Abstract
Leucine-rich repeats and immunoglobulin-like domains 3 (Lrig3) was identified by microarray analysis among genes that show differential expression during gastrulation in Xenopus laevis. Lrig3 was expressed in the neural plate and neural crest (NC) at neurula stages, and in NC derivatives and other dorsal structures during tailbud stages. A prominent consequence of the morpholino-induced inhibition of Lrig3 expression was impaired NC formation, as revealed by the suppression of marker genes, including Slug, Sox9 and Foxd3. In the NC induction assay involving Chordin plus Wnt3a-injected animal caps, Lrig3 morpholino inhibited expression of Slug, Sox9 and Foxd3, but not of Pax3 and Zic1. In line with this, Lrig3 knockdown prevented NC marker induction by Pax3 and Zic1, suggesting that Lrig3 acts downstream of these two genes in NC formation. Injection of Lrig3 and Wnt3a led to low-level induction of NC markers and enhanced induction of Fgf3, Fgf4 and Fgf8 in animal caps, suggesting a positive role for Lrig3 in Wnt signaling. Lrig3 could attenuate Fgf signaling in animal caps, did interact with Fgf receptor 1 in cultured cells and, according to context, decreased or increased the induction of NC markers by Fgf. We suggest that Lrig3 functions in NC formation in Xenopus by modulating the Wnt and Fgf signaling pathways.
Keywords: Fgf8, MAPK, Neural crest, Slug, Wnt3a, Xenopus laevis, Leucine-rich repeats protein, Animal cap, DNA microarray
INTRODUCTION
The neural crest (NC) arises at the border between the neural plate and the epidermis, and represents multipotent cells that migrate to various parts of the embryo and differentiate into multiple cell types. Induction of the NC requires precisely balanced combinations of signals generated by the BMP, Wnt, Fgf, retinoic acid and Notch/Delta pathways (Cornell and Eisen, 2005; Huang and Saint-Jeannet, 2004; Steventon et al., 2005; Yanfeng et al., 2003). These signals lead to the activation of multiple transcription factors such as Msx1, Pax3 (Monsoro-Burq et al., 2005), Zic1 (Sato et al., 2005), Foxd3 (Kos et al., 2001; Pohl and Knochel, 2001; Sato et al., 2005), Sox9 (Spokony et al., 2002), Slug (Mayor et al., 1995), Snail (Essex et al., 1993; Mayor et al., 1993), Myc (Bellmeyer et al., 2003) and Ap2 (Luo et al., 2003). Gain- and loss-of-function experiments suggested tentative regulatory networks containing many of these factors (Monsoro-Burq et al., 2005; Saint-Jeannet et al., 1997; Sato et al., 2005).
In addition to NC formation, Fgf family members have crucial roles in at least three steps in embryogenesis, neural specification (Curran and Grainger, 2000; Hongo et al., 1999; Ribisi, Jr et al., 2000; Streit et al., 2000), posterior mesoderm formation (Amaya et al., 1993; Umbhauer et al., 1995) and cell migration (Chung et al., 2007; Yokota et al., 2003). Fgf signaling involves binding to one of four Fgf receptors (FGFRs), inducing activation of at least three downstream cascades: the PI3K-Akt, Raf-MEK1/2-ERK1/2 and PLCγ pathways (Eswarakumar et al., 2005; Schlessinger, 2004; Tsang and Dawid, 2004), the first two having been linked to mesoderm formation (Carballada et al., 2001; Umbhauer et al., 1995). Inhibition of the Fgf signaling by dominant negative Fgfr1 (Amaya et al., 1993), Sef (Tsang et al., 2002) or Mkp3 (Tsang et al., 2004) results in defects of axis formation. Knockdown of Fgf8 by antisense morpholino (MO) impairs mesoderm and NC formation (Fletcher et al., 2006; Hong and Saint-Jeannet, 2007; Monsoro-Burq et al., 2005). It is notable that in NC formation, Fgf signaling is essential, but an excess of Fgf signaling is inhibitory (Hong and Saint-Jeannet, 2007).
The Wnt/β-catenin pathway has been implicated in NC formation by gain- and loss-of-function studies in vivo and in explants (Abu-Elmagd et al., 2006; Garcia-Castro et al., 2002; Monsoro-Burq et al., 2005; Saint-Jeannet et al., 1997; Sato et al., 2005). In NC formation, Wnt signaling cooperates with other signal cascades, notably an attenuated BMP signal, in the regulation of the specification and differentiation process (LaBonne and Bronner-Fraser, 1998; Saint-Jeannet et al., 1997).
In this paper, we report that Lrig3, a single-pass transmembrane protein, is involved in NC formation by modulating FGF and Wnt signaling. We identified Lrig3 as a gene preferentially expressed in dorsal marginal zone explants from Xenopus gastrulae, using a DNA microarray approach. Among three human Lrig family members, Lrig1 has been shown to function as an EGF signaling inhibitor by enhancing EGFR ubiquitination (Gur et al., 2004; Laederich et al., 2004), and has also been linked to HGF signaling as a negative regulator (Shattuck et al., 2007). However, little is known about the function of Lrig family proteins in embryonic development. In this study, we determined that Lrig3 is required for NC formation in the Xenopus embryo.
MATERIALS AND METHODS
Embryo dissection and animal cap assay
Xenopus eggs were fertilized in vitro as described (Sive, 2000), embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1975). Animal caps were dissected at stage 9 in 1×MMR, and cultured in L-15 medium [67% L-15, 7.5 mM Tris-HCl (pH 7.5), 1 mg/ml BSA]. For microarray analysis, stage 10 Xenopus embryos were dissected into four explants. The dorsal marginal zone, composed mostly of endomesoderm, ventral marginal zone, animal cap and vegetal explant. To avoid possible cross contamination that could blur the microarray data, the junction regions between the explants were removed.
Microarray and data analysis
The dissected explants were homogenized in Stat 60 (TEL TEST), RNA was treated with DNase I and purified using RNeasy kit (Qiagen). Biotinylated probe was prepared from 100 ng total RNA using the OVATION RNA amplification system (Nugen Technologies). Probes were hybridized to Xenopus genome arrays (Affymetrix) according to the manufacture's instructions. Hybridized arrays were processed by the GeneChip Fluidics system (Affymetrix), and scanned in the GeneChip Scanner (Affymetrix). Gene expression profiles were analyzed by the GCOS software (Affymetrix).
DNA constructs
The ORF of Lrig3 was cloned into pCS2+ (Turner and Weintraub, 1994) or into pCS2flag, pCS2myc and pCS2GFP. The tags were located c-terminal to Lrig3. Deletion mutants of Lrig3 were generated by PCR and subcloned into pCS2myc.
Morpholino oligo
The splicing morpholino (Genetools) against Lrig3 (L3MO) recognizes both pseudo-alleles is GGGTTTCTGAAAGATAAAAACAAGC, and the Control-MO is CCTCTTACCTCAGTTACAATTTATA.
lacZ staining, whole-mount in situ hybridization, Alcian Blue staining
lacZ staining was performed as described (Zhao et al., 2001). Whole-mount in situ hybridization was performed as described (Harland, 1991). The following probes were used: Sox2 (Kishi et al., 2000), Rx2a (Yoshitake et al., 1999), Krox20 (Bradley et al., 1993), Slug (Snail2) (Mayor et al., 1995), Sox9 (Spokony et al., 2002), Ap2a (Luo et al., 2002), Inca (Luo et al., 2007), Myc (Bellmeyer et al., 2003), Twist (Hopwood et al., 1989) and Traf4 (this laboratory) were examined. Alcian Blue cartilage staining was performed as described (Pasqualetti et al., 2000).
Cell culture and transfection
HEK293T and COS7 cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's manual.
RT-PCR assay
Superscript III (Invitrogen) was used for cDNA synthesis. PCR primers are listed in Table 1.
Table 1.
