Abstract
Metabolic flexibility is the capacity for skeletal muscle to shift reliance between lipids and glucose during fasting or in response to insulin. We hypothesized that body fat, adipose tissue characteristics, e.g. larger adipocytes, presence of inflammatory gene markers and impaired suppression of non-esterified fatty acids (NEFAs) during insulin infusion might be related to metabolic flexibility.
We measured changes in respiratory quotient (ΔRQ) before and during euglycemic-hyperinsulinemic clamp in healthy young males. Body fat by DXA, laboratory measurements, abdominal subcutaneous adipose tissue biopsies and fat cell size (FCS) were obtained after an overnight fast. Gene expression for 17 adipose tissue genes related to lipid synthesis, uptake, oxidation and storage, lipolysis and inflammation were measured.
Reduced metabolic flexibility was associated with higher body fat, larger FCS and impaired insulin suppression of NEFAs. Metabolic flexibility was associated with higher serum adiponectin levels. Lower adipose tissue gene expression for inflammation markers was associated with greater NEFA suppression by insulin and metabolic flexibility.
Combined, these results indicate that body fat, larger adipocytes, failure of insulin to suppress NEFAs, decreased adiponectin levels and inflammation markers in adipose tissue are associated with decreased insulin-stimulated glucose uptake and oxidation, which is an important component of reduced metabolic flexibility.
Keywords: Fat cell size, Non-esterified free fatty acids, Euglycemic-hyperinsulinemic, clamp, Adipose tissue, Metabolic flexibility, Inflammatory markers, Adiponectin
1. Introduction
Metabolic flexibility is the capacity for skeletal muscle to acutely shift its reliance between lipids and glucose during fasting or in response to insulin, such as in postprandial conditions. Two fundamental features of reduced metabolic flexibility are decreased fat oxidation in the fasting state (i.e. higher respiratory quotient (RQ)) and decreased insulin-stimulated glucose oxidation. Another important feature of reduced metabolic flexibility is an impaired suppression of non-esterified free fatty acid (NEFA) release (lipolysis) in response to insulin [1]. Impaired insulin stimulation of glucose uptake by skeletal muscle [2], impaired skeletal muscle mitochondrial biogenesis and decreased capacity for oxidation of dietary fat are all involved in reducing metabolic flexibility [3]. Furthermore, these physiologic characteristics are enriched in healthy young men with a family history of type 2 diabetes (T2D) [4]. The cause of the derangements in skeletal muscle of type 2 diabetic patients remains to be elucidated. Impaired mitochondrial function is a likely candidate. Evidence from both in vivo and ex vivo studies supports the idea that an impaired skeletal muscle mitochondrial function is related to the development of insulin resistance and type 2 diabetes. A decreased mitochondrial oxidative capacity in skeletal muscle was revealed in diabetic patients, using in vivo 31-Phosphorus Magnetic Resonance Spectroscopy (31P-MRS) [5]. Insulin resistance is associated with metabolic inflexibility, impaired switching of substrate oxidation from fatty acids to glucose in response to insulin. Ukropcova et al. recently demonstrated that muscle mitochondrial content was higher in flexible subjects with high fat oxidation after a high fat diet (HFD) and contributed 49% of the variance. Subjects with a family history of diabetes were inflexible and had reduced HFD-induced fat oxidation and muscle mitochondrial content but did not differ in the amount of body or visceral fat. Metabolic inflexibility, lower adaptation to an HFD and reduced muscle mitochondrial mass cluster together in subjects with a family history of diabetes, supporting the role of an intrinsic metabolic defect of skeletal muscle in the pathogenesis of insulin resistance [4].
Fuel selection is a key component of insulin sensitivity. Skeletal muscle accounts for 80–90% of insulin-stimulated glucose disposal and is the primary tissue responsible for peripheral insulin sensitivity and glucose homeostasis [6]; however, the role of adipose tissue has been unexplored. White adipose tissue (WAT) is a major site of energy storage, and it is important for energy homeostasis. WAT stores energy in the form of triglycerides when in positive energy balance and releases energy as NEFAs when energy expenditure exceeds energy intake [7,8]. While WAT provides a survival advantage in times of starvation, excess WAT and larger adipocytes are now recognized as links to the health problems associated with obesity of developed countries. Furthermore, while adipocytes are traditionally known as fat storage cells, adipose tissue is also an endocrine organ that secretes hormones, chemokines and cytokines. Increased basal/fasted lipolysis in white adipose tissue leads to increase fasting NEFA levels [9]. Increased chemotaxis and macrophage content in WAT are characteristics of the obese state. While there are very few manuscript that measure both body composition and adipose tissue gene expression, several of these are from our own group and no paper, to our knowledge, exists that compares inflammatory markers to high quality gene expression. One paper from our group demonstrated that the combined activation of peroxisome proliferator-activated receptor-gamma and beta-adrenergic receptors has beneficial effects on body weight, plasma triglycerides and lipid metabolism in subcutaneous fat by increasing the expression of genes required for fatty acid catabolism [10].
We hypothesized that the quantity and the characteristics of the adipose tissue (larger adipocytes, expression of macrophage genes and disordered fatty acid oxidation) might contribute to the state of reduced metabolic flexibility. To explore this hypothesis, we studied 56 healthy young men under carefully controlled conditions, examining how adipose tissue mass and fat cell size (FCS) influence metabolic flexibility during a euglycemic-hyperinsulinemic clamp (EHC).
2. Research design and methods
2.1. Study population and design
All procedures were approved by the PBRC IRB board. After providing written informed consent, a cohort of 56 healthy young men, aged 22.6 ± 3.2 years with a BMI of 26.4 ± 4.1 kg/m2 underwent physical examination, medical laboratory tests and measurement of body fat by dual energy X-ray absorptiometry (DXA). Thirteen of the 56 healthy young men had a BMI > 30, but less than 35 kg/m2, and 16 of the 56 participants had a family history of T2D. These clinical characteristics were taken into account in the analyses. All participants presented to the Pennington inpatient unit and ate a weight-maintaining diet (35% fat, 16% protein and 49% carbohydrate) as a "control" prepared by the metabolic kitchen. After 48 h of this “control” diet, a euglycemic-hyperinsulinemic clamp was performed the next morning following a 10 h fast by the study participant. Earlier studies describe this cohort in terms of skeletal muscle oxidative phosphorylation and metabolic flexibility [3,11]. The present investigation only involves measures made prior to a 3-day dietary intervention; thus, no analysis was performed on the data collected after the 3-day dietary intervention. All measurements were made after the 48-h “control” diet, and all data reported in this paper relates to baseline.
2.2. Euglycemic-hyperinsulinemic clamp
Glucose disposal rate (GDR; mg/kg FFM/min) and metabolic flexibility (change in respiratory quotient (ΔRQ) from fasting to the insulin-stimulated state were measured by a euglycemic-hyperinsulinemic clamp) [12]. Briefly, after an overnight fast intravenous catheters were inserted in an antecubital vein for infusions and in a vein on the dorsum of the contra-lateral hand for sampling of arterialized blood. After baseline sampling, a primed-continuous insulin infusion (80 mIU/m2/min] was continued for 3–4 h. Insulin was infused for at least 1 h after reaching a concentration of glucose ∼90 mg/dl. Plasma glucose was measured every 5 min and maintained by a variable 20% glucose infusion. The mean rate of exogenous glucose infusion during steady state (last 30 min) was divided by fat free mass (FFM) as determined by DXA to assess insulin sensitivity. GDR = sum of glucose infused during steady state/30 min × kg FFM. Delta RQ (ΔRQ) was calculated as post-insulin steady-state RQ-fasting RQ.
Fasting RQ was measured at fasting state, just before the clamp in resting conditions. Post-insulin steady-state RQ-fasting RQ was measured duringthe steady state of the clamp.
2.3. Maximal aerobic capacity (VO2max)
Maximal oxygen uptake was determined by a progressive treadmill test to exhaustion [13]. Oxygen consumption (VO2) and CO2 production (VCO2) were measured continuously using a breath-by-breath indirect calorimeter (V-Max29 Series, SensorMedics, Yorba Linda, California).
2.4. Body composition
Body fat mass and lean body mass were measured on a Hologic Dual Energy X-Ray Absorptiometer (DXA; QDR 4500, Hologic, Inc. Waltham, MA). Visceral adipose tissue (VAT) mass by multi-slice CT as previously described [14].
