Abstract
Background/Aims
To determine breast cancer metastasis suppressor 1 (BRMS1) expression in breast cancers and the efficacy of BRMS1 as a prognostic indicator, BRMS1 expression was assessed in two sets of breast cancer tissues.
Methods
Epithelial cells from 36 frozen samples of breast cancers and corresponding normal breast were collected by laser capture microdissection and assessed for BRMS1 by quantitative RT-PCR and immunohistochemistry. BRMS1 was also evaluated by immunohistochemistry in a tissue microarray of 209 breast cancers and correlated with indicators of prognosis [estrogen receptor (ER), progesterone receptor (PR), ErbB2, p53, p27Kip1, Bcl2 and Ki-67].
Results
BRMS1 mRNA and protein were higher in 94 and 81%, respectively, of breast cancers than in corresponding normal epithelium. BRMS1 localization was predominantly nuclear, but 60–70% of cancers also exhibited cytoplasmic immunostaining. Breast cancers with lower nuclear than cytoplasmic BRMS1 (nuclear score – cytoplasmic score ≤0; 11% of cancers) had lower ER, lower PR and higher Ki-67 expression. There was also a trend toward poorer overall survival in this group of cancers, but this was only of borderline significance (p = 0.073). In Cox proportional hazards models, loss of nuclear BRMS1 was not a significant predictor of overall survival.
Conclusions
Loss of nuclear BRMS1 was associated with ER-negative cancers and a high rate of proliferation, but was not an independent indicator of prognosis.
Key Words: Breast cancer progression, Immunohistochemistry, Prognosis, Breast cancer metastasis suppressor 1
Introduction
Of the women diagnosed with early-stage, apparently localized breast cancer, approximately one third will develop life-threatening, systemic disease. Unfortunately, it is not yet possible to discern in which patients the disease will recur and in which it will not. As a result, the majority of women receiving adjuvant treatments do not really need it. The costs – economic, anxiety, quality of life – are enormous. Thus, a high priority is to develop tests that will predict which patients are at higher risk for developing metastases so that physicians can be objectively guided toward the most appropriate treatment course.
Metastasis suppressors are a growing family of molecules that are functionally defined by their ability to suppress metastasis without blocking orthotopic, primary tumor growth when re-expressed [1]. The first metastasis suppressor, Nm23, was discovered in the 1980s and there are now more than 25 published metastasis suppressor genes [1]. They have diverse mechanisms of action and many appear to function in multiple tumor types. Breast cancer metastasis suppressor 1 (BRMS1) was discovered by comparing parental to metastasis-suppressed chromosome 11 microcell hybrids using subtractive hybridization [2]. The BRMS1 gene maps to chromosome 11q13, a gene-dense region in which several nonrandom amplification and deletion events associated with breast cancer progression and metastasis have been described.
BRMS1 appears to exert its antimetastatic effect by regulating gene expression as a result of its participation in SIN3:histone deacetylase complexes [3, 4]. BRMS1 selectively reduces prometastatic genes and signaling pathways, including phosphoinositide (4,5)-bisphosphate levels and associated downstream signaling [5, 6], NFκB transcriptional activity [7, 8], epidermal growth factor receptor expression and downstream signaling [5], urokinase-type plasminogen activator expression [7] and osteopontin expression [9,10,11], and alters gap junctional intercellular communication in tumor cells [12,13,14]. Likewise, BRMS1 re-expression selectively increases expression of antimetastatic microRNA [15]. Microarray [10, 16] and proteomic [17, 18] analyses also showed multiple changes in gene and protein expression when BRMS1 was introduced into metastatic cells. Since the ability to successfully metastasize requires coordinated expression of multiple genes [19], the cumulative data describe a process in which BRMS1 selectively regulates multiple metastasis-associated genes.
Because of their roles in regulating metastasis, the metastasis suppressors would seem to offer great promise as prognostic or predictive markers. However, relatively few studies have been performed to test this potential, largely due to the lack of antibodies. A limited number of prior studies assessing BRMS1 in breast and pheochromocytoma cancer specimens have examined mRNA levels in tumor contaminated with stroma [20,21,22,23,24], which can cause complications with interpretation since BRMS1 is ubiquitously expressed [2]. In general, prior studies have not found statistically significant correlations between BRMS1 mRNA expression and survival, development of metastasis or other well-established prognostic markers. Two studies actually reported that BRMS1 expression was a poor prognostic marker [20, 21].
In addition to the complications arising because of contaminating stromal cells, recent data from our laboratory suggest that mRNA and protein levels for BRMS1 do not always correlate [25], that BRMS1 expression is post-transcriptionally regulated [26] and that splice variants of BRMS1 can be detected using RT-PCR [25]. Thus, the development of specific antibodies recognizing BRMS1 was essential in order to study primary breast cancer specimens for protein expression. Using an antibody recognizing an N-terminal epitope, Hicks et al. [27] recently reported that BRMS1 protein expression was lost in approximately 25% of cases and that there was no overall correlation with disease-free survival. However, subsets of patients who were estrogen receptor (ER) or progesterone receptor (PR) negative or human epidermal growth factor receptor 2 (HER2) overexpressing did show correlation of loss of BRMS1 with disease-free survival. These data suggested a linkage between BRMS1 expression and hormone and HER2 status. Moreover, the data suggested that BRMS1 staining could be helpful as a prognostic marker in conjunction with other prognostic markers. More recently, using an antibody for which the epitope has not been defined, Smith et al. [28] measured BRMS1 expression in non-small cell lung carcinoma and found that BRMS1 expression correlates with improved patient survival, suggesting that BRMS1 expression may have prognostic value in other types of cancers.
Our laboratory has identified several protein interaction domains that are important for the metastasis suppressor and/or corepressor activities of BRMS1 [3, 29]. As a result, other antibodies recognizing a C-terminal epitope were developed that provided important tools for dissecting the mechanism(s) of action. In the present study, we utilized antibodies recognizing the C terminus of BRMS1 to measure BRMS1 protein expression in breast cancer tissues. As found previously, BRMS1 was not an independent prognostic marker; however, the data from this study revealed, for the first time, that the cellular localization of BRMS1 protein may be important in controlling metastasis.
Materials and Methods
Breast Tissues and Tissue Microarray
Thirty-six snap-frozen breast cancer tissues, corresponding histologically and grossly normal breast tissue from the same breast resection specimen and corresponding axillary lymph node metastases (in 7 cases) were collected from the University of Alabama at Birmingham (UAB) Tissue Procurement Facility. The cancer and corresponding normal samples, although derived from the same resection specimen, were separate tissue samples, and information about the distance of the normal sample from the breast cancer sample within the resection specimen was not provided. Tissue microarrays were constructed in collaboration with Applied Genomics Inc. after collecting formalin-fixed, paraffin-embedded infiltrating ductal carcinomas, acquired between 1988 and 1996, from the archives of the UAB Department of Pathology. Two tissue microarrays were constructed, each containing a single 1-mm-diameter core from the same 209 infiltrating ductal carcinomas. Clinical data corresponding to each breast cancer were retrieved from the Department of Surgery and the UAB Tumor Registry. Demographic data for the patients are summarized in table 1. Tissue collection, tissue microarray construction and retrieval of clinical data were performed after Institutional Review Board approval.