Primers used for RT-PCR
| Gene name | Sequence (upstream and downstream, respectively) | Reference |
|---|---|---|
| Ap2 | TCCGACCCGAACGAGCAAGTA | |
| ATCAGGACCGGGCAATGTTCTA | This work | |
| Bmp4 | GATCTCAGACTCAACGGCAC GCATGTACGGATAAGTCGATC | De Robertis* |
| Chordin | CCTCCAATCCAAGACTCCAGAG | |
| GGAGGAGGAGGAGCTTTGGGACAAG | De Robertis* | |
| Cerberus | GCTGAACTATTTGATTTCACC | |
| ATGGCTTGTATTCTGTGGGGCG | De Robertis* | |
| Fgf3 | GTCATTTGTTTCCAGACTTC | |
| TATCTGTAGGTGGTACTTAG | This work | |
| Fgf4 | TTACCGGACGGAAGGATA | |
| CCTCGATTCGTAAGCGTT | This work | |
| Fgf8 | CTGGTGACCGACCAACTAAG | |
| ACCAGCCTTCGTACTTGACA | This work | |
| Foxd3 | GGAGGGAGGGGGCAATGCAC | |
| CCCCGAGCTCGCCTACT | (Pohl and Knochel, 2001) | |
| Frzb-1 | GACCACTGAATGTAGCCAGGAC | |
| GGAGATGCAGACTCCTCTGTCA | De Robertis* | |
| Lrig3 | GTGCAGTGTACAAGGCCTCA | |
| TTGGAATGTACACGGACTGC | This work | |
| Lrig3A intron | CGGCTAGACTAGAGTGTGCT | |
| AGCATCTTCAAGGTCTGTGT | This work | |
| Lris3B intron | CAGCTAGATTAGAATGTGCAGCG | |
| CCATTAATGGCCGTAGAAACGATG | This work | |
| Msx1 | GCTAAAAATGGCTGCTAA | |
| AGGTGGGCTGTGTAAAGT | De Robertis* | |
| Odc | CAGCTAGCTGTGGTGTGG | |
| CAACATGGAAACTCACACC | De Robertis* | |
| Pax3 | CTACCTCGGTTTCTTGACTG | |
| TGGTCAATCCTTCTTAATGG | This work | |
| Siamois | CAGGTTTGGTTTCAGAACAG | |
| TTCAGGCCAGTTGAATGTAT | This work | |
| Slug | TCCCGCACTGAAAATGCCACGATC | |
| CCGTCCTAAAGATGAAGGGTATCCTG | (LaBonne and Bronner-Fraser, 1998) | |
| Snail | AAGCACAATGGACTCCTT | |
| CCAATAGTGATACACACC | (LaBonne and Bronner-Fraser, 1998) | |
| Sox2 | GAGGATGGACACTTATGCCCAC | |
| GGACATGCTGTAGGTAGGCGA | De Robertis* | |
| Sox3 | TGATGCAGGACCAGTTGGGC | |
| TGAAGTGAAGGGTCGCTGGC | (Ishibashi and Yasuda, 2001) | |
| Sox9 | AAGCAGAATGTCCTCTGTGA | |
| AAGGCCAGATTCAGTTCTTC | This work | |
| Sox17β | GTCATGGTAGGAGAGAAC | |
| ATCTGTTTAGCCATCACTG | De Robertis* | |
| VegT | CAAGTAAATGTGAGAAACCG | |
| CAA ATACACACACATTTCCC | De Robertis* | |
| Xbra | TTAAGTGCTGTAATCTCTTCA | |
| GCTGGAAGTATGTGATGGAG | De Robertis* | |
| Xlim-1 | GAAGGATGAGACCACTGGTGG | |
| CACTGCCGTTTCGTTCATTTC | De Robertis* | |
| XTwist | AGTCCGATCTCAGTGAAGCGCA | |
| TGTGTGTGGCCTGAGCTGTAG | (LaBonne and Bronner-Fraser, 1998) | |
| XVent-1 | TTCCCTTCAGCATGGTTCAAC | |
| GCATCTCCTTGGCATATTTGG | (Gawantka et al., 1995) | |
| Zic1 | ATGAAGGTCCACGAAGCATC | |
| CGTGCTGTGATTGGACGTGT | (Nakata et al., 1998) |
Immunoprecipitation and immunoblotting
Cells were lysed in lysis buffer [137 mM NaCl, 5 mM EDTA, 1% TrionX-100, 10 mM Tris-HCl (pH 7.5)]. Immune complexes were captured by Fastflow protein G beads (GE Health), washed in lysis buffer five times, separated on NuPage Norvex 4-12% Bis-Tris gels (Invitrogen), and transferred to PVDF membranes (Invitrogen) for blotting.
ERK phosphorylation
Animal caps were lysed in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA and 5 mM sodium orthovanadate (Bottcher et al., 2004). The equivalent of 10 animal caps was blotted using monoclonal anti-diphospho-ERK antibody (clone MAPK-YT, Sigma) at 1:1000, and monoclonal anti-pan-ERK (BD) at 1:4000.
Immunofluorescence
Cells were plated on chambered microslides (Nunc). The cells were washed by cold 1× PBS, fixed by 2% PFA in 1× PBS, blocked with 20% serum in 1× PBS, and incubated with antibodies for 1 hour. Alexa Fluor 488- or 568-conjugated secondary antibodies were used. Slides were mounted in VECTASHIELD (Vector Laboratories) and viewed using a Zeiss LSM510 confocal microscope.
Luciferase assay
HEK293T cells were plated into 24-well plates for 1 day, a total of 0.8 μg/well of plasmid DNA including Topflash-luciferase (0.25 μg/well) and Renilla luciferase pRL-CMV (0.025 μg/well) were added, using pCS2+ DNA to adjust the total amount. After 1 day, mouse Wnt3a (100 ng/ml) or LiCl (30 mM) was added. Luciferase activity was measured using the Dual luciferase system (Promega) after 24 hours.
RESULTS
Properties and expression of Xenopus and zebrafish Lrig3
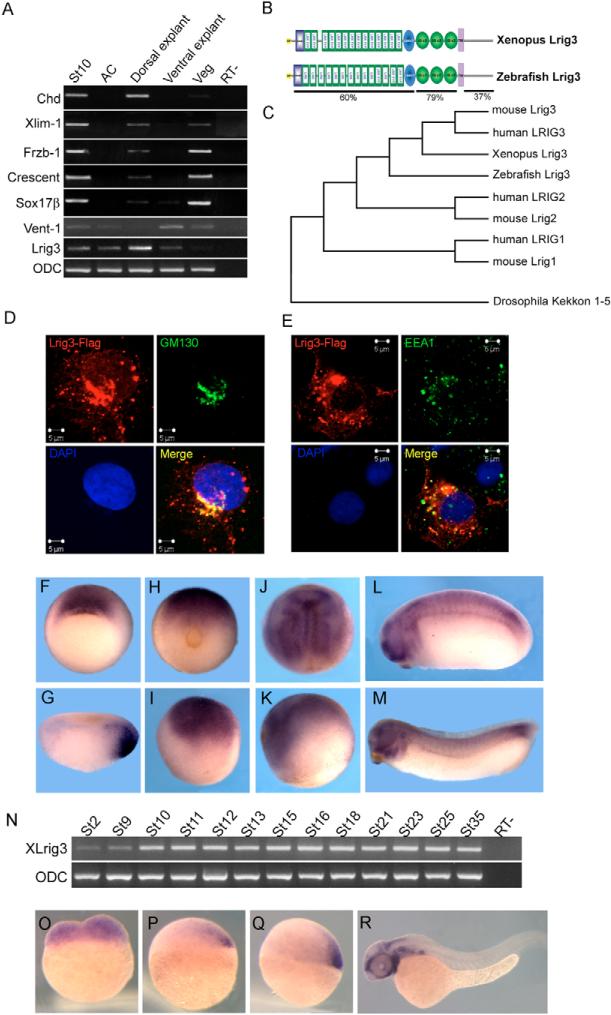
To study genome-wide gene expression during embryogenesis, we used DNA microarrays to investigate spatial differences in gene expression. Stage 10 Xenopus embryos were dissected into four explants (see Materials and methods). Several known genes were used to test the dissection (Fig. 1A). The original results of these microarray experiments were deposited at GEO and will be discussed elsewhere. The Accession Numbers are GSM227883 (animal cap explant), GSM227884 (dorsal explant), GSM227963 (vegetal explant) and GSM227964 (ventral explant). Here, we focus on one gene identified in this study that encodes a protein most similar to human LRIG3; we named this gene Xenopus leucine-rich repeats and immunoglobulin-like domains 3 (Lrig3). Lrig3 contains a signal peptide, 14 tandem leucine-rich repeats, N- and C-terminal specialized leucine-rich repeats, three immunoglobulin C2 domains, a single transmembrane domain and a cytoplasmic region with no identified domain structure (Fig. 1B). To determine whether a similar gene is expressed in different vertebrate embryos, we isolated zebrafish lrig3 by 5′ RACE (Fig. 1B). Accession numbers are EU126151 (Xenopus) and EU126152 (zebrafish). The phylogenetic tree in Fig. 1C presents the relationships within the vertebrate Lrig family, also including Drosophila Kekkon as the closest homolog.
Fig. 1. Characterization of the Lrig3 gene.
(A) Regional expression of Lrig3 in stage 10 embryos. Lrig3 was expressed strongly in dorsal explants, but barely detected in other regions. The indicated genes were assayed to test quality of dissection. St10, stage 10 embryo; AC, animal cap; Veg, vegetal explant; RT-, control without reverse transcriptase. (B) Schematic drawing of the protein structure of Xenopus and zebrafish Lrig3. SP (yellow), signal peptide; LRRNT (blue box), leucine-rich repeat N-terminal domain; LRR TYP, leucine-rich repeats, typical; LRR, leucine-rich repeats; LRRCT (blue oval), leucine rich repeat C-terminal domain; IG C2 (green oval), immunoglobulin C-2 Type; TM (purple), transmembrane domain. Sequence identity between zebrafish and Xenopus Lrig3 is indicated. (C) Dendrogram of the Lrig3 family including Kekkon of Drosophila. (D,E) Subcellular distribution of Lrig3 after transfection into COS7 cells. (D) Transfected Lrig3-Flag (red) co-localized with the cis Golgi apparatus marker GM130 (green); the nucleus was stained with DAPI (blue). (E) A small proportion of Lrig3-Flag co-localized with the early endosome marker EEA1 (green). (F-M) Expression pattern of Lrig3 in Xenopus. Vegetal view at stage 10, expression in the organizer (F); stage 10 section (G). (H,I) Stage 12 (H, posterior view; I, lateral view). (J,K) Stage 15 (J, dorsal view; K, lateral view). Tailbud (stage 24, L) and tadpole (stage 32, M); expression is seen in brain, eye, somites and branchial arches. (N) Temporal expression of Lrig3 during Xenopus development. (O-R) Expression of lrig3 in zebrafish. Transcripts are present maternally (O), become localized in the forming organizer at 30% epibody (P) and subsequently in the shield (Q), and were found in brain, eye and branchial arches at 24 hours (R). (P-R) Lateral views.
Lrig3 contains a putative signal peptide and transmembrane domain, and is thus predicted to be a transmembrane protein. To investigate the subcellular localization of Lrig3, we expressed epitope tagged protein in COS7 or CHO cells. A fraction of Lrig3-Flag was localized at the cell membrane, while a majority localized to the Golgi apparatus, as seen by colocalization with the cis Golgi marker GM130 (Fig. 1D). In addition, punctate distribution of Lrig3-Flag was observed in the cytosol. Only a small proportion of these puncta overlapped with the early endosome marker EEA1 (Fig. 1E). The nucleus was essentially devoid of Lrig3-Flag staining.
In the Xenopus embryo, Lrig3 is expressed around the dorsal blastopore lip, including the prospective neural ectoderm, involuting dorsal mesoderm and anterior dorsal endoderm (Fig. 1F,G). Dorsally restricted expression was verified by RT-PCR (Fig. 1A). As gastrulation progressed, the future neural plate, including the NC anlagen, was stained (Fig. 1H-K). In tailbud and tadpole stages, Lrig3 was highly expressed in the brain, branchial arches and paraxial mesoderm (Fig. 1L,M). RT-PCR indicated that Lrig3 is a maternal factor, increases by gastrulation and continues to be expressed through embryogenesis (Fig. 1N). This pattern is consistent with Lrig3 induction by Chordin or a truncated BMP receptor in animal caps (see Fig. S2 in the supplementary material).
The expression pattern of zebrafish lrig3 was investigated to ask whether the Xenopus pattern is conserved. Zebrafish lrig3 is present maternally, localizes to the shield at gastrula and later shows a complex pattern with strong expression in the branchial arches (Fig. 1O-R).
Lrig3 is required for neural crest differentiation
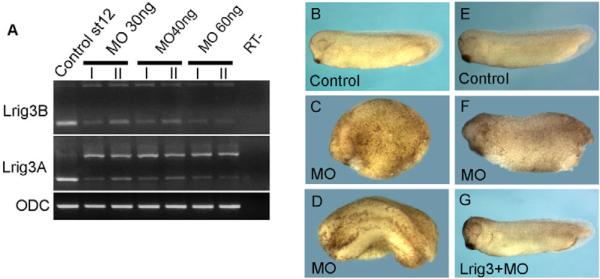
To study the function of Lrig3 in Xenopus development, we used a splicing morpholino antisense oligonucleotide (hereafter L3MO) to reduce expression of the endogenous protein. The effectiveness of L3MO was checked by RT-PCR, showing that it greatly reduced the level of mature mRNA while leading to the appearance of a 1 kb PCR product corresponding to unspliced RNA (Fig. 2A). Injection of L3MO allowed completion of gastrulation but led to repression of anterior development and bent axis (Fig. 2B-D, Table 2). These phenotypes could be rescued by co-injection with Lrig3 mRNA, indicating that the effect of L3MO is specific (Fig. 2E-G, Table 3).
Fig. 2. Suppression of Lrig3 by a splicing morpholino disturbed embryonic development.
(A) RT-PCR shows that injection of L3MO decreased the mature Lrig3 mRNA while inducing the predicted ~1 kb unspliced band, using intron-spanning primers for pseudoalleles Lrig3A and Lrig3B. ODC was loading control. (B-D) Phenotypes induced by L3MO. The morphants showed severe anterior defects, delayed or failed neural fold closure, and shortened axis. (C,D) Embryos were injected with 30 ng L3MO; uninjected control in B. (E-G) The phenotype was rescued by co-injection of Lrig3 mRNA. (E) Uninjected control; (F) L3MO (30 ng); (G) 30 ng of L3MO plus 20 pg Lrig3 mRNA.
Table 2.
Phenotype induced by L3MO
| Head repression, short and bent axis |
||||
|---|---|---|---|---|
| L3MO dose | n | Normal | Mild | Severe |
| 10 ng | 48 | 46 (95.8%) | 2 (4.2%) | 0 |
| 20 ng | 32 | 17 (53.1%) | 15 (46.8%) | 0 |
| 30 ng | 97 | 7 (7.2%) | 13 (13.4%) | 77 (79.4%) |
| 40 ng | 68 | 0 | 9 (13.2%) | 59 (86.7%) |
Table 3.
Rescue of L3MO phenotype by co-injection of Lrig3 mRNA
| Head repression, short and bent axis |
||||
|---|---|---|---|---|
| n | Normal | Mild | Severe | |
| L3MO 30 ng | 82 | 7 (8.5%) | 24 (29.3%) | 51 (62.2%) |
| L3MO+Lrig3 (30 ng + 20 pg) | 80 | 50 (62.5%) | 24 (30%) | 4 (5%) |
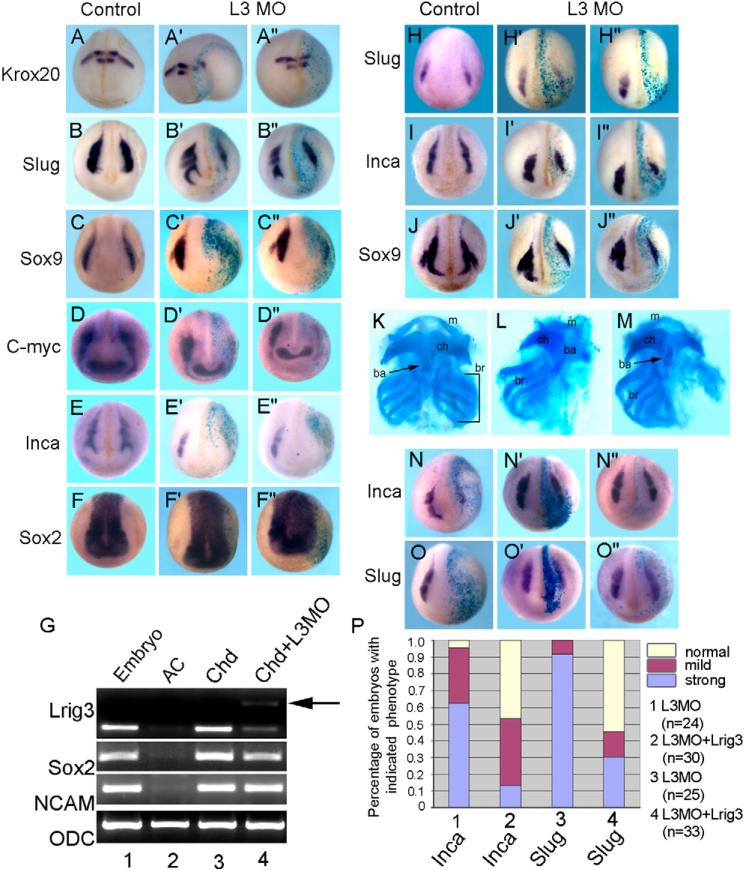
Examination of the expression of various marker genes at different stages in L3MO-injected embryos showed that NC formation was impaired in these embryos. This is illustrated by in situ hybridization with embryos injected with L3MO in one side, the other side acting as control. When examined at around stage 19, Krox20 is still expressed within rhombomeres 3 and 5, but the stripe of Krox20-positive cells that marks the NC stream emanating from rhombomere 5 was strongly inhibited (Fig. 3A-A″), while the retinal marker Rx2a was only mildly inhibited (see Fig. S3A-A″ in the supplementary material). Thus, Lrig3 does not appear to be required for neural induction, as also seen by the lack of inhibition of the pan-neural marker Sox2 (Fig. 3F-F″), and the resistance of neural induction by Chordin in animal caps to the morpholino (Fig. 3G). However, compensation by maternal Lrig3 or by other Lrig family members has not been excluded. By contrast, severe reduction up to a complete loss of expression was seen in the great majority of L3MO-injected embryos for a panel of NC markers (Fig. 3B-E″; see Fig. S3B-D in the supplementary material; see legend for numbers of embryos affected). Knock-down of Lrig3 had little effect on the anterior expression domain of Myc which marks the border of the neural plate, whereas expression in the NC anlage was strongly reduced (Fig. 3D-D″); this emphasizes the differential nature of the effect. The loss of NC markers could be rescued by co-injection of L3MO with Lrig3 mRNA, as seen in Fig. 3N-O″ for Slug and Inca; quantification is shown in Fig. 3P.
Fig. 3. Knockdown of Lrig3 results in defects of the NC and its derivatives.
(A-F″) In situ hybridization with the indicated markers for uninjected or two examples (′,″) of L3MO-injected embryos. One dorsal blastomere at the four-cell stage was injected with 15 ng L3MO and 100 pg lacZ mRNA, and fixed at about stage 19. The expression of hindbrain marker Krox20 (rhombomeres 3 and 5) (A-A″), NC markers Slug (B-B″), Sox9 (C-C″), Myc (D-D″) and Inca (E-E″), and pan-neural marker Sox2 (F-F″) were examined. The injected side was traced by lacZ staining. The stream of Krox20-positive cells extending from the hindbrain was absent in 97% (28 of 29 embryos) on the injected side. The expression of NC markers was reduced on the injected side in the following percentage of embryos: Slug (58%, 14 of 24), Sox9 (79%, 15 of 19), Myc (83%, 20 of 24) and Inca (65%, 17 of 26). (G) Neural induction by overexpression of Chordin (Chd) in animal caps was barely affected by L3MO. Chd (100 pg) or Chd (100 pg) plus L3MO (30 ng) were injected, caps dissected at stage 9, and assayed at equivalent stage 22. Pan neural markers Sox2 and Ncam were examined, and interference with Lrig3 splicing was verified (see also Fig. 2A); arrow indicates the predicted unspliced band (lane 4). (H-J″) NC markers, including Slug, Inca and Sox9, were examined in embryos injected with 7.5 ng L3MO in one animal dorsal blastomere at the eight-cell stage. Strong inhibition was observed for Slug (95%, 18 of 19), Inca (89%, 24 of 27) and Sox9 (73%, 19 of 26). (K-M) Cranial cartilages were reduced by knockdown of Lrig3. Ventral views of Alcian Blue stained cartilage from embryos injected with control morpholino (K) or L3MO (L,M). ba, basihyal; br, branchial; ch, ceratohyal; m, Meckel's. Percentages of embryos with reduced head cartilage are 82% (38 of 47) at 7.5 ng L3MO and 97% (31 of 32) at 15 ng L3MO. (N-P) Rescue of L3MO effect. All embryos were injected with 15 ng L3MO, and embryos in N-O″ were co-injected with 20 pg Lrig3 mRNA; in situ hybridization with Inca (N-N″) and Slug (O-O″). Percentages of strongly affected (examples in N,O), mildly affected (N′,O′) and normal (N″,O″) are shown in P.
The inhibition of NC might possibly be explained by suppression of mesoderm formation; therefore, we examined the expression of Xbra, Chordin, Cerberus and goosecoid in embryos targeted dorsally with L3MO at the four-cell stage (see Fig. S4 in the supplementary material). The expression domain and expression level of these mesodermal markers were not substantially affected. Furthermore, we checked the expression of Slug, Inca and Sox9 in embryos in which L3MO was injected into one animal-dorsal blastomere at the eight-cell stage to restrict the range of cells that are affected. Strong inhibition of NC was again observed in these embryos (Fig. 3H-J″).
Next, we checked for the presence of differentiated NC derivatives at swimming tadpole stages in Lrig3-depleted embryos. NC cells of the branchial arches differentiate into the cartilages of the embryonic facial skeleton (Le Douarin and Kalcheim, 1999). Cartilage formation in Lrig3 morphants, as visualized by Alcian Blue staining, was strongly inhibited in the injected side, the phenotype ranging from missing a single branchial arch to loss of the entire cranial cartilage (Fig. 3K-M, see legend for the percentage of affected embryos). As NC precursors also give rise to ganglion cells of the peripheral nervous system (Huang and Saint-Jeannet, 2004; Le Douarin and Kalcheim, 1999) we examined trigeminal nerve formation in Lrig3 morphants by staining with Xenopus Synuclein γ, and found it to be strongly reduced (see Fig. S5 in the supplementary material).
Lrig3 is required for NC induction in animal caps
To investigate the mechanism of Lrig3 involvement in NC formation, we used animal caps, in which NC can be induced by the combined action of a canonical Wnt ligand and a BMP antagonist (Saint-Jeannet et al., 1997). Animal caps were dissected from embryos injected with the indicated mRNAs with or without L3MO, cultured to stage 16, and gene expression was examined by RT-PCR. In agreement with previous findings, the neural makers Otx2, Ncam, Sox3 and Sox2 were induced by the injection of Chordin, but were suppressed by the co-injection of Wnt3a, while the NC markers Slug, Sox9, Foxd3, Ap2, Twist and Snail were induced by the combination of Wnt3a and Chordin (Fig. 4A,A′). Induction of Slug, Sox9, Foxd3 and Twist was decreased by co-injection of L3MO, while Snail was affected to a lesser extent and Ap2 was unaffected. Ap2 is expressed in epidermis in addition to the NC (Luo et al., 2003; Luo et al., 2002), possibly explaining the reduced effect. In contrast to NC markers, the neural genes Sox3 and Sox2, but not Otx2 or Ncam, were up-regulated moderately by the morpholino, suggesting that the neural fate that had been suppressed by adding Wnt3a to Chordin-expressing animal caps was partially recovered (Fig. 4A,A′). Overexpression of Lrig3 alone could not induce neural or NC makers in animal caps, nor could addition of Lrig3 increase the level of Slug, Sox9 and Foxd3 over that achieved by Wnt3a plus Chordin alone (Fig. 4A). These experiments also confirmed that Lrig3 itself is strongly induced by injection of Chordin or a combination of Chordin and Wnt3a, and that L3MO reduced the level of mature Lrig3 mRNA (Fig. 4A′). Thus, Lrig3 is required for the induction of NC in explants as well as in whole embryos.
Fig. 4. Lrig3 is required for NC formation in animal caps downstream of Pax3 and Zic1.