2.5. Indirect calorimetry
After an overnight fast, the RQ was measured at baseline and RQ was measured again using 20-min time intervals during the euglycemic-hyperinsulinemic clamp by indirect calorimetry using a Deltatrac II indirect calorimeter (DATEX-Ohmeda, Helsinki, Finland). Oxidative and non-oxidative glucose disposal were calculated as described by Livesey and Elia [15].
2.6. Fat cell size
Fat cell size was determined as previously described [16]. Briefly, subcutaneous adipose tissue was fixed in osmium tetrachloride/collidine-HCl followed by disassociation via urea digestion. Cells were counted on a Multisizer-3 (Beckman Coulter, Fullerton, CA) using a 400-µm aperture (dynamic linear range, 12–320µm) and reported as the mean of all adipocytes >22.5 µm.
2.7. Laboratory measures
Fasting serum glucose and non-esterified free fatty acids were assayed on a Beckman Synchron CX7 (Beckman Coulter, Brea, CA). Wako (Richmond, VA) reagents were used for the NEFA determination, except the NEFA obtained during the period of insulin infusion during the clamp. Because of their low level they were measured in triplicate by high-performance liquid chromatography (HPLC) as previously described [17]. Fasting plasma insulin and C-peptide were measured on an Immulite autoanalyzer (DPC, Los Angeles, CA). Following insulin infusion, serum glucose and insulin were assayed during the euglycemic-hyperinsulinemic clamp in the same manner as the fasting samples.
2.8. Adipose tissue biopsy
Adipose tissue was obtained from subcutaneous abdominal adipose tissue using the technique of Bergstrom. This technique has advantages over the liposuction or small needle sampling of adipose tissue as the tissue fragments are larger and relatively more intact making utilization for tissue blocks and immunohistochemistry a more realistic endeavour. Participants were placed in a supine position and the skin lateral to the umbilicus cleansed with povidone-iodine solution. After a sterile drape was placed, the skin and adipose tissue were anesthetized using <5 ml of a 50%/50% mixture of bupivicaine and lidocaine (final concentrations 1.0% and 0.125%). The site was chosen taking care to avoid the line under belt and lateral to the small subcutaneous arteries that lie parallel to the midline by 2–4 cm. The skin was incised (approximately 0.75 cm) with a #11 scalpel. The Bergstrom needle (6 mm) was inserted into the adipose tissue away from large fibrous structures. After suction was applied from the proximal port, the sample (approximately 50 mg) was cut and the needle removed.
2.9. RNA and DNA extraction
Total RNA from ∼200 mg of human subcutaneous abdominal adipose tissue was isolated with Trizol reagent (Invitrogen, Carlsbad, CA). The quantity and integrity of the RNA was confirmed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
2.10. Real-time qRT-PCR for RNA
All primers and probes were designed using Primer Express version 2.1 (Applied Biosystems, Roche, Branchburg, NJ). Sequences of primers and probes are shown in Table 1. Realtime qRT-PCR reactions [18] were performed as one-step reactions in ABI PRISM 7900 (Applied, Biosystems, Branchburg, NJ) using the following parameters: one cycle of 48 °C for 30 min, then 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. 18S was used as the internal control. All expression data were normalized by dividing the target gene quantity in nanograms by the internal control gene (Cyclophilin B for adipose tissue and ribosomal protein, large, P0 for skeletal muscle tissue) quantity in nanograms. Both internal control genes were tested for interindividual variability with a coefficient of variance to ensure precision in the measurements.
Table 1.
Oligonucleotide sequences for primer/probe sets used for qRT-PCR and qPCR.
| Official gene symbol |
Common gene name |
Accession number |
Forward primer | Probe | Reverse primer |
|---|---|---|---|---|---|
| PPARGC1A | PGC1a | NM_0013261 | AGGTGAAAGTGTAATACTGTTGGTTGA | TGCTGAAGAGGGAAAGTGAGCGATTAGTTGA | CATGTAGAATTGGCAGGTGGAA |
| ND1 | mtDNA | AF346985 | CCCTAAAACCCGCCACATCT | CCATCACCCTCTACATCACCGCCC | GAGCGATGGTGAGAGCTAAGGT |
| genomicLPL | NC_000008 | CGAGTCGTCTTTCTCCTGATGAT | ACATTCACCAGAGGGTC | TTCTGGATTCCAATGCTTCGA | |
| CD36 | CD36 | NM_000072 | AGTCACTGCGACATGATTAATGGT | CAGATGCAGCCTCATTTCCACCTTTTG | CTGCAATACCTGGCTTTTCTC |
| LPL | LPL | NM_000237 | TATCCGCGTGATTGCAGAGA | CTAGCTGGTCCACATCTCCAAGTCCT | AGAGAGTCGATGAAGAGATGAATGG |
| FAS | FAS | NM_000043 | TATGCTTCTTCGTGCAGCAGTT | AGCGCCTCCAGCACCCTGTTGT | GCTGCCACACGCTCCTCTAG |
| PCK1 | PCK1 | NM_002591 | CAGGCGGCTGAAGAAGTATGA | AACTGCTGGTTGGCTCTCACTGACCC | AACCGTCTTGCTTTCGATCCT |
| SCD | SCD1 | NM_005063 | TGGCATTCCAGAATGATGTCTATG | CGTGACCACCGTGCCCACCACA | GGAATTATGAGGATCAGCATGTGT |
| ACADM | MCAD | NM_000016 | TGCCCTGGAAAGGAAAACTTT | TGTAGAGCACCAAGCAATATCATTTATG | GTTCAACTTTCATTGCCATTTCAG |
| PPARA | PPARα | NM_001001928 | GCTTTGGCTTTACGGAATACCA | AGCCATCTGAGCCAGGACAGCTTCCTAA | TGAAAGCGTGTCCGTGATGA |
| PLIN | perilipin | NM_002666 | CACCGTGGCCATGTGGAT | CCCCTGAGCAGCCTGGCCC | GCCTGCATGGCCACTGAG |
| SORBS1 | CAP | NM_001034954 | CAAATTCCCTGAACTTCCTGAAA | CCAGCAAACTTCCGAAGAGGACAATCC | AGGAAACTGGTAGGTGGGAGTGTA |
| PNPLA2 | ATGL | NM_020376 | CCACGGCGCTGGTCA | TTGGCACCAGCCTCACCCAGG | GGGCCTCTTTAGATACCTCAATGA |
| LIPE | HSL | NM_005357 | GACTTCCTCCGGGAGTATGTC | TGCATAAGGGATGCTTCTATGGCC | GCGTGAACTGGAAGCCCA |
| PPARG | PPARγ1 | NM_005037 | GTCAAACGAGAGTCAGCCTTTAACG | AGAGATGCCATTCTGGCCCACCAACTT | CCACGGAGCTGATCCCAA |
| PPARG | PPARγ2 | NM_015869 | GATACACTGTCTGCAAACATATCACAA | AGAGATGCCATTCTGGCCCACCAACTT | CCACGGAGCTGATCCCAA |
| CD68 | CD68 | NM_001040059 | GCTTCTCTCATTCCCCTATGGA | CAGCTTTGGATTCATGCAGGACCTCC | ATGTAGCTCAGGTAGACAACCTTCTG |
| CD163 | MAC-2 | NM_004244 | TGCAGAAAACCCCACAAAAAG | CACAACAGGTCGCTCATCCCGTCA | CAAGGATCCCGACTGCAATAA |
| CCL2 | MCP-1 | NM_002982 | GATCTCAGTGCAGAGGCTCG | AGCTATAGAAGAATCACCAGCAGCAAGTGTCCC | AATGGTCTTGAAGATCACAGCTTCT |
| CCL3 | MIP-1α | NM_002983 | ACAGAATTTCATAGCTGACTACTTTGAGA | AGTGCTCCAAGCCCGGTGTCATCTTC | GCCGGCTTCGCTTGGT |
| RPS18 | 18S | NM_022551 | CGCCGCTAGAGGTGAAATTC | ACCGGCGCAAGACGGACCAGA | CATTCTTGGCAAATGCTTTCG |
2.11. Statistical analysis
Population characteristics are represented as means ± S.D. Gene expression and clinical data were correlated using regression analysis. Gene expression data are represented as means ± S.E. The change in respiratory quotient (ΔRQ) was divided into quartiles (quartile 1 = ΔRQ < 0.06; quartile 2 = 0.06 < ΔRQ ≤ 0.08; quartile 3 = 0.08 < ΔRQ ≤ 0.11; quartile 4 = ΔRQ > 0.11) to characterize the range in metabolic flexibility within the cohort. ANOVA was used to test for differences in biopsy and blood parameters across quartiles of metabolic flexibility (ΔRQ), with post hoc testing by mean equality contrast between different groups using the Tukey-Kramer HSD; alpha = 0.05. Type I error rate was set a priori at p < 0.05. Analysis was performed using JMP version 5.0 (SAS, Cary, NC). Correlations were preformed on continuous variables for the analysis in Table 2 and Table 3. The categorical variable quartiles of metabolic flexibility (factors) were used for the ANOVA analysis presented in Fig. 1–Fig. 3. Repeat analyses in 40 subjects give coefficients of variation (CV) for body composition measurements of 0.6%, 1.1% and 1.1% as lean mass, fat mass and percentage of body fat, respectively.