Table 1.
Clinicopathologic features of the 209 patients with infiltrating ductal carcinomas
| Age range, years | 25–89 |
| Race, % | |
| Caucasian | 73 |
| African-American | 25 |
| Other | 2 |
| Clinical stage, % | |
| I | 22 |
| II | 38 |
| III/IV | 23 |
| Unknown | 17 |
| Mean follow-up, years | 8.03 |
Laser Capture Microdissection and Quantitative RT-PCR
Frozen sections (8 μm thick) were prepared from each tissue and rapidly stained with hematoxylin and eosin. Reagents were prepared with diethylpyrocarbonate-treated water, when possible. Laser capture microdissection (Molecular Devices/Arcturus Engineering PixCell II) was used to isolate 2,000–5,000 histologically normal or neoplastic epithelial cells from each tissue. RNA was isolated and treated with DNase (RNAqueous-Micro Kit, Ambion, Austin, Tex., USA). Prior to cDNA synthesis, all RNA samples were diluted to 4 ng/μl using RNase-free water containing 12.5 ng/μl of total yeast RNA (Ambion) as a carrier. cDNA was prepared using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, Calif., USA) according to the manufacturer's instructions. The forward, reverse and probe oligonucleotides were synthesized and purified by HPLC (Applied Biosystems). Fluorescent signal data were collected using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). The log-linear phase of amplification was monitored to obtain the threshold cycle values for each RNA sample. Ribosomal S9 was used as the internal reference and was selected because it exhibits minimal variability in tissues of different origins [30]. The comparative threshold cycle method was employed to determine BRMS1 expression levels in each sample relative to expression of ribosomal S9. Each sample was run in triplicate.
Immunohistochemistry
A rabbit polyclonal antiserum (S07050) was produced to a peptide corresponding to the C terminus of BRMS1 (231KARAAVSPQKRKSDGP246). Specificity of the antibodies was verified by Western blotting, immunoprecipitation and mass spectroscopy and electrospray sequencing (electrospray ionization mass spectrometry-mass spectrometry, data not shown). For immunohistochemistry, optimal primary antibody incubation and concentration were determined by serial dilutions on placental tissue (positive control). For immunostaining of frozen tissues, frozen sections (5 μm thickness) were fixed in 70% ethanol overnight. Subsequently, sections were incubated with an aqueous solution of 3% hydrogen peroxide for 5 min followed by 1% goat serum for 1 h at room temperature. Sections were then incubated with primary antibodies diluted 1:1,000 in phosphate-buffered saline (pH 7.6) containing 1% bovine serum albumin, 1 mM ethylenediamine tetraacetic acid and 1.5 mM sodium azide for 1 h at room temperature. Secondary detection was achieved with a streptavidin-biotin-horseradish peroxidase system (USA Ultrastreptavidin Multi-Species Detection System, Signet/Covance Laboratories). Diaminobenzidine tetrachloride was the chromogen (Super Sensitive Substrate Kit, Biogenex, San Ramon, Calif., USA). Negative controls consisted of histologic sections processed without the addition of primary antibody. After immunostaining, sections were counterstained with hematoxylin.
For immunostaining of formalin-fixed, paraffin-embedded tissue microarrays, 5-μm sections were deparaffinized in xylene, followed by hydration in graded alcohols. Sections were incubated in anti-BRMS1 (S07050, 1:1,000), and subsequent immunostaining was carried out as described above. Immunostaining for Bcl2, ErbB2, p53, ER, PR, p27Kip1 and Ki-67 was performed as previously described [31, 32]. Briefly, immunostaining for Bcl2 required antigen retrieval consisting of incubation in boiling 10 mM citrate buffer (pH 6.0) for 5 min. Immunostaining for ER, PR, p27Kip1 and Ki-67 required low-temperature antigen retrieval with enzymatic pretreatment (0.1% trypsin) followed by incubation in 10 mM citrate buffer, pH 6, for 2 h at 80°C. The antibodies included anti-ErbB2 (clone 3B5, Oncogene Research Products, San Diego, Calif., USA; 0.25 μg/ml), anti-Bcl-2 (clone 124, Genosys Biotechnologies Inc., Cambridge, UK; 12.5 μg/ml), anti-p53 (clone BP53.12, Oncogene Research Products; 0.25 μg/ml), anti-ER (clone ER88, Biogenex; 0.33 mg/ml total protein), anti-PR (clone PR88, Biogenex; 0.33 mg/ml total protein), anti-Ki-67 (clone MIB-1, Biogenex; 0.37 mg/ml total protein) and anti-p27Kip1 (clone 1B4, Novocastra Laboratories Ltd., Newcastle upon Tyne, UK; 8.0 μg/ml IgG). Secondary detection was similar to that described for BRMS1.
All slides were examined and scored by 2 observers concurrently. The intensity of cytoplasmic (anti-BRMS1 and anti-Bcl2), cell membrane (anti-ErbB2) and nuclear immunostaining (anti-BRMS1) of individual cells was scored on a scale of 0 (no staining) to 4+ (strongest possible intensity), and the percentage of cells staining at each intensity was estimated. The proportion of cells at each intensity was multiplied by the corresponding intensity value and these products were added to obtain an immunostaining score (immunoscore) ranging from 0 to 4 [33, 34]. For ER, PR, p53, p27Kip1 and Ki-67, the percentage of cancer cells with nuclei that stained positively at any intensity was determined. To quantify total cellular BRMS1 protein for comparison with BRMS1 mRNA levels, BRMS1 cytoplasmic and nuclear immunoscores were summed. To quantify a shift in BRMS1 protein from nucleus to cytoplasm, the cytoplasmic BRMS1 immunoscore was subtracted from the nuclear BRMS1 immunoscore. Using this ‘cytoplasmic shift’ score, a negative value indicates a greater level of cytoplasmic BRMS1 than nuclear BRMS1.
Statistical Analyses
Mean mRNA expression of BRMS1 relative to S9, immunoscores, percentages of positively staining cells and tumor sizes were compared between groups by the Mann-Whitney test or Wilcoxon signed rank test, as appropriate. Lymph node status, clinical stage and low BRMS1 expression in ER-positive versus ER-negative cancers were compared between groups by Fisher's exact test. Kaplan-Meier methods were used to estimate the survival function for overall survival for patients with high-BRMS1 versus low-BRMS1 cancers [35]. Overall survival was defined as the time from the date of diagnosis to the date of death. Patients who were alive at the date of last contact, died from unknown causes or died from a disease other than breast cancer were censored at the date of last contact. Differences in overall survival functions were assessed using the log-rank test. Cox proportional hazards models were fitted to the data to assess the effect of BRMS1 on overall survival after adjusting for the effects of other covariates [36]. Interaction effects between covariates were also considered by including their cross-products in the Cox models.