(A,A′) L3MO inhibits NC marker induction by Chordin (Chd) plus Wnt3a in animal caps. Embryos were injected with 300 pg Chd, 1 ng Lrig3, 30 ng L3MO, 300 pg Chd + 300 pg Wnt3a, 300 pg Chd + 300 pg Wnt3a + 30 ng L3MO, or with 300 pg Wnt3a + 300 pg Chd +1 ng Lrig3. Gene expression was assayed by RT-PCR. (B) NC induced by co-injection of Pax3 and Zic1 was inhibited by L3MO. Pax3 (200 pg) and Zic1 (200 pg) each, alone or with 30 ng L3MO were injected, and assayed as in A. AC, uninjected animal caps. RT-, without reverse transcriptase.
The Zic1 and Pax3 genes play an important role in NC specification by inducing NC markers such as Slug and Foxd3 in the presence of a Wnt signal (Monsoro-Burq et al., 2005; Sato et al., 2005) (for a review, see Steventon et al., 2005). L3MO did not block the expression of Zic1 and Pax3 in animal caps injected with Chordin and Wnt3a (Fig. 4A), suggesting that Lrig3 functions between Pax3/Zic1 and Slug/Foxd3 in the regulatory hierarchy of NC formation. To address this possibility, we examined the L3MO effect on the NC inducing activity of Pax3 plus Zic1. As reported, co-injection of Pax3 and Zic1 induced NC markers Slug, Sox9 and Twist (Hong and Saint-Jeannet, 2007; Monsoro-Burq et al., 2005); this induction was inhibited by L3MO (Fig. 4B). We therefore conclude that Lrig3 functions downstream of Pax3 and Zic1 in NC formation.
Lrig3 acts in NC specification in coordination with a Wnt signal
Lrig3 is required for NC formation in animal caps exposed to a Wnt signal and BMP inhibition. We attempted to determine which of these two pathways interacts with Lrig3. Induction of the Wnt target genes Xnr3 and Siamois in animal caps was only slightly enhanced by co-injection of Lrig3 (Fig. 5A), and Lrig3 transfection stimulated Wnt-dependent Topflash reporter activity in cultured cells to a modest but significant extent (see Fig. S6 in the supplementary material). In addition to activating dorsal genes such as Siamois, canonical Wnt signaling is also capable of inducing mesoderm formation (Schohl and Fagotto, 2003). We found that the slight induction of Xbra by Wnt3a was greatly enhanced by co-injection of Lrig3 (Fig. 5B). The combination of Wnt3a and Lrig3 could also induce the NC markers Slug, Foxd3 and Sox9, but to a lower level than that obtained after injection of a combination of Wnt3a, Chordin and Lrig3 (Fig. 5B). These results indicate that Lrig3 can enhance canonical Wnt signaling, with the strength of the effect dependent on context.
Fig. 5. Lrig3 is involved in NC formation by modulating Wnt and Fgf signaling.
(A) Lrig3 slightly enhanced Xnr3 and Siamois induction by Wnt3a. Animal cap assay at stage 10, embryos injected with 300 pg Wnt3a or 300 pg Wnt3a + 1 ng Lrig3. (B) NC markers were moderately induced by the combination of Lrig3 and Wnt3a. Embryos were injected with Lrig3 (1 ng), Wnt3a (300 pg), Wnt3a and Lrig3, Chd (300 pg), Chd and Lrig3, and Chd, Wnt3a and Lrig3, and animal caps were assayed at equivalent stage 16. (C) Lrig3 enhanced the induction of Fgf3, Fgf4 and Fgf8 by Wnt3a. RNAs were injected at the concentrations listed in B, and animal caps were assayed at stages indicated.
The role of Wnt signaling in mesoderm formation is correlated with its induction of Fgf3 (Schohl and Fagotto, 2003). As Xbra is a direct target of Fgf signaling (Smith et al., 1991), we tested whether the enhanced Xbra expression induced by Wnt and Lrig3 was accompanied by upregulation of Fgf3, Fgf4 (also known as eFgf) or Fgf8, three genes known to be expressed in early Xenopus embryos (Fletcher et al., 2006; Isaacs et al., 1995; Lombardo et al., 1998; Tannahill et al., 1992). Injection of Lrig3 alone did not induce any of these genes. When harvested at stage 10, animal caps co-injected with Lrig3 and Wnt3a expressed Fgf8, while Wnt3a alone did not induce any of the three Fgf genes (Fig. 5C). By stage 16, animal caps injected with Wnt3a induced a low level of Fgf3, but co-injection of Wnt3a with Lrig3 led to the clear increase in the induction of all three Fgf genes tested, consistent with the induction of Xbra under the same conditions (Fig. 5C). We conclude that Lrig3 strongly enhances the ability of canonical Wnt signaling to induce three Fgf genes in Xenopus embryonic tissues.
In contrast to the interaction seen between Lrig3 and Wnt3a, no interaction was observed with Chordin. Chordin alone induces neural markers but does not induce NC markers, and addition of Lrig3 did not change this behavior (Fig. 5B). Likewise, Chordin induces Fgf8 but not the other Fgf genes, and again addition of Lrig3 had no obvious effect (Fig. 5C). Thus, the role of Lrig3 in NC induction appears to be related to the Wnt rather than the BMP antagonist arm of the signaling cascade.
Lrig3 can modulate Fgf signaling in mesoderm and NC induction
We investigated the relationship of Lrig3 to the Fgf pathway for three reasons. First, modulation of Wnt signaling in the context of NC induction in explants results in enhanced expression of Fgf genes. Second, mammalian Lrig1 affects EGF signaling, which shares several signal transduction components with the Fgf pathway. Third, Fgf signaling has been implicated in NC formation. We first checked the effect of Lrig3 on Fgf-induced ERK (MAP kinase) phosphorylation in animal caps harvested at an early stage where the effects of Fgf have been well studied. Injection of active Ras or Fgf4 mRNA into animal caps stimulates ERK phosphorylation (Gupta and Mayer, 1998; Suga et al., 2006; Whitman and Melton, 1992), and this could also be achieved by treatment with bFgf (Fig. 6A). ERK phosphorylation was inhibited by injection of Lrig3; similar inhibition was observed with Sef or Mkp3, known inhibitors of Fgf signaling that were used as controls (Kovalenko et al., 2003; Tsang et al., 2002; Tsang et al., 2004) (Fig. 6A). Using induction of Xbra by Fgf (Smith et al., 1991) as an assay for Fgf signaling also showed inhibition by injection of Lrig3 (Fig. 6B). This inhibition is specific because Lrig3 could not inhibit Xbra induction by activin, while Smad7, an inhibitor of activin/nodal signaling (Bhushan et al., 1998), blocked the activin effect (Fig. 6C). Multiple Fgf ligands are expressed in the Xenopus embryo, and while the signaling pathways downstream of these ligands are similar, varying consequences of different Fgfs have been reported (Fletcher et al., 2006; Hardcastle et al., 2000). Therefore, we checked the effect of Lrig3 on Fgf8-induced gene activation in animal caps, injecting increasing doses of Fgf8a RNA with or without a constant level of Lrig3 RNA. Induction of Xbra, Msx1 and Wnt8 increased with the dose of Fgf8a, peaked at 100 pg/embryo, and decreased at higher doses. It has been shown previously that overexpression of Fgf8a inhibits Xbra expression in the whole embryo (Hardcastle et al., 2000). Co-injection of Lrig3 with Fgf8a led to a decrease of the expression of Xbra, Msx1 and Wnt8 at 100 pg Fgf8a RNA, while at higher doses, Lrig3 had less effect (Fig. 6D). Thus, the level of Fgf at which peak induction is obtained shifted to higher values with the addition of Lrig3. These results suggest that (1) induction of the three mesodermal genes by Fgf8 is dose dependent with a discrete maximum; and (2) Lrig3 attenuates the Fgf signal under these conditions.
Fig. 6. Lrig3 attenuates Fgf signaling.
(A) Lrig3 inhibited ERK phosphorylation. Embryos were injected with active Ras (HaRas, 100 pg), eFgf (1 pg), Lrig3 (1 ng), zebrafish sef (500 pg) or mkp3 (500 pg). Animal caps were treated with bFgf (20 ng/ml) for 3 hours as indicated. Cell lysates were analyzed for diphosphorylated (dp)-ERK and total (pan)-ERK. AC, untreated. (B) Overexpression of Lrig3 inhibits Xbra induction by bFgf. Animal caps were treated with bFgf (50 ng/ml) for 4 hours. Xbra expression was induced by bFgf treatment in uninjected (lane 4), but not in Lrig3 (1 ng) injected caps (lane 5). (C) Lrig3 did not attenuate Xbra expression induced by activin (250 pM). Injection of Smad7 (500 pg), but not of Lrig3 (1 ng), inhibited Xbra induction. (D) Lrig3 inhibits Fgf8 activity in animal caps. Embryos were injected with the indicated dose of Fgf8a alone or with 1 ng of Lrig3. Expression of Xbra, Xwnt8 and Xmsx1 was examined at equivalent stage 11.5. (E) Differential effect of Lrig3 on Slug and Twist induction by different doses of Fgf8a. Fgf8a at increasing doses, as indicated, was injected alone or with a constant amount (1 ng) of Lrig3, and induction of Slug and Twist was assayed at equivalent stage 22.
We argued that modulation of Fgf signaling might have a role in Lrig3 function during NC induction. Fgf8 is required for NC formation as reduction of Fgf8 expression by a morpholino inhibits NC formation (Hong and Saint-Jeannet, 2007; Monsoro-Burq et al., 2005), while injection of Fgf4 (LaBonne and Bronner-Fraser, 1998) or Fgf8 (Monsoro-Burq et al., 2003) RNA can induce NC markers in animal caps. Titration experiments in whole embryos showed that Fgf8 has a dual role in this process as a modest increase enhanced, but an excess of Fgf8 inhibited, NC formation (Hong and Saint-Jeannet, 2007). To examine the role of Lrig3 in this system, we injected different levels of Fgf8a with or without Lrig3 into animal caps and tested for induction of NC genes (Fig. 6E). At the lowest dose used, Fgf8a RNA induced both Slug and Twist, and addition of Lrig3 inhibited the induction. At higher doses, NC marker induction by Fgf8a alone was reduced, but it was strongly enhanced by co-injection of Lrig3. This is most obvious in the case of Slug induction at 1 ng of Fgf8a, which changes from undetectable to a robust induction upon addition of Lrig3. Even more pronounced than in the case of mesodermal gene induction, addition of Lrig3 shifts the peak of NC gene induction to higher concentrations of Fgf. These results are compatible with the conclusion that an optimal level of Fgf signaling is required for NC induction, and that Lrig3 attenuates Fgf signal transduction to bring the output into optimal range at the higher levels of Fgf signaling.
Lrig3 interacts with Fgfr1 and decreases its level of expression
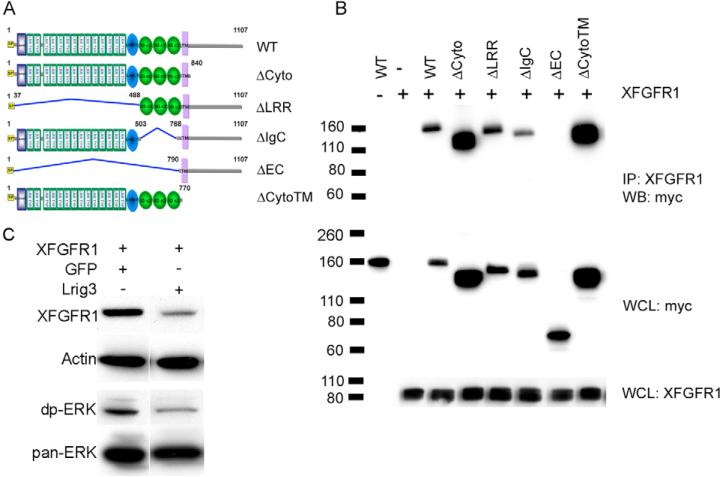
Lrig3 contains a signal peptide and a transmembrane domain, and a fraction of Lrig3 is localized on the cell membrane (Fig. 1). Lrig1 interacts with all four ErbB receptors as well as Met receptors (Gur et al., 2004; Shattuck et al., 2007). Therefore, we investigated the possible interaction of Lrig3 with Fgf receptors. Lrig3 was co-immunoprecipitated with Xfgfr1 when both were co-expressed in 293T cells, showing that these two membrane proteins can bind each other (Fig. 7B; see Fig. S7 in the supplementary material). To determine the Fgfr1-binding domains of Lrig3, we constructed a series of deletion mutants, ΔCyto (deleting the cytoplamic domain), ΔLRR (deleting the LRR domains), ΔIgC (deleting the IG C2 domains), ΔEC (deleting the extracellular portion including LRR and IG C2 domains) and ΔCytoTM (deleting the transmembrane and cytoplasmic domains), and performed co-immunoprecipitation assays. Interaction with Xfgfr1 was retained in constructs that contained all or even a part of the ectodomains, but was lost when the entire ectodomain was deleted (Fig. 7B). The intracellular and transmembrane domains were not involved in the binding. We conclude that the extracellular region of Lrig3 is responsible for binding of Fgfr1.
Fig. 7. Lrig3-Xfgfr1 interactions.
(A) The domain structure of wild-type Lrig3 and its deletion mutants; all constructs were tagged with Myc. LRRNT (blue box), leucine-rich repeat N-terminal domain; LRR TYP, leucine-rich repeats, typical; LRRCT (blue oval), leucine-rich repeat C-terminal domain; IG C2 (green oval), immunoglobulin C-2 Type; SP (yellow), signal peptide;. TM (purple), transmembrane domain. (B) Lrig3 binds to Xfgfr1 through its ectodomains. Co-immunoprecipitation was carried out in extracts of 293T cells co-transfected with Xfgfr1 and wild type or mutants of Lrig3 using anti-Xfgfr1 antibody, and blotted with anti-Myc antibody. (C) Lrig3 decreases Xfgfr1 levels. 293T cells were co-transfected with Xfgfr1 and Lrig3 or GFP, which was used to adjust the amount of DNA. Xfgfr1 and endogenous diphospho-ERK and Pan-ERK were detected by western blotting. Actin was employed as a loading control. IP, immunoprecipitate; WCL, whole cell lysate; WB, western blot.
As Lrig1 negatively regulates the stabilities of EGFR and the Met receptor (Gur et al., 2004; Shattuck et al., 2007), we next asked whether Lrig3 affects Fgf receptor metabolism. Xfgfr1 protein was expressed at a much lower level in cells co-transfected with Lrig3 when compared with controls (Fig. 7C). This result correlates with the reduced ERK phosphorylation in the presence of Lrig3 that was seen in these cells (Fig. 7C) and in animal caps (Fig. 6A). These results suggest a role for Lrig3 in the regulation of FGFR1 availability in the cell.
DISCUSSION
In this paper, we describe the transmembrane protein Lrig3 as a novel factor required for the induction of the NC in Xenopus embryos. The Lrig3 gene is expressed in several regions of the early Xenopus and zebrafish embryo, including the NC precursor domain. A splice morpholino that strongly reduces the level of mature Lrig3 mRNA inhibits the expression of multiple NC marker genes and the formation of NC derivatives such as the cartilages of the branchial arches in the Xenopus embryo. We suggest that Lrig3 function is required downstream of Pax3 and Zic1 in NC formation, and that Lrig3 exerts its role by modulating signal transduction in the embryo. Lrig3 thus joins the factors known to be required for NC specification, which so far have included transcription factors such as Slug and Myc, and signaling factors such as Wnt ligands, an attenuated BMP signal and Fgf proteins.
