Abstract
In Saccharomyces cerevisiae, SAL1 encodes a Ca2+-binding mitochondrial carrier. Disruption of SAL1 is synthetically lethal with the loss of a specific function associated with the Aac2 isoform of the ATP/ADP translocase. This novel activity of Aac2 is defined as the V function (for Viability of aac2 sal1 double mutant), which is independent of the ATP/ADP exchange activity required for respiratory growth (the R function). We found that co-inactivation of SAL1 and AAC2 leads to defects in mitochondrial translation and mitochondrial DNA (mtDNA) maintenance. Additionally, sal1Δ exacerbates the respiratory deficiency and mtDNA instability of ggc1Δ, shy1Δ and mtg1Δ mutants, which are known to reduce mitochondrial protein synthesis or protein complex assembly. The V function is complemented by the human Short Ca2+-binding Mitochondrial Carrier (SCaMC) protein, SCaMC-2, a putative ATP-Mg/Pi exchangers on the inner membrane. However, mitochondria lacking both Sal1p and Aac2p are not depleted of adenine nucleotides. The Aac2R252I and Aac2R253I variants mutated at the R252-254 triplet critical for nucleotide transport retain the V function. Likewise, Sal1p remains functionally active when the R479I and R481I mutations were introduced into the structurally equivalent R479-T480-R481 motif. Finally, we found that the naturally occurring V-R+ Aac1 isoform of adenine nucleotide translocase partially gains the V function at the expense of the R function by introducing the mutations P89L and A96V. Thus, our data support the view that the V function is independent of adenine nucleotide transport associated with Sal1p and Aac2p and this evolutionarily conserved activity affects multiple processes in mitochondria.
Keywords: Mitochondria, adenine nucleotide translocase, Sal1, Ca2+-binding, transport
Introduction
Mitochondria produce cellular energy by oxidative phosphorylation. The assembly of the oxidative phosphorylation apparatus requires a coordinated expression of both nuclear and mitochondrial genes (Tzagoloff and Dieckmann 1990; Attardi and Schatz 1988; Wallace 2005). The expression of mitochondrial genes is affected by a plethora of nuclear-encoded genes that are implicated in processes such as mtDNA replication, transcription and translation. In addition, these cellular processes require a continuous supply of substrates from the cytosol. Some of the substrate molecules could directly regulate mtDNA transactions. For instance, recent studies have shown that mtDNA copy number in yeast is regulated by the size of mitochondrial dNTP pools (Taylor et al. 2005). The ATP level in the mitochondrial matrix is also an important signal, which is sensed by mitochondrial RNA polymerase to activate transcription (Amiott and Jaehning 2006).
The homeostasis of substrates and regulatory molecules inside mitochondria is ensured by proteins of the mitochondrial carrier family (MCF). The yeast and human proteomes have 34 and at least 65 MCF members, respectively (del Arco and Satrustegui 2005; Kaplan 2001; Palmieri 2004; Wohlrab 2005). A typical MCF protein contains about 300-320 amino acid residues, and is predicted to have six transmembrane α-helices forming a barrel of pseudo-3-fold symmetry with a conspicuous central cavity for substrate translocation (Pebay-Peyroula et al. 2003). Recently, a subfamily of MCF proteins, called Ca2+-binding mitochondrial carriers (CaMC), has been identified. CaMC proteins contain a hydrophilic amino terminal extension of ∼350 amino acids harboring Ca2+-binding EF-hand motifs (del Arco et al. 2000; del Arco and Satrustegui 1998) and are proposed to fulfill their transport function in a Ca2+-regulated manner. The prototypical members of the CaMC family are the human citrin and aralar 1 proteins, which have been identified as aspartate/glutamate exchangers, which play a role in the urea cycle and the aspartate/malate NADH shuttle system (Palmieri et al. 2001). Mutations in citrin are associated with adult-onset type II citrullinemia in humans (del Arco et al. 2000).
Several CaMC proteins have a shorter Ca2+-binding domain of ∼200 residues, as represented by the yeast Sal1 protein. SAL1 (for Suppressor of aac2-lethality) was initially identified as a strain-polymorphic gene that maintains the viability of cells disrupted in AAC2, a gene that encodes the major isoform of the adenine nucleotide translocase (Chen 2004). However, SAL1 does not complement the respiratory deficiency of aac2 mutants. We have proposed that Aac2p is bifunctional. In addition to its function in catalyzing ATP/ADP exchange required for respiratory growth (the R function), it also promotes a novel transport activity essential for cell viability (the V function) in concert with Sal1p. In addition, we have found that an isoform of the yeast adenine nucleotide translocase, Aac1p, is unable to suppress the lethality of sal1 aac2 double mutant, despite the fact that Aac1p is capable of catalyzing ATP/ADP exchange and complementing the respiratory-deficient phenotype of the aac2 mutant. Aac1p is therefore naturally V-R+, in contrast to Aac2p and Sal1p which are V+R+ and V+R- respectively.
Although Sal1p cannot replace Aac2p in supporting respiratory growth, it displays significant sequence similarity to adenine nucleotide translocases. This led to the proposal that Sal1p may be a novel type of adenine nucleotide transporter (Robinson and Kunji 2006). Furthermore, Sal1p has three orthologs in humans, namely SCaMC-1, -2 and -3 (for Short CaMC) (del Arco and Satrustegui 2004), or APC1, APC3 and APC2 (for ATP-Mg/Pi Carrier) (Fiermonte et al. 2004), respectively. In reconstituted proteoliposomes, SCaMC-1 and -3 have been shown to promote the exchange between ATP, ADP, AMP or ATP-Mg, and phosphate (Fiermonte et al. 2004). It was assumed that SCaMC-2 may have similar activity. This transport activity is insensitive to carboxyatractyloside (CATR), a specific inhibitor of adenine nucleotide translocase. In the present study, we have found that sal1 mutant can be complemented by the human SCaMC-2 protein. The data demonstrate that the loss of Sal1 and Aac2 functions have profound implications for several fundamental processes including mitochondrial protein synthesis, mtDNA stability and the biogenesis of membrane protein complexes. These effects are not caused by depletion of adenine nucleotides in mitochondria. Mutagenic analysis of AAC2, SAL1 and AAC1 revealed that the structural requirement for the V and R functions can be genetically dissected.
Experimental procedures
Growth media and strains
Complete (YP) and minimal medium (YNB) were prepared with 2% dextrose (D), 2% galactose (Gal), 2% raffinose (Raf), 3% glycerol (G), or 2% lactate (L). Strains used in this study are listed in Table 1.
TABLE 1. Genotype and sources of yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| W303-1B | MATα, leu2-3, 112 his3-1,1 15 ura3-1 ade2-1 trp1-1 can1-100 SAL1 | R. Rothstein |
| CS415 | as W303-1B, but sal1Δ∷kan | (Chen 2004) |
| CS341 | as W303-1B, but aac2Δ∷kan | (Chen 2004) |
| CS523/3 | as W303-1B, but aac2Δ∷LEU2 sal1Δ∷kan ura3∷pURA3-GAL10-AAC2 | (Chen 2004) |
| BK3 | as CS415, but [pRS426-ADH1-SAL1] | This study |
| BK18 | as CS523/3, but [HS40] | This study |
| CS494-2B | MATa, leu2 his4 aac2Δ∷kan ura3∷GAL10-AAC2 sal1-1 | (Chen 2004) |
| CS277/2 | MATa/α, leu2/leu2 ura3/ura3 +/ade2 +/his4 sal1-1/sal1-1 +/aac2Δ∷LEU2 | (Chen 2004) |
| CS1345 | as W303-1B, but diploid, +/aac2Δ∷LEU2, +/sal1Δ∷kan | This study |
| BK16 | as W303-1B, but diploid, +/ggc1Δ∷kan +/sal1Δ∷kan | This study |
| BK16-1C | BK16 segregant, ggc1Δ∷kan | This study |
| BK16-1B | BK16 segregant, ggc1Δ∷kan sal1Δ∷kan | This study |
| BK37 | as W303-1B, but diploid, +/shy1Δ∷kan +/sal1Δ∷kan | This study |
| BK37-1B | BK37 segregant, shy1Δ∷kan | This study |
| BK37-1C | BK37 segregant, shy1Δ∷kan sal1Δ ∷kan | This study |
| BK38 | as W303-1B, but diploid, +/mtg1Δ∷kan +/sal1Δ∷kan | This study |
Isolation of mitochondria
Crude mitochondria were isolated from cells grown to late exponential phase as previously described (Diekert et al. 2001). Mitochondria used for HPLC analysis and determination of dATP levels were immediately frozen at -70°C after isolation. Those used for nucleotide transport experiments were further purified by Histodenz (Sigma) density gradient centrifugation.
Measurement of adenine nucleotide content by HPLC
Preparation of HClO4 mitochondrial extracts (2-3 mg of proteins) and the determination of nucleotide concentrations were performed as described previously (Giannattasio et al. 2003).
ATP uptake
Assays for intramitochondrial ATP accumulation were performed at 25°C as previously described (Aprille and Austin 1981). Mitochondria (0.4 mg of proteins) were added to 0.5 ml of incubation mixture containing 0.6 M mannitol, 2 mM K2PO4/KH2PO4, pH 6.8, 10 mM Tris-maleate, pH 6.8, 5 mM MgCl2, 10 mM KCl and 0.1% ethanol, with or without 5 μM carboxyatractyloside (Calbiochem) and CaCl2. 20 seconds after the addition of mitochondria, 4 mM [8-14C]-ATP (∼50 nCi/μmol, Amersham) was added. The reaction was stopped after 2 min by the addition of 10 ml ice cold 0.3 M NaCl and was applied to Millipore HNWP filters under vacuum. Collected mitochondria were washed with 10 ml of 0.3 M NaCl. The radioactivity retained on the filters was counted using scintillation counter LS 6500 (Beckman).
Determination of mitochondrial dATP concentration
The mitochondrial dATP concentrations were determined by an enzymatic assay as previously described (Roy et al. 1999). Crude mitochondria isolated from cells grown in YPD were extracted with 60% cold methanol ethanol at the protein concentration of 3 mg/ml. Just before use, dried extracts were dissolved in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2 to concentrations that would correspond to 100 mg of proteins/ml. Oligonucleotides p13 (5′-TCGCAGCCGTCCA) and tA (3′-AGCGTCGGCAGGTAATAATAATAA) were used for the preparation of the primer/template duplex. dATP determination assays ware performed using unlabelled and labelled duplexes mixed at a 97:3 molar ratio. The polymerase reaction, which was performed for 20 min at 37°C in total volume of 10 μl, contained 1.5 pmol of duplex, 5 μM dTTP, 10 mM Tris-HCl, pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiotreitol, 0.25U of Klenow fragment, and 1 μl of mitochondrial extract. The reaction was stopped by the addition of an equal volume of formamide loading buffer (80% formamide, 10 mM EDTA, pH 8.0, 0.1% xylene cyanol, 0.1% bromophenol blue). The DNA products were separated by PAGE electrophoresis in 12% polyacrylamide 8M urea gel. Autoradiography of the gel was performed using phosphorimaging plates and analyzed by a STORM 820 phopshoimager (Molecular Dynamics). Densitometric analysis of each band was performed using ImageQuant software. The amount of dATP in pmol was calculated according to the formula [pmol of primer × (1 × PR1 + 2 × PR2 + 3 × PR3)]/total signal in lane, where PR1 represents primer elongated for TTA, PR2 for TTATTA and PR3 for TTATTATTATT.
Mitochondrial translation
Mitochondrial translation products were labeled in vivo as previously described (Rodeheffer and Shadel 2003; Westermann et al. 2001) with minor modifications. Prior to labelling, collected cells were suspended to an OD600=6-12 and incubated in YNBGal or YNBD at 30° for 1 hour. Mitochondria were isolated as described (Fox et al. 1991) and resuspended in 10 μl of sample buffer (40 mM Tris-Cl, pH 6.8, 8 M urea, 5% SDS, 0.1 mM EDTA, pH 7.5, 1% β-mercaptoethanol, 0.01% bromophenol blue). Proteins were denatured at 92°C for 2 min, separated by electrophoresis in a 17% SDS-PAGE gel, blotted onto a nitrocellulose membrane and visualized by autoradiography using phosphorimaging plates.
Expression of human SCaMC-1, -2 and -3
The ORFs of the human cDNA clones containing SCaMC-1, -2 and -3 (kindly provided by A. del Arco, Universidad Autónoma, Spain) were amplified by PCR and cloned into the vector pRS415-ADH1. The coding sequence of SCaMC-1 corresponds to variant 1 of SLC25A24 in the NCBI database, except that the N- and C-termini are shortened by 6 and 13 amino acids, respectively. SCaMC-2 corresponds to variant A of SLC25A25, and SCaMC-3 matches SLC25A23. When compared to the cDNA clones used by Palmieri's group for in vitro transport assay (Fiermonte et al. 2004), SCaMC-1 matches APC1 except for the first 54 residues. SCaMC-3 is identical to APC2 and SCaMC-2 matches APC3 except for the first 53 residues. For the c-Myc tagged variants of the human genes, the epitope was added on their N-terminus and cloned into pRS415-ADH1.
Analysis of transcripts from HS40
For detection of mitochondrial transcripts from the HS40 ρ- genome, [32P]-labeled oligonucleotide 5′-GTGCTTTGTATTTATTGAATATTCTGG was used as probe for ori5.
Random mutagenesis of AAC1
Hydroxylamine mutagenesis was performed to introduce random mutations into AAC1. Briefly, 10 μg of the plasmid pCXJ24-AAC2-1 (LEU2), in which the AAC1 coding sequence was placed under the control of the AAC2 promoter, was mixed with 500 μl of freshly made hydroxylamine solution (1 M hydroxylamine-HCl, 0.45 N NaOH). After incubation for 20 hours at 56°C, the DNA was purified on a Quick-Spin column (QIAGEN). The mutagenized plasmids were transformed into CS494-2B on YNBGal medium. Approximately 600 Leu+ transformants were screened for the formation of viable colonies on YPD medium. The plasmids in the viable colonies were rescued and re-transformed into CS494-2B to test the ability to complement the V function. The positive plasmids were sequenced to identify the mutations.
Other methods
A HIS6-tagged truncated SAL1 allele comprising the first 220 codons was expressed in E. coli. The purified protein was used to raise a polyclonal antibody in rabbit through a commercial service. For transmission electron microscopy (TEM), cells grown aerobically for 24 hours in YPGal+Raf medium were fixed with glutaraldehyde/KMnO4 and stained by uranyl acetate as described (Wright 2000). TEM was performed using JEOL 1200 EX microscope. The aac2R252I, aac2R253I, aac2R254I, sal1R479I and sal1R481I alleles were generated by changing the AGA codons for arginine to ATA that codes for isoleucine using site-directed in vitro mutagenesis (Stratagene).
Results
Membrane topology and expression of Sal1p
Sal1p is a bipartite protein. The predicted transmembrane domain is preceded by a 220-residue amino-terminal extension which harbors two recognizable Ca2+-binding EF-hand motifs. Sal1 is localized in mitochondria (Cavero et al. 2005). To determine the membrane topology of Sal1p, we prepared a polyclonal antibody that specifically targets the 220-residue N-terminal segment. Like Aac2p and Ilv5p, markers for the inner membrane and matrix proteins respectively, Sal1p is protected from proteinase K digestion in intact isolated mitochondria (Figure 1A). The outer membrane protein Mmm1p is sensitive to proteinase K digestion under these conditions. Upon osmotic swelling, both Aac2p and Sal1p became accessible to proteinase K while Ilv5p remains largely intact. The hypersensitivity to proteolysis suggests that Sal1p is an inner membrane protein that likely adopts the conventional Nout-Cout configuration on the inner membrane with the Ca2+-binding domain facing to the intermembrane space (Figure 1B), as is the case with the human CaMC proteins (Palmieri et al. 2001). Immunoblot analysis showed that cells grown in media containing glucose, glycerol or galactose maintain a comparable steady state level of Sal1p, indicating that expression of SAL1 is not subject to glucose repression (data not shown). Compared with the wild-type, the sal1Δ mutant has only a mildly reduced growth rate in complete medium containing a non-fermentable carbon source (data not shown).
Fig. 1.
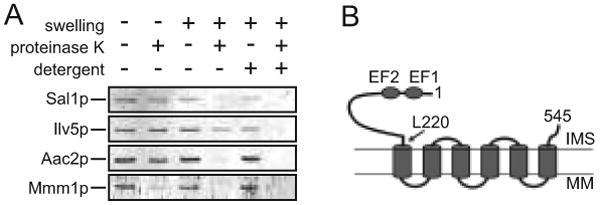
Membrane topology of Sal1p and growth characteristics of the sal1Δ mutant. (A) Immunoblot analysis using the anti-Sal11-220 antibody. Isolated mitochondria were incubated in 0.6 M or 0.06 M sorbitol to induce mitochondrial swelling, with or without the addition of proteinase K and NP40. Ilv5p was used as a marker for mitochondrial matrix proteins and Mmm1p and Aac2p were used as markers for the outer and inner membrane proteins, respectively. (B) Schematic representation of Sal1p on the mitochondrial inner membrane. Indicated are the first and last codons, and the codon L220 separating the Ca2+-binding and the transmembrane domains. EF1 and EF2, Ca2+-binding EF-hand motifs; IMS, intermembrane space; MM, mitochondrial matrix.
Loss of ρ+ but not ρ- mtDNA in the sal1 aac2 double mutant
To evaluate the mitochondrial defects in the sal1 aac2 double mutant, we developed a procedure for preparing cells lacking both Sal1p and Aac2p. After sporulation and dissection of the diploid strain CS277 (sal1-1/sal1-1, +/aac2Δ) on YPD, the spores were germinated under anaerobic conditions to derepress AAC3 that encodes a V+R+ isoform of adenine nucleotide translocase. Viable meiotic segregants carrying both aac2Δ and sal1-1 were then returned to aerobic conditions, where they can only divide for ∼12 generations before a complete cessation of cell division (Figure 2A). Examination by electron microscopy revealed that mitochondria from the sal1-1 aac2Δ double mutant are grossly swollen, deprived of cristae structure and accumulate materials of low electron density in the matrix (Figure 2B).
Fig. 2.
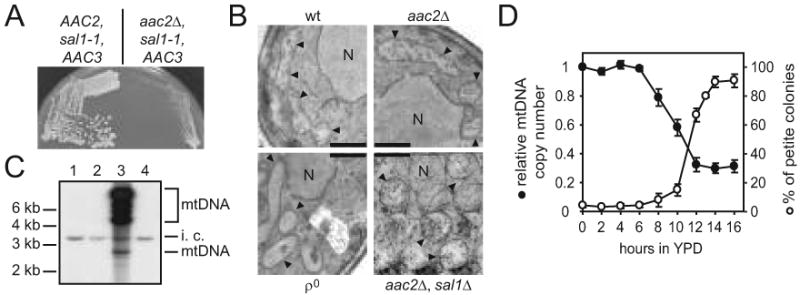
The sal1 aac2 double mutant is unable to maintain ρ+ mtDNA. (A) Anaerobically grown meiotic segregants from CS277/2 (sal1-1/sal1-1 +/aac2Δ) were streaked onto YPD medium and incubated aerobically for three days. Left half panel, growth of a sal1-1 segregant; right half panel, growth of a segregant carrying both the aac2Δ and sal1-1 alleles. (B) Transmission electron microscopy of aerobically-grown W303-1B (wt), CS341 (aac2Δ), CS1345 (+/aac2Δ, +/sal1Δ) segregant (aac2Δ, sal1Δ) and W303-1B ρ° (ρ°). Scale bar = 0.5 μm; N, nucleus, arrowheads, mitochondria. (C) Southern blot analysis of CfoI-digested total cellular DNA isolated from cells grown aerobically in YPD. Total mtDNA was used as probe. Lane 1 and 2, representative meiotic segregants of CS277/2 carrying both the sal1-1 and aac2Δ alleles; lane 3, wild type control; lane 4, ρ° control. i.c., internal control for DNA loading that results from cross-hybridization of mtDNA probe to nuclear ribosomal DNA.(D) Petite production and mtDNA levels in CS523/3 (aac2Δ, sal1Δ, GAL10-AAC2) upon depletion of Aac2p. Cells were collected after a shift from YPGal to YPD. The frequency of petite colony formation (open circles) was estimated in 3-6 independent cultures for each time point after plating cells onto YPGal. The relative mtDNA copy number (full circles) in two of these cultures was calculated as mtDNA/nuclear DNA ratio estimated by Southern-blot analysis of HaeIII digested total DNA hybridized with probes for mitochondrial 21S rRNA and for the nuclear gene ACT1. Both plots represent average values with average deviations.
The lack of cristae is reminiscent of phenotypes associated with ρ° and ρ- conditions or with cells impaired in membrane organization (Stevens 1981; Lefebvre-Legendre et al. 2005). Indeed, Southern-blot analysis failed to detect mtDNA in the aerobically arrested cells lacking both Sal1p and Aac2p (Figure 2C). In a parallel experiment, we depleted Aac2p in the strain CS523/3 (sal1Δ, aac2Δ, GAL10-AAC2) by repressing the expression of the GAL10-AAC2 cassette in glucose medium and monitored petite formation (Figure 2D). After 14 hours of glucose repression, over 75% of the cells were capable of forming viable colonies when returned to YPGal. However, 90% of these colonies were petite. Thus, cells lacking both Sal1p and Aac2p are unable to maintain a functional mitochondrial genome, which may consequently affect cell viability because of the ρ°-lethal nature of aac2 cells (see Discussion).
A possible explanation for the mtDNA instability is that Sal1p and Aac2p may play a role in maintaining the level of a substrate essential for mtDNA replication. One such candidate molecule is dATP, which is likely imported from the cytosol since yeast mitochondria are not thought to have the ribonucleotide reductase and nucleoside diphospho-kinase, enzymes required to convert ADP into dADP and dATP. Using an enzymatic assay, we found no significant difference in dATP levels in mitochondria isolated from CS415 (sal1Δ), CS341 (aac2Δ), CS523/3 (sal1Δ, aac2Δ, GAL10-AAC2) and wild-type cells grown in YPD medium (Figure 3A and 3B).
Fig. 3.
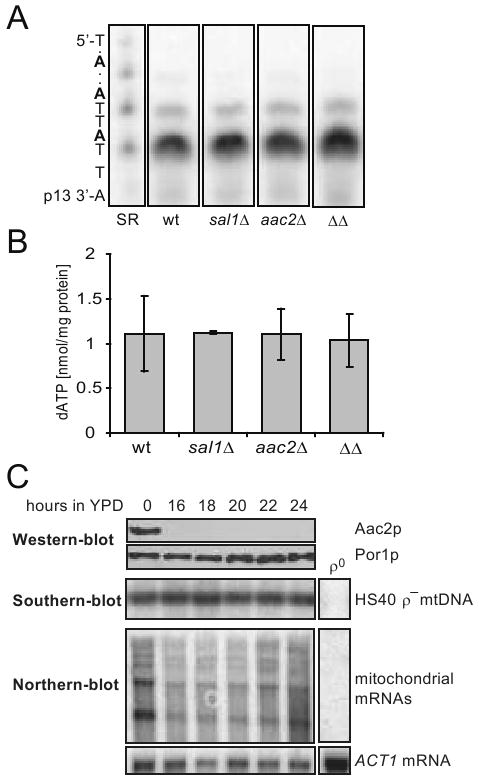
dATP content in purified mitochondria and the stability of HS40 ρ- mtDNA in cells lacking Sal1p and Aac2p. (A) dATP in mitochondria isolated from YPD-grown W303-1B (wt), CS415 (sal1Δ), CS341 (aac2Δ), and CS523/3 (aac2Δ, sal1Δ, GAL10-AAC2; ΔΔ). Mitochondria were prepared from cells grown in YPD after approximately 10 doubling times. Representative 10% polyacrylamide-urea gel with elongated products containing one and two incorporated dATPs. SR, standard reaction with 1 pmole of dATP. (B) dATP content in mitochondrial extracts calculated after signal quantification. Plot represents average of two independent experiments for each strain, error bars represent average deviations. (C) BK18 cells grown in YPGal (aac2Δ, sal1Δ, GAL10-AAC2, [HS40] were shifted to YPD. Aliquots were collected in 2 hour intervals and subjected to western (upper panel), Southern (middle panel) and northern-blot (bottom panel) analysis for monitoring the level of Aac2p, HS40 mtDNA and mitochondrial transcripts, respectively. The HS40 mtDNA was used as probe for Southern-blot analysis. The probe for Northern analysis is described in Materials and Methods. The transcripts are multiple lengths because the ρ- genome is organized in the form of tandem repeats.
Further evidence excluding a direct role of Sal1p and Aac2p in mtDNA replication came from the observation that cells lacking both proteins are capable of maintaining ρ- mtDNA. By cytoduction, we replaced the wild-type mitochondrial genome in the strain CS523/3 (sal1Δ, aac2Δ, GAL10-AAC2) with the 760-bp ρ- mtDNA, HS40 (Parikh et al. 1989). The resulting strain, BK18, is viable on YPGal and can stably maintain HS40. After incubation for 16-24 hours in YPD, Aac2p was completely depleted (Figure 3C, upper panel). Interestingly, in contrast to the ρ+ genome, no depletion of the HS40 ρ- genome was observed (Figure 3C, middle panel). Sal1p and Aac2p are therefore not required to maintain the ρ- genome.
Effect on mitochondrial protein synthesis
The most common conditions that permit cells to preferentially maintain ρ- but not ρ+ genomes are nuclear mutations that affect mitochondrial transcription and translation (Fangman et al. 1990; Myers et al. 1985; Weislogel and Butow 1970). The retention of the HS40 ρ- genome in BK18 (sal1Δ, aac2Δ, GAL10-AAC2, [HS40]) cells after Aac2p-depletion allowed us first to examine whether mitochondrial transcription is affected. By Northern-blot analysis, we found that in cells lacking both Sal1p and Aac2p, transcription from the ori5 promoter was initially reduced during incubation in YPD because of glucose repression. The transcripts are more abundant in later time points during incubation and ultimately reach a level comparable to that in the YPGal-grown pre-cultures (Figure 3C, lower panel). Thus, in the sal1 aac2 double mutant, active mitochondrial transcription is retained.
We then tested whether the mitochondrial protein synthesis machinery is affected in cells in which SAL1 and AAC2 are co-inactivated. Using the strain CS523/3 (sal1Δ, aac2Δ, GAL10-AAC2), we simultaneously followed the steady state levels of mitochondrial translation products, mtDNA and mtRNA during Aac2p-depletion. It was found that Aac2p was depleted after 4-6 hours of glucose repression. This was followed by the disappearance of mitochondrially encoded Cox2p with a lag of 2-4 hours (Figure 4A). Loss of the mitochondrial translation product clearly occurs before mtDNA instability which becomes significant only after 10 hours of glucose repression (Figure 2D). In these cells, COX2 mRNA persisted even after 16 hours of cultivation despite a relatively low mtDNA copy number.
Fig. 4.