Results
Expression of BRMS1 Is Higher in Breast Cancer Epithelial Cells than in Corresponding, Histologically Normal Breast Epithelium
BRMS1 is ubiquitously expressed in normal tissues, including inflammatory and stromal cells [2]. Therefore, to measure the expression of BRMS1 in normal breast epithelium and corresponding breast carcinoma cells without the presence of contaminating stromal or inflammatory cells, we isolated each of these epithelial cell types by laser microdissection. Normal or cancer epithelial cells were isolated from histologic sections of each frozen tissue. All cancers were infiltrating ductal adenocarcinomas. BRMS1 mRNA content was quantified by real-time quantitative PCR, and expression was normalized to expression of the housekeeping gene ribosomal S9. Mean expression of BRMS1 was 5 times higher in cancer cells than in matching normal epithelial cells (p < 0.001; fig. 1A). BRMS1 was increased in cancer epithelium in 34 of 36 matching cancer/normal pairs (94%). In 1 cancer/normal pair, BRMS1 was decreased, while in another pair, expression of BRMS1 was not detected in either normal or cancer epithelium. In addition, tissue from axillary lymph node metastases was available for 7 cancers. Carcinoma cells in the lymph nodes were also microdissected, and BRMS1 mRNA levels were determined by real-time quantitative PCR. BRMS1 expression was similar in primary tumor cells and their corresponding lymph node metastases, and expression was higher in both the primary carcinoma and the metastasis than in the matching normal epithelium (p = 0.016 and p = 0.031, respectively; fig. 1A).
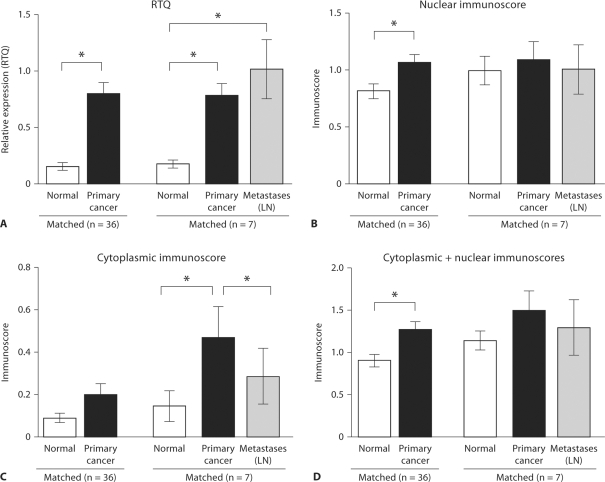
Fig. 1.
BRMS1 expression is higher in breast cancer than in normal breast epithelium. A Epithelium from infiltrating ductal carcinomas (n = 36), corresponding normal tissue (n = 36) and axillary lymph nodes (LN) with metastases (n = 7) was collected by laser capture microdissection. RNA was isolated, cDNA was prepared and expression of BRMS1 and ribosomal S9 was measured by real-time quantitative PCR (RTQ). Expression of BRMS1 was normalized to ribosomal S9. The mean and standard error are presented. BRMS1 expression was significantly greater in cancer than in matched normal epithelium (p < 0.001). BRMS1 expression was also greater in breast cancer epithelium (p = 0.016) and in metastatic cancer epithelial cells (p = 0.031) than in normal breast epithelium in the 7 breast cancers with matched primary and metastatic cancer. B–D The carcinomas and corresponding normal breast and lymph nodes presented in A were immunostained for expression of BRMS1. Nuclear expression of BRMS1 was greater than cytoplasmic expression. Both nuclear and cyto plasmic BRMS1 in epithelial cells was semiquantified (immunoscore). Reflecting BRMS1 mRNA, nuclear BRMS1 protein was significantly greater in cancer than in normal epithelial cells (p = 0.011; B), but this was not the case for cytoplasmic BRMS1 (C). The total of the nuclear and cytoplasmic immunoscores (representing the total cellular BRMS1) was significantly higher in cancer than in normal epithelium (p = 0.002; D). In the 7 breast cancers with corresponding lymph node metastases, BRMS1 protein was again higher in cancer epithelium than in corresponding normal epithelium, but this was statistically significant only for cytoplasmic BRMS1 (p = 0.036; C). In these 7 cases, the metastatic cancer cells had similar nuclear (B) and total (D) BRMS1 expression to the primary cancer epithelial cells, but cytoplasmic BRMS1 was lower in the metastatic cells than in the primary cancer cells (p = 0.033; C). The mean and standard error are presented. Statistical significance is indicated by asterisks.
To identify BRMS1 protein, the frozen breast tissues were also immunostained using S07050. Consistent with the known nuclear functions and location of BRMS1 protein, immunostaining was primarily nuclear in the breast tissue. However, 49% of normal epithelium and 57% of cancer epithelium also exhibited weak to moderate cytoplasmic staining. BRMS1 expression (either nuclear or both cytoplasmic and nuclear) was detected in all breast carcinomas. Both cytoplasmic and nuclear staining were semiquantified using a previously described scoring system [32] that incorporates the number of cells that stain and the intensity of the staining. Similar to the mRNA data, the nuclear and cytoplasmic immunoscores were higher in cancer than in the corresponding normal epithelium (n = 36), but only the difference in the nuclear immunoscores was statistically significant (p = 0.011; fig. 1B, C). The total (nuclear + cytoplasmic) immunoscore (representing total cellular BRMS1 protein) was also significantly higher in cancer than in normal epithelium (p = 0.002; fig. 1D). In the 7 breast cancers with corresponding lymph node metastases, there was a decrease in cytoplasmic BRMS1 in the lymph node metastases (p = 0.033) compared to the primary cancers (fig. 1C), but this decrease was not reflected by a difference in total or nuclear BRMS1 protein (fig. 1B, D). Higher expression of BRMS1 protein in the primary cancer than in the corresponding normal epithelium was also seen in this small group of 7 cases, but this was statistically significant only for cytoplasmic expression (p = 0.036; fig. 1B–D). In addition, the number of normal/cancer pairs in which BRMS1 protein expression was higher in cancer tissue was 29 of 36 (81%), which is a slightly lower percentage than for BRMS1 mRNA. This result supports prior findings showing that BRMS1 mRNA and protein levels do not necessarily correlate [25].
