The hepatitis B virus (HBV) is a major cause of liver disease in humans[1] and its non-infectious capsid is of interest for nanotechnology, including for drug-delivery applications. A precise biophysical characterization of these particles is of importance not only for these applications, but also because it may provide further insight into the replication cycle and assembly pathway of the virus, and thus contribute to the future development of drugs.[2,3] The HBV capsid protein (cp) forms icosahedral capsids of two sizes in vivo and in vitro (with triangulation numbers of T = 3 and T = 4[4] that contain 180 and 240 subunits, respectively[5–8]).The capsid protein has two domains—a core domain (amino acids 1–140) and a “protamine domain” (amino acids 150–183)—connected by a 10-residue linker;[9] of these, the core domain is necessary and sufficient for assembly of the capsid. The length of the linker and the conditions under which assembly take place determine the ratio of the T = 3 and T = 4 capsids obtained.[10] Capsid protein dimers are stabilized by an intermolecular four-helix bundle[11–13] and a disulfide bond within the bundle (Cys61); these dimers represent the building blocks for the formation of the capsid. However, the disulfide bond is not required for dimerization or assembly, as Cys61 can be replaced with Ala[9, 10] without affecting either process. The interfaces of the dimers display protruding spikes that result in an uneven surface.[6, 13] Although extensive structural studies of the HBV capsid structure have been performed by electron microscopy (EM) and X-ray crystallography,[14] knowledge of its biophysical properties is limited.[15, 16] Here, we present data from macromolecular tandem and ion mobility mass spectrometry (MS)[17–19] that have a bearing on the stability and conformational diversity of HBV capsids in vacuo.
Multiple charged ions of a protein or protein complex are generated in mass spectrometry by electrospray ionization (ESI), which typically gives rise to approximately Gaussian-shaped charge-state distributions of ion signals for each species present in solution (Figure 1A).The charge and, thus, the mass of the analyte may be determined from such spectra with very high precision. Impressive progress in MS has enabled measurement of the accurate mass of macromolecular particles as large as ribosomes[20] and transcriptosomes,[21] and has contributed unique insights into their compositions, structures, and stabilities. Even whole viruses have been introduced into mass spectrometers.