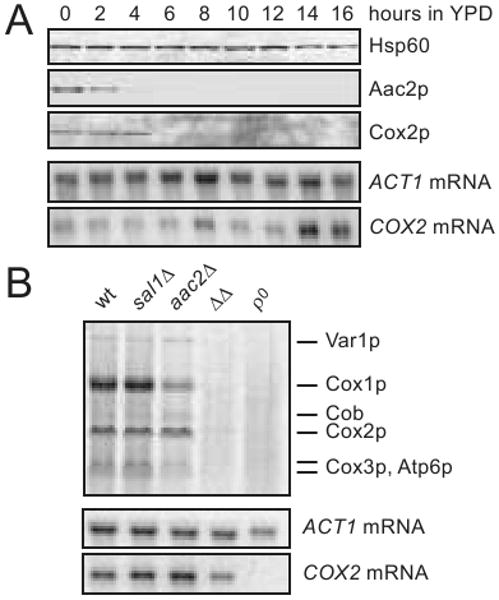
Defects in mitochondrial protein synthesis in cells lacking Sal1p and Aac2p. (A) Complete loss of the mitochondrially encoded Cox2p following the depletion of Aac2p in CS523/3 (aac2Δ, sal1Δ, GAL10-AAC2) during growth in YPD. Hsp60 was used as loading control in the western-blot analysis. COX2 and ACT1 mRNA levels were monitored by northern-blot analysis. (B) In vivo labeling of mitochondrial translation products in W303-1B (wt), CS415 (sal1Δ), CS341 (aac2Δ), CS523/3 (ΔΔ) and W303-1B ρ° (ρ°) cells grown for 15 hours in YPD. [35S]-methionine incorporation proceeded in YNBD for 15 min at 30°C (Rodeheffer and Shadel 2003). Cells were collected in parallel for northern-blot analysis of COX2 and ACT1 mRNAs.
[35S]-methionine labeling of mitochondrial translation products provided further evidence that co-inactivation of SAL1 and AAC2 affects mitochondrial protein synthesis. After repression of AAC2 expression for 16 hours in CS523/3 (sal1Δ, aac2Δ, GAL10-AAC2), COX2 mRNA levels were moderately reduced (Figure 4B, lower panel). However, the incorporation of [35S]-methionine into mitochondrial translation products was globally reduced to a barely detectable level (Figure 4B, upper panel). Incorporation of [35S]-methionine was severely reduced but remained detectable with mitochondria isolated after depletion of AAC2 expression for 10 hours (data not shown).
The possible effect of Sal1p on mitochondrial protein synthesis was further supported by examining the genetic interactions between sal1Δ and mutations known to affect mitochondrial protein translation or biogenesis, even in the presence of a functional AAC2. GGC1 encodes the mitochondrial GTP/GDP exchanger required for GTP homeostasis and protein synthesis (Vozza et al. 2004). After sporulation and dissection of BK16, heterozygous for both sal1Δ and ggc1Δ, the meiotic segregants were replica-plated onto YPGly to test for respiratory competency (Figure 5A). We found that both ggc1Δ and sal1Δ single mutants are capable of forming colonies, whereas all the double mutants failed to grow.
Fig. 5.
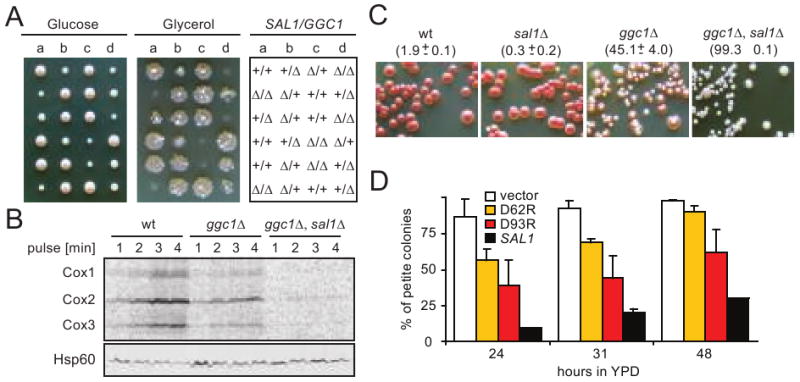
Disruption of SAL1 exacerbates the mtDNA instability and mitochondrial translation defects of the ggc1 mutant. (A) Cells carrying both sal1Δ and ggc1Δ form respiratory deficient petite colonies on YPD. BK16 (+/ggc1Δ +/sal1Δ) was sporulated and dissected on YPD (left panel). After four days of incubation at 30°C, the meiotic segregants were replica-plated onto YPG (middle panel). The genotypes of the segregants are indicated (right panel). (B) In vivo mitochondrial translation in YPG-grown W303-1B (wt), BK16-1C (ggc1Δ), and BK16-1B (ggc1Δ sal1Δ) performed in YNBGal at 25°C (Rodeheffer and Shadel 2003; Westermann et al. 2001). Hsp60 was used as a loading control (lower palnel). (C) Growth phenotype of ggc1 and sal1 mutants on YPD. Cells were shifted from YPG to YPD, grown for 48 hours and plated onto YPD. The petite frequencies (%), scored after incubation at 30° for 4 (wt and sal1Δ) and 10 (ggc1Δ, sal1Δ ggc1Δ) days are indicated in the parentheses. The values represent the average of two experiments with average deviations. (D) Calcium binding domains are important for Sal1p function in vivo. The formation of petite colonies was estimated in strain BK16-1B (ggc1Δ, sal1Δ) transformed with empty pCXJ24 (vector) and pCXJ24 bearing wild type SAL1 (SAL1), or alleles with single amino acid substitutions in putative calcium binding EF-hand motifs sal1[D62R] (D62R) and sal1[D93R] (D93R) after a shift from YNBGly to YNBD medium. Plot represents average of two independent experiments for each strain; error bars represent average deviations.
By dissecting BK16 on complete glycerol medium, we obtained respiratory competent sal1Δ and ggc1Δ double mutants capable of retaining mtDNA, which permitted us to measure mtDNA instability and the rate of mitochondrial protein synthesis. In comparison to wild-type, the ggc1Δ mutant had a significantly reduced translation activity. This defect was further exacerbated in the sal1Δ ggc1Δ double mutant (Figure 5B). The synergistic interaction between sal1Δ and ggc1Δ can also be deduced from the mtDNA instability phenotype observed after cultivation in glucose medium. The sal1Δ mutant has a stable mitochondrial genome, whereas mtDNA stability is significantly compromised in the ggc1Δ mutant (Figure 5C). More importantly, sal1Δ ggc1Δ double mutants have much higher levels of petite formation. mtDNA instability in the sal1Δ ggc1Δ double mutant is largely suppressed by the introduction of SAL1, but poorly suppressed by the sal1D62R and sal1D93R alleles mutated in the Ca2+-binding EF-hand motifs (Figure 5D), suggesting that the function of Sal1p in maintaining mitochondrial protein synthesis is Ca2+-dependent. The sal1D62R and sal1D93R alleles have previously been shown to affect Sal1 function (Chen 2004). By using a more robust in vivo functional test, we have recently found that the sal1D62R and sal1D93R alleles retain a residual Sal1 function in vivo (data not shown). This activity may explain the partial suppression of petite formation in the sal1Δ ggc1Δ double mutant.
The genetic interaction with ggc1Δ suggested that Sal1p, and possibly also Aac2p, affect mitochondrial protein synthesis by directly importing GTP. However, our epistasis analysis indicated that this is an unlikely scenario. We found that ggc1Δ, which has a much more severe defect in mitochondrial protein synthesis and mtDNA maintenance than sal1Δ, is not synthetically lethal with aac2Δ (data not shown). Thus, SAL1, but not GGC1, is epistatic to AAC2. This suggests that Sal1p and Aac2p do not promote the same transport process as Ggc1p.
sal1Δ was also found to synergize the defect in mitochondrial protein synthesis or biogenesis in shy1Δ and mtg1Δ mutants. Shy1p is required for efficient synthesis and/or assembly of cytochrome oxidase subunit I. shy1Δ mutants retain only 10-15% fully assembled and functional cytochrome c oxidase activity (Barrientos et al. 2002; Mashkevich et al. 1997; Nijtmans et al. 2001). Mtg1p is a putative GTPase that likely functions in the assembly of the large ribosomal subunit in mitochondria. mtg1Δ mutants exhibit low respiratory activity (Barrientos et al. 2003). We found that although the shy1Δ mutant exhibits a detectable growth defect on YPD plates within 3-5 days (not illustrated), it becomes undistinguishable from wild type and sal1Δ single mutant by forming large red colonies after extended incubation period (Figure 6A). However, the shy1Δ sal1Δ double mutant forms either light pink or white colonies, which is indicative of a severe respiratory deficiency. Thus, sal1Δ is synergistic to shy1Δ for respiratory deficiency. This is further supported by a quantitative assay showing that shy1Δ sal1Δ double mutants produce petite colonies at a frequency significantly higher than that for shy1Δ or sal1Δ single mutants (Figure 6B).
Fig. 6.
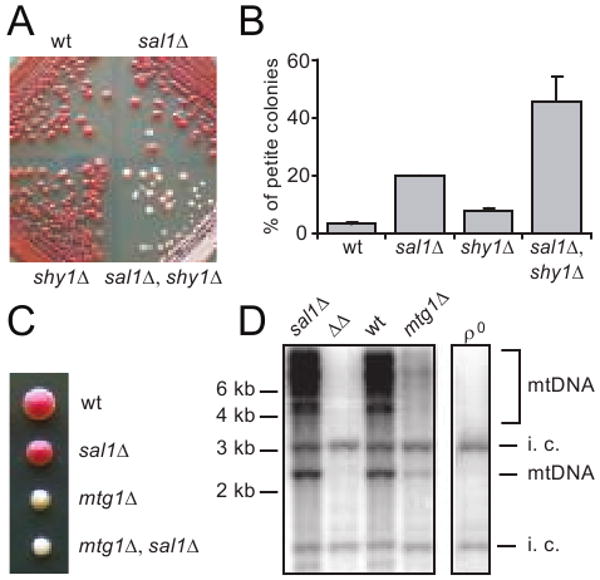
Disruption of SAL1 exacerbates the respiratory deficient and mtDNA instability phenotypes of shy1Δ and mtg1Δ mutants. (A) Growth phenotype of W303-1B (wt), CS415 (sal1Δ), BK37-1B (shy1Δ) and BK37-1C (sal1Δ shy1Δ) on YPD after 10 days at 30°. (B) Petite frequency in strains shown in (A) after 31 hrs of growth in YP supplemented with 10% glucose. The values represent the average of two independent experiments with average deviations. (C) A representative tetrad from BK38 (+/mtg1Δ, +/sal1Δ) dissected on YPD. (D) Southern-blot analysis of CfoI-digested total cellular DNA extracted from a complete tetrad grown in YPGal. Total mtDNA was used as probe. i.c. internal control for loading. ΔΔ, sal1Δ mtg1Δ mutant.
Likewise, we found that mtg1Δ sal1Δ double mutants form completely white colonies after dissection on YPD medium, in contrast to mtg1Δ single mutants that form colonies of light but yet visible pink color (Figure 6C). Southern blot analysis showed that mtg1Δ single mutants have reduced mtDNA levels, whereas mtDNA is undetectable in mtg1Δ sal1Δ double mutants (Figure 6D). Taken together, these data strongly suggest that loss of Sal1 function further decreases mitochondrial protein synthesis or protein complex biogenesis in ggc1Δ, shy1Δ and mtg1Δ mutants.
Complementation of the sal1 mutant by the human SCaMC-2 protein
The membrane regions of Sal1p display 31-35% sequence identity to those in the human SCaMC-1, -2 and -3 proteins. SCaMC-1 and -3 have been assigned as ATP-Mg/Pi exchangers (Fiermonte et al. 2004). For the complementation test, we expressed the three human genes in the strain CS494-2B (sal1-1, aac2Δ, GAL10-AAC2) in which the sal1-1 allele produces a truncated nonfunctional protein. The transformants carrying an empty vector failed to form colonies on glucose medium, since AAC2 expression is repressed (Figure 7A). We found that the expression of SCaMC-2, but not SCaMC-1 and -3, suppresses cell lethality. SCaMC-2 is therefore a functional homolog of Sal1p. As expected, none of the human proteins supports respiratory growth on medium containing glycerol when introduced into a yeast aac2 mutant strain (but SAL1) (Figure 7B), indicating that these proteins do not catalyze ATP/ADP exchange in vivo. The three human genes are all expressed and targeted into mitochondria in yeast (Figure 7C).
Fig. 7.
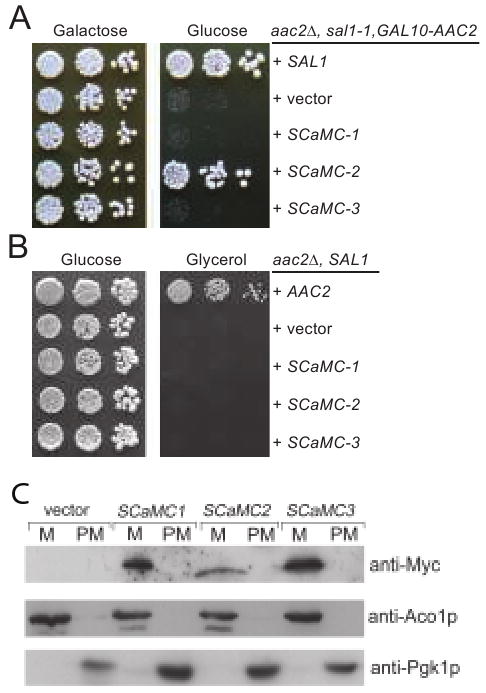
Expression of human SCaMC-1, -2 and -3 in yeast. (A) SCaMC-2 suppresses lethality in cells inactivated for both SAL1 and AAC2. Serial dilutions of CS494-2B (aac2Δ, GAL10-AAC2, sal1-1) expressing SCaMC-1, -2 and -3 from the ADH1 promoter were spotted onto YPGal and YPD. (B) SCaMC-1, -2 and -3 are unable to complement the respiratory deficiency of aac2 mutant. CS341 (aac2Δ, but SAL1) cells expressing SCaMC-1, -2 and -3 from the ADH1 promoter were diluted in water and spotted on YPD and YPG. Plates were incubated for five days at 30°C before being photographed. C. The expression levels and mitochondrial targeting of Myc-SCaMC-1, Myc-SCaMC-2 and Myc-SCaMC-1 in yeast. Yeast transformants expressing the human genes from the ADH1 promoter were grown in YPD and fractionated into mitochondrial pellet (M) and post-mitochondrial supernatant (PM). Proteins were separated by SDS-PAGE and immunobloted with the anti-Myc antibody. The Aco1 and Pgk1 proteins were used as markers for mitochondrial and cytosolic proteins respectively.
sal1 aac2 double mutant is not depleted of adenine nucleotides
To test whether the V function is related to the maintenance of adenine nucleotide homeostasis in mitochondria, we directly measured a possible role of Sal1p in ATP import. When isolated mitochondria were incubated in medium containing 2 mM phosphate as a counterion, CATR-sensitive ATP uptake mediated by Aac2p was readily detectable (Figure 8A). However, in the presence of CATR, mitochondria from the strains CS415 and BK3 do not exhibit decreased or increased ATP accumulation regardless the presence or absence of Ca2+. CS415 carries a null mutation for SAL1 and BK3 overexpresses Sal1p at a level ∼11 fold higher than that in the wild type (Figure 8A, insert), Thus, Sal1p does not seem to confer a detectable ATP-Mg(ext)/Pi(int) exchange activity under these experimental conditions.
Fig. 8.
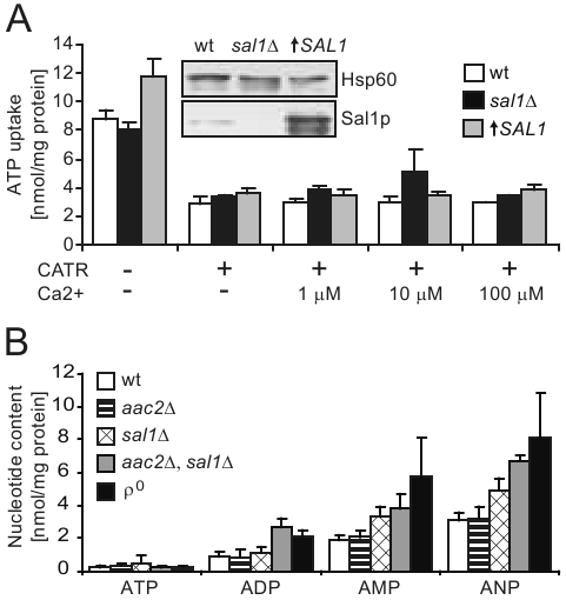
Mitochondrial ATP uptake and adenine nucleotide content. (A) Uptake of ATP by mitochondria isolated from YPL-grown cells with disruption or overexpression of SAL1, in the presence or absence of Ca2+ and carboxyatractyloside (CATR). The values represent an average of 2-5 independent experiments for each strain. Error bars represent average deviations. The steady state level of Sal1p in W303-1B (wt), CS415 (sal1Δ), and BK3 (↑SAL1) in shown by western-blot analysis (insert). (B) HPLC analysis of adenine nucleotide content in mitochondria isolated from YPD-grown W303-1B (wt), CS341 (aac2Δ), CS415 (sal1Δ), CS523/3 (aac2Δ, sal1Δ, GAL10-AAC2), and W303-1B ρ° (ρ°). Values represent averages from 3-4 independent measurements for each strain. Error bars represent average deviations.
If Sal1p affects mtDNA stability and mitochondrial protein synthesis by affecting the net accumulation of adenine nucleotides, we would also expect a depletion of the nucleotides in the matrix of sal1 aac2 double mutants. To test this, we analyzed mitochondrial adenine nucleotide contents by using a HPLC-based procedure. In contrary to our prediction, we found that mitochondria from CS523/3 (sal1Δ, aac2Δ, GAL10-AAC2) grown in YPD accumulate adenine nucleotides, with the total adenine nucleotide content (ANP) 2.2 fold higher than wild-type levels (Figure 8B). As CS523/3 is depleted of mtDNA under these conditions, mitochondria from ρ° cells were used as control. A comparable elevation of adenine nucleotide levels was found. This suggests that the noticeable elevation of ANP in the cells lacking both Sal1p and Aac2p may be an attribute of the ρ° conditions. Taken together, our data strongly indicate that there is no adenine nucleotide depletion associated with the co-inactivation of SAL1 and AAC2. A third molecular entity may contribute to maintain the adenine nucleotide levels in mitochondria in the absence of these two genes.