Loss of BRMS1 Expression Occurs in a Small Percentage of Breast Cancers
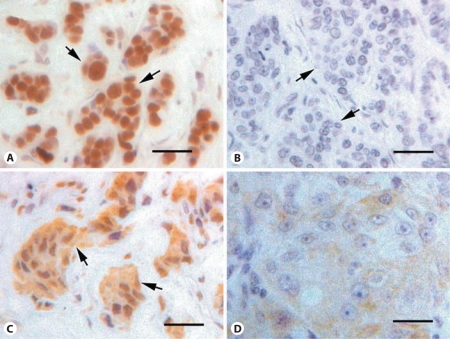
To analyze BRMS1 expression in a larger number of breast cancers, tissue microarrays containing 209 infiltrating ductal adenocarcinomas were immunostained with S07050 antiserum. Two tissue microarrays were stained, each containing a different core sample of the same 209 breast cancers. The mean immunoscore for each cancer was determined after histologic evaluation, as described above. BRMS1 protein was detected in the majority of breast cancers (205/209, 98%) and was primarily, but not exclusively, nuclear in location. BRMS1 staining was located in the nucleus in 203 of 209 cancers (97%). Cytoplasmic staining was present in 141 of 209 cancers (67%). Four cancers (2%) expressed neither cytoplasmic nor nuclear BRMS1 (fig. 2).
Fig. 2.
BRMS1 expression in breast cancers evaluated by immunohistochemistry. A tissue microarray of 209 infiltrating ductal carcinomas was immunostained for BRMS1 and scored. The cytoplasmic shift score is a means of quantifying the distribution of BRMS1 in the nucleus and cytoplasm and consists of the cytoplasmic immunoscore subtracted from the nuclear immunoscore. A A carcinoma with strong nuclear BRMS1 (cytoplasmic shift score = 2.53). B A carcinoma with no cytoplasmic or nuclear BRMS1 staining (cytoplasmic shift score = 0). C A carcinoma with greater cytoplasmic than nuclear staining (cytoplasmic shift score = −0.96). D A carcinoma with only cytoplasmic staining (cytoplasmic shift score = −0.93). Arrows indicate clusters of cancer epithelial cells. Original magnification: ×200. Scale bar = 50 μm.
Low Nuclear Expression of BRMS1 Correlates with Indicators of Poor Prognosis
The breast cancers constituting the microarrays were also immunostained for detection of expression of ER, PR, ErbB2, p53, Bcl2, p27Kip1 and Ki-67, as previously described [31, 32]. Each of these protein biomarkers functions in the growth and progression of breast cancer and is a prognostic indicator or is associated with other indicators of prognosis. There is a general consensus that ER and PR content is a weak prognosticator, with most studies conferring a more favorable prognosis on ER-positive and PR-positive tumors [37, 38]. Overexpression of ErbB2, decreased expression of Bcl2 and mutation of p53 (identified by nuclear accumulation of p53 protein) [39] correlate with other indicators of a poor prognosis, such as a negative ER status [39,40,41,42,43,44,45,46]. A high rate of cancer cell proliferation is also a poor prognostic indicator, irrespective of the method used to measure proliferation, including identifying the incidence of cells that immunostain for the proliferation marker Ki-67 [47]. p27Kip1 is a member of the Cip/Kip family of cyclin-dependent kinase inhibitors which function to inhibit the transition from G1 to the S phase of the cell cycle. Low or absent nuclear expression of p27Kip1 detected by immunohistochemistry is a clinical marker of disease progression in several types of tumors, including breast cancer [47,48,49]. Additionally, clinical data for the 209 cases, including tumor size, axillary lymph node status, clinical stage and clinical follow-up data, were obtained. Clinicopathologic features of the 209 cases are presented in table 1.
Expression of ErbB2 at the cell membrane and cytoplasmic expression of Bcl2 were semiquantified by the same immunoscoring system used for BRMS1 and as previously described [31]. For ER, PR, p53, Ki-67 and p27Kip1, the percentage of cells with nuclear staining was determined. Nuclear expression of BRMS1 (immunoscore) was positively correlated with expression of Bcl2, ER, PR and p27Kip1, but negatively correlated with Ki-67 (p = 0.008, p < 0.001, p = 0.001, p = 0.003 and p = 0.007, respectively; table 2). In other words, lower nuclear BRMS1 correlates with lower levels of Bcl2, ER, PR and p27Kip1 and greater proliferation, all indicators of a more aggressive breast cancer phenotype. Interestingly, cytoplasmic expression of BRMS1 was inversely correlated with expression of ER and positively correlated with Ki-67 (p = 0.007 and p = 0.033, respectively), which was opposite to the results for nuclear BRMS1 (table 2). In addition, cytoplasmic BRMS1 was positively correlated with nuclear accumulation of p53 and expression of ErbB2. Taken together, these correlations suggest an association of cytoplasmic BRMS1 with more aggressive breast cancer, which is again opposite to the findings for nuclear BRMS1. These findings raise the possibility that nuclear and cytoplasmic BRMS1 expression are inversely correlated. Indeed, there was an inverse correlation between nuclear and cytoplasmic BRMS1, but it was not statistically significant (r = −0.127, p = 0.066).
Table 2.
Correlation of BRMS1 nuclear and cytoplasmic staining with other biomarkers
| r | p | |
| Nuclear score | ||
| ErbB2 | 0.13 | 0.071 |
| Bcl2 | 0.19 | 0.008 |
| p53 | −0.02 | 0.814 |
| ER | 0.29 | <0.001 |
| PR | 0.25 | 0.001 |
| p27Kip1 | 0.22 | 0.003 |
| Ki-67 | −0.20 | 0.007 |
| BRMS1 cytoplasmic | −0.13 | 0.066 |
| Cytoplasmic score | ||
| ErbB2 | 0.15 | 0.038 |
| Bcl2 | −0.13 | 0.071 |
| p53 | 0.15 | 0.034 |
| ER | −0.20 | 0.007 |
| PR | −0.07 | 0.367 |
| p27Kip1 | −0.04 | 0.603 |
| Ki-67 | 0.16 | 0.033 |
| BRMS1 nuclear | −0.13 | 0.066 |
Breast Cancers with Equal or Higher Cytoplasmic than Nuclear Expression of BRMS1 Have Low ER and PR Expression and a High Rate of Proliferation
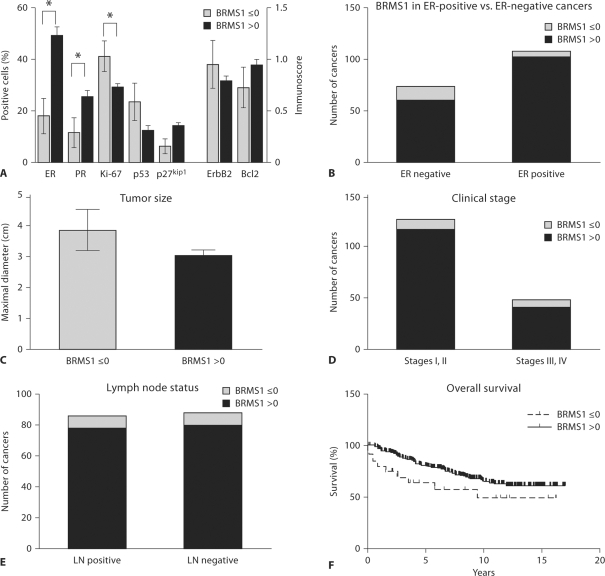
