Abstract
Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis of multiple tumor types without blocking tumorigenesis. BRMS1 forms complexes with SIN3, histone deacetylases and selected transcription factors that modify metastasis-associated gene expression (e.g., EGFR, OPN, PI4P5K1A, PLAU). microRNA (miRNA) are a recently discovered class of regulatory, noncoding RNA, some of which are involved in neoplastic progression. Based on these data, we hypothesized that BRMS1 may also exert some of its antimetastatic effects by regulating miRNA expression. Micro-RNA arrays were done comparing small RNAs that were purified from metastatic MDA-MB-231 and MDA-MB-435 and their non-metastatic BRMS1-transfected counterparts. miRNA expression changed by BRMS1 were validated using SYBR Green RT-PCR. BRMS1 decreased metastasis-promoting (miR-10b, -373 and -520c) miRNA, with corresponding reduction of their downstream targets (e.g., RhoC which is downstream of miR-10b). Concurrently, BRMS1 increased expression of metastasis suppressing miRNA (miR-146a, -146b and -335). Collectively, these data show that BRMS1 coordinately regulates expression of multiple metastasis-associated miRNA and suggests that recruitment of BRMS1-containing SIN3:HDAC complexes to, as yet undefined, miRNA promoters might be involved in the regulation of cancer metastasis.
Keywords: metastasis suppression, microRNA, BRMS1
Gene regulation by microRNA (miRNA) is a conserved mechanism in animals and plants.1 Endogenous, miRNA range from 15 to 28 nucleotides (nt) in Homo sapiens and, most commonly, negatively regulate gene expression, although some gene expression is positively regulated by miRNA. miRNA acts as templates for RNA-induced silencing complexes (RISC) to target mRNA. Animal miRNA differs functionally from plant miRNA in that they have imperfect base pairing with their target mRNA and more commonly inhibit protein translation than degrade mRNA.2 Imperfect or promiscuous base pairing allows animal miRNA to target multiple mRNA or even entire cellular pathways.3,4 To date, miRNA has been shown to regulate multiple cellular processes or pathways critical for neoplastic transformation and progression,5 including apoptosis,6,7 cell cycle regulation,8 differentiation,4,9,10 immune function11 and metabolism.12,13 Up- and down-regulation of miRNA expression are correlated with development of multiple cancers,5,14 including breast carcinoma,15–17 and a growing number of miRNA also contribute to promotion and suppression of cancer invasion and metastasis.16,18–27
Metastasis suppressors, defined by their ability to suppress metastasis without blocking orthotopic tumor growth, are an expanding family of >25 molecules.28,29 Breast cancer metastasis suppressor 1 (BRMS1) inhibits breast, melanoma, nonsmall cell lung and ovarian cancer metastasis in xenograft and syngeneic models.30–36 Expression of BRMS1 protein,30,37 but not necessarily mRNA,38,39 expression generally correlates inversely with survival and development of metastasis. BRMS1 associates with SIN3:histone deacetylase complexes36,40,41 which are involved in chromatin structure and selective regulation of gene expression. Collectively, these factors lead to the hypothesis that BRMS1 suppresses metastasis by altering expression of metastasis-associated genes. Previous studies have identified selective regulation of the epidermal growth factor receptor,42 osteopontin,35,36,43,44 connexins 45 and urokinase plasminogen activator.46 We hypothesized that BRMS1 might also regulate recently discovered, metastasis-associated miRNA.
To determine whether BRMS1 regulates miRNA expression, we compared miRNA expression patterns in nonexpressing and BRMS1 re-expressing breast carcinoma cells. miRNA expression was compared using microarrays imprinted with 328 known human miR probes and a selected common subset was further validated using quantitative real-time PCR (RTQ). In this article, we report that BRMS1 alters miRNA expression in metastatic breast carcinoma cells and notably down-regulated 3 of the 4 published metastasis-promoting miRNA22,23,47 and up-regulated all 3 of the known metastasis-suppressing miRNA.16,24
Material and methods
Cell lines and cell culture
MDA-MB-231 (231) and MDA-MB-435 (435) are human estrogen receptor- and progesterone receptor-negative cell lines derived from pleural effusions of metastatic infiltrating ductal breast carcinomas.48,49 Both cell lines form progressively growing tumors when injected into the mammary fat pads of immunocompromised mice. MDA-MB-435 cells develop macroscopic metastases in lungs and regional lymph nodes by 10–12 weeks postinoculation, but infrequently metastasize after direct injection into the lateral tail vein or following subcutaneous injection. In contrast, MDA-MB-231 cells form macroscopic metastases when injected intravenously, but less commonly following injection into an orthotopic site. Both lines form osteolytic metastases following injection into the left ventricle of the heart.31,50–52 The origin of 435 has been questioned,53 however, this does not affect the interpretation of the results since BRMS1 suppresses metastasis of tumor cell lines from multiple tissue origins.33,34
Parental cell lines were cultured in a mixture (1:1 v:v) of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s-F12 medium (DMEM/F12; Invitrogen, Carlsbad, CA) supplemented with 2 mM L-glutamine, 0.02 mM nonessential amino acids and 5% fetal bovine serum (Invitrogen). All cultures were maintained without antibiotics or antimycotics on 100-mm tissue culture dishes (Corning, Corning, NY) at 37°C with 5% CO2 in a humidified atmosphere. When cultures reached 80–90% confluence they were passaged using a solution of 2 mM EDTA in Ca2+/Mg2+-free Dulbecco’s phosphate buffered saline (CMF-DPBS; Invitrogen). All cultures were regularly tested and confirmed negative for Mycoplasma spp. infection using a PCR-based test (TaKaRa, Shiga, Japan).