The role of Lrig3 in NC formation
Studies in Xenopus have suggested that an intermediate level of BMP signaling is necessary but not sufficient for NC formation, and that activation of the Wnt and Fgf pathways are involved in this process (Steventon et al., 2005). Inhibition of Wnt/β-catenin signaling by either Frizzled 7 MO (Abu-Elmagd et al., 2006) or Gsk3b (Saint-Jeannet et al., 1997) blocks NC formation, while loss of Wnt1 and Wnt3a in mice causes NC deficiencies (Ikeya et al., 1997). Furthermore, activation of Wnt signaling by overexpression of Lrp6 expands the expression of Slug in the embryo (Tamai et al., 2000). In Xenopus animal caps, canonical Wnt ligands, including Wnt1, Wnt3a, Wnt7b and Wnt8, together with a BMP antagonist, such as Chordin or Noggin, induce NC markers, but Wnt alone is ineffective (Chang and Hemmati-Brivanlou, 1998; Deardorff et al., 2001; Saint-Jeannet et al., 1997). The importance of Wnt signaling is emphasized by the presence of a TCF-binding site in the Slug promoter and its response to Wnt signaling (Vallin et al., 2001). Lrig3 is able to enhance Wnt signaling in animal caps, as most clearly seen in the enhanced induction of several Fgf genes (Fig. 5C). As Fgf signaling can induce NC markers under certain conditions (Fig. 6E), these observations may account for the modest induction of NC genes in Wnt3a plus Lrig3-injected animal caps (Fig. 5B).
A requirement for Lrig3 in NC formation was seen by knockdown of Lrig3 using a splicing MO in whole embryos or animal caps (Fig. 3B-E″,H-J″, Fig. 4A-B). In animal caps we find that the expression of Zic1 and Pax3 is not inhibited by L3MO, while the expression of Slug, Foxd3 and Sox9 is. In a proposed network of NC formation, Zic1 and Pax3 are upstream of the other markers tested (Monsoro-Burq et al., 2005; Steventon et al., 2005), and thus our results can be interpreted by placing Lrig3 function downstream of Zic1 and Pax3. Indeed, L3MO prevented the expression of NC markers induced by Zic1 and Pax3 (Fig. 4B). A Wnt signal is required in combination with Zic1 and Pax3 in NC induction, suggesting that Lrig3 has a role in modulating this signal at this position of the regulatory hierarchy.
In addition to the Wnt pathway and an attenuated BMP signal, Fgf signaling is important in NC induction. Gain-of-function experiments indicate that the ectopic expression of Fgf8 at a level just above physiological leads to expansion of the NC, while an excess of Fgf8 expression is inhibitory (Hong and Saint-Jeannet, 2007). Furthermore, knockdown of Fgf8 by MO inhibits NC formation (Hong and Saint-Jeannet, 2007; Monsoro-Burq et al., 2005), while Fgf8 is able to induce NC markers in animal cap assays (Monsoro-Burq et al., 2003). We showed in animal caps that Fgf8 was able to induce NC markers at low but not at high doses, and addition of Lrig3 shifts peak induction to higher Fgf levels (Fig. 6E). We suggest that Lrig3 has a role in optimizing the Fgf signal during NC induction in Xenopus.
Lrig3 as a modulator of different signaling cascades during embryonic development
The proposed role of Lrig3 in modulating the Fgf signal is compatible with the observations that Lrig3 can inhibit Fgf-dependent ERK phosphorylation and mesodermal gene induction in animal caps (Fig. 6). As a transmembrane protein containing leucine-rich repeats, Lrig3 might be expected to interact with Fgf receptors, and we find that such an interaction can be observed after expressing Lrig3 and Fgfr1 in cultured cells (Fig. 7; see Fig. S7 in the supplementary material). Lrig3 bears some resemblance to Xflrt3 (Bottcher et al., 2004), which also contains leucine-rich repeats and interacts with Fgfr1, but functions as a Fgf signaling enhancer. Human LRIG1 has been reported as an inhibitor of EGF signaling (Gur et al., 2004), and it is possible that Lrig3 also affects EGF signaling. EGF signaling and the ErbB receptor family regulate mesoderm formation (Nie and Chang, 2006) and gastrulation movements (Nie and Chang, 2007) in Xenopus, although a role in NC formation has not been identified. Human LRIG1 negatively regulates the stability of EGFR and Met receptors through different mechanism. Lrig1 destabilizes EGFR by enhancing its ubiquitination in a Cbl-dependant manner (Gur et al., 2004), while it destabilizes the Met receptor in a Cbl-independent manner (Shattuck et al., 2007). Our data suggest a negative influence of Lrig3 on Xfgfr1 expression levels, expanding the range of apparent interactions between Lrig family members and RTK receptors.
Although Lrig3 inhibited the function of Fgf in animal caps, it enhanced the transcription of Fgf3, Fgf4 and Fgf8 induced by Wnt3a. Wnt/β-catenin signaling has at least two distinct functions in very early Xenopus development. First, maternal Wnt signaling is involved in the establishment of the DV axis (Kofron et al., 2007; Tao et al., 2005). Second, Wnt/β-catenin signaling is required for mesoderm induction through Fgf and Nodal, and promotes ventral/lateral but restricts dorsal development (Schohl and Fagotto, 2003) (Christian and Moon, 1993; Hoppler et al., 1996; Hoppler and Kavanagh, 2007). We observed an effect of Lrig3 on Wnt signaling mostly in the context of the latter process. Induction of Siamois and Xnr3 by Wnt, representing its role in axis formation, was only slightly affected by Lrig3, and overexpression of Lrig3 in ventral blastomeres did not induce a secondary axis (data not shown). However, induction of Xbra, and notably of Fgf3, Fgf4 and Fgf8 by Wnt was stimulated by Lrig3 (Fig. 5B), and Topflash reporter activity was moderately enhanced by Lrig3 (see Fig. S6 in the supplementary material). These observations suggest that Lrig3 can enhance some but not all outcomes of the Wnt signal transduction pathway.
In this study, we show that Lrig3 is required for NC formation during early embryonic development by modulating Wnt and Fgf signaling, and we placed Lrig3 downstream of Zic1 and Pax3 in the NC regulatory network. As is well established, an attenuated BMP signal in concert with a canonical Wnt signal is crucial in NC formation. Lrig3 was induced in this context and stimulated some aspects of Wnt signaling. One of the outputs of the enhanced Wnt signal is the induction of Fgf genes, providing an additional signal required in NC specification. Similar to BMP, which is required in NC formation but must be kept at a low level, an optimum Fgf level must be maintained for effective NC induction. Lrig3, through its ability to attenuate Fgf signaling, may assure achievement of this optimal level. In this model, its ability to modulate different signaling cascades is the basis for the requirement for Lrig3 in NC induction in the embryo.
Supplementary Material
Acknowledgments
We thank Dr Eddy M. De Robertis, Roberto Mayor, Tom Sargent, Jan L. Christian, Jean-Pierre Saint-Jeannet, Carole LaBonne, Yoshiki Sasai, Naoto Ueno and Robert Friesel for reagents, and members of the Dawid laboratory for discussions. This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health, USA.