This trend toward an inverse correlation between cytoplasmic and nuclear BRMS1 suggests that a shift from nuclear to cytoplasmic localization of BRMS1 in breast cancer may indicate a worse prognosis. To quantify this shift, we subtracted the cytoplasmic BRMS1 immunoscore from the nuclear BRMS1 immunoscore. A negative ‘cytoplasmic shift’ score indicates a greater level of cytoplasmic BRMS1 than nuclear BRMS1, which occurred in 19 of 209 cancers (9%). Four cancers had a cytoplasmic shift score of 0, with no cytoplasmic or nuclear BRMS1 staining. Breast cancers with no BRMS1 staining or greater cytoplasmic than nuclear staining (i.e. a cytoplasmic shift score ≤0) constituted a minority of cases (23/209, 11%), similar to the findings of a prior report of loss of nuclear BRMS1 expression in breast cancer (25% of cases) [27]. In addition, this group of breast carcinomas with a cytoplasmic shift score ≤0 had significantly lower expression of ER and PR and a higher rate of proliferation than those breast cancers with a cytoplasmic shift score greater than 0 (fig. 3A). When comparing the number of ER-positive versus ER-negative cancers with a cytoplasmic shift score ≤0, ER-negative cancers were significantly more likely than ER-positive cancers to have low BRMS1. Fourteen of 74 ER-negative cancers (18.9%) versus 6 of 108 ER-positive cancers (5.6%) had a cytoplasmic shift score ≤0 (p = 0.007, Fisher's exact test; fig. 3B). ER-negative status was defined as those cancers with <10% of cells staining positively for ER. Corresponding to the higher rate of proliferation, tumor size was also larger and p27Kip1 expression was lower in breast cancers with a cytoplasmic shift score ≤0, but these differences were of only borderline significance (p = 0.075 and p = 0.085, respectively; fig. 3C, A). There was no significant difference in ErbB2, Bcl2 or p53 expression in breast cancers with cytoplasmic shift scores ≤0 versus >0 (fig. 3A). From the cases of a high clinical stage at presentation (stages III and IV), a larger proportion had a cytoplasmic shift score ≤0 (7/48, 15%) compared to cases of a lower clinical stage (stages I and II; 10/126, 8%), but this difference was not statistically significant (fig. 3D). There was also no difference in the proportion of lymph node-positive (i.e. cases with metastases to axillary lymph nodes) versus lymph node-negative cases with a cytoplasmic shift score ≤0 [8/86 (9%) versus 8/88 (9%); fig. 3E]. Patients with breast cancers with a cytoplasmic score ≤0 also had poorer overall survival, although this was only of borderline significance (p = 0.079; fig. 3F). In the Cox proportional hazards model for overall survival with BRMS1 cytoplasmic shift score and all covariates (i.e. age, race, tumor size, lymph node metastasis, ErbB2, Bcl2, p53, ER, PR, Ki-67 and p27Kip1), BRMS1 was not a significant predictor of survival. Only age was significant. After removing all nonsignificant covariates from the model for predictors of longer overall survival, only 1 covariate, age, stays in the Cox model (hazard ratio 0.982, 95% confidence interval 0.967–0.998; p = 0.0249) and BRMS1 is still not significant (hazard ratio 0.734, 95% confidence interval 0.513–1.050; p = 0.0903).
Fig. 3.
Breast cancers with lower nuclear than cytoplasmic BRMS1 expression or no BRMS1 expression have lower expression of ER and PR and a higher rate of proliferation. A Expression of ER, PR, Ki-67, p53 and p27Kip1 was assessed by immunohistochemistry [percentage of positively staining (nuclear) cancer epithelial cells] in the breast cancers included in the tumor microarray. ErbB2 (membrane expression) and Bcl2 (cytoplasmic expression) were also assessed by immunohistochemistry in cancer epithelial cells and semiquantified by an immunoscore. Cancers with a cytoplasmic shift score (i.e. nuclear minus cytoplasmic expression) ≤ 0 (n = 23) had lower ER and PR expression and a higher rate of proliferation (i.e. Ki-67 expression; p = 0.002, p = 0.023 and p = 0.046, respectively) than those with a cytoplasmic shift score 1 0 (n = 186). Significant differences are indicated by asterisks. The mean and standard error are presented. B The proportion of breast cancers with a cytoplasmic shift score ≤ 0 was greater in ER-negative than in ER-positive cancers. Fourteen of 74 ER-negative cancers (18.9%) versus 6 of 108 ER-positive cancers (5.6%) had a cytoplasmic shift score ≤ 0 (p = 0.007, Fisher's exact test). C There was no statistically significant difference in tumor size in breast cancers with a BRMS1 cytoplasmic shift score ≤ 0 versus > 0. The mean size and standard error are presented. D There was no significant difference in the proportion of cancers with a BRMS1 cytoplasmic shift score ≤ 0 in cases of clinical stages I and II (10/126, 8%) versus those of clinical stages III and IV (7/48, 15%). E There was no significant difference in the proportion of cancers with a BRMS1 cytoplasmic shift score ≤ 0 in cases with axillary lymph node metastases (8/86, 9%) versus those without axillary lymph node metastases (8/88, 9%). F There was a trend for overall survival, estimated by the Kaplan-Meier method, to be lower in patients with breast cancers with a cytoplasmic shift score ≤ 0, but this was not statistically significant (p = 0.079, log-rank test).
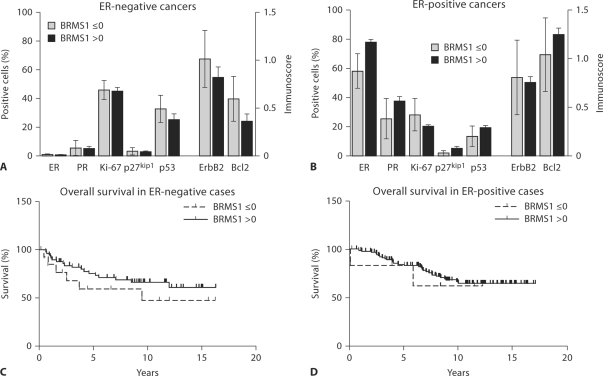
To determine whether BRMS1 may be prognostic independently of ER status, we assessed those cancers with a BRMS1 cytoplasmic shift score ≤0 versus >0 in both ER-positive and ER-negative breast cancers separately. There was no difference in biomarker expression in those cancers with low (≤0) versus high (>0) BRMS1 cytoplasmic shift scores in either ER-negative or ER-positive cancers (fig. 4A, B). However, there was a trend toward statistically significantly lower expression of ER in low-BRMS1, ER-positive cancers (p = 0.081), underscoring the strong association between BRMS1 and ER expression (fig. 4B). There was also no significant difference in tumor size or in the proportion of cases with a BRMS1 cytoplasmic shift score ≤0 in patients with low-stage (stages I, II) versus high-stage (stages III, IV) disease or in those with axillary lymph node metastases versus those without (data not shown). The overall survival was slightly poorer in patients with low BRMS1, ER-negative cancers, but this was not statistically significant (p = 0.273; fig. 4C). The BRMS1 cytoplasmic shift score had no impact on overall survival in patients with ER-positive cancers (fig. 4D). These results indicate that loss of BRMS1 is not an independent prognostic indicator but is closely associated with negative or low ER status.
Fig. 4.
BRMS1 in ER-negative and ER-positive breast cancers. A, B When ER-negative and ER-positive carcinomas were analyzed separately, there was no significant difference in biomarker expression in ER-negative (A) or ER-positive (B) carcinomas with a BRMS1 cytoplasmic shift score ≤ 0 versus > 0. However, there was a trend toward statistically significantly lower expression of ER in low-BRMS1, ER-positive cancers (p = 0.081). The mean biomarker expression and standard error are presented. C There was no significant difference in overall survival in patients with ER negative breast carcinomas with a BRMS1 cytoplasmic shift score ≤0 (n = 14) versus > 0 (n = 60; p = 0.273). D There was no significant difference in overall survival in patients with ER-positive breast cancers with a BRMS1 cytoplasmic shift score ≤ 0 (n = 6) versus > 0 (n = 102; p = 0.705). Kaplan-Meier methods were employed to estimate overall survival.
Discussion
In general, the data reported here are consistent with the only other published report examining BRMS1 protein expression in breast cancer [27]. Similar to this previous report, we found that loss of nuclear BRMS1 protein expression occurred in a minority of cancers (11%) but correlated with ER and PR expression and that there was a trend toward worse overall survival in patients with cancers with loss of nuclear BRMS1 expression. Hicks et al. [27] also found a loss of nuclear BRMS1 in a minority of cases (25%), showed a correlation between BRMS1 and hormone receptor status and found that loss of BRMS1 expression was associated with decreased disease-free survival in the context of hormone receptor-negative tumors as well as loss of BRMS1 with HER2/ErbB2 overexpression. Hicks et al. [27] did not report cytoplasmic localization of BRMS1, whereas in this study, we found that 60–70% of the breast cancers exhibited non-nuclear staining of BRMS1 in addition to nuclear localization, which still predominated. We report for the first time that BRMS1 can be found in readily measurable amounts in the cytoplasm. Moreover, if the cytoplasmic BRMS1 levels exceeded the nuclear levels, the cancers generally exhibited patterns associated with a worse prognosis (e.g. lower/negative ER, lower PR and higher Ki-67 expression), but the correlation with overall survival was of only borderline significance (p = 0.073).
Our ability to detect cytoplasmic BRMS1 could be due to utilization of an antibody that recognizes the C terminus of BRMS1. The nuclear localization sequences for BRMS1 are located at the C terminus. That the antibodies still recognize the epitope argues that gross deletions of the C terminus are not responsible for loss of nuclear localization. Rather, associations of BRMS1 with chaperones or other proteins may be, at least partially, responsible for the change in localization. Nonetheless, the data presented here suggest that BRMS1 localization within the cell appears to be as important as total expression. The contribution of protein localization within cells to pathologies is not unprecedented. For example, the nuclear to cytoplasmic ratios of β-catenin [50,51,52], p63 [53], p21 [54], p27Kip1[55, 56] and p53 [57] contribute to cellular transformation.
Recent studies examining BRMS1 mRNA expression in clinical human breast cancer samples failed to show any correlation between BRMS1 expression and parameters of local dissemination such as tumor size and lymph node metastasis. However, BRMS1 protein is expressed in all normal mammary epithelium and in unaffected breast tissues from patients with cancer as well as in host inflammatory cells. Therefore the measurement of BRMS1 mRNA levels in non-microdissected clinical samples could potentially be confounded by the expression of the BRMS1 gene by normal host cells within tumor tissue.
The mechanism underlying loss of BRMS1 expression in cancer cells is not known. However, previous reports indicate that mutations are not common and that epigenetic mechanisms (e.g. methylation) are responsible for loss of BRMS1 expression [13, 25, 58]. Coupled with prior reports that BRMS1 protein stability can be regulated by chaperones [26], altered localization and protein stability issues take on increased significance with regard to biological behavior and prognostic value. Using Cox proportional hazards models, we found that loss of nuclear BRMS1 was not a significant predictor of survival. Thus, while loss of nuclear BRMS1 was associated with ER-negative status and a high rate of proliferation, both of which are indicators of a poorer prognosis, loss of nuclear BRMS1 was not an independent indicator of prognosis. Nonetheless, BRMS1 expression and localization may have some utility in combination with other breast cancer prognostic biomarkers. Additional studies with larger data sets will be required.
Erratum
In the article ‘A Shift from Nuclear to Cytoplasmic Breast Cancer Metastasis Suppressor 1 Expression Is Associated with Highly Proliferative Estrogen Receptor-Negative Breast Cancers’ by Frolova et al. (Tumor Biol 2009; 30: 148–159), the following Acknowledgments were erroneously left out:
Acknowledgements
This work was supported by the NIH SPORE in Breast Cancer CA91421 (N.F., M.R.J., D.R.W. and A.R.F.), NIH CA87728 (M.D.E., T.M.B. and D.R.W.), US Army Medical Research Defense Command W81-XWH-08-1-0786 (M.D.E. and D.R.W) and W81-XWH- 08-1-0779 (T.M.B. and D.R.W.), and the National Foundation for Cancer Research (A.R.F. and D.R.W.).
References
- 1.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- 3.Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC, Welch DR. Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–7444. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- 5.Vaidya KS, Harihar S, Stafford LJ, Hurst DR, Hicks DG, Casey G, DeWald DB, Welch DR. Breast cancer metastasis suppressor-1 differentially modulates growth factor signaling. J Biol Chem. 2008;283:28354–28360. doi: 10.1074/jbc.M710068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeWald DB, Torabinejad J, Samant RS, Johnston D, Erin N, Shope JC, Xie Y, Welch DR. Metastasis suppression by breast cancer metastasis suppressor 1 involves reduction of phosphoinositide signaling in MDA-MB-435 breast carcinoma cells. Cancer Res. 2005;65:713–717. [PubMed] [Google Scholar]
- 7.Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-kB activity. Cancer Res. 2005;65:3586–3595. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol. 2006;26:8683–8696. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedley BD, Welch DR, Allan AL, Al-Katib W, Dales DW, Postenka CO, Casey G, MacDonald IC, Chambers AF. Downregulation of osteopontin contributes to metastasis suppression by breast cancer metastasis suppressor 1. Int J Cancer. 2008;123:526–534. doi: 10.1002/ijc.23542. [DOI] [PubMed] [Google Scholar]
- 10.Shevde LA, Samant RS, Paik JC, Metge BJ, Chambers AF, Casey G, Frost AR, Welch DR. Osteopontin knockdown suppresses tumorigenicity of human metastatic breast carcinoma. Clin Exp Metastasis. 2006;23:123–133. doi: 10.1007/s10585-006-9013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR, Shevde LA. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor P, Saunders MM, Li Z, Zhou Z, Schaeffer N, Kunze EL, Samant RS, Welch DR, Donahue HJ. Breast cancer metastatic potential: correlation with increased heterotypic gap junctional intercellular communication between breast cancer cells and osteoblastic cells. Int J Cancer. 2004;111:693–697. doi: 10.1002/ijc.20318. [DOI] [PubMed] [Google Scholar]
- 13.Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- 14.Saunders MM, Seraj MJ, Li ZY, Zhou ZY, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–1767. [PubMed] [Google Scholar]
- 15.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champine PJ, Michaelson J, Weimer B, Welch DR, DeWald DB. Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exp Metastasis. 2007;24:551–565. doi: 10.1007/s10585-007-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006–4018. doi: 10.1021/pr0703167. [DOI] [PubMed] [Google Scholar]
- 18.Cicek M, Samant RS, Kinter M, Welch DR, Casey G. Identification of metastasis-associated proteins through protein analysis of metastatic MDA-MB-435 and metastasis-suppressed BRMS1 transfected-MDA-MB-435 cells. Clin Exp Metastasis. 2004;21:149–157. doi: 10.1023/b:clin.0000024729.19084.f0. [DOI] [PubMed] [Google Scholar]
- 19.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombardi G, Di Cristofano C, Capodanno A, Iorio MC, Aretini P, Isola P, Tancredi M, Collecchi P, Naccarato AG, Porta RP, Bevilacqua G, Caligo MA. High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int J Cancer. 2006;120:1169–1178. doi: 10.1002/ijc.22379. [DOI] [PubMed] [Google Scholar]
- 21.Kelly LM, Buggy Y, Hill A, O'Donovan N, Duggan C, McDermott EW, O'Higgins NJ, Young L, Duffy MJ. Expression of the breast cancer metastasis suppressor gene, BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumor Biol. 2005;26:213–216. doi: 10.1159/000086955. [DOI] [PubMed] [Google Scholar]
- 22.Ohta S, Lai EW, Pang ALY, Brouwers FM, Chan WY, Eisenhofer G, deKrijger R, Ksinantova L, Breza J, Blazicek P, Kvetnansky R, Wesley RA, Pacak K. Downregulation of metastasis suppressor genes in malignant pheochromocytoma. Int J Cancer. 2005;114:139–143. doi: 10.1002/ijc.20670. [DOI] [PubMed] [Google Scholar]
- 23.Kaushik N, Fear D, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, Kerr JR. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol. 2005;58:826–832. doi: 10.1136/jcp.2005.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Yamashita H, Toyama T, Yamamoto Y, Kawasoe T, Iwase H. Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res. 2006;12:6410–6414. doi: 10.1158/1078-0432.CCR-06-1347. [DOI] [PubMed] [Google Scholar]
- 25.Hurst DR, Xie Y, Edmonds MD, Welch DR. Multiple forms of BRMS1 are differentially expressed in the MCF10 isogenic breast cancer progression model. Clin Exp Metastasis. 2009;26:89–96. doi: 10.1007/s10585-008-9216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA, Shevde LA, Hopper JE, Xie Y, Welch DR, Samant RS. Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Commun. 2006;348:1429–1435. doi: 10.1016/j.bbrc.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri T, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost AR, Welch DR, Casey G. Loss of BRMS1 protein expression predicts reduced disease-free survival in hormone receptor negative and HER2 positive subsets of breast cancer. Clin Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith PW, Liu Y, Siefert SA, Moskaluk CA, Petroni GR, Jones DR. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276:196–203. doi: 10.1016/j.canlet.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silveira AC, Hurst DR, Vaidya KS, Ayer DE, Welch DR. Over-expression of the BRMS1 family member SUDS3 does not suppress metastasis of human cancer cells. Cancer Lett. 2009;276:32–37. doi: 10.1016/j.canlet.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanquicett C, Johnson MR, Heslin M, Diasio RB. Housekeeping gene variability in normal and carcinomatous colorectal and liver tissues: applications in pharmacogenomic gene expression studies. Anal Biochem. 2002;303:209–214. doi: 10.1006/abio.2001.5570. [DOI] [PubMed] [Google Scholar]
- 31.Talley L, Chhieng DC, Bell WC, Grizzle WE, Frost AR. Immunohistochemical detection of EGFR, p185(erbB-2), Bcl-2 and p53 in breast carcinomas in pre-menopausal and post-menopausal women. Biotech Histochem. 2008;83:5–14. doi: 10.1080/10520290701822436. [DOI] [PubMed] [Google Scholar]
- 32.Talley LI, Grizzle WE, Waterbor JW, Brown D, Weiss H, Frost AR. Hormone receptors and proliferation in breast carcinomas of equivalent histologic grades in pre- and postmenopausal women. Int J Cancer. 2002;98:118–127. doi: 10.1002/ijc.10171. [DOI] [PubMed] [Google Scholar]
- 33.Grizzle WE, Myers RB, Manne U, Stockard CR, Harkins LE, Srivastava S. Factors affecting immunohistochemical evaluation of biomarker expression in neoplasia. In: Hanausek M, Walaszek Z, editors. John Walker's Methods in Molecular Medicine – Tumor Marker Protocols. Totowa: Humana Press; 1998. pp. 161–179. [Google Scholar]
- 34.Frost AR, Sparks D, Grizzle WE. Methods of antigen recovery vary in their usefulness in unmasking specific antigens in immunohistochemistry. Appl Immunohistochem Mol Morphol. 2000;8:236–243. doi: 10.1097/00129039-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 36.Cox D. Regression models and life tables. J R Stat Soc. 1972;34:187–220. [Google Scholar]
- 37.Pichon MF, Broet P, Magdelenat H, Delarue JC, Spyratos F, Basuyau JP, Saez S, Rallet A, Courriere P, Millon R, Asselain B. Prognostic value of steroid receptors after long-term follow-up of 2257 operable breast cancers. Br J Cancer. 1996;73:1545–1551. doi: 10.1038/bjc.1996.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher B, Redmond C, Fisher ER, Caplan R. Relative worth of estrogen or progesterone receptor and pathologic characteristics of differentiation as indicators of prognosis in node negative breast cancer patients: findings from National Surgical Adjuvant Breast and Bowel Project Protocol B-06. J Clin Oncol. 1988;6:1076–1087. doi: 10.1200/JCO.1988.6.7.1076. [DOI] [PubMed] [Google Scholar]
- 39.Zellars RC, Hilsenbeck SG, Clark GM, Allred DC, Herman TS, Chamness GC, Elledge RM. Prognostic value of p53 for local failure in mastectomy-treated breast cancer patients. J Clin Oncol. 2000;18:1906–1913. doi: 10.1200/JCO.2000.18.9.1906. [DOI] [PubMed] [Google Scholar]
- 40.Stefano R, Agostara B, Calabro M, Campisi I, Ravazzolo B, Traina A, Miele M, Castagnetta L. Expression levels and clinical-pathological correlations of HER2/neu in primary and metastatic human breast cancer. Ann NY Acad Sci. 2004;1028:463–472. doi: 10.1196/annals.1322.055. [DOI] [PubMed] [Google Scholar]
- 41.Barbareschi M, Doglioni C, Veronese S, Bonzanini M, Dalla PP, Harris AL, Caffo O. p21WAF1 and p53 immunohistochemical expression in breast carcinoma may predict therapeutic response to adjuvant treatment. Eur J Cancer. 1996;32A:2182–2183. doi: 10.1016/s0959-8049(96)00222-5. [DOI] [PubMed] [Google Scholar]
- 42.Barbareschi M, Caffo O, Veronese S, Leek RD, Fina P, Fox S, Bonzanini M, Girlando S, Morelli L, Eccher C, Pezzella F, Doglioni C, Dalla PP, Harris A. Bcl-2 and p53 expression in node-negative breast carcinoma: a study with long-term follow-up. Hum Pathol. 1996;27:1149–1155. doi: 10.1016/s0046-8177(96)90307-x. [DOI] [PubMed] [Google Scholar]
- 43.Barbareschi M, Caffo O, Doglioni C, Fina P, Marchetti A, Buttitta F, Leek R, Morelli L, Leonardi E, Bevilacqua G, Dalla PP, Harris AL. p21WAF1 immunohistochemical expression in breast carcinoma: correlations with clinicopathological data, oestrogen receptor status, MIB1 expression, p53 gene and protein alterations and relapse-free survival. Br J Cancer. 1996;74:208–215. doi: 10.1038/bjc.1996.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jalava PJ, Collan YU, Kuopio T, Juntti-Patinen L, Kronqvist P. Bcl-2 immunostaining: a way to finding unresponsive postmenopausal N+ breast cancer patients. Anticancer Res. 2000;20:1213–1219. [PubMed] [Google Scholar]
- 45.Nakopoulou LL, Alexiadou A, Theodoropoulos GE, Lazaris AC, Tzonou A, Keramopoulos A. Prognostic significance of the co-expression of p53 and c-erbB-2 proteins in breast cancer. J Pathol. 1996;179:31–38. doi: 10.1002/(SICI)1096-9896(199605)179:1<31::AID-PATH523>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 46.Horita K, Yamaguchi A, Hirose K, Ishida M, Noriki S, Imamura Y, Fukuda M. Prognostic factors affecting disease-free survival rate following surgical resection of primary breast cancer. Eur J Histochem. 2001;45:73–84. doi: 10.4081/1616. [DOI] [PubMed] [Google Scholar]
- 47.Van Diest PJ, van der Wall E, Baak JP. Prognostic value of proliferation in invasive breast cancer: a review. J Clin Pathol. 2004;57:675–681. doi: 10.1136/jcp.2003.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbareschi M, van Tinteren H, Mauri FA, Veronese S, Peterse H, Maisonneuve P, Caffo O, Scaioli M, Doglioni C, Galligioni E, Dalla Palma P, Michalides R. P27Kip1 expression in breast carcinomas: an immunohistochemical study on 512 patients with long-term follow-up. Int J Cancer. 2000;89:236–241. [PubMed] [Google Scholar]
- 49.Tsuchiya A, Zhang GJ, Kanno M. Prognostic impact of cyclin-dependent kinase inhibitor p27kip1 in node-positive breast cancer. J Surg Oncol. 1999;70:230–234. doi: 10.1002/(sici)1096-9098(199904)70:4<230::aid-jso6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 50.Kielhorn E, Provost E, Olsen D, D'Aquila TG, Smith BL, Camp RL, Rimm DL. Tissue microarray-based analysis shows phospho-β-catenin expression in malignant melanoma is associated with poor outcome. Int J Cancer. 2003;103:652–656. doi: 10.1002/ijc.10893. [DOI] [PubMed] [Google Scholar]
- 51.Johnson JP. Cell adhesion molecules in the development and progression of malignant melanoma. Cancer Metastasis Rev. 1999;18:345–357. doi: 10.1023/a:1006304806799. [DOI] [PubMed] [Google Scholar]
- 52.Nakopoulou L, Mylona E, Papadaki I, Kavantzas N, Giannopoulou I, Markaki S, Keramopoulos A. Study of phospho-beta-catenin subcellular distribution in invasive breast carcinomas in relation to their phenotype and the clinical outcome. Mod Pathol. 2006;19:556–563. doi: 10.1038/modpathol.3800562. [DOI] [PubMed] [Google Scholar]
- 53.Narahashi T, Niki T, Wang T, Goto A, Matsubara D, Funata N, Fukayama M. Cytoplasmic localization of p63 is associated with poor patient survival in lung adenocarcinoma. Histopathology. 2006;49:349–357. doi: 10.1111/j.1365-2559.2006.02507.x. [DOI] [PubMed] [Google Scholar]
- 54.Ohata M, Nakamura S, Fujita H, Isemura M. Prognostic implications of p21 (Waf1/Cip1) immunolocalization in multiple myeloma. Biomed Res. 2005;26:91–98. doi: 10.2220/biomedres.26.91. [DOI] [PubMed] [Google Scholar]
- 55.Singh SP, Lipman J, Goldman H, Ellis FHJ, Aizenman L, Cangi MG, Signoretti S, Chiaur DS, Pagano M, Loda M. Loss or altered subcellular localization of p27 in Barrett's associated adenocarcinoma. Cancer Res. 1998;58:1730–1735. [PubMed] [Google Scholar]
- 56.Ogino S, Kawasaki T, Ogawa A, Kirkner GJ, Loda M, Fuchs CS. Cytoplasmic localization of p27 (cyclin-dependent kinase inhibitor 1B/KIP1) in colorectal cancer: inverse correlations with nuclear p27 loss, microsatellite instability, and CpG island methylator phenotype. Hum Pathol. 2007;38:585–592. doi: 10.1016/j.humpath.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sembritzki O, Hagel C, Lamszus K, Deppert W, Bohn W. Cytoplasmic localization of wild-type p53 in glioblastomas correlates with expression of vimentin and glial fibrillary acidic protein. Neuro Oncol. 2002;4:171–178. doi: 10.1093/neuonc/4.3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metge BJ, Frost AR, King JA, Dyess DL, Welch DR, Samant RS, Shevde LA. Epigenetic silencing contributes to the loss of BRMS1 expression in breast cancer. Clin Exp Metastasis. 2008;25:753–763. doi: 10.1007/s10585-008-9187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]