Constructs and transductions
Cells (231 and 435) were transduced with GFP to facilitate tracking of cells in vivo (231GFP/435GFP) as previously described.50 Full-length BRMS1 cDNA was cloned into lentiviral constructs and transduced into 231GFP and 435GFP cells. Single cell clones (231BRMS1 and 435BRMS1) were isolated and BRMS1 mRNA and protein expression were verified. Transduced cells were initially selected with puromycin (500 μg/ml) and maintained in puromycin (100 μg/ml) to ensure stable transduction. For routine culture, no antibiotic selection was used and expression has been verified to be stable for more than 2 years.
MicroRNA arrays
Cells were grown to 90% confluence, media aspirated, washed in ice-cold PBS and lysed in acid phenol:chloroform (Ambion, Austin, TX). Small RNA species (≤200 nt) were isolated using mirVana PARIS kit (Ambion) according to manufacturer’s instructions and immediately stored at −80°C. RNA quantification was performed with use of a DU 800 spectrophotometer (Beckman Coulter, Fullerton, CA). Three independently isolated samples were collected for every cell line.
miRNA array profiling was performed at the Vanderbilt Microarray Shared Resource according to the core’s standard operating procedures (http://array.mc.vanderbilt.edu/microarray/expr/protocol.vmsr) as summarized later. RNA species (10–40 nt) were enriched using the flashPAGE fractionater (Ambion). The 3′ ends were polyadenylated with modified amines; RNA from 231GFP and 435GFP were labeled with Cy5 whereas RNA from 231BRMS1 and 435BRMS1 cells were labeled with Cy3. The yields on the coupling reactions were typical of miRNA labeling reactions with no detectable CyDye, but a large increase in measurable RNA indicated that poly A polymerase was effective in adding the modified dNTPs to the miRNA (data not shown). RNA samples were suspended in 3X Hyb™ buffer (Ambion), heated for 95°C for 2 min, allowed to cool at room temperature for 1 min and then loaded onto mirVana miRNA bioarrays v2 (Ambion) with Maui DC cover slips (BioMicro Systems, Salt Lake City, UT). Hybridizations were performed using the Maui hybridization station at 42°C for 16 hr with Mix D settings in Maui A1-A3. Arrays were washed once in Ambion SlideHyb™ low-stringency buffer for 30 sec with mixing, followed by 2 Ambion SlideHyb™ high-stringency buffer washes for 30 sec with mixing. Arrays were spun dry and scanned on the AXON 4000B scanner (Molecular Devices, Sunnyvale, CA).
Raw GeneChip files from GeneChip Operating Software (GCOS, Affymetrix, CA) were uploaded and background was subtracted. Expression changes were normalized to the respective control cells (231GFP or 435GFP) to calculate the intensity ratio/fold changes of the BRMS1-expressing counterparts (Cy5/Cy3). Data were sorted from greatest to least intensity. miR spots with foreground intensities less than 150 were disregarded as signals were too near background median. miR with values of −50 and −100 were also excluded as spot irregularity-affected fluorescence.
Raw data from Axon were background-subtracted, normalized to the respective control cells (231GFP or 435GFP) to calculate the intensity ratio/fold changes of the BRMS1-expressing counterparts (Cy5/Cy3). miR spots with foreground intensities less than 150 were disregarded as signals were too near background median. miR with values of −50 and −100 were also excluded as spot irregularity-affected fluorescence.
Antibodies and immunoblotting
BRMS1 monoclonal antibody clone 1a5.7 was used at 1:2,500 and was previously described.36,54 Other antibodies used in this study were purchased and used at the titer indicated: anti-Twist1 (1:1,000), anti-α tubulin (1:1,000) and anti-EGFR (1:1,000; Cell Signaling Technology, Danvers, MA), anti-HoxD10 E20 (1:200; Santa Cruz Biotechnology, Santa Cruz, CA), anti-RhoC (1:1,000; Abcam, Cambridge, MA). Western blotting was performed as previously described.36,54
Validation using real-time quantitative PCR
miRNA expression was determined by collecting total RNA from 70 to 90% confluent cell cultures using Qiazol (Qiagen Inc, Valencia, CA). RNA was purified using miRNeasy (Qiagen Inc). MiScript primers (Qiagen Inc), designed to amplify specifically mature miRNA (i.e., not pre-miR or pri-miR), were as follows: hsa-miR-10b_2, miR-96_1, hsa-miR-30e3p_1, hsa-miR-30e5p, hsa-miR-151, hsa-miR-320, hsa-miR-335_1, hsa-miR-373_1 and hsa-miR-520c. All samples were normalized to small nuclear RNA U6 and fold changes were calculated as previously described.55
Results and discussion
Small RNAs (≤40 nt) were enriched from metastatic 231GFP, 435GFP and metastasis-suppressed BRMS1-transduced isogenic counterparts. BRMS1-associated mature miRNA expression changes were determined using miR-microarrays in 3 independent experiments. The top 25 consistently increased and decreased mature miRNA as a result of BRMS1 re-expression from both cell line comparisons are listed in Table I. The arrays revealed changes in mature miRNA expression that could contribute to BRMS1 metastasis suppression. For example, the oncogenic miR-155 decreased in 435 cells and tumor suppressing let-7a increased in 231 cells; however, as the arrays were used as a screen for miRNA changes observed in both cell lines, many miRNA were excluded from further study in this report. In general, the direction of miRNA expression change was consistent during the validations, but the magnitude of the changes was often under-appreciated on the arrays.
TABLE I.