Amino acids essential for adenine nucleotide transport are not necessarily required for the V function
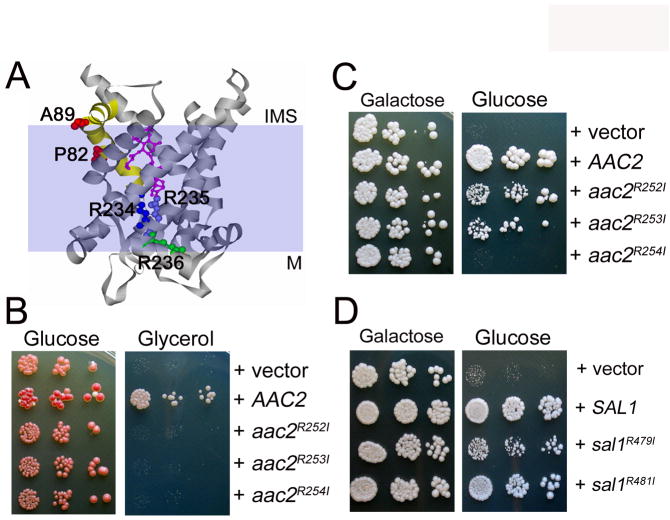
Further experiments supported the idea that the V function is likely independent of adenine nucleotide transport. We generated the aac2R252I and aac2R253I and aac2R254I alleles. These arginine residues are equivalent to Arg234, Arg235 and Arg236 in the bovine Ant1, which are conserved across all species. The arginine triplet is proposed to act as a transport switch, with the first two residues facing outside for the binding of adenine nucleotides from the cytosolic side. The third arginine residue faces inside, and is likely required for the binding of nucleotides from the matrix (Pebay-Peyroula et al. 2003) (see Figure 9A). Mutations to any of the arginine residues lead to a respiratory deficiency (Figure 9B) and have barely detectable nucleotide transport activity in all possible modes including ADP/ADP, ADP/ATP, ATP/ADP and ATP/ATP exchanges (Heidkamper et al. 1996). Interestingly, we found that aac2R252I and aac2R253I retain a significant activity in supporting cell viability (or V+) in the absence of Sal1p (Figure 9C). In contrast, aac2R254I was found to be V-.
Fig. 9.
Mutagenic bisection of Aac2p and Sal1p. (A) Projected localization of Pro82 and Ala89 (red) on the transmembrane helix 2 (yellow), and the nucleotide-binding Arg234 (dark blue), Arg235 (light blue) and Arg236 (green) in the crystal structure of bovine Ant1 bound by carboxyatractyloside (magenta) (Pebay-Peyroula et al. 2003). These amino acids correspond to Pro89 and Ala96 of Aac1p and to Arg252, 253 and 254 of Aac2p respectively. M, matrix; IMS, inter-membrane space. (B) Complementation of the aac2 (but SAL1) mutant for respiratory growth by mutant aac2 alleles. CS341 was transformed with the centromeric pCXJ24 (LEU2) expressing AAC2, aac2R252I, aac2R253I and aac2R254I respectively and the transformants were tested for respiratory growth on YPGly. (C) aac2R252I and aac2R253I, but not aac2R254I, retain the V function. Transformants of CS494-2B (aac2Δ, GAL10-AAC2, sal1-1) were tested on YPD to deplete the expression of AAC2. (D) Arg479 and Argh481 are not essential for Sal1 function. Transformants of CS494-2B expressing the sal1R479I and sal1R481I alleles were tested for cell viability on YPD.
In place of the Arg252-254 triplet in Aac2p, Sal1p has the Arg479-Thr480-Arg481 motif which is conserved among numerous non-nucleotide transporters (Nelson et al. 1998). We introduced the R479I and R481I mutations and found that that R481I has little effect on the Sal1 function, whereas R479I partially retains the capability to support cell growth (Figure 9D). Thus, none of these positively charged residues is essential for Sal1 function.
Aac1p partially gains the V function by specific mutations in a region putatively controlling substrate entrance from the cytosolic side
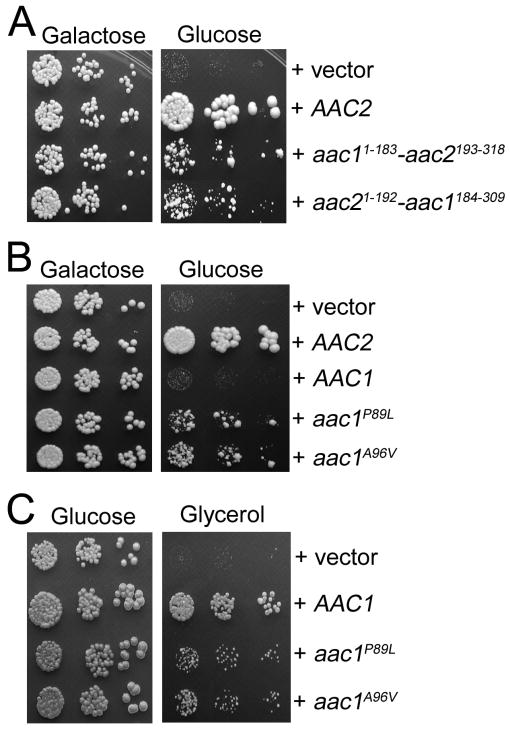
We previously reported that unlike Aac2p, the Aac1 isoform of adenine nucleotide translocase, which is otherwise active in promoting ADP/ATP exchange, intrinsically lacks the V function (Chen 2004). We reasoned that understanding the molecular basis for the difference between Aac1p and Aac2p could provide insight into the nature of the V function. Although Aac1p and Aac2p share 78.2% identity in amino acid sequence, the two proteins are most divergent in the first cytosolic loop and the second matrix loop, which are involved in substrate recognition from both sides of the membrane. Using an in vivo functional assay, we found that conversion of these regions of Aac2p to those found in Aac1p does not reduce the V function of Aac2p (data not shown). This suggests that these regions are not essential for the V function. We subsequently constructed the two chimeras, Aac11-183-Aac2193-318 and Aac21-192-Aac1184-309, each containing three trans-membrane domains of Aac1p and Aac2p on the N- or C-termini respectively. Interestingly, both chimeras retained partial activity for the V function (Figure 10A). Therefore, it appears that both moieties of Aac2p contribute to the V function.
Fig. 10.
Analysis of Aac1-Aac2 chimeras and the gain-of-function P89L and A96V mutants of Aac1p. (A) Serial dilutions of CS494-2B (aac2Δ, GAL10-AAC2, sal1-1) transformants expressing Aac11-183-Aac2193-318 and Aac21-192-Aac1184-309 from the AAC2 promoter were spotted onto YPGal (Galactose) and YPD (Glucose). (B) Serial dilutions of CS494-2B transformants expressing aac1P89L and aac1A96V from the AAC2 promoter were spotted onto YPGal and YPD. (C) aac1P89L and aac1A96V are partially respiratory deficient. CS341 (aac2Δ, but SAL1) cells expressing aac1P89L and aac1A96V were diluted in water and spotted on YPD and YPGly (Glycerol).
In a reciprocal approach, we randomly mutagenized AAC1 under the control of the AAC2 promoter and screened for mutants that may gain the V function. We identified the P89L and A96V mutations that confer partial but significant activity to Aac1p in supporting cell viability in the absence of both Aac2 and Sal1p (Figure 10B). Intriguingly, these two alleles severely reduce the R function as manifested by decreased growth on glycerol medium (Figure 10C). Thus the gain of V function appears to occur at the expense of the R function. Pro89 and Ala96, equivalent to Pro82 and Ala89 in bovine Ant1, are located in a region that acts as a gate for substrate entrance from the cytosolic side (see Figure 9A, discussed below).
Discussion
We investigated how Sal1p, the only identified CaMC protein in yeast, acts in concert with the Aac2 isoform of the adenine nucleotide translocase to promote mitochondrial biogenesis. These two proteins were found to be required for the maintenance of a functional mitochondrial genome. The inability to maintain the mitochondrial genome provides a plausible explanation for the lethality of the sal1 aac2 double mutant. Yeast aac2 cells are known to be intolerant to ρ°/ρ- conditions (Kovacova et al. 1968), because in the absence of an active electron transport chain, the electrogenic exchange between cytosolic ATP4- and matrix ADP3- catalyzed by Aac2p is required to maintain the electrochemical gradient across the mitochondrial inner membrane and for cell viability (Giraud and Velours 1997; Kominsky et al. 2002; Chen and Clark-Walker 2000). Thus, ρ°/ρ--lethality may be responsible for the lethal phenotype associated with the co-inactivation of SAL1 and AAC2.
How might Sal1p and Aac2p affect mtDNA stability? Although mutations in a plethora of genes can affect the stability of the mitochondrial genome, only a subset is essential for mtDNA maintenance. These genes include those directly involved in mtDNA replication, transcription, translation and mitochondrial morphogenesis (Chen and Butow 2005; Contamine and Picard 2000; Scott et al. 2003). As the HS40 ρ- genome can be stably maintained in cells lacking both Sal1p and Aac2p, it is unlikely that the two proteins directly affect the levels of nucleotide precursors essential for mtDNA replication. We also showed that mitochondria from sal1 aac2 double mutants maintain a wild-type level of dATP, which is thought to be imported from the cytosol in yeast. Furthermore, we demonstrated that mitochondrial transcripts from ori5 in the HS40 ρ- genome remain robust in cells lacking both Sal1p and Aac2p. Sal1p and Aac2p are therefore not essential for mtDNA replication and mitochondrial transcription.
Several lines of evidence support the idea that Sal1p and Aac2p affect the accumulation of mitochondrial translation products. First, co-inactivation of SAL1 and AAC2 leads to a reduction of mitochondrial translation products before the loss of mtDNA and mtRNA becomes noticeable. This observation suggests that mitochondrial protein translation is severely impaired, or that the translation products are rapidly degraded. Pulse-chase experiments showed a slightly reduced, rather than accelerated turnover of mitochondrial translation products in aac2 sal1 double mutants, compared with the single mutants or wild type cells (data not shown). Thus, the loss of translation products is likely caused by defects in protein synthesis. Secondly, the loss of Sal1 and Aac2 functions destabilizes ρ+ but not ρ- mtDNA. By an as yet not fully understood mechanism, a preferential effect on the stability of ρ+ versus ρ- mtDNA is a phenotype consistently associated with conditions compromising mitochondrial protein synthesis (Myers et al. 1985; Weislogel and Butow 1970). Finally, our genetic experiments clearly demonstrate that sal1Δ exacerbates the respiratory deficiency and mtDNA instability phenotypes of the ggc1Δ and mtg1Δ mutants, which are known to partially compromise mitochondrial protein synthesis. In the case of ggc1Δ, which affects mitochondrial GTP homeostasis required for protein translation, the genetic interaction with sal1 was validated by a synergistic defect in the incorporation of [35S]-methionine into mitochondrial translation products. We speculate that Sal1p may directly promote the transport of a substrate required for mitochondrial protein synthesis. Alternatively, Sal1p and Aac2p may be implicated in a fundamental process in mitochondrial biogenesis, which secondarily affects protein synthesis and mtDNA stability. In fact, in yeast, mitochondrial translation requires a coordinated assembly of mt-ribosomes and coupled transcription and translation at the mitochondrial inner membrane (Fox 1996; Bryan et al. 2002; Rodeheffer et al. 2001). In addition to translational activators that are directly required for efficient translation of specific mRNAs (Barrientos et al. 2004; Fox 1996; Krause et al. 2004; Green-Willms et al. 2001; Steele et al. 1996), defects in other processes (e.g., membrane biogenesis) can also be expected to affect ribosomal assembly and ultimately, mitochondrial protein synthesis.
Sal1p is closely related to the human SCaMC-1, -2 and -3 proteins. SCaMC-1 and -3 transport adenine nucleotides by exchange with phosphate and several other substrates (Fiermonte et al. 2004). The closely related SCaMC-2 is assumed to have a similar transport activity. A Ca2+-dependent ATP-Mg(ext)/Pi(int) exchange activity has been proposed to be necessary for maintaining the adenine nucleotide pool size in the matrix during mitochondrial proliferation (Aprille 1993). An optimal adenine nucleotide level is also required for effective mitochondrial protein synthesis (McKee and Poyton 1984). It is speculated that the SCaMC proteins may fulfill such a function in vivo. This raises the possibility that the yeast Sal1p, together with Aac2p, may be required for the maintenance of adenine nucleotide homeostasis in the matrix, thereby affecting mitochondrial protein synthesis and other processes. In the present study, only SCaMC-2 was confirmed to functionally complement sal1Δ among the three human genes. Although the SCaMC-1 cDNA used in our study is different from the variant used by Fiermonte and co-workers in in vitro transport assays in the first 54 amino acids, the SCaMC-3 variant used in our complementation test is identical to the one which is otherwise active in nucleotide transport (Fiermonte et al. 2004). Thus, a plausible explanation would be that only Sal1p and SCaMC-2 have the V function in addition to their activity in promoting ATP-Mg/Pi exchange. As mitochondria lacking both Sal1p and Aac2p are not depleted of adenine nucleotides, it is unlikely that the lethality of the sal1 aac2 double mutant is caused by the loss of adenine nucleotide homeostasis. In the double mutant, additional molecular entities may function to maintain adenine nucleotide homeostasis. In our in organello assays, we were unable to detect any appreciable ATP import activity in isolated mitochondria over-expressing Sal1p, although a rather weak activity to uptake radioactive ATP has previously been reported with wild-type mitochondria in response to high levels of Ca2+ (Cavero et al. 2005). We postulate that Sal1p may possess a low nucleotide transport activity in vivo, which is beyond a reproducible detection by in organello assays.
Additional experiments support the notion that the V function of Aac2p and Sal1p is independent of adenine nucleotide transport. We found that the aac2R252I and aac2R253I alleles, having a nucleotide transport activity as low as 1-6% of the wild type protein (Heidkamper et al. 1996), retain significant V function activity. In this regard, it may be argued that the residual transport activity is sufficient for maintaining cell viability. However, we demonstrated that the P89L and A96V mutants of the naturally occurring V-R+ Aac1p isoform gain a significant activity in the V function at the expense of the R function (i.e., adenine nucleotide transport). Furthermore, Sal1p mutated in Arg479 and Arg481 in the RTR motif remains functionally active in vivo. The equivalent arginine triplet in all known adenine nucleotide transporters is essential for adenine nucleotide transport. Thus, it remains possible that like Aac2p, Sal1p is also bifunctional.
Another relevant observation is that the gain-of-function mutations in Aac1 occur at Pro89 and Ala96. Although Pro89 is conserved among approximately half of the 34 members of mitochondrial carriers, Ala96 is specific to adenine nucleotide translocases with a few exceptions (Nelson et al. 1998). These two amino acids are located at the cytosolic end of the transmembrane domain 2 (see Figure 9A), in which several amino acids face to the substrate translocation pathway in the crystallized bovine Ant1 (Pebay-Peyroula et al. 2003). This particular region undergoes dynamic structural changes during nucleotide transport and has been proposed to contribute to the recognition of substrates from the cytosolic side (Kihira et al. 2004). Although Pro89 and Ala96 are invariant amino acids in Aac2p, our currently available data favor the model that the P89L and A96V mutations confer or increase the capability of recognizing a novel substrate other than ATP and ADP from the cytosolic side. Transport of multiple substrates by mitochondrial carriers is a rather common phenomenon. For instance, the human ATP-Mg(ext)/Pi(int) exchangers, SCaMC-1 and -3, are capable of mediating transport of a wide range of substrates that include adenine nucleotides, Pi, 3′-AMP, 3′,5′-ADP, deoxyadenine nucleotides, GMP, GDP and TDP, among others (Fiermonte et al. 2004). As Pro89 and Ala96 in Aac1p face to the interface with the phospholipid bilayer, it remains an open question whether or not the transport activity associated with the V function is dependent on the central nucleotide translocation pathway that implicates Arg252 and Arg253. Adenine nucleotide translocase has been previously linked to several activities unrelated to nucleotide exchange, which include membrane uncoupling via the proton conductance on the protein-phospholipid interface and co-transportation of fatty acid anions (Brand et al. 2005; Brustovetsky and Klingenberg 1994).
In summary, our data demonstrate a role of the evolutionarily conserved Ca2+-dependent proteins of Sal1-family and Aac2p in supporting several fundamental processes including mitochondrial protein synthesis and mtDNA stability. These effects are likely through an activity other than adenine nucleotide transport. Identifying the nature of this novel activity remains a challenge for future studies. Sal1p, together with other SCaMC proteins, provide an interesting link between cellular Ca2+-signalling and mitochondrial function. In mammals, it is known that direct flux of cytosolic Ca2+ into mitochondria stimulates the rate of NADH production and ATP output by the allosteric activation of matrix dehydrogenases (McCormack et al. 1990), so that energy production is synchronized with the energy demands of Ca2+-activated processes in extramitochondrial compartments. The Sal1-family of proteins therefore provides an example in which cytosolic Ca2+ could affect mitochondrial function by processes other than the activation of metabolic dehydrogenases. This novel pathway does not involve a direct influx of Ca2+ into the organelle but is essential for mitochondrial biogenesis. On the other hand, our finding of the bifunctionality of Aac2p could also have implications for fully understanding the biology of the adenine nucleotide translocase, especially when considering the fact that this evolutionarily conserved protein not only contributes to oxidative phosphorylation, but also plays an important role in cell death in higher eukaryotes.
Acknowledgments
We thank Martin Kucej for constant discussions and comments on the work, Araceli del Arco (Universidad Autónoma, Spain) for kindly providing the human SCaMC cDNA clones, Kelly Salinas for critical reading of the manuscript and Tom Januszewski for help with TEM. This work is dedicated to the memory of Ronald Butow who provided unreserved encouragement and support. This work was supported by grants from the American Heart Association (0435047N) and NIA/NIH (AG023731) to X.J.C.
References
- Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol Cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Aprille JR. Mechanism and regulation of the mitochondrial ATP-Mg/P(i) carrier. J Bioenerg Biomembr. 1993;25:473–481. doi: 10.1007/BF01108404. [DOI] [PubMed] [Google Scholar]
- Aprille JR, Austin J. Regulation of the mitochondrial adenine nucleotide pool size. Arch Biochem Biophys. 1981;212:689–699. doi: 10.1016/0003-9861(81)90413-6. [DOI] [PubMed] [Google Scholar]
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Barwell KJ, Sjulsen C, Gajewski CD, Manfredi G, Ackerman S, Tzagoloff A. MTG1 codes for a conserved protein required for mitochondrial translation. Mol Biol Cell. 2003;14:2292–2302. doi: 10.1091/mbc.E02-10-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Pakay JL, Ocloo A, Kokoszka J, Wallace DC, Brookes PS, Cornwall EJ. The basal proton conductance of mitochondria depends on adenine nucleotide translocase content. Biochem J. 2005;392:353–362. doi: 10.1042/BJ20050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustovetsky N, Klingenberg M. The reconstituted ADP/ATP carrier can mediate H+ transport by free fatty acids, which is further stimulated by mersalyl. J Biol Chem. 1994;269:27329–27336. [PubMed] [Google Scholar]
- Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavero S, Traba J, Del Arco A, Satrustegui J. The calcium-dependent ATP-Mg/Pi mitochondrial carrier is a target of glucose-induced calcium signalling in Saccharomyces cerevisiae. Biochem J. 2005;392:537–544. doi: 10.1042/BJ20050806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ. Sal1p, a calcium-dependent carrier protein that suppresses an essential cellular function associated with the Aac2 isoform of ADP/ATP translocase in Saccharomyces cerevisiae. Genetics. 2004;167:607–617. doi: 10.1534/genetics.103.023655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Clark-Walker GD. The petite mutation in yeasts: 50 years on. Int Rev Cytol. 2000;194:197–238. doi: 10.1016/s0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Arco A, Agudo M, Satrustegui J. Characterization of a second member of the subfamily of calcium-binding mitochondrial carriers expressed in human non-excitable tissues. Biochem J. 2000;345(Pt 3):725–732. doi: 10.1042/0264-6021:3450725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Arco A, Satrustegui J. Molecular cloning of Aralar, a new member of the mitochondrial carrier superfamily that binds calcium and is present in human muscle and brain. J Biol Chem. 1998;273:23327–23334. doi: 10.1074/jbc.273.36.23327. [DOI] [PubMed] [Google Scholar]
- del Arco A, Satrustegui J. Identification of a novel human subfamily of mitochondrial carriers with calcium-binding domains. J Biol Chem. 2004;279:24701–24713. doi: 10.1074/jbc.M401417200. [DOI] [PubMed] [Google Scholar]
- del Arco AD, Satrustegui J. New mitochondrial carriers: an overview. Cell Mol Life Sci. 2005;62:2204–2227. doi: 10.1007/s00018-005-5197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert K, de Kroon AIPM, Kispal G, Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. In: Pon LA, Schon EA, editors. Mitochondrion. San Diego: Academic Press; 2001. pp. 41–44. [DOI] [PubMed] [Google Scholar]
- Fangman WL, Henly JW, Brewer BJ. RPO41-independent maintenance of [rho-] mitochondrial DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa FM, Palmieri F. Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2004;279:30722–30730. doi: 10.1074/jbc.M400445200. [DOI] [PubMed] [Google Scholar]
- Fox TD. Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia. 1996;52:1130–1135. doi: 10.1007/BF01952112. [DOI] [PubMed] [Google Scholar]
- Fox TD, Folley LS, Mulero JJ, McMullin TW, Thorsness PE, Hedin LO, Costanzo MC. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Giannattasio S, Gagliardi S, Samaja M, Marra E. Simultaneous determination of purine nucleotides, their metabolites and beta-nicotinamide adenine dinucleotide in cerebellar granule cells by ion-pair high performance liquid chromatography. Brain Res Brain Res Protoc. 2003;10:168–174. doi: 10.1016/s1385-299x(02)00215-5. [DOI] [PubMed] [Google Scholar]
- Giraud MF, Velours J. The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho- yeast cells by a lack of assembly of the catalytic sector F1. Eur J Biochem. 1997;245:813–818. doi: 10.1111/j.1432-1033.1997.00813.x. [DOI] [PubMed] [Google Scholar]
- Green-Willms NS, Butler CA, Dunstan HM, Fox TD. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J Biol Chem. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- Heidkamper D, Muller V, Nelson DR, Klingenberg M. Probing the role of positive residues in the ADP/ATP carrier from yeast. The effect of six arginine mutations on transport and the four ATP versus ADP exchange modes. Biochemistry. 1996;35:16144–16152. doi: 10.1021/bi960668j. [DOI] [PubMed] [Google Scholar]
- Kaplan RS. Structure and function of mitochondrial anion transport proteins. J Membr Biol. 2001;179:165–183. doi: 10.1007/s002320010046. [DOI] [PubMed] [Google Scholar]
- Kihira Y, Iwahashi A, Majima E, Terada H, Shinohara Y. Twisting of the second transmembrane alpha-helix of the mitochondrial ADP/ATP carrier during the transition between two carrier conformational states. Biochemistry. 2004;43:15204–15209. doi: 10.1021/bi0494222. [DOI] [PubMed] [Google Scholar]
- Kominsky DJ, Brownson MP, Updike DL, Thorsness PE. Genetic and biochemical basis for viability of yeast lacking mitochondrial genomes. Genetics. 2002;162:1595–1604. doi: 10.1093/genetics/162.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacova V, Irmlerova J, Kovac L. Oxidative phosphorylatiion in yeast. IV. Combination of a nuclear mutation affecting oxidative phosphorylation with cytoplasmic mutation to respiratory deficiency. Biochim Biophys Acta. 1968;162:157–163. doi: 10.1016/0005-2728(68)90097-2. [DOI] [PubMed] [Google Scholar]
- Krause K, Lopes de Souza R, Roberts DG, Dieckmann CL. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol Biol Cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Salin B, Schaeffer J, Brethes D, Dautant A, Ackerman SH, di Rago JP. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J Biol Chem. 2005;280:18386–18392. doi: 10.1074/jbc.M410789200. [DOI] [PubMed] [Google Scholar]
- Mashkevich G, Repetto B, Glerum DM, Jin C, Tzagoloff A. SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J Biol Chem. 1997;272:14356–14364. doi: 10.1074/jbc.272.22.14356. [DOI] [PubMed] [Google Scholar]
- McCormack JG, Halestrap AP, Denton RM. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev. 1990;70:391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- McKee EE, Poyton RO. Mitochondrial gene expression in Saccharomyces cerevisiae. I. Optimal conditions for protein synthesis in isolated mitochondria. J Biol Chem. 1984;259:9320–9331. [PubMed] [Google Scholar]
- Myers AM, Pape LK, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Felix CM, Swanson JM. Highly conserved charge-pair networks in the mitochondrial carrier family. J Mol Biol. 1998;277:285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Artal Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 2001;498:46–51. doi: 10.1016/s0014-5793(01)02447-4. [DOI] [PubMed] [Google Scholar]
- Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Pardo B, Lasorsa FM, del Arco A, Kobayashi K, Iijima M, Runswick MJ, Walker JE, Saheki T, Satrustegui J, Palmieri F. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001;20:5060–5069. doi: 10.1093/emboj/20.18.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Conrad-Webb H, Docherty R, Butow RA. Interaction between the yeast mitochondrial and nuclear genomes influences the abundance of novel transcripts derived from the spacer region of the nuclear ribosomal DNA repeat. Mol Cell Biol. 1989;9:1897–1907. doi: 10.1128/mcb.9.5.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- Robinson AJ, Kunji ER. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci U S A. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J Biol Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Beuneu C, Roux P, Buc H, Lemaire G, Lepoivre M. Simultaneous determination of pyrimidine or purine deoxyribonucleoside triphosphates using a polymerase assay. Anal Biochem. 1999;269:403–409. doi: 10.1006/abio.1999.4051. [DOI] [PubMed] [Google Scholar]
- Scott SV, Cassidy-Stone A, Meeusen SL, Nunnari J. Staying in aerobic shape: how the structural integrity of mitochondria and mitochondrial DNA is maintained. Curr Opin Cell Biol. 2003;15:482–488. doi: 10.1016/s0955-0674(03)00070-x. [DOI] [PubMed] [Google Scholar]
- Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci U S A. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B. Mitochondrial structure. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces - Life cycle and inheritance. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1981. pp. 471–504. [Google Scholar]
- Taylor SD, Zhang H, Eaton JS, Rodeheffer MS, Lebedeva MA, O'Rourke TW, Siede W, Shadel GS. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozza A, Blanco E, Palmieri L, Palmieri F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weislogel PO, Butow RA. Low temperature and chloramphenicol induction of respiratory deficiency in a cold-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1970;67:52–58. doi: 10.1073/pnas.67.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Herrmann JM, Neupert W. Analysis of mitochondrial translation products in vivo and in organello in yeast. In: Pon LA, Schon EA, editors. Mitochondrion. San Diego: Academic Press; 2001. pp. 429–438. [DOI] [PubMed] [Google Scholar]
- Wohlrab H. The human mitochondrial transport protein family: identification and protein regions significant for transport function and substrate specificity. Biochim Biophys Acta. 2005;1709:157–168. doi: 10.1016/j.bbabio.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Wright R. Transmission electron microscopy of yeast. Microsc Res Tech. 2000;51:496–510. doi: 10.1002/1097-0029(20001215)51:6<496::AID-JEMT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]