Table 2.
Relationships between adipose tissue gene expression, body fatness (FFAs) and metabolic flexibility (ΔRQ) (R2).
| Functional category |
Gene expression (mRNA) |
% Fat | VAT | FCS | Fasting FFAs |
Steady-state FFAsa |
ΔRQ |
|---|---|---|---|---|---|---|---|
| Lipid uptake | CD36 | −0.02 | 0.00 | −0.01 | −0.06 | 0.00 | 0.00 |
| LPL | 0.00 | 0.00 | 0.08 | −0.03 | 0.00 | −0.06 | |
| Lipid synthesis | FAS | −0.06 | −0.13* | −0.05 | 0.00 | −0.13** | 0.05 |
| PCK1 | −0.07 | −0.06** | −0.06 | −0.08 | 0.00 | 0.00 | |
| PPARγ1 | −0.05 | −0.02 | −0.05 | −0.16* | −0.01 | 0.00 | |
| PPARγ2 | 0.06 | −0.00 | 0.05 | 0.04 | 0.00 | 0.01 | |
| Lipid storage | SCD1 | −0.05 | −0.05 | −0.01 | 0.00 | −0.06 | 0.06 |
| Lipid oxidation | MCAD | −0.08** | −0.08** | −0.06 | −0.14** | −0.01 | 0.01 |
| PPARα | −0.10** | −0.08** | −0.07 | −0.10** | −0.01 | 0.00 | |
| Lipolysis | perilipin | −0.01 | −0.01 | 0.00 | −0.02 | −0.01 | 0.00 |
| CAP | −0.02 | −0.03 | 0.00 | −0.03 | −0.03 | 0.01 | |
| ATGL | −0.02 | −0.03 | −0.02 | −0.01 | −0.01 | 0.00 | |
| HSL | −0.08** | −0.06** | −0.05 | −0.08** | −0.03 | 0.00 | |
| Inflammation | CD68 | 0.07 | 0.12* | 0.10** | 0.04 | 0.38* | −0.10** |
| MAC-2 | 0.08 | 0.10** | 0.07 | 0.01 | 0.20* | −0.10** | |
| MCP-1 | 0.04 | 0.06** | 0.04 | 0.01 | 0.23* | −0.1l** | |
| MIP-1α | 0.20* | 0.21* | 0.18* | 0.10** | 0.20* | −0.16* |
All data are presented as R2. VAT, visceral adipose tissue; FCS, fat cells size; FFA, free fatty acids; ΔRQ, change in respiratory quotient; LPL, lipoprotein lipase; FAS, fatty acid synthase; PCK1, phosphoenolpyruvate carboxykinase 1; SCD1, stearoyl-CoA desaturase; MCAD, acyl-Coenzyme A dehydrogenase C-4 to C-12 straight chain; CAP, cbl-associated protein; ATGL, adipose triglyceride lipase; HSL, hormone-sensitive lipase; MCP-1, monocyte chemoattractant protein-1; MIP-1α, macrophage inflammatory protein 1, alpha subunit.
During insulin infusion (80 mIU/m2 BSA/min).
p < 0.01.
p ≤ 0.05.
Table 3.
Relationships among transcriptional markers of chemotaxis and macrophage content (R2).
| Gene expression (mRNA) |
CD68 | MAC-2 | MCP-1 | MIP-1α |
|---|---|---|---|---|
| CD68 | 1.00 | 0.78 | 0.45 | 0.58 |
| MAC-2 | 0.78 | 1.00 | 0.48 | 0.42 |
| MCP-1 | 0.45 | 0.48 | 1.00 | 0.35 |
| MIP-1α | 0.58 | 0.42 | 0.35 | 1.00 |
All data are presented as R2, all p < 0.01. FFA, free fatty acids; MCP-1, monocyte chemoattractant protein-1; MIP-1α, macrophage inflammatory protein 1, alpha subunit.
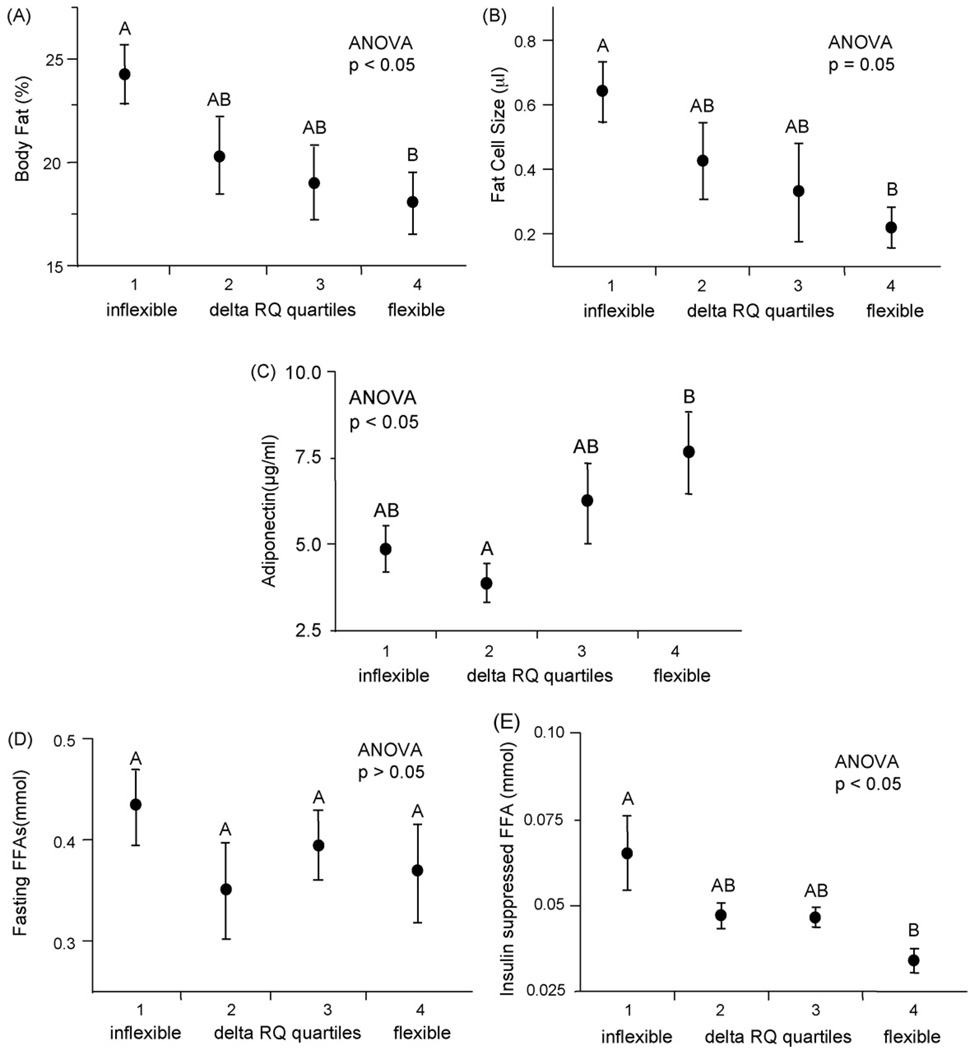
Fig. 1.
Body fatness, NEFAs after insulin infusion and adiponectin are related to reduced metabolic flexibility (ΔRQ) in healthy young men. Change in respiratory quotient (ΔRQ; metabolic flexibility) was measured before and during a euglycemic-hyperinsulinemic clamp (EHC) in the population of 56 healthy young men. ΔRQ was subdivided into quartiles and ANOVA was used to test for differences in biopsy and blood parameters across quartiles of metabolic flexibility (ΔRQ), with post hoc testing by mean equality contrast between different groups using the Tukey-Kramer HSD; alpha = 0.05. Type I error rate was set a priori at p < 0.05. Data are shown as means ± S.E. Levels which do not share the same letter are significantly different. (A) Percent body fat, (B) fat cell size, (C) serum adiponectin, (D) fasting non-esterified free fatty acids (NEFAs) as measured by enzyme assay and (E) insulin-suppressed NEFAs during insulin infusion of the EHC as measured by high-performance liquid chromatography (HPLC).