TOP 25 MOST COMMONLY REGULATED MIR FOLLOWING RE-EXPRESSION OF BRMS1
| Cell line | Direction change | microRNA | Cy5:Cy3 ratio | Locus | Accession no. Sanger database |
|---|---|---|---|---|---|
| MDA-MB-231 | DOWN | hsa_miR_200a_AS | 62.190 | 1p36 | MIMAT0001620 |
| hsa_miR_200b | 16.213 | 1p36 | MIMAT0000318 | ||
| hsa_miR_380_3p | 12.441 | 14q32 | MIMAT0000735 | ||
| hsa_miR_326 | 8.200 | 11q13 | MIMAT0000756 | ||
| hsa_miR_483 | 6.864 | 11p15 | MIMAT0002173 | ||
| hsa_miR_210 | 3.964 | 11p15 | MIMAT0000267 | ||
| hsa_miR_99b | 3.580 | 19q13 | MIMAT0000689 | ||
| hsa_miR_330 | 2.945 | 19q13 | MIMAT0000751 | ||
| hsa_miR_324_5p | 2.860 | 17p13 | MIMAT0000761 | ||
| hsa_miR_324_3p | 2.769 | 17p13 | MIMAT0000762 | ||
| hsa_miR_320 | 2.684 | 8p21 | MIMAT0000510 | ||
| hsa_miR_520h | 2.621 | 19q13 | MIMAT0002867 | ||
| hsa_miR_149 | 2.614 | 2q37 | MIMAT0000450 | ||
| hsa_miR_423 | 2.553 | 17q11 | MIMAT0001340 | ||
| hsa_miR_425 | 2.552 | 3p21 | MIMAT0001343 | ||
| hsa_miR_501 | 2.478 | Xp11 | MIMAT0002872 | ||
| hsa_miR_452 | 2.430 | Xq28 | MIMAT0001635 | ||
| hsa_miR_511 | 2.414 | 10p12 | MIMAT0002808 | ||
| hsa_miR_339 | 2.336 | 7p22 | MIMAT0000764 | ||
| hsa_miR_151 | 2.299 | 8q24 | MIMAT0000757 | ||
| hsa_miR_491 | 2.126 | 9p21 | MIMAT0002807 | ||
| hsa_miR_362 | 2.064 | Xp11 | MIMAT0000705 | ||
| hsa_miR_17_3p | 2.031 | 13q31 | MIMAT0000071 | ||
| hsa_miR_18a_AS | 1.969 | 13q31 | MIMAT0002891 | ||
| MDA-MB-231 | UP | hsa_let_7a | 0.295 | 22q13 | MIMAT0000062 |
| hsa_let_7f | 0.328 | 9q22 | MIMAT0000067 | ||
| hsa_miR_128b | 0.358 | 3p22 | MIMAT0000676 | ||
| hsa_miR_30c | 0.370 | 6q13 | MIMAT0000244 | ||
| hsa_let_7g | 0.384 | 3p21 | MIMAT0000414 | ||
| hsa_miR_30a_3p | 0.385 | 6q13 | MIMAT0000088 | ||
| hsa_miR_21 | 0.386 | 17q22 | MIMAT0000076 | ||
| hsa_miR_10a | 0.396 | 17q21 | MIMAT0000253 | ||
| hsa_miR_128a | 0.408 | 2q21 | MIMAT0000424 | ||
| hsa_miR_146b | 0.429 | 10q21 | MIMAT0002809 | ||
| hsa_miR_181a | 0.434 | 1q31 | MIMAT0000256 | ||
| hsa_miR_182 | 0.434 | 7q32 | MIMAT0000259 | ||
| hsa_miR_96 | 0.435 | 7q32 | MIMAT0000095 | ||
| hsa_miR_20a | 0.436 | 13q31 | MIMAT0000075 | ||
| hsa_miR_200c | 0.436 | 12p13 | MIMAT0000617 | ||
| hsa_miR_26a | 0.463 | 3p22 | MIMAT0000082 | ||
| hsa_let_7i | 0.470 | 12q14 | MIMAT0000415 | ||
| hsa_miR_15b | 0.504 | 3q25 | MIMAT0000417 | ||
| hsa_miR_148a | 0.510 | 7p15 | MIMAT0000243 | ||
| hsa_miR_30a_5p | 0.515 | 6q13 | MIMAT0000087 | ||
| hsa_miR_15a | 0.518 | 13q14 | MIMAT0000068 | ||
| hsa_miR_27a | 0.520 | 19p13 | MIMAT0000084 | ||
| hsa_miR_30e_5p | 0.527 | 1p34 | MIMAT0000692 | ||
| hsa_miR_16 | 0.540 | 13q14 | MIMAT0000069 | ||
| hsa_miR_142_5p | 0.541 | 17q22 | MIMAT0000433 | ||
| MDA-MB-435 | DOWN | hsa_miR_34a | 102.208 | 1p36 | MIMAT0000255 |
| hsa_miR_126 | 99.692 | 9q34 | MIMAT0000445 | ||
| hsa_miR_224 | 53.970 | Xq28 | MIMAT0000281 | ||
| hsa_let_7b | 51.088 | 22q13 | MIMAT0000063 | ||
| hsa_miR_155 | 50.785 | 21q21 | MIMAT0000646 | ||
| hsa_miR_139 | 31.486 | 11q13 | MIMAT0000250 | ||
| hsa_miR_199a | 28.600 | 19p13 | MIMAT0000231 | ||
| hsa_miR_452 | 25.500 | Xq28 | MIMAT0001635 | ||
| hsa_miR_148a | 24.778 | 7p15 | MIMAT0000243 | ||
| hsa_miR_200b | 20.446 | 1p36 | MIMAT0000318 | ||
| hsa_miR_222 | 16.220 | Xp11 | MIMAT0000279 | ||