Footnotes
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/135/7/1283/DC1
References
- Abu-Elmagd M, Garcia-Morales C, Wheeler GN. Frizzled7 mediates canonical Wnt signaling in neural crest induction. Dev. Biol. 2006;298:285–298. doi: 10.1016/j.ydbio.2006.06.037. [DOI] [PubMed] [Google Scholar]
- Amaya E, Stein PA, Musci TJ, Kirschner MW. FGF signalling in the early specification of mesoderm in Xenopus. Development. 1993;118:477–487. doi: 10.1242/dev.118.2.477. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev. Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Chen Y, Vale W. Smad7 inhibits mesoderm formation and promotes neural cell fate in Xenopus embryos. Dev. Biol. 1998;200:260–268. doi: 10.1006/dbio.1998.8965. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat. Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- Bradley LC, Snape A, Bhatt S, Wilkinson DG. The structure and expression of the Xenopus Krox-20 gene: conserved and divergent patterns of expression in rhombomeres and neural crest. Mech. Dev. 1993;40:73–84. doi: 10.1016/0925-4773(93)90089-g. [DOI] [PubMed] [Google Scholar]
- Carballada R, Yasuo H, Lemaire P. Phosphatidylinositol-3 kinase acts in parallel to the ERK MAP kinase in the FGF pathway during Xenopus mesoderm induction. Development. 2001;128:35–44. doi: 10.1242/dev.128.1.35. [DOI] [PubMed] [Google Scholar]
- Chang C, Hemmati-Brivanlou A. Neural crest induction by Xwnt7B in Xenopus. Dev. Biol. 1998;194:129–134. doi: 10.1006/dbio.1997.8820. [DOI] [PubMed] [Google Scholar]
- Christian JL, Moon RT. Interactions between Xwnt-8 and Spemann organizer signaling pathways generate dorsoventral pattern in the embryonic mesoderm of Xenopus. Genes Dev. 1993;7:13–28. doi: 10.1101/gad.7.1.13. [DOI] [PubMed] [Google Scholar]
- Chung HA, Yamamoto TS, Ueno N. ANR5, an FGF target gene product, regulates gastrulation in Xenopus. Curr. Biol. 2007;17:932–939. doi: 10.1016/j.cub.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin. Cell Dev. Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Curran KL, Grainger RM. Expression of activated MAP kinase in Xenopus laevis embryos: evaluating the roles of FGF and other signaling pathways in early induction and patterning. Dev. Biol. 2000;228:41–56. doi: 10.1006/dbio.2000.9917. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Tan C, Saint-Jeannet JP, Klein PS. A role for frizzled 3 in neural crest development. Development. 2001;128:3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- Essex LJ, Mayor R, Sargent MG. Expression of Xenopus snail in mesoderm and prospective neural fold ectoderm. Dev. Dyn. 1993;198:108–122. doi: 10.1002/aja.1001980205. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. Antagonizing the Spemann organizer: role of the homeobox gene Xvent-1. EMBO J. 1995;14:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RW, Mayer BJ. Dominant-negative mutants of the SH2/SH3 adapters Nck and Grb2 inhibit MAP kinase activation and mesoderm-specific gene induction by eFGF in Xenopus. Oncogene. 1998;17:2155–2165. doi: 10.1038/sj.onc.1202158. [DOI] [PubMed] [Google Scholar]
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle Z, Chalmers AD, Papalopulu N. FGF-8 stimulates neuronal differentiation through FGFR-4a and interferes with mesoderm induction in Xenopus embryos. Curr. Biol. 2000;10:1511–1514. doi: 10.1016/s0960-9822(00)00825-3. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of pax3 and zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo I, Kengaku M, Okamoto H. FGF signaling and the anterior neural induction in Xenopus. Dev. Biol. 1999;216:561–581. doi: 10.1006/dbio.1999.9515. [DOI] [PubMed] [Google Scholar]
- Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J. Cell Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- Hoppler S, Brown JD, Moon RT. Expression of a dominant-negative Wnt blocks induction of MyoD in Xenopus embryos. Genes Dev. 1996;10:2805–2817. doi: 10.1101/gad.10.21.2805. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A, Gurdon JB. A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev. Biol. 2004;275:1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Isaacs HV, Pownall ME, Slack JM. eFGF is expressed in the dorsal midline of Xenopus laevis. Int. J. Dev. Biol. 1995;39:575–579. [PubMed] [Google Scholar]
- Ishibashi S, Yasuda K. Distinct roles of maf genes during Xenopus lens development. Mech. Dev. 2001;101:155–166. doi: 10.1016/s0925-4773(00)00585-2. [DOI] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, Yamazaki H, Shiota K, Nakanishi S, Sasai Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Kofron M, Birsoy B, Houston D, Tao Q, Wylie C, Heasman J. Wnt11/beta-catenin signaling in both oocytes and early embryos acts through LRP6-mediated regulation of axin. Development. 2007;134:503–513. doi: 10.1242/dev.02739. [DOI] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Kovalenko D, Yang X, Nadeau RJ, Harkins LK, Friesel R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J. Biol. Chem. 2003;278:14087–14091. doi: 10.1074/jbc.C200606200. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J. Biol. Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The Neural Crest. Cambridge University Press; Cambridge: 1999. [Google Scholar]
- Lombardo A, Isaacs HV, Slack JM. Expression and functions of FGF-3 in Xenopus development. Int. J. Dev. Biol. 1998;42:1101–1107. [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Thomas ML, Weeks DL, Sargent TD. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev. Biol. 2002;245:136–144. doi: 10.1006/dbio.2002.0621. [DOI] [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc. Natl. Acad. Sci. USA. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Xu Y, Hoffman TL, Zhang T, Schilling T, Sargent TD. Inca: a novel p21-activated kinase-associated protein required for cranial neural crest development. Development. 2007;134:1279–1289. doi: 10.1242/dev.02813. [DOI] [PubMed] [Google Scholar]
- Mayor R, Essex LJ, Bennett MF, Sargent MG. Distinct elements of the xsna promoter are required for mesodermal and ectodermal expression. Development. 1993;119:661–671. doi: 10.1242/dev.119.3.661. [DOI] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Fletcher RB, Harland RM. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic family and its role in neural and neural crest development. Mech. Dev. 1998;75:43–51. doi: 10.1016/s0925-4773(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Nie S, Chang C. Regulation of early Xenopus development by ErbB signaling. Dev. Dyn. 2006;235:301–314. doi: 10.1002/dvdy.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Chang C. Regulation of Xenopus gastrulation by ErbB signaling. Dev. Biol. 2007;303:93–107. doi: 10.1016/j.ydbio.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North-Holland; Amsterdam: 1975. [Google Scholar]
- Pasqualetti M, Ori M, Nardi I, Rijli FM. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development. 2000;127:5367–5378. doi: 10.1242/dev.127.24.5367. [DOI] [PubMed] [Google Scholar]
- Pohl BS, Knochel W. Overexpression of the transcriptional repressor FoxD3 prevents neural crest formation in Xenopus embryos. Mech. Dev. 2001;103:93–106. doi: 10.1016/s0925-4773(01)00334-3. [DOI] [PubMed] [Google Scholar]
- Ribisi S, Jr, Mariani FV, Aamar E, Lamb TM, Frank D, Harland RM. Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev. Biol. 2000;227:183–196. doi: 10.1006/dbio.2000.9889. [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet JP, He X, Varmus HE, Dawid IB. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc. Natl. Acad. Sci. USA. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- Schohl A, Fagotto F. A role for maternal beta-catenin in early mesoderm induction in Xenopus. EMBO J. 2003;22:3303–3313. doi: 10.1093/emboj/cdg328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, Sweeney C. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol. Cell. Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Smith JC, Price BM, Green JB, Weigel D, Herrmann BG. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Semin. Cell Dev. Biol. 2005;16:647–654. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Suga A, Hikasa H, Taira M. Xenopus ADAMTS1 negatively modulates FGF signaling independent of its metalloprotease activity. Dev. Biol. 2006;295:26–39. doi: 10.1016/j.ydbio.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- Tannahill D, Isaacs HV, Close MJ, Peters G, Slack JM. Developmental expression of the Xenopus int-2 (FGF-3) gene: activation by mesodermal and neural induction. Development. 1992;115:695–702. doi: 10.1242/dev.115.3.695. [DOI] [PubMed] [Google Scholar]
- Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci. STKE. 20042004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- Tsang M, Maegawa S, Kiang A, Habas R, Weinberg E, Dawid IB. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131:2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Umbhauer M, Marshall CJ, Mason CS, Old RW, Smith JC. Mesoderm induction in Xenopus caused by activation of MAP kinase. Nature. 1995;376:58–62. doi: 10.1038/376058a0. [DOI] [PubMed] [Google Scholar]
- Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J. Biol. Chem. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- Whitman M, Melton DA. Involvement of p21ras in Xenopus mesoderm induction. Nature. 1992;357:252–254. doi: 10.1038/357252a0. [DOI] [PubMed] [Google Scholar]
- Yanfeng W, Saint-Jeannet JP, Klein PS. Wnt-frizzled signaling in the induction and differentiation of the neural crest. BioEssays. 2003;25:317–325. doi: 10.1002/bies.10255. [DOI] [PubMed] [Google Scholar]
- Yokota C, Kofron M, Zuck M, Houston DW, Isaacs H, Asashima M, Wylie CC, Heasman J. A novel role for a nodal-related protein; Xnr3 regulates convergent extension movements via the FGF receptor. Development. 2003;130:2199–2212. doi: 10.1242/dev.00434. [DOI] [PubMed] [Google Scholar]
- Yoshitake Y, Howard TL, Christian JL, Hollenberg SM. Misexpression of Polycomb-group proteins in Xenopus alters anterior neural development and represses neural target genes. Dev. Biol. 1999;215:375–387. doi: 10.1006/dbio.1999.9473. [DOI] [PubMed] [Google Scholar]
- Zhao H, Cao Y, Grunz H. Isolation and characterization of a Xenopus gene (XMLP) encoding a MARCKS-like protein. Int. J. Dev. Biol. 2001;45:817–826. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.