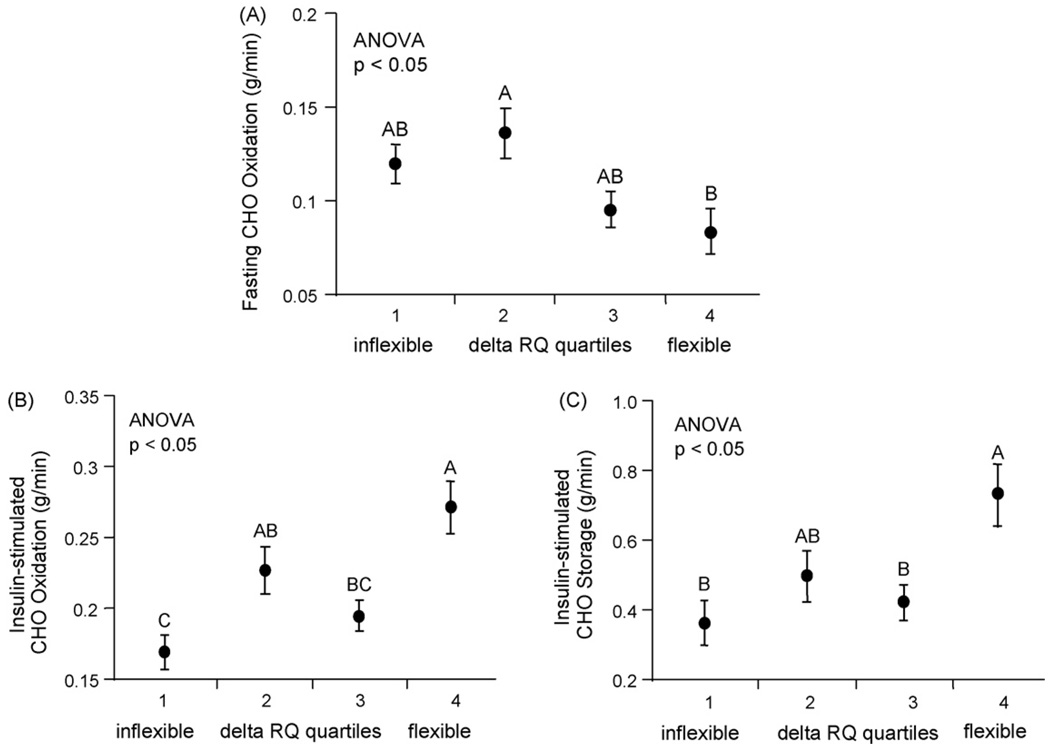
Fig. 3.
Oxidative and non-oxidative carbohydrate (CHO) disposal are related to reduced metabolic flexibility (ΔRQ) in healthy young men. Higher fasting carbohydrate (CHO) oxidation (A) was associated with lower ΔRQ. Lower levels of insulin-suppressed CHO oxidation (B) and storage (C) were associated with lower ΔRQ. Delta RQ (ΔRQ) was subdivided into quartiles. ANOVA was used to test for differences in biopsy and blood parameters across quartiles of metabolic flexibility (ΔRQ), with post hoc testing by mean equality contrast between different groups using the Tukey-Kramer HSD; alpha = 0.05. Type I error rate was set a priori at p < 0.05. Data are shown as means ± S.E. All levels not connected by same letter are significantly different.
3. Results
3.1. Body fat, fat cell size and serum adiponectin are associated with metabolic flexibility (ΔRQ) in healthy young men
Subject characteristics are listed in Table 4. All subjects were sedentary healthy young men ranging widely in BMI (20.1–34.7 kg/m2) and percent body fat (8.4–32.3%). RQ was measured before and during the euglycemic-hyperinsulinemic clamp to determine metabolic flexibility (ΔRQ). ΔRQ varied greatly, but was normally distributed, ranging from inflexible (ΔRQ= 0.03) to flexible (ΔRQ= 0.25). We divided metabolic flexibility (ΔRQ) into quartiles (quartile 1 = ΔRQ < 0.06; quartile 2 = 0.06 < ΔRQ ≤ 0.08; quartile 3 = 0.08 < ΔRQ ≤ 0.11; quartile 4 = ΔRQ > 0.11) and compared quartiles to adipose tissue chemokine and macrophage gene expression. ΔRQ was positively correlated with GDR (R2 = 0.23, p < 0.01; data not shown) and VO2max (R2 = 0.10, p < 0.05; data not shown), demonstrating the relationship between metabolic flexibility, insulin sensitivity and aerobic fitness. The lowest quartile of ΔRQ was associated with higher percent body fat (ANOVA, p < 0.05; Fig. 1A), larger fat cell size (ANOVA, p = 0.05; Fig. 1B) and lower serum adiponectin levels (ANOVA, p < 0.05; Fig. 1C). None of these associations were influenced by family history of T2D or BMI.
Table 4.
Characteristics of the study population.
| Mean ± S.D. | Range | |
|---|---|---|
| Age (years) | 22.6 ± 3.2 | 18.0–29.0 |
| Height (cm) | 176.9 ± 5.8 | 163.0–189.5 |
| Weight (kg) | 82.5 ± 13.2 | 59.2–118.3 |
| BMI (kg/m) | 26.4 ± 4.1 | 20.1–34.7 |
| WHR (au) | 0.87 ± 0.07 | 0.7–1.0 |
| Body fat (%) | 20.3 ± 6.5 | 8.4–32.3 |
| Visceral adipose tissue mass (kg) | 2.1 ± 1.3 | 0.5–5.7 |
| Fasting RQ (au) | 0.84 ± 0.04 | 0.74–0.95 |
| RQ after insulin infusiona (au) | 0.93 ± 0.04 | 0.85–1.03 |
| ΔRQ (au) | 0.09 ± 0.04 | 0.03–0.25 |
| VO2max (ml/kg/min) | 41.2 ± 7.2 | 23.5–59.2 |
| Fasting glucose (mg/dl) | 80.5 ± 5.4 | 66.0–90.0 |
| Glucose after insulin infusiona (mg/dl) | 89.0 ± 4.5 | 78.0–102.0 |
| Fasting insulin (µU/ml) | 8.2 ± 4.6 | 2.6–22.4 |
| Insulin after insulin infusiona (µU/ml) | 160.1 ± 35.7 | 103.9–251.3 |
| C-peptide (ng/ml) | 1.93 ± 0.70 | 0.9–3.5 |
| GDR (mg/kg FFM/min) | 11.1 ± 4.1 | 4.0–24.5 |
| Fasting FFA (mmol) | 0.6 ± 0.2 | 0.07–0.84 |
| FFA after insulin infusiona (mmol) | 0.05 ± 0.02 | 0.02–0.16 |
| Fat cell size (µl) | 0.6 ± 0.2 | 0.22–0.95 |
n = 16 with a family history of diabetes; n = 40 without a family history of diabetes. BMI, body mass index; GDR, glucose disposal rate; WHR, waist to hip ratio; FFA, free fatty acids. All male subjects were chosen from the larger mixed gender study population.
During insulin infusion (80 mIU/m2 BSA/min).
3.2. Insulin suppression of non-esterified free fatty acid is correlated with metabolic flexibility
We next explored the relationship between metabolic flexibility and plasma NEFAs in the fasting state and after insulin suppression during the euglycemic-hyperinsulinemic clamp. ΔRQ was not related to fasting NEFA levels (ANOVA, p > 0.05; Fig. 1D) but was related to NEFA levels after insulin infusion. Greater suppression of NEFAs (i.e. lower concentration of insulin-suppressed NEFAs after a 2-h insulin infusion) was associated with metabolic flexibility (higher ΔRQ) (ANOVA, p < 0.05; Fig. 1E). FCS was positively correlated with both fasting and insulin-suppressed NEFAs during the clamp (R2 = 0.20, p < 0.01 and R2 = 0.19, p < 0.01; data not shown).
3.3. Metabolic flexibility and gene expression in adipose tissue
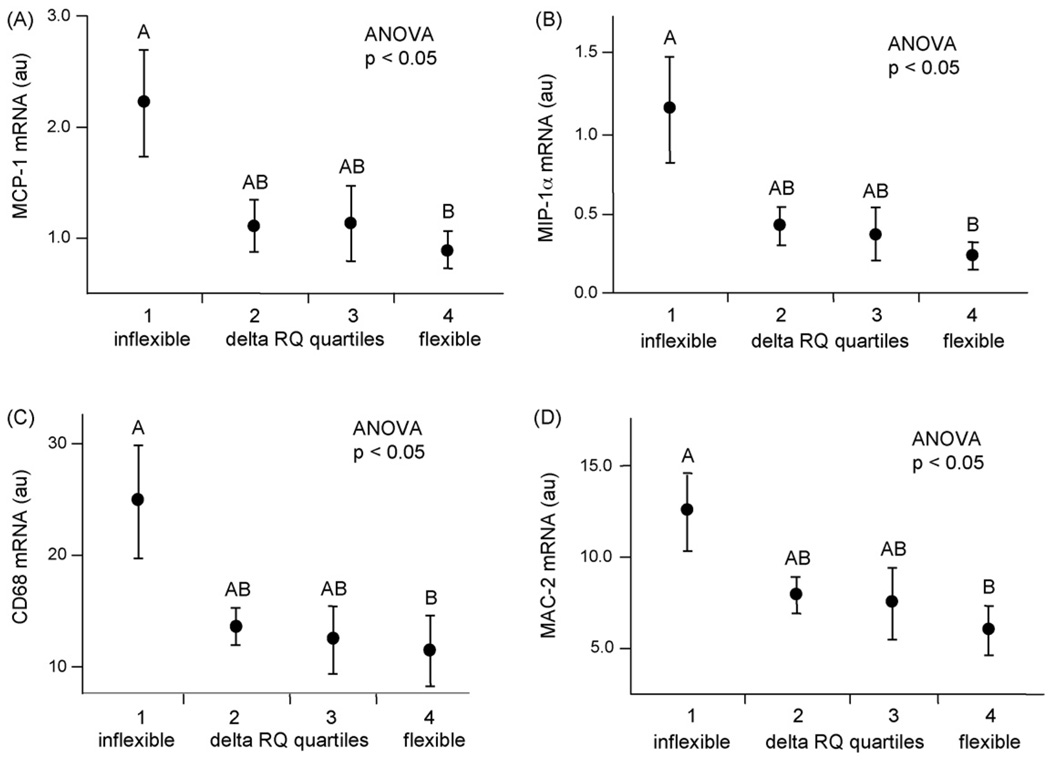
We next examined mRNA expression of candidate genes involved in fatty acid metabolism (synthesis, uptake, oxidation and storage, as well as lipolysis) and inflammation(chemokines and macrophage markers) and related these to insulin-suppressed NEFAs and metabolic flexibility (change in respiratory quotient; ΔRQ) (Table 2). We previously correlated the protein and mRNA expressions of all four inflammation genes in two distinct populations of lean and obese men. Insulin-suppressed NEFAs were significantly correlated with the expression of genes involved in chemotaxis (MCP-1 and MIP-1α) and markers of macrophage content (CD68 and MAC-2) (Table 2). Metabolic flexibility (ΔRQ) was negatively correlated with MCP-1, MIP-1α, CD68 and MAC-2 (Table 2). The expressions of these four genes were highly intercorrelated with R2 ranging from 0.35 to 0.78 (data not shown). Percent body fat was positively correlated with only MIP-1α (Table 2); however, total adipose tissue mass is positively correlated with all four gene markers of inflammation (data not shown). Fat cell size was positively correlated with gene expression of the chemokine MIP-1α (Table 2) and the macrophage marker CD68 (Table 2). There was no relationship between fasting NEFAs and the chemokines MCP-1 and MIP-1α; however, there was a strong positive correlation between insulin-suppressed NEFAs and both MCP-1 and MIP-1α (Table 2). There was no relationship between the macrophage markers CD68 and MAC-2 and fasting NEFAs. Insulin-suppressed NEFAs were strongly correlated with CD68 and MAC-2 (Table 2). The gene expression for chemokines MCP-1 and MIP-1α was lower in metabolically flexible subjects (i.e. those with lower ΔRQ) (ANOVA, p < 0.05; Fig. 2A and ANOVA, p < 0.05; Fig. 2B, respectively), as were the macrophage markers CD68 and MAC-2 (ANOVA, p < 0.05; Fig. 2C and ANOVA, p < 0.05; Fig. 2D).
Fig. 2.
Relationships between reduced metabolic flexibility (ΔRQ) and expressions of chemokines and macrophage markers. Change in respiratory quotient (ΔRQ; metabolic flexibility) was measured before and during a euglycemic-hyperinsulinemic clamp (EHC) in the population of 56 healthy young men. ΔRQ was subdivided into quartiles and was associated with gene expressions of chemokines and macrophage markers, with post hoc testing by mean equality contrast between different groups using the Tukey-Kramer HSD; alpha = 0.05. Type I error rate was set a priori at p < 0.05. mRNA expression data were normalized to 18S. Data are shown as means ± S.E. Levels which do not share the same letter are significantly different. (A) MCP-1 expression, (B) MIP-1α expression, (C) CD68 expression and (D) MAC-2 expression.
3.4. Oxidative and non-oxidative glucose disposal are related to metabolic flexibility (ΔRQ) in healthy young men
To explore the contribution of the fasting and insulin-stimulated sides of glucose disposal, we calculated fasting glucose oxidation, as well as oxidative and non-oxidative glucose disposal during insulin infusion. Metabolic flexibility (higher ΔRQ) was associated with lower fasting glucose oxidation (ANOVA, p < 0.05; Fig. 3A). As expected, higher ΔRQ was associated with higher insulin-stimulated glucose oxidation (ANOVA, p < 0.05; Fig. 3B); and with higher non-oxidative glucose disposal (glucose storage) (ANOVA, p < 0.05; Fig. 3C). Upon insulin stimulation, glucose storage was significantly greater than glucose oxidation for both flexible (higher ΔRQ) and inflexible subjects (lower ΔRQ) (0.57 ± 0.06 g/min vs. 0.22 ± 0.02 g/min, p < 0.05 and 0.43 ± 0.05 g/min vs. 0.20 ± 0.01 g/min, p < 0.05, respectively; data not shown).
3.5. Correlates of metabolic flexibility (ΔRQ) in healthy young men
To identify the dominant factors that contribute to metabolic flexibility, we used a combination of univariate and stepwise linear regression analyses in our cohort of healthy youngmen. We first performed univariate correlations between metabolic flexibility (ΔRQ) and clinical characteristics, as well as adipose tissue gene expression. All factors significantly correlated with ΔRQ (fat cell size, total adipose tissue mass, visceral adipose tissue mass, deep subcutaneous adipose tissue mass, subcutaneous adipose tissue mass, fasting triglycerides, fasting high-density lipoprotein (HDL), percent body fat, VO2max, fasting glucose, insulin-suppressed NEFAs, serum adiponectin, glucose disposal rate, CD68, MAC-2, MCP-1 and MIP-1α) were loaded into the stepwise linear regression analysis. The only three adipose tissue characteristics that correlated with metabolic flexibility in these men were insulin-suppressed NEFAs, HDL and serum adiponectin (model R2 = 0.46, p < 0.05; data not shown).
4. Discussion
Metabolic flexibility (ΔRQ) is influenced by both skeletal muscle and adipose tissue. The contribution of skeletal muscle to metabolic flexibility is related to mitochondrial content and function [3,19]. The aim of this study was to determine how adipose tissue might relate to metabolic flexibility. We found that metabolic flexibility is negatively associated with percent body fat, fat cell size and insulin-suppressed NEFAs; that is, those who were metabolically flexible were less fat, had smaller fat cells and had greater suppression of plasma fatty acids when insulin was infused. We also found that family history of T2D did not affect the relationships between metabolic flexibility and body fatness, fat cell size and insulin suppression of NEFAs.
Gene markers for chemokines and macrophages in adipose tissue were also positively correlated with reduced metabolic flexibility. Metabolically flexible subjects expressed lower levels of the chemokine genes for MCP-1 and MIP-1α and lower levels of macrophage markers CD68 and MAC-2. In contrast, they had higher circulating adiponectin. Civitarese et al. reported that adiponectin signaling regulates mitochondrial bioenergetics in skeletal muscle and adiponectin levels strongly correlate with mitochondrial DNA (mtDNA) content. These findings define a pathway by which adiponectin increases mitochondrial number and function and exerts antidiabetic effects [20]. Both lower glucose oxidation (higher fat oxidation) during fasting and higher glucose oxidation/storage after insulin stimulation were associated with the metabolically flexible phenotype. This is in accordance with findings from Kelley and Mandarino, using the leg balance technique, that glucose oxidation was increased in leg muscle of type 2 diabetic subjects studied post-absorptively under conditions of fasting hyperglycemia. In fact, leg RQs in individuals with type 2 diabetes averaged 0.92 under fasting conditions [21].
In this study, metabolic flexibility varied greatly (0.03–0.25), but was normally distributed, across the cohort of 56 healthy young men; however, those with a lower percentage of body fat were more metabolically flexible (i.e. had a higher ΔRQ during insulin infusion). It is important to note that the risk of metabolic complications is increased not only by the amount and location of adipose tissue, but also by the size of the fat cells [22,23]. Human fat cells can change by 20-fold in diameter and several thousand-fold in volume. Jernas et al. identified genes with markedly higher expression in large, compared with small, human adipocytes and they concluded that these genes may link hypertrophic obesity to T2D [24]. Enlarged adipocytes have both increased lipolysis and impaired insulin-mediated antilipolysis in the post-absorptive state and are associated with hyperglycemia and predict the onset of T2D [25]. In contrast, the stimulating effect of insulin on the rate of glucose metabolism is inversely related to the size of the fat cell [26,27]. Our results show that small adipocytes are associated with metabolic flexibility in these healthy young men and that larger fat cells are more associated with a reduced metabolic flexibility. The transcriptional link between larger adipocytes and reduced metabolic flexibility (low ΔRQ) is complementary to our data showing a physiological link between larger adipocytes and impaired insulin suppression of lipolysis, which is associated with reduced metabolic flexibility. Therefore, increased body fatness, coupled with larger adipocytes, is a determinant of reduced metabolic flexibility.
Another strong determinant for metabolic flexibility is fuel availability [1]. NEFA levels are pivotal in complex disease states such as T2D and its associated reduced metabolic flexibility. Recent studies by Bajaj et al. demonstrate an improvement in peripheral insulin action in response to a 7-day reduction of plasma NEFAs in type 2 diabetics. Lipid infusion inhibits glucose metabolism [28]. Impaired insulin suppression of lipolysis leads to excess circulating non-esterified free fatty acids when glucose uptake and oxidation/storage should be maximal. Given the high fractional uptake of NEFAs and the obligatory oxidation of these NEFAs, this inappropriate lipid supply might be an important contributor to reduced metabolic flexibility.
Accumulation of excess fatty acids in the form of triacylglycerol (TAG) in skeletal muscle and in liver is associated with reduced metabolic flexibility [21,29,30]. Here we report that reduced metabolic flexibility is strongly related to lower suppression of NEFAs by insulin. These data are consistent with a model whereby the failure to suppress NEFA (after a meal) leads to reduced metabolic flexibility in the skeletal muscle which is then unable to switch substrates from fatty acid oxidation to glucose oxidation in the presence of insulin. It is important to note that a lower fatty acid suppression during insulin infusion might be a manifestation of decreased insulin sensitivity. However, while it is not clear whether it is a failure to turn off lipolysis or a failure of peripheral tissues to take up fatty acids, we do think it is related to the size of the fat cell as demonstrated by the negative association between metabolic flexibility and fat cell size, as well as the positive association between fat cell size and insulin-suppressed fatty acids. The link between adipose tissue and metabolic flexibility emphasizes the important role of adipose tissue with its inflammatory cells in buffering the daily influx of dietary fat entering the circulation and preventing excessive exposure of other tissues to fatty acids. In addition, we demonstrated that fat cell size was positively correlated with fasting and insulin-suppressed NEFAs during the hyperinsulinemic clamp. These findings suggest a relationship among the characteristics of adipose tissue, NEFAs and reduced metabolic flexibility.
The next step was to determine if other characteristics of adipose tissue were related to metabolic flexibility. Macrophage infiltration [31] and the expression of genes involved in chemotaxis and macrophage content are higher in obesity and related metabolic disorders [32–37]. Chronic activation of the inflammatory pathway might be a mechanism for certain obesity-related metabolic disorders, such as T2D [38]. Recent attention has focused on the potential role of macrophages in this process [39]. Weisberg et al. and Xu et al. have shown that in obesity, adipose tissue contains an increased number of resident macrophages and that, in some circumstances, macrophages can constitute almost half of the cell population within an adipose tissue depot [40,41]. Macrophages have been shown to be a potential source of secreted proinflammatory factors, and these data have led to the concept that macrophages in adipose tissue can directly influence adipocyte biology and perhaps lead to a state of reduced metabolic flexibility. More than this, immunohistochemical analysis from Weisberg et al. of human subcutaneous adipose tissue revealed that the percentage of cells expressing the macrophage antigen CD68 was significantly and positively correlated with both adipocyte size and body fat mass [40]. The total contribution of adipose tissue to metabolic flexibility is still unclear. While our results show that percent body fat is positively correlated with only MIP-1α gene expression (Table 2), total adipose tissue mass is positively correlated with all four gene markers of inflammation. All analyses were performed in subcutaneous abdominal adipose tissue since this depot is the most accessible, but discussions are ongoing regarding the pertinence of this tissue in metabolic aspects. Our results also reveal that metabolically inflexible subjects had higher gene expression of both macrophage markers (CD68 and MAC-2), as well as both chemokine markers (MCP-1 and MIP-1α). Failure of insulin to suppress NEF As adequately was associated with higher gene expression for the macrophage markers CD68 and MAC-2 and for the chemokine markers MCP-1 and MIP-1α; however, fasting NEFAs were not related to expression of inflammatory genes. This is in contrast to the findings of Kelley et al. where fasting NEFAs significantly contribute to reduced metabolic flexibility in an older, more obese population [2].
Cytokine release within adipose tissue is correlated with adipocyte size [42–45], and hypertrophic adipocytes may contribute to the excess accumulation of FAs in other tissues [46]. This concept of “lipotoxicity” has been related to insulin resistance and compromised flexibility in substrate utilization [47–51]. In this study, fat cell size was positively correlated with the macrophage CD68 and the chemokine MIP-1α. The correlation of macrophage markers and chemokines with insulin-suppressed NEFA levels supports a relationship between macrophage infiltration/chemotaxis and a reduced ability of the adipose tissue to respond to insulin’s signal to suppress lipolysis. Even more striking was that mRNA expression of both the macrophage markers and the chemokine markers were coordinately increased, suggesting an interdependent relationship. These data are consistent with a model where larger adipocytes lead to chemotaxis and macrophage infiltration, resulting in impaired insulin suppression of NEFAs.
To further identify the role of adipose tissue as an endocrine organ in metabolic flexibility, we correlated serum levels of adiponectin to metabolic flexibility. We found that lower serum adiponectin levels were associated with reduced metabolic flexibility. Recent studies by Civitarese et al. demonstrated that low adiponectin levels are associated with low mtDNA copy number [20]. Given a tight relationship between mitochondrial content and insulin sensitivity (Ukropcova [4]), adiponectin could influence metabolic flexibility.
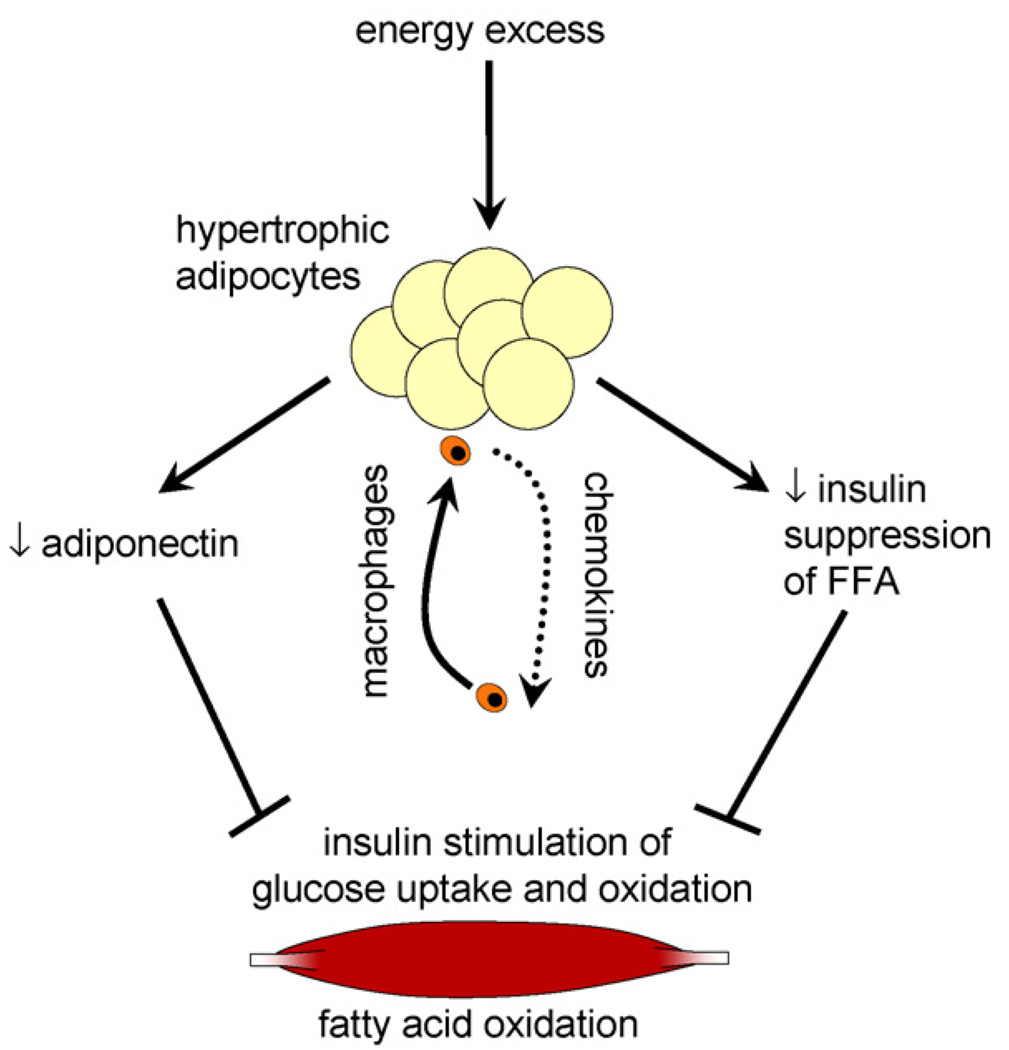
Body fat, larger adipocytes, reduced adiponectin, increased chemokines and macrophage infiltration and impaired insulin suppression of non-esterified free fatty acids all contribute to reduced metabolic flexibility in healthy young men (Fig. 4). Future studies that isolate each of these factors will be required to determine causality. Taken together, these results suggest that both the quantity and the quality of the adipose organ are important in metabolic flexibility. The clinical implications for this paper are twofold. First, this suggests that metabolic defects namely metabolic flexibility and adipose tissue inflammation occur in young healthy men without overt disease. This suggests that early interventions to reverse these defects might prevent diabetes and cardiovascular disease. Second, metabolic inflexibility is fundamental and early component of the dysmetabolic syndrome and provides new insight into why some people develop diabetes and some do not.
Fig. 4.
A model for the role of adipose tissue in metabolic flexibility. Energy excess leads to increased body fatness (%) and hypertrophic adipocytes. Hypertrophic adipocytes secrete chemokines and lead to macrophage infiltration. The macrophage-infiltrated hypertrophic adipocytes decrease insulin-stimulated suppression of lipolysis, as well as decrease adiponectin secretion. Elevated levels of NEFAs during insulin infusion, coupled with decreased serum adiponectin levels, lead to an inhibition of insulin-stimulated glucose uptake and oxidation in skeletal muscle and a decreased capacity for fat oxidation.
Acknowledgments
This work was supported by USDA grant #2003-34323-14010 and USPHS grant HL67933. We also thank the study participants.
Footnotes
Conflict of interest
None.
REFERENCES
- 1.Storlien L, Oakes ND, Kelley DE. Metabolic flexibility. Proc. Nutr. Soc. 2004;63:363–368. doi: 10.1079/PNS2004349. [DOI] [PubMed] [Google Scholar]
- 2.Kelley DE, Goodpaster B, Wing RR, Simoneau JA. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am. J. Physiol. 1999;277:E1130–E1141. doi: 10.1152/ajpendo.1999.277.6.E1130. [DOI] [PubMed] [Google Scholar]
- 3.Ukropcova B, McNeil M, Sereda O, de Jonge L, Xie H, Bray GA, et al. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J. Clin. Invest. 2005;115:1934–1941. doi: 10.1172/JCI24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ukropcova B, Sereda O, de Jonge L, Bogacka I, Nguyen T, Xie H, et al. Family history of diabetes links impaired substrate switching and reduced mitochondrial content in skeletal muscle. Diabetes. 2007;56:720–727. doi: 10.2337/db06-0521. [DOI] [PubMed] [Google Scholar]
- 5.Phielix E, Mensink M. Type 2 diabetes mellitus and skeletal muscle metabolic function. Physiol. Behav. 2008;94:252–258. doi: 10.1016/j.physbeh.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 6.DeFronzo RA, Gunnarsson R, Bjorkman O, Olsson M, Wahren J. Effects of insulin on peripheral and splanchnic, glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J. Clin. Invest. 1985;76:149–155. doi: 10.1172/JCI111938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahn CR. Triglycerides and toggling the tummy. Nat. Genet. 2000;25:6–7. doi: 10.1038/75610. [DOI] [PubMed] [Google Scholar]
- 8.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 9.Boden G, Shulman GI. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. Eur. J. Clin. Invest. 2002;32 Suppl. 3:14–23. doi: 10.1046/j.1365-2362.32.s3.3.x. [DOI] [PubMed] [Google Scholar]
- 10.Bogacka I, Gettys TW, de Jonge L, Nguyen T, Smith JM, Xie H, et al. The effect of beta-adrenergic and peroxisome proliferator-activated receptor-gamma stimulation on target genes related to lipid metabolism in human subcutaneous adipose tissue. Diabetes Care. 2007;30:1179–1186. doi: 10.2337/dc06-1962. [DOI] [PubMed] [Google Scholar]
- 11.Sparks LM, Xie H, Koza RA, Mynatt RL, Hulver MW, Bray G, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 12.Heilbronn LK, Rood J, Janderova L, Albu JB, Kelley DE, Ravussin E, et al. Relationship between serum resistin concentrations and insulin resistance in nonobese, obese, and obese diabetic subjects. J. Clin. Endocrinol. Metab. 2004;89:1844–1848. doi: 10.1210/jc.2003-031410. [DOI] [PubMed] [Google Scholar]
- 13.Carter H, Jones AM, Barstow TJ, Burnley M, Williams CA, Doust JH. Oxygen uptake kinetics in treadmill running and cycle ergometry: a comparison. J. Appl. Physiol. 2000;89:899–907. doi: 10.1152/jappl.2000.89.3.899. [DOI] [PubMed] [Google Scholar]
- 14.Smith SR, Lovejoy JC, Greenway F, Ryan D, Dejonge L, De La Bretonne J, et al. Contributions of total body fat, abdominal subcutaneous adipose tissue compartments, and visceral adipose tissue to the metabolic complications of obesity. Metabolism. 2001;50:425–435. doi: 10.1053/meta.2001.21693. [DOI] [PubMed] [Google Scholar]
- 15.Livesey G, Elia M. Food energy values of artificial feeds for man. Clin. Nutr. 1985;4:99–111. doi: 10.1016/0261-5614(85)90051-2. [DOI] [PubMed] [Google Scholar]
- 16.Smith SR, Xie H, Baghian S, Needham A, McNeil M, Bogacka I, et al. Pioglitazone changes the distribution of adipocyte size in type 2 diabetics. Adipocytes. 2006;2:11–22. [Google Scholar]
- 17.Miles JM, Judd RL, Persson M, Coppack SW. Methods for estimating the kinetics of lipid fuels in vivo. In: Standl W, Mogensen CE, editors. Research Methodologies in Human Diabetes. Diabetes Forum V. New York: Walter de Gruyter; 1995. pp. 345–359. [Google Scholar]
- 18.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 19.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 20.Civitarese AE, Ukropcova B, Carling S, Hulver M, DeFronzo RA, Mandarino L, et al. Role of adiponectin in human skeletal muscle bioenergetics. Cell Metab. 2006;4:75–87. doi: 10.1016/j.cmet.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a re-examination. Diabetes. 2000;49:677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 22.Weyer C, Foley JE, Bogardus C, Tataranni PA, Pratley RE. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia. 2000;43:1498–1506. doi: 10.1007/s001250051560. [DOI] [PubMed] [Google Scholar]
- 23.Xie W, Hamilton JA, Kirkland JL, Corkey BE, Guo W. Oleate-induced formation of fat cells with impaired insulin sensitivity. Lipids. 2006;41:267–271. doi: 10.1007/s11745-006-5096-4. [DOI] [PubMed] [Google Scholar]
- 24.Jernas M, Palming J, Sjoholm K, Jennische E, Svensson PA, Gabrielsson BG, et al. Separation of human adipocytes by size: hypertrophic fat cells display distinct gene expression. FASEB J. 2006;20:1540–1542. doi: 10.1096/fj.05-5678fje. [DOI] [PubMed] [Google Scholar]
- 25.Sharma AM, Staels B. Peroxisome proliferator-activated receptor γ (PPARγ) and adipose tissue understanding obesity-related changes in regulation of lipid and glucose metabolism. J. Clin. Endocrinol. Metab. 2006;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- 26.Salans LB, Knittle JL, Hirsch J. The role of adipose cell size and adipose tissue insulin sensitivity in the carbohydrate intolerance of human obesity. J. Clin. Invest. 1968;47:153–165. doi: 10.1172/JCI105705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salans LB, Zarnowski MJ, Segal R. Effect of insulin upon the cellular character of rat adipose tissue. J. Lipid Res. 1972;13:616–623. [PubMed] [Google Scholar]
- 28.Bajaj M, Suraamornkul S, Romanelli A, Cline GW, Mandarino LJ, Shulman GI, et al. Effect of a sustained reduction in plasma free fatty acid concentration on intramuscular long-chain fatty acyl-CoAs and insulin action in type 2 diabetic patients. Diabetes. 2005;54:3148–3153. doi: 10.2337/diabetes.54.11.3148. [DOI] [PubMed] [Google Scholar]
- 29.Coppack SW, Evans RD, Fisher RM, Frayn KN, Gibbons GF, Humphreys SM, et al. Adipose tissue metabolism in obesity lipase action in vivo before and after a mixed meal. Metabolism. 1992;41:264–272. doi: 10.1016/0026-0495(92)90269-g. [DOI] [PubMed] [Google Scholar]
- 30.Kelley DE, Goodpaster BH. Skeletal muscle triglyceride. An aspect of regional adiposity and insulin resistance. Diabetes Care. 2001;24:933–941. doi: 10.2337/diacare.24.5.933. [DOI] [PubMed] [Google Scholar]
- 31.Permana PA, Menge C, Reaven PD. Macrophage-secreted factors induce adipocyte inflammation and insulin resistance. Biochem. Biophys. Res. Commun. 2006;341:507–514. doi: 10.1016/j.bbrc.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Bruun JM, Helge JW, Richelsen B, Stallknecht B. Diet and exercise reduce low-grade inflammation and macrophage infiltration in adipose tissue but not in skeletal muscle in severely obese subjects. Am. J. Physiol. Endocrinol. Metab. 2006;290:E961–E967. doi: 10.1152/ajpendo.00506.2005. [DOI] [PubMed] [Google Scholar]
- 33.Neels JG, Olefsky JM. Inflamed fat: what starts the fire? J. Clin. Invest. 2006;116:33–35. doi: 10.1172/JCI27280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neels JG, Pandey M, Hotamisligil GS, Samad F. Autoamplification of tumor necrosis factor-alpha: a potential mechanism for the maintenance of elevated tumor necrosis factor-alpha in male but not female obese mice. Am. J. Pathol. 2006;168:435–444. doi: 10.2353/ajpath.2006.050699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sell H, Dietze-Schroeder D, Eckardt K, Eckel J. Cytokine secretion by human adipocytes is differentially regulated by adiponectin, AICAR, and troglitazone. Biochem. Biophys. Res. Commun. 2006;343:700–706. doi: 10.1016/j.bbrc.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Sell H, Dietze-Schroeder D, Kaiser U, Eckel J. Monocyte chemotactic protein-1 is a potential player in the negative cross-talk between adipose tissue and skeletal muscle. Endocrinology. 2006;147:2458–2467. doi: 10.1210/en.2005-0969. [DOI] [PubMed] [Google Scholar]
- 37.Weisberg SP, Hunter D, Huber R, Lemieux J, Slaymaker S, Vaddi K, et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J. Clin. Invest. 2006;116:115–124. doi: 10.1172/JCI24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellen KE, Hotamisligil GS. Inflammation, stress and diabetes. J. Clin. Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouloumie A, Curat CA, Sengenes C, Lolmede K, Miranville A, Busse R. Role of macrophage tissue infiltration in metabolic diseases. Curr. Opin. Clin. Nutr. Metab. Care. 2005;8:347–354. doi: 10.1097/01.mco.0000172571.41149.52. [DOI] [PubMed] [Google Scholar]
- 40.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sopasakis VR, Sandqvist M, Gustafson B, Hammarstedt A, Schmelz M, Yang X, et al. High local concentrations and effects on differentiation implicate interleukin-6 as a paracrine regulator. Obes. Res. 2004;12:454–460. doi: 10.1038/oby.2004.51. [DOI] [PubMed] [Google Scholar]
- 43.Van Harmelen V, Reynisdottir S, Eriksson P, Thorne A, Hoffstedt J, Lonnqvist F, et al. Leptin secretion from subcutaneous and visceral adipose tissue in women. Diabetes. 1998;47:913–917. doi: 10.2337/diabetes.47.6.913. [DOI] [PubMed] [Google Scholar]
- 44.Winkler G, Kiss S, Keszthelyi L, Sapi Z, Ory I, Salamon F, et al. Expression of tumor necrosis factor (TNF)-alpha protein in the subcutaneous and visceral adipose tissue in correlation with adipocyte cell volume, serum TNF-alpha, soluble serum TNF-receptor-2 concentrations and C-peptide level. Eur. J. Endocrinol. 2003;149:129–135. doi: 10.1530/eje.0.1490129. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Guo KY, Diaz PA, Heo M, Leibel RL. Determinants of leptin gene expression in fat depots of lean mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;282:R226–R234. doi: 10.1152/ajpregu.00392.2001. [DOI] [PubMed] [Google Scholar]
- 46.Lelliott C, Vidal-Puig AJ. Lipotoxicity, an imbalance between lipogenesis de novo and fatty acid oxidation. Int. J. Obes. Relat. Metab. Disord. 2004;28 Suppl. 4:S22–S28. doi: 10.1038/sj.ijo.0802854. [DOI] [PubMed] [Google Scholar]
- 47.Goodpaster BH, Stenger VA, Boada F, McKolanis T, Davis D, Ross R, et al. Skeletal muscle lipid concentration quantified by magnetic resonance imaging. Am. J. Clin. Nutr. 2004;79:748–754. doi: 10.1093/ajcn/79.5.748. [DOI] [PubMed] [Google Scholar]
- 48.Goodpaster BH, Thaete FL, Kelley DE. Composition of skeletal muscle evaluated with computed tomography. Ann. N.Y. Acad. Sci. 2000;904:18–24. doi: 10.1111/j.1749-6632.2000.tb06416.x. [DOI] [PubMed] [Google Scholar]
- 49.Greco AV, Mingrone G, Giancaterini A, Manco M, Morroni M, Cinti S, et al. Insulin resistance in morbid obesity: reversal with intramyocellular fat depletion. Diabetes. 2002;51:144–151. doi: 10.2337/diabetes.51.1.144. [DOI] [PubMed] [Google Scholar]
- 50.Kelley DE, Williams KV, Price JC, McKolanis TM, Goodpaster BH, Thaete FL. Plasma fatty acids, adiposity, and variance of skeletal muscle insulin resistance in type 2 diabetes mellitus. J. Clin. Endocrinol. Metab. 2001;86:5412–5419. doi: 10.1210/jcem.86.11.8027. [DOI] [PubMed] [Google Scholar]
- 51.Perseghin G, Scifo P, De Cobelli F, Pagliato E, Battezzati A, Arcelloni C, et al. Intramyocellular triglyceride content is a determinant of in vivo insulin resistance in humans: a 1H–13C nuclear magnetic resonance spectroscopy assessment in offspring of type 2 diabetic parents. Diabetes. 1999;48:1600–1606. doi: 10.2337/diabetes.48.8.1600. [DOI] [PubMed] [Google Scholar]