| hsa_miR_200c | 12.161 | 12p13 | MIMAT0000617 | ||
| hsa_miR_380_3p | 11.759 | 14q32 | MIMAT0000735 | ||
| hsa_miR_503 | 7.627 | Xq26 | MIMAT0002874 | ||
| hsa_miR_138 | 7.141 | 3p21 | MIMAT0000430 | ||
| hsa_miR_485_5p | 6.773 | 14q32 | MIMAT0002175 | ||
| hsa_let_7f | 6.644 | 9q22 | MIMAT0000067 | ||
| hsa_miR_21 | 6.037 | 17q22 | MIMAT0000076 | ||
| hsa_miR_152 | 5.913 | 17q21 | MIMAT0000438 | ||
| hsa_miR_125b | 5.000 | 11q24 | MIMAT0000423 | ||
| hsa_miR_100 | 4.076 | 11q24 | MIMAT0000098 | ||
| hsa_miR_210 | 4.040 | 11p15 | MIMAT0000267 | ||
| hsa_miR_520h | 4.031 | 19q13 | MIMAT0002867 | ||
| hsa_miR_221 | 3.980 | Xp11 | MIMAT0000278 | ||
| MDA-MB-435 | UP | hsa_miR_146a | 0.001 | 5q33 | MIMAT0000449 |
| hsa_miR_363 | 0.001 | Xq26 | MIMAT0000707 | ||
| hsa_miR_506 | 0.002 | Xq27 | MIMAT0002878 | ||
| hsa_miR_508 | 0.003 | Xq27 | MIMAT0002880 | ||
| hsa_miR_509 | 0.003 | Xq27 | MIMAT0002881 | ||
| hsa_miR_513 | 0.003 | Xq27 | MIMAT0002877 | ||
| hsa_miR_31 | 0.004 | 9p21 | MIMAT0000089 | ||
| hsa_miR_105 | 0.006 | Xq25 | MIMAT0000102 | ||
| hsa_miR_211 | 0.009 | 15q13 | MIMAT0000268 | ||
| hsa_miR_510 | 0.013 | Xq27 | MIMAT0002882 | ||
| hsa_miR_20b | 0.016 | Xq26 | MIMAT0001413 | ||
| hsa_miR_146b | 0.025 | 10q24 | MIMAT0002809 | ||
| hsa_miR_9_AS | 0.028 | 1q22 | MIMAT0000442 | ||
| hsa_miR_204 | 0.032 | 9q21 | MIMAT0000265 | ||
| hsa_miR_501 | 0.034 | Xp11 | MIMAT0002872 | ||
| hsa_miR_500 | 0.038 | Xp11 | MIMAT0002871 | ||
| hsa_miR_502 | 0.042 | Xp11 | MIMAT0002873 | ||
| hsa_miR_362 | 0.045 | Xp11 | MIMAT0000705 | ||
| hsa_miR_188 | 0.061 | Xp11 | MIMAT0000457 | ||
| hsa_miR_185 | 0.070 | 22q11 | MIMAT0000455 | ||
| hsa_miR_497 | 0.079 | 17p13 | MIMAT0002820 | ||
| hsa_miR_196a | 0.087 | 17q21 | MIMAT0000226 | ||
| hsa_miR_195 | 0.089 | 17p13 | MIMAT0000461 | ||
| hsa_miR_96 | 0.182 | 7q32 | MIMAT0000095 |
Small RNAs were isolated from cell lines not expressing (MDA-MB-231 or MDA-MB-435) or expressing BRMS1, labeled with Cy5 or Cy3 and hybridized to arrays containing 328 human microRNA. Arrays were performed 3 times in independent experiments. Only consistent changes are shown in this table. Sanger database v12: http://microrna.sanger.ac.uk/sequences/. Subsequent to the evaluation of miR expression on the microarrays, homologs of some miR have been submitted to the Sanger databases. The chromosomal locations provided in this table correspond to the first miR deposited into the database.
The criteria for prioritizing candidates were as follows: (i) technical replicates on the arrays were consistent; (ii) the direction of miRNA expression change was consistent in both 231/231BRMS1 and 435/435BRMS1 cell pairs; (iii) miRNA that target metastasis-associated mRNA (or proteins); and (iv) miRNA previously demonstrated to alter phenotypes associated with invasion and/or metastasis. Based on these criteria 6 miRNA were initially selected for further follow-up: miR-10b, -30e-3p, -30e-5p, -96, -151, -339 (Fig. 1).
Figure 1.
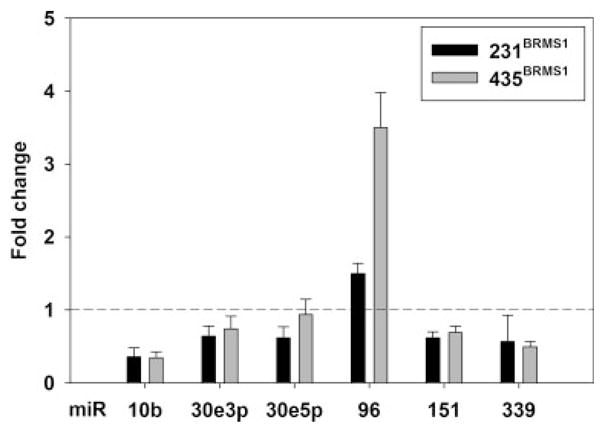
BRMS1 re-expression changes miR levels in metastasis suppressed breast cancer cells. Total RNA was extracted from 231/231BRMS1 and 435/435BRMS1 cell lines, reverse transcribed and miR assessed using SYBR-green real-time RT-PCR.