[22] We recently used MS to determine the accurate masses (±0.1%) of HBV capsids assembled in vitro.[8] Herein we further investigate the two HBV cp149 constructs (consisting of the core domain plus a nearly complete linker), namely the cysteine-free mutant (3C→A) and a mutant containing a single cysteine residue (61C, containing the intradimer disulfide bond).T he mass spectra of both the 3C→A and 61C capsids showed two well-resolved sets of ion signals around m/z 22 000 and 25 000 (Figure 1A). The high resolution enabled us to determine the masses of the capsids (Figure 1A),[8] which led us to conclude that the capsids are composed of exactly 90 and 120 dimers.
Figure 1.
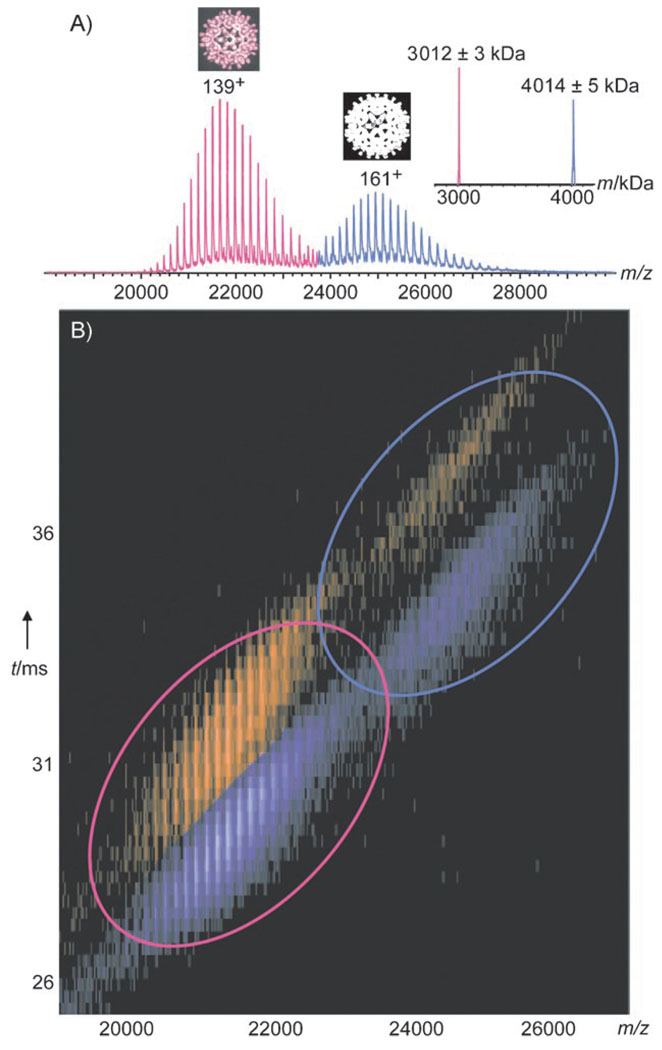
Ion mobility MS on HBV cp149 61C capsids. A) Mass spectrum of 61C, adapted from Uetrecht et al., 2008.[8] The distributions around m/z 22000 and 25000 represent the T = 3 and T = 4 capsids (pink and blue), respectively. For each distribution, the most abundant charge state is indicated. The inset on the right shows a spectrum deconvolved to uncharged species. The calculated monomer mass is 16706 Da. B) Contour plot of the ion mobility drift time versus m/z for 61C capsids at 0.04 µm (ca. 8 µm cp monomer) in 200 mm ammonium acetate, pH 6.8, without fragmentation (150 V accelerating voltage). Both the T = 3 (pink ellipse) and T = 4 (blue ellipse) capsids show small (purple) and large (orange) conformers.
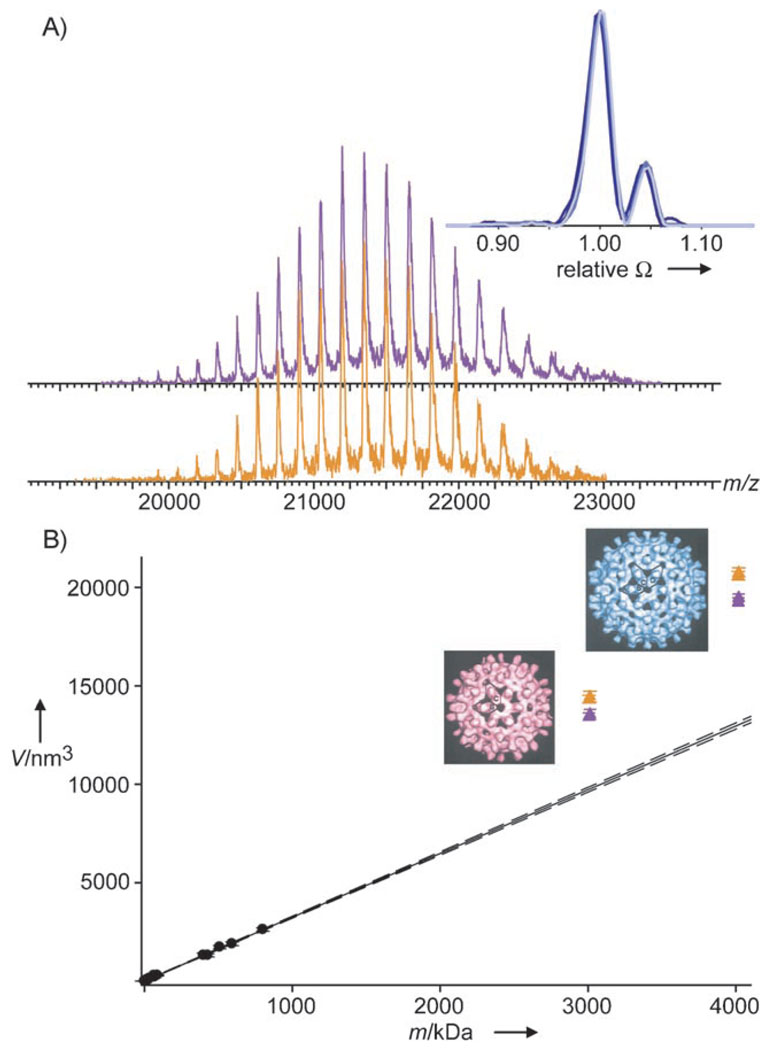
These data already proved that capsids can remain intact in vacuo. We further analyzed cp149 capsids by ion mobility MS (IMS) to investigate the capsid structure/shape under in vacuo conditions.[18, 23] Here, ions are separated not only on the basis of their m/z ratio but also, inside a gas-filled ion mobility chamber, according to their drift time, which is shape-dependent. Typically, molecules with larger cross-sections, that is, larger volumes, exhibit longer drift times. The cross-section or average projected area of a molecule can be determined from the drift time.[18] A contour plot of IM drift time versus m/z for the 61C capsids is shown in Figure 1B. Strikingly, two significantly different drift times are observed for both the T = 3 and T = 4 capsids, and even for each charge state thereof. This finding suggests that at least two different conformers are present. The ratio between the two conformers is independent of the charge state and applied acceleration voltage (Figure 2A), thus indicating that the conformers are either already present in solution or, more likely, formed during the ESI ionization/desolvation process. For each charge state and for both T = 3 and T = 4, the smaller conformer is more abundant. Only a quarter (22±6%) of the capsids correspond to the conformer with the larger cross-section (by 4.4±0.5%; Figure 2A inset; see also Table S1 in the Supporting Information).The results for 3C→A and 61C were very similar (see Tables S1 and S2 in the Supporting Information).The capsid radii calculated from the measured cross-sections are 14.8 and 15.1 nm for the two different T = 3 conformers; these values are in excellent agreement with radii ranging from 13.1 to 15.9 nm as determined from cryo-EM reconstructions.[10] The volumes of the capsids, as calculated from the cross-sections measured in vacuo, are plotted versus particle mass in Figure 2B, together with similar data obtained for a number of unrelated proteins and protein complexes under comparable experimental conditions (see also the Supporting Information). A linear relationship is observed between the volume and mass, except for all of the analyzed HBV capsids, which seem to occupy larger volumes in vacuo than do globular proteins of similar mass. These data suggest that the capsids retain their hollow-sphere character to a large extent in vacuo.
Figure 2.
Shape of HBV capsids in vacuo. A) Charge state distributions of the small (purple) and large (orange) T = 3 conformers as extracted from Figure 1B normalized to the intensity of their individual base signal. The inset shows the unchanged abundance and relative size of conformers, which is independent of the charge state and accelerating voltage (165+ ion at 100 V and the 169+, 165+, and 161+ ions at 150 V (dark to light blue) of 61C T = 4). The cross-sections (Ω) and intensities are normalized on the base signal of a particular charge state and overlay almost perfectly. B) Volumes derived from the cross-sections for a large set of proteins and protein complexes (circles, see also Table S2 in the Supporting Information) and the 3C→A and 61C capsids (triangles). Error bars represent the standard deviation. The small and large conformers are depicted in purple and orange, respectively. The solid line represents a linear regression in the set of globular proteins. The dashed lines correspond to a 99% confidence interval of the linear regression. The EM reconstructions[10] of T = 3 and T = 4 are shown to the left of the triangles.
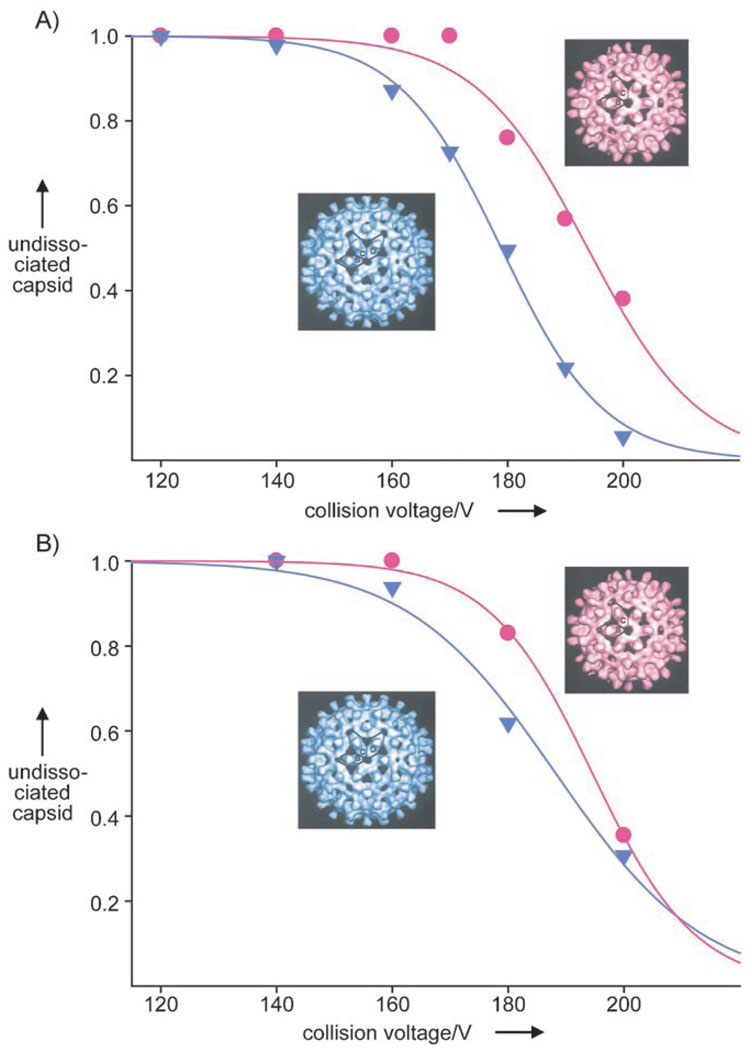
Little is known about the relative stabilities of the T = 3 and T = 4 capsids.[8, 24] We thus studied both capsids by tandem MS,[17, 19, 25] by using a quadrupole analyzer to select the T = 3 or T = 4 capsids. In general, single highly charged subunits are ejected from the precursor ions of a protein complex in tandem MS, with the concomitant formation of lower charged counter-complexes. The masses and charges of the fragments should add up to the mass and charge of the precursor.[17, 26] This dissociation is induced in the collision cell, where an additional, accelerating voltage activates the selected precursor ions through collisions. In line with this scenario, the T = 3 and T = 4 3C→A capsid ions fragment through ejection of cp monomers, but the 61C capsids fragment through the loss of dimers, presumably because of the presence of the covalent disulfide bond.[8] To directly compare the T = 3 and T = 4 capsids, we gradually increased the collisional activation and selected ions of both species around the same m/z value which should then acquire similar internal energies upon collisional activation.[25, 26] The energy-resolved breakdown curves of the T = 3 and T = 4 capsids are plotted for both constructs in Figure 3. These data reveal that the T = 3 3C→A and 61C capsids have a higher resistance to dissociation in vacuo than their T = 4 counterparts. These results are in agreement with data on the other charge states present in the mass spectra (data not shown).
Figure 3.
Curves showing the breakdown of HBV cp149 capsids in vacuo. Abundance of undissociated T = 3 (circles) and T = 4 (triangles) capsids from: A) m/z 24 500 3C→A and B) m/z 24000 61C precursor selection at different collision voltages. The solid lines correspond to sigmoidal fits. The curve progressions are indistinguishable irrespective of whether the laboratory frame or center of mass energy is displayed in the plots.
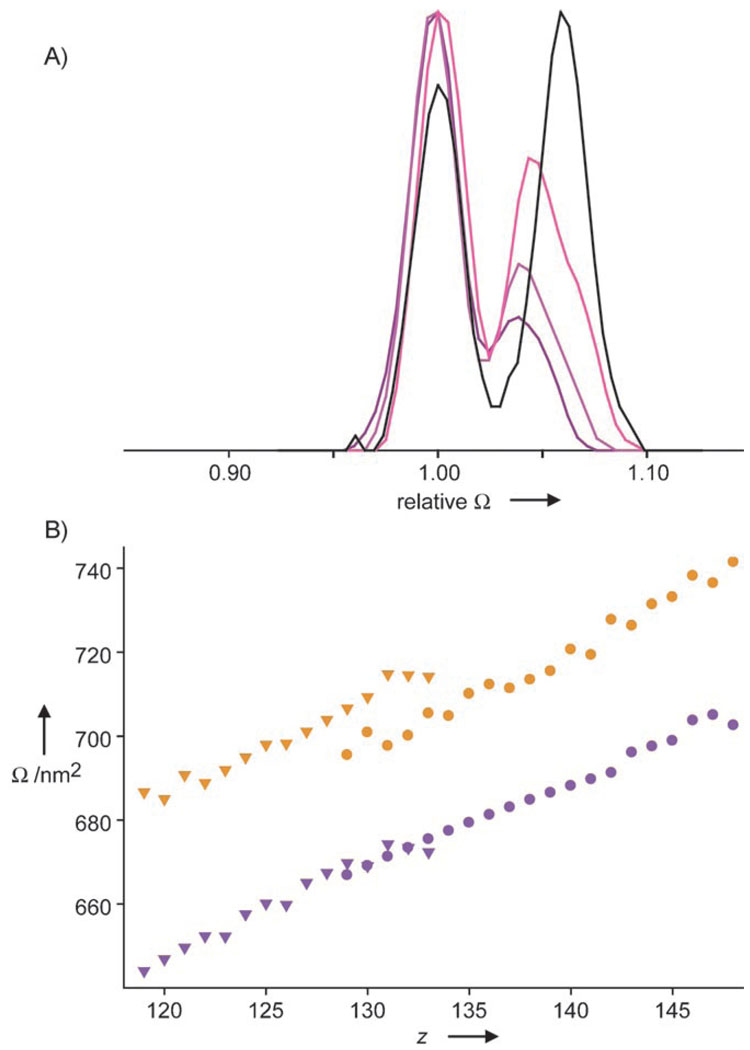
It should be noted that the data in Figure 3 are averaged over the whole ensemble of conformations present at one m/z value, as the signals for the precursors are above the m/z range that can be selected in the instrument used for separation of the conformers. However, distinctive behavior could still be observed for the two conformers upon collision-induced dissociation in the IMS experiments. In detail, the relative abundance of the smaller 61C T = 3 conformer became significantly reduced (from 74% to 60%) when the collision voltage was increased from 150 to 200 V (Figure 4A and see Table S1 in the Supporting Information), which indicates that this smaller conformer dissociates more readily. We also observed that after elimination of a dimer the capsids remain similar in size to the intact precursors (Figure 4B), thus indicating the high stability of its quaternary structure even in vacuo. Interestingly, the larger conformer was more abundant (57%) than the smaller conformer in the capsid-derived fragment ions missing a dimer, as the smaller conformer undergoes another fragmentation step to give a product whose high-mass ions were barely detectable. Our results indicate that the large conformer is, in general, more stable than the small one for T = 3.Fragmentation of T = 4 capsids was not detected in the IMS experiments because of the lower signal intensity and high m/z values of the ions. As far as the overall shape of the capsid particles in vacuo is concerned, we can exclude a distorted open structure, although a slight flattening is compatible with the data. We therefore conclude that a spherical structure is indeed retained.
Figure 4.
Dissociation and cross-sections (Ω) of capsids in vacuo. A) Relative abundance of the conformers of a 141+ ion (undissociated T = 3 61C) without (150 V; dark purple) and with fragmentation (175 and 200 V; purple and pink, respectively) compared to the 126+ counterion from which one dimer was ejected (200 V, black). Cross-sections were normalized on the small conformation and intensities on the base signal. B) Cross-sections of61 C capsids for large (orange) and small (purple) conformers of undissociated T = 3 (circles) and T = 3 missing a dimer (triangles) for all the detected charge states. The linear correlation between the cross-section and charge indicates similar structures for the undissociated precursor and the fragmentation products.
Our IMS data reveal that both the T = 3 and T = 4 capsids exhibit two distinct conformers in vacuo. All conformers have a much larger apparent volume than would be expected for “mass-identical” nonhollow globular proteins. Moreover, the cross-sections of all the conformers are reasonably consistent with published cryo-EM data. Both observations imply that the overall hollow spherical shell structure of the capsids is retained in vacuo. Notably, the “water-free”, but uncharged, HBV particles also preserve their spherical shape under the high-vacuum conditions applied for scanning tunneling electron microscopy (STEM).[9]
Two slightly different conformations have been reported in a cryo-EM study of RNA- and DNA-containing HBV capsids.[27] However, we suspect that this conformational distinction differs from what we observe here in vacuo. In the first place, our capsids contain neither RNA nor DNA. Second, the calculated size difference between the conformers as determined by MS (4.4%) appears too large to go undetected in cryo-EM.[28] Rather, it appears more likely that the conformers we observe in vacuo represent alternative, approximately isoenergetic, states that are populated in the ionization/desolvation process. This difference might become amplified or clamped when water is removed in vacuo, as this treatment is thought to reduce interconversion between conformations.[29]
Recycling of cp149 capsids (dissociation into dimers and reassembly) results in a progressive increase in the T = 3 to T = 4 ratio, which suggests that T = 3 assembly in vitro is thermodynamically favored, since external factors such as nucleic acids are absent.[30] In this context, tandem MS studies revealed that the T = 3 capsids had a higher stability than the T = 4 capsids in vacuo. The question remains whether the observed difference correlates with their stability in vitro, as the capsids may be affected by the ionization process and in vacuo (that is, water-free) conditions in the spectrometer. Despite the use of different conditions, the present results are consistent with studies that revealed that the T = 3 capsids had a higher resistance than T = 4 capsids to mutation-induced distortion (for example, insertion of additional amino acids).[24] However, atomic force microscopy studies could not detect a significant difference in the elasticity between the two geometries.[8]
In the specific context of HBV assembly, tandem MS analysis of capsids assembled from subunits of different sizes (that is, different linker lengths) should yield information about changes in stability. Therefore, these measurements could reveal whether the discrimination between the assembly of T = 3 and T = 4 capsids is thermodynamically or kinetically driven, irrespective of whether there is a direct correlation to their behavior in solution.
In general, IMS enhances the resolving power by adding a shape-related dimension to the mass separation. This offers the possibility to identify and differentiate states in a viral assembly not only by mass but also by conformation. This possibility enables the detection of structural variations and transitions within and between protein complexes, which will have possible implications for their function.[31, 32] The coupling of IMS to macromolecular MS provides unprecedented mass accuracy and tolerance towards sample heterogeneity compared to alternative techniques used to probe the size and structural integrity of viruses in vacuo; these alternative methods are limited to particles larger than 50 nm in diameter, and only an indirect mass determination can be performed.[33]
Experimental Section
The detailed experimental approach is described in the Supporting Information.
Tandem MS experiments were carried out on a modified Q-ToF 1 spectrometer (Waters, UK).[34] IMS was performed on a Synapt HDMS (Waters, UK).[35]
Supplementary Material
Footnotes
This research was supported by the Netherlands Proteomics Centre and the NIAMS Intramural Research Program of the NIH.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200802410.
Contributor Information
Charlotte Uetrecht, Biomolecular Mass Spectrometry and Proteomics Group, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Sorbonnelaan 16, 3584 CA Utrecht (The Netherlands) Fax: (+31) 30-251-8219.
Cees Versluis, Biomolecular Mass Spectrometry and Proteomics Group, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Sorbonnelaan 16, 3584 CA Utrecht (The Netherlands) Fax: (+31) 30-251-8219.
Norman R. Watts, Protein Expression Laboratory, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892 (USA)
Paul T. Wingfield, Protein Expression Laboratory, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892 (USA)
Alasdair C. Steven, Laboratory of Structural Biology Research, National Institute of Arthritis, Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892 (USA)
Prof. Dr. Albert J. R. Heck, Email: a.j.r.heck@uu.nl, Biomolecular Mass Spectrometry and Proteomics Group, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Sorbonnelaan 16, 3584 CA Utrecht (The Netherlands), Fax: (+31) 30-251-8219.
References
- 1.Blumberg BS. Proc. Natl. Acad. Sci. USA. 1997;94:7121. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. Science. 2003;299:893. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 3.Singh P, Gonzalez MJ, Manchester M. Drug Dev. Res. 2006;67:23. [Google Scholar]
- 4.Caspar DL, Klug A. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 5.Kenney JM, von Bonsdorff CH, Nassal M, Fuller SD. Structure. 1995;3:1009. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 6.Crowther RA, