Tzeng S, Horng J. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acid Res. 2005;33:W226–229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmahl J, Raymond C, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat Genet. 2006 doi: 10.1038/ng1922. Ahead of print. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Wada R, Yamashita T, Mi Y, Deng CX, Hobson JP, Rosenfeldt HM, Nava VE, Chae SS, Lee MJ, Liu CH, Hla T, Spiegel S, Proia RL. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J Clin Invest. 2000;106:951–961. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobrosotskaya I, Seegmiller A, Brown M, Goldstein J, Rawson R. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 73.Kariya Y, Kihara A, Ikeda M, Kikuchi F, Nakamura S, Hashimoto S, Choi CH, Lee YM, Igarashi Y. Products by the sphingosine kinase/sphingosine 1-phosphate (S1P) lyase pathway but not S1P stimulate mitogenesis. Genes Cells. 2005;10:605–615. doi: 10.1111/j.1365-2443.2005.00862.x. [DOI] [PubMed] [Google Scholar]
- 74.Kume H, Sasaki H. Ethanolamine modulates DNA synthesis through epidermal growth factor receptor in rat primary hepatocytes. In Vitro Cell Dev Biol Anim. 2006;42:20–26. doi: 10.1007/s11626-006-0007-9. [DOI] [PubMed] [Google Scholar]
- 75.Crilly K, Benyhe S, Kiss Z. Promitogenic effects of ethanol, methanol, and ethanolamine in insulin-treated fibroblasts. Biochem Pharmacol. 2000;60:1391–1398. doi: 10.1016/s0006-2952(00)00456-1. [DOI] [PubMed] [Google Scholar]
- 76.Kennedy E, Weiss S. The function of cytidine coenzymes in the biosynthesis of phospholipids. J Biol Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 77.Mou Z, Wang X, Fu Z, Dai Y, Han C, Ouyang J, Bao F, Hu Y, Li J. Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell. 2002;14:2031–2043. doi: 10.1105/tpc.001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gottlieb D, Heideman W, Zhou J, Oskouian B, Saba J. The DPL1 gene is involved in mediating the response to nutrient deprivation in Saccharomyces cerevisiae. Mol Cell Biol Res Comm. 1999;1:66–71. doi: 10.1006/mcbr.1999.0109. [DOI] [PubMed] [Google Scholar]
- 79.Li G, Alexander H, Schneider N, Alexander S. Molecular basis for resistance to the anticancer drug cisplatin in Dictyostelium. Microbiology. 2000;146(Pt 9):2219–2227. doi: 10.1099/00221287-146-9-2219. [DOI] [PubMed] [Google Scholar]
- 80.Min J, Stegner A, Alexander H, Alexander S. Overexpression of sphingosine-1-phosphate lyase or inhibition of sphingosine kinase in Dictyostelium discoideum results in a selective increase in sensitivity to platinum-based chemotherapy drugs. Eukaryotic Cell. 2004;3 doi: 10.1128/EC.3.3.795-805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Min J, Van Veldhoven PP, Zhang L, Hanigan MH, Alexander H, Alexander S. Sphingosine-1-phosphate lyase regulates sensitivity of human cells to select chemotherapy drugs in a p38-dependent manner. Mol Cancer Res. 2005;3:287–296. doi: 10.1158/1541-7786.MCR-04-0197. [DOI] [PubMed] [Google Scholar]
- 82.Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, Alexander S. (Dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Mol Cancer Res. 2007;5:801–812. doi: 10.1158/1541-7786.MCR-07-0100. [DOI] [PubMed] [Google Scholar]
- 83.Phan V, Herr D, Panton D, Fyrst H, Saba J, Harris G. Disruption of sphingolipid metabolism elicits apoptosis-associated reproductive defects in Drosophila. Dev Biol. 2007;309:329–341. doi: 10.1016/j.ydbio.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fyrst H, Zhang X, Herr D, Byun H, Bittman R, Phan V, Harris G, Saba J. Identification and characterization by electrospray mass spectrometry of endogenous Drosophila sphingadienes. J Lipid Res. 2008;49:597–606. doi: 10.1194/jlr.M700414-JLR200. [DOI] [PubMed] [Google Scholar]
- 85.Chiba K, Yanagawa Y, Masubuchi Y, Kataoka H, Kawaguchi T, Ohtsuki M. FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. J Immunol. 1998;160:5037–5044. [PubMed] [Google Scholar]
- 86.Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, Thornton R, Shei GJ, Card D, Keohane C, Rosenbach M, Hale J, Lynch CL, Rupprecht K, Parsons W, Rosen H. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 87.Sanchez T, Estrada-Hernandez T, Paik JH, Wu MT, Venkataraman K, Brinkmann V, Claffey K, Hla T. Phosphorylation and action of the immunomodulator FTY720 inhibits vascular endothelial cell growth factor-induced vascular permeability. J Biol Chem. 2003;278:47281–47290. doi: 10.1074/jbc.M306896200. [DOI] [PubMed] [Google Scholar]
- 88.Graler MH, Goetzl EJ. The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G protein-coupled receptors. Faseb J. 2004;18:551–553. doi: 10.1096/fj.03-0910fje. [DOI] [PubMed] [Google Scholar]
- 89.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 90.Zhi L, Leung B, Melendez A. Sphingosine kinase 1 regulates pro-inflammatory responses triggered by TNFalpha in primary human monocytes. J Cell Physiol. 2006;208:109–115. doi: 10.1002/jcp.20646. [DOI] [PubMed] [Google Scholar]
- 91.Pettus BJ, Bielawski J, Porcelli AM, Reames DL, Johnson KR, Morrow J, Chalfant CE, Obeid LM, Hannun YA. The sphingosine kinase 1/sphingosine-1-phosphate pathway mediates COX-2 induction and PGE2 production in response to TNF-alpha. Faseb J. 2003;17:1411–1421. doi: 10.1096/fj.02-1038com. [DOI] [PubMed] [Google Scholar]
- 92.Seo EY, Park GT, Lee KM, Kim JA, Lee JH, Yang JM. Identification of the target genes of atopic dermatitis by real-time PCR. J Invest Dermatol. 2006;126:1187–1189. doi: 10.1038/sj.jid.5700234. [DOI] [PubMed] [Google Scholar]
- 93.Visentin B, Vekich J, Sibbald B, Cavalli A, Moreno K, Matteo R, Garland W, Lu Y, Yu S, Hall H, Kundra V, Mills G, Sabbadini R. Validation of an anti-sphingosine-1-phosphate antibody as a potential therapeutic in reducing growth, invasion, and angiogenesis in multiple tumor lineages. Cancer Cell. 2006;9:225–238. doi: 10.1016/j.ccr.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 94.LaMontagne K, Littlewood-Evans A, Schnell C, O’Reilly T, Wyder L, Sanchez T, Probst B, Butler J, Wood A, Liau G, Billy E, Theuer A, Hla T, Wood J. Antagonism of sphingosine-1-phosphate receptors by FTY720 inhibits angiogenesis and tumor vascularization. Cancer Res. 2006;66:221–231. doi: 10.1158/0008-5472.CAN-05-2001. [DOI] [PubMed] [Google Scholar]
- 95.Schmid G, Guba M, Ischenko I, Papyan A, Joka M, Schrepfer S, Bruns CJ, Jauch KW, Heeschen C, Graeb C. The immunosuppressant FTY720 inhibits tumor angiogenesis via the sphingosine 1-phosphate receptor 1. J Cell Biochem. 2007;101:259–270. doi: 10.1002/jcb.21181. [DOI] [PubMed] [Google Scholar]
- 96.Xia P, Gamble JR, Wang L, Pitson SM, Moretti PA, Wattenberg BW, D’Andrea RJ, Vadas MA. An oncogenic role of sphingosine kinase. Curr Biol. 2000;10:1527–1530. doi: 10.1016/s0960-9822(00)00834-4. [DOI] [PubMed] [Google Scholar]
- 97.Sukocheva OA, Wang L, Albanese N, Vadas MA, Xia P. Sphingosine Kinase Transmits Estrogen Signaling in Human Breast Cancer Cells. Mol Endocrinol. 2003;24:24. doi: 10.1210/me.2003-0119. [DOI] [PubMed] [Google Scholar]
- 98.Johnson KR, Johnson KY, Crellin HG, Ogretmen B, Boylan AM, Harley RA, Obeid LM. Immunohistochemical distribution of sphingosine kinase 1 in normal and tumor lung tissue. J Histochem Cytochem. 2005;53:1159–1166. doi: 10.1369/jhc.4A6606.2005. [DOI] [PubMed] [Google Scholar]
- 99.Sarkar S, Maceyka M, Hait NC, Paugh SW, Sankala H, Milstien S, Spiegel S. Sphingosine kinase 1 is required for migration, proliferation and survival of MCF-7 human breast cancer cells. FEBS Lett. 2005;579:5313–5317. doi: 10.1016/j.febslet.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 100.Kawamori T, Osta W, Johnson KR, Pettus BJ, Bielawski J, Tanaka T, Wargovich MJ, Reddy BS, Hannun YA, Obeid LM, Zhou D. Sphingosine kinase 1 is up-regulated in colon carcinogenesis. Faseb J. 2006;20:386–388. doi: 10.1096/fj.05-4331fje. [DOI] [PubMed] [Google Scholar]
- 101.Cuvillier O. Sphingosine kinase-1--a potential therapeutic target in cancer. Anticancer Drugs. 2007;18:105–110. doi: 10.1097/CAD.0b013e328011334d. [DOI] [PubMed] [Google Scholar]
- 102.French K, Upson J, Keller S, Zhuang Y, Yun J, Smith C. Antitumor activity of sphingosine kinase inhibitors. J Pharmacol Exp Ther. 2006;318:596–603. doi: 10.1124/jpet.106.101345. [DOI] [PubMed] [Google Scholar]
- 103.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 104.Hibbs K, Skubitz K, Pambuccian S, Casey R, Burleson K, Oegema TJ, Thiele J, Grindle S, Bliss R, Skubitz A. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hong G, Baudhuin LM, Xu Y. Sphingosine-1-phosphate modulates growth and adhesion of ovarian cancer cells. FEBS Lett. 1999;460:513–518. doi: 10.1016/s0014-5793(99)01400-3. [DOI] [PubMed] [Google Scholar]
- 106.Xu Y, Xiao Y, Baudhuin L, Schwartz B. The role and clinical applications of bioactive lysolipids in ovarian cancer. J Soc Gynecol Investig. 2001;8:1–13. [PubMed] [Google Scholar]
- 107.Paris F, Perez G, Fuks Z, Haimovitz-Friedman A, Nguyen H, Bose M, Ilagan A, Hunt P, Morgan W, Tilly J, Kolesnick R. Sphingosine-1-phosphate preserves fertility in irradiated female mice without propagating genomic damage in offspring. Nature Medicine. 2002;8:901–902. doi: 10.1038/nm0902-901. [DOI] [PubMed] [Google Scholar]
- 108.Huang L, Mivechi N, Moskophidis D. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Mol Cell Biol. 2001;21:8575–8591. doi: 10.1128/MCB.21.24.8575-8591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]