Our original goals were to determine whether BRMS1 regulates miRNA and, if so, whether BRMS1-regulated miRNAs are downstream mediators of BRMS1 metastasis suppression. However, as these results were being collected, other laboratories reported on miRNA regulation of invasion and/or metastasis,16,22,23 compelling more detailed analysis of our data with regard to BRMS1 regulation of those miRNA (Table II).
TABLE II.
SUMMARY OF METASTASIS-ASSOCIATED MIR CHANGES FOLLOWING BRMS1 RE-EXPRESSION
| miR | Accession no. | Change |
Reported role(s) of miR in tumor progression | References | |
|---|---|---|---|---|---|
| 231BRMS1:231 | 435BRMS1:435 | ||||
| Decrease | |||||
| 10b | MI0000267 | 0.36 | 0.35 | Increased migration, invasion, metastasis | 23 |
| 373 | MI0000781 | 0.53 | 0.69 | Increased migration, invasion, metastasis | 22 |
| 520c | MI0007801 | 0.59 | 0.65 | Increased migration, invasion, metastasis | 22 |
| Increase | |||||
| 146a | MI0000477 | 6 | 64 | Decreased invasion, metastasis | 24,25 |
| 146b | MI0003129 | 0.6 | 42 | Decreased invasion, metastasis | 24,25 |
| 335 | MI0000816 | 6 | 11 | Decreased migration, metastasis | 16 |
| 21 | MI0000077 | 2.1 | 1.1 | Decreased migration, metastasis | 46,59 |
The first miRNA validated to promote metastasis was miR-10b.23 Knockdown of miR-10b in 231 cells decreased in vitro migration and invasion. Additionally, ectopic expression of miR-10b in HMEC and SUM149 cells promoted in vitro migration and invasion. In addition, there was increased dissemination to the lung and formation of macroscopic foci in the peritoneum by SUM159 cells.23 Correspondingly, miR-10b expression was suppressed in 231BRMS1 and 435BRMS1 cells by greater than 50%, which is consistent with prior data showing that BRMS1 suppresses invasion and metastasis. Furthermore, when miR-10b expression was examined in a panel of cells denoting breast cancer progression, BRMS1 reduced miR-10b levels toward those found in nonmetastatic cells (Fig. 2).
Figure 2.
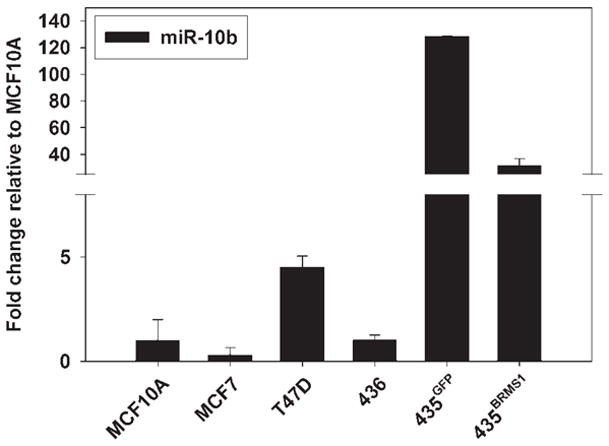
BRMS1 restores miR-10b levels to those found in non-metastatic cells. A panel of breast cancer cell lines was evaluated for miR-10b expression. Total RNA was extracted, reverse transcribed and miR assessed using SYBR-green real-time RT-PCR.
Ma et al.23 further showed that miR-10b down-regulated HoxD10 protein which led to an increased expression of RhoC, which is a positive regulator of metastasis.56–58 Although RhoC mRNA expression decreased in both 231BRMS1 and 435BRMS1 cells (Fig. 3), HoxD10 mRNA increased in 435BRMS1 cells but decreased in 231BRMS1 cells. The differences observed when comparing cell lines suggest a nonlinear pathway and/or other (i.e., non-HoxD10) mediators of miR-10b. Because we were unable to detect HoxD10 with the commercially available antibodies, we measured mRNA. The inconsistency of mRNA levels between cell lines is in agreement with the findings reported by Ma et al.23
Figure 3.
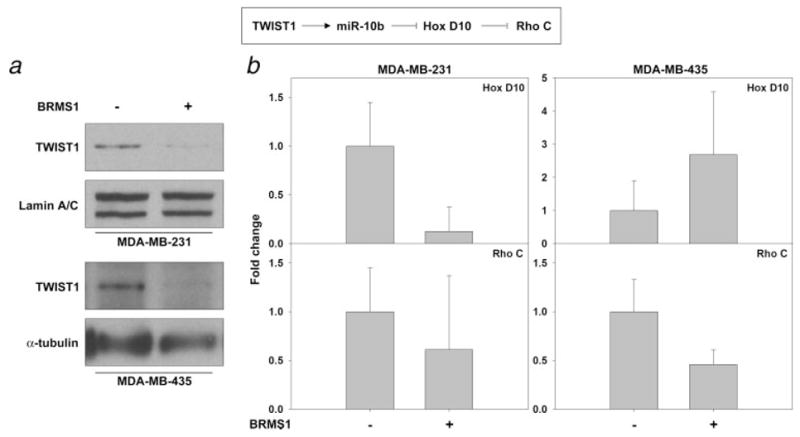
BRMS1 decreases a prometastatic pathway. (a) Whole cell and nuclear fractions were obtained and probed for TWIST1. α-tubulin and lamin A/C were used as loading controls for the whole cell and nuclear fractions, respectively. (b) Total RNA was collected from 231/231BRMS1 and 435/435BRMS1 cell lines. RNA (3 μl) was reverse transcribed and messages detected using Taqman primer probes for RhoC and HoxD10. Results are given as fold change relative to non-BRMS1 expressing 231 and 435 cells ± S.E.M. Ribosomal S9 was used as a normalization control for equal loading.
Relatedly, Huang et al.22 utilized a library of miR-transduced into human breast cancer cells to identify which miRNA promoted invasion in vitro. miR-373 and miR-520c stimulated migration and invasion in vitro and metastasis in vivo. Expression of miR-373 and -520c was decreased in 231BRMS1 and 435BRMS1 cells (Fig. 4 and Table II).
Figure 4.
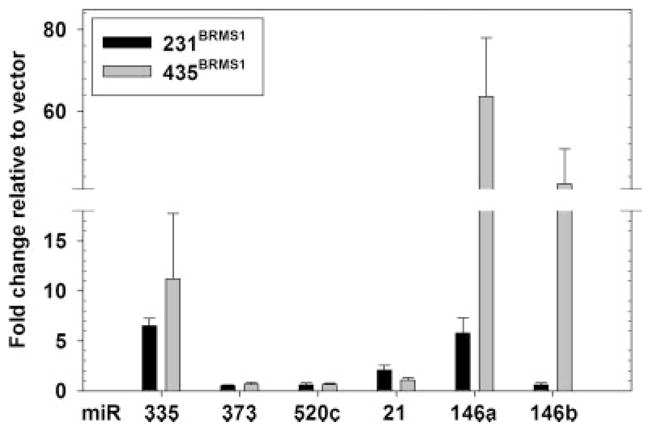
BRMS1 decreases prometastatic and increases metastasis-suppressing miR in breast cancer cells. RNA was collected using Qiazol and the miRNeasy kits. Purified RNA (6 μl) was reverse transcribed and then amplified usin miR specific primer probes. Relative amounts were calculated using the comparative cycle threshold (CT) method. The miR amounts were normalized to small nuclear RNA-U6. Results are given as fold change relative to non-BRMS1 expressing 231 and 435 cells ± S.E.M.
Tavazoie et al.16 compared miRNA expression patterns in paired sets of 231 variants which showed preferential organ-selective metastasis patterns. miR-335, when over-expressed by 250-fold, significantly suppressed lung and bone metastasis. Re-expression of BRMS1 suppressed metastasis to lungs and bone31,34,36 and increased expression of the metastasis-suppressing miR-335 by 6-fold (Fig. 4 and Table II). In the same report, miR-126 was described as metastasis suppressing16; however, it also suppressed orthotopic tumor growth, thereby excluding it as a metastasis suppressor.
Classically metastasis suppressor genes have encoded proteins; however, the demonstration that miR-335 and miR-146a/b suppress metastasis without blocking primary tumor growth shows that the types of molecules considered to be regulating metastasis needs to be more inclusive. Thus far, few pathways have been linked between known metastasis suppressors. In this article, we show for the first time that BRMS1 coordinately regulates expression of multiple miRNA and their corresponding downstream targets [e.g., RhoC (this report) and epidermal growth factor receptor42]. The findings are consistent with the emerging awareness that metastasis involves simultaneous control of multiple genes. Our observations beg the question as to whether other metastasis suppressors also mediate some or all of their affects through miRNA regulation.
Prior studies relating miRNA and metastasis have focused attention primarily toward identifying miRNA targets. Comparatively little is known about miRNA regulation.59,60 Ma et al.23 identified TWIST1, a major regulator of epithelial-mesenchymal transition,61 as a promoter of miR-10b expression.23 We discovered that BRMS1 decreases TWIST expression in whole cell lysates as well as nuclear fractions (Fig. 3). Moreover, BRMS1 mutants that do not suppress metastasis fail to down-regulate TWIST1 in 231 cells. Whether BRMS1 directly binds to the miR-10b promoter is still not known. But it is conceivable that BRMS1 might be part of a corepressor complex(es) recruited to miR-10b because BRMS1 is a component of several SIN3:histone deacetylase complexes.36,41 Similarly, the mechanism(s) by which regulation of other miRNA occurs will be the object of intense future investigation. Given the decrease in such a prominent epithelial-to-mesenchymal transition (EMT) regulator (Twist1), one could speculate that BRMS1 suppresses metastasis by invoking the reverse process (mesenchymal-to-epithelial transition, MET) in metastatic cells; however, some published and unpublished data argue against this possibility. Several recent publications indicate that miR-200 family are associated with increased expression of E-cadherin,20,21,26,62,63 which is typically lost during EMT.64 Data presented in Table I show decreases in miR-200 family members when BRMS1 is expressed. This is the opposite of what would be predicted. Moreover, changes in miR-200c are not consistent between 231 and 435 cells when BRMS1 is re-expressed. Also, we have previously reported that BRMS1 re-expression has no effect on E-cadherin mRNA or protein expression.65 Finally, BRMS1 consistently increases the vimentin expression, which is opposite of what one would expect if BRMS1 were regulating metastasis solely by regulating EMT.
Some miRNA expression changes observed in BRMS1-expressing 231 and 435 cells are large (miR-10b; 0.35/0.36, respectively), whereas others are more subtle (miR-373; 0.69/0.53 and miR-520c; 0.65/0.59, respectively). Nonetheless, it is interesting that nearly all (6 of 7; 86%) of the known metastasis-associated miRNA are regulated in the direction predicted for a metastasis suppressor. The exception was miR-21 which promoted metastasis in breast47 and colorectal cancer66 cells. BRMS1 increased miR-21 expression in 231 by ~ 2-fold, but did not change in 435. It is quite possible that in 231 cells, the combined effects changing expression of the other metastasis-associated miRNA may overwhelm the miR-21 up-regulation or the miR-21 expression change may be an attempt by cells to compensate for other changes.
Among the many miRNA changes observed in BRMS1-expressing cells, miR-146a and -146b expression were increased. Coupled with our prior reports that BRMS1 inhibits NFκB activity46 and EGFR expression,42 NFκB regulation by miR-146a/b25,67 and demonstration that EGFR is a target sequence for miR-146a/b, we tested the hypothesis that miR-146 was a downstream mediator of BRMS1 metastasis suppression. Transfection of miR-146a or miR-146b into 231 cells resulted in a significant suppression of lung metastases in experimental xenograft models.24 We emphasize that, unlike prior studies testing the role of miRNA in the processes of invasion and metastasis, the data reported here are based upon near physiologic expression levels of BRMS1 and the corresponding downstream miRNA. This point is key because off-target effects are more likely if experiments involve gross over-expression.
In summary, we demonstrate that BRMS1 regulates virtually all of the known miRNA regulating invasion and/or metastasis. Given that a single metastasis suppressor regulates several key metastasis-associated miRNA, future studies should focus on which step(s) of the metastatic cascade are dysregulated in human disease and the specific miRNA expression regulatory elements that are regulated by BRMS1 directly.
Acknowledgments
This work was supported by U.S. Public Health Service Grants CA87728 (D.R.W.), CA13148 (D.C.), UL1RR025777 (D.C.) and F32CA113037 (D.R.H.); a predoctoral fellowship from the U.S. Army Medical Research and Materiel Command W81-XWH-08-1-0786 (M.D.E.); a postdoctoral fellowship from Susan G. Komen for the Cure PDF1122006 (K.S.V.) and a grant from the National Foundation for Cancer Research—Center for Metastasis Research (D.R.W.). This work is submitted in partial fulfilment of the requirements of the Molecular and Cellular Pathology Graduate Program (M.D.E.). The authors thank Dr. Janet Price (University of Texas M.D. Anderson Cancer Center) for providing the MDA-MB-231 and -435 cell lines. They also thank the Vanderbilt Microarray Core Facility for superb technical assistance with the microRNA arrays. Author contributions: M.D.E. and D.R.W. designed research; M.D.E., D.R.H., K.S.V. and L.J.S. performed research; M.D.E., D.R.H., K.S.V., D.C. and D.R.W. analyzed data; D.C. contributed new analytical tools and M.D.E., D.R.H., K.S.V. and D.R.W. wrote the article.
Grant sponsor: U.S. Public Health Service Grants; Grant numbers: CA87728, CA13148, UL1RR025777, F32CA113037; Grant sponsor: U.S. Army Medical Research and Materiel Command; Grant number: W81-XWH-08-1-0786; Grant sponsor: Susan G. Komen for the Cure; Grant number: PDF1122006; Grant sponsor: National Foundation for Cancer Research—Center for Metastasis Research
Abbreviations
- BRMS1
breast cancer metastasis suppressor 1
- CMF-DPBS
calcium- and magnesium-free Dulbecco’s phosphate buffered saline
- Cy
cyanine
- DMEM
Dulbecco’s modified Eagle’s medium
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- hsa
Homo sapiens
- MET
mesenchymal-to-epithelial transition (reverse of EMT)
- miR
microRNA
- miRNA
microRNA
- nt
nucleotide
- RTQ
real-time quantitative PCR
- TTBS
Tween-20 containing Tris buffered saline
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
References
- 1.Tang GL, Tang XQ, Mendu V, Tang XH, Jia XY, Chen QJ, He LH. The art of microRNA: various strategies leading to gene silencing via an ancient pathway. BBA-Gene Regul Mech. 2008;1779:655–62. doi: 10.1016/j.bbagrm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Van den Berg A, Mols J, Han JH. RISC-target interaction: cleavage and translational suppression. BBA-Gene Regul Mech. 2008;1779:668–77. doi: 10.1016/j.bbagrm.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T, Mor G. Regulation of IKK beta by miR-199a affects NF-κ B activity in ovarian cancer cells. Oncogene. 2008;27:4712–23. doi: 10.1038/onc.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 8.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–32. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 9.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 10.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–51. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Shyu AB, Wilkinson MF, Van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–81. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–89. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 14.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–77. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 15.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AWM, Klijn JGM, Wiemer EAC, Martens JWM. Four miR-NAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–6. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang QQ, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 18.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–84. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 20.Korpal M, Kang YB. The emerging role of miR-200 family of micro-RNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–19. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The mir-200 family and mir-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 22.Huang QH, Gumireddy K, Schrier M, LeSage C, Nagel R, Nair S, Egan DA, Li AP, Huang GH, Klein-Szanto AJ, Gimotty PA, Katsaros D, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 24.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 BRMS1 up-regulates miR-146 that suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–83. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-κB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;42:5643–7. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 27.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. Micro-RNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 28.Bodenstine TM, Welch DR. Metastasis suppressors and the tumor microenvironment. Cancer Microenviron. 2008;1:1–11. doi: 10.1007/s12307-008-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–91. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Smith PW, Liu Y, Siefert SA, Moskaluk CA, Petroni GR, Jones DR. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276:196–203. doi: 10.1016/j.canlet.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phadke PA, Vaidya KS, Nash KT, Hurst DR, Welch DR. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol. 2008;172:809–17. doi: 10.2353/ajpath.2008.070772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene. BRMS1. Int J Gynecol Cancer. 2006;16:522–31. doi: 10.1111/j.1525-1438.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- 33.Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene. BRMS1. Exp Cell Res. 2002;273:229–39. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- 34.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor. BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–9. [PubMed] [Google Scholar]
- 35.Hedley BD, Welch DR, Allan AL, Al-Katib W, Dales DW, Postenka CO, Casey G, MacDonald IC, Chambers AF. Down-regulation of osteopontin contributes to metastasis suppression by breast cancer metastasis suppressor 1. Int J Cancer. 2008;123:526–34. doi: 10.1002/ijc.23542. [DOI] [PubMed] [Google Scholar]
- 36.Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC, Welch DR. Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–44. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri T, Tubbs RR, et al. Loss of BRMS1 protein expression predicts reduced disease-free survival in hormone receptor negative and HER2 positive subsets of breast cancer. Clin Cancer Res. 2006;12:6702–8. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lombardi G, Di Cristofano C, Capodanno A, Iorio MC, Aretini P, Isola P, Tancredi M, Collecchi P, Naccarato AG, Porta RP, Bevilacqua G, Caligo MA. High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int J Cancer. 2006;120:1169–78. doi: 10.1002/ijc.22379. [DOI] [PubMed] [Google Scholar]
- 39.Kelly LM, Buggy Y, Hill A, O’Donovan N, Duggan C, McDermott EW, O’Higgins NJ, Young L, Duffy MJ. Expression of the breast cancer metastasis suppressor gene. BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumor Biol. 2005;26:213–16. doi: 10.1159/000086955. [DOI] [PubMed] [Google Scholar]
- 40.Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA, Shevde LA, Hopper JE, Xie Y, Welch DR, Samant RS. Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Commun. 2006;348:1429–35. doi: 10.1016/j.bbrc.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–9. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- 42.Vaidya KS, Harihar S, Stafford LJ, Hurst DR, Hicks DG, Casey G, DeWald DB, Welch DR. Breast cancer metastasis suppressor-1 differentially modulates growth factor signaling. J Biol Chem. 2008;283:28354–60. doi: 10.1074/jbc.M710068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR, Shevde LA. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-κB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shevde LA, Samant RS, Paik JC, Metge BJ, Chambers AF, Casey G, Frost AR, Welch DR. Osteopontin knockdown suppresses tumorigenicity of human metastatic breast carcinoma. Clin Exp Metastasis. 2006;23:123–33. doi: 10.1007/s10585-006-9013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saunders MM, Seraj MJ, Li ZY, Zhou ZY, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–7. [PubMed] [Google Scholar]
- 46.Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-kB activity. Cancer Res. 2005;65:3586–95. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- 47.Zhu SM, Wu HL, Wu FT, Nie DT, Sheng SJ, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 48.Cailleau R, Young R, Olive M, Reeves WJ. Breast tumor cell lines from pleural effusions. J Natl Cancer Inst. 1974;53:661–74. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cailleau R, Olive M, Cruciger QVJ. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14:911–15. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- 50.Phadke PA, Mercer RR, Harms JF, Jia Y, Frost AR, Jewell J, Bussard KM, Nelson S, Moore C, Kappes JC, Gay CV, Mastro AM, et al. Kinetics of metastatic breast cancer cell trafficking in bone. Clin Cancer Res. 2006;12:1431–40. doi: 10.1158/1078-0432.CCR-05-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koblinski JE, Kaplan-Singer BR, VanOsdol SJ, Wu M, Engbring JA, Wang S, Goldsmith CM, Piper JT, Vostal JG, Harms JF, Welch DR, Kleinman HK. Endogenous osteonectin/SPARC/BM-40 expression inhibits MDA-MB-231 breast cancer cell metastasis. Cancer Res. 2005;65:7370–7. doi: 10.1158/0008-5472.CAN-05-0807. [DOI] [PubMed] [Google Scholar]
- 52.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-β signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 Melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2006;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 54.Hurst DR, Xie Y, Edmonds MD, Welch DR. Multiple forms of BRMS1 are differentially expressed in the MCF10 isogenic breast cancer progression model. Clin Exp Metastasis. 2009;26:89–96. doi: 10.1007/s10585-008-9216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iiizumi M, Bandyopadhyay S, Pai SK, Watabe M, Hirota S, Hosobe S, Tsukada T, Miura K, Saito K, Furuta E, Liu W, Xing F, et al. RhoC promotes metastasis via activation of the Pyk2 pathway in prostate cancer. Cancer Res. 2008;68:7613–20. doi: 10.1158/0008-5472.CAN-07-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu M, Wu ZF, Kumar-Sinha C, Chinnaiyan A, Merajver SD. RhoC induces differential expression of genes involved in invasion and metastasis in MCF10A breast cells. Breast Cancer Res Treat. 2004;84:3–12. doi: 10.1023/B:BREA.0000018426.76893.21. [DOI] [PubMed] [Google Scholar]
- 58.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 59.Ozsolak F, Poling LL, Wang ZX, Liu H, Liu XS, Roeder RG, Zhang XM, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson PT, Wang WX, Wilfred BR, Tang GL. Technical variables in high-throughput miRNA expression profiling: Much work remains to be done. BBA-Gene Regul Mech. 2008;1779:758–65. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Korpal M, Lee ES, Hu GH, Kang YB. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 65.Samant RS, Seraj MJ, Saunders MM, Sakamaki T, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon LR, Meehan WJ, Winter CR, Christensen ND, et al. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2001;18:683–93. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- 66.Asangani IA, Rasheed SAK, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 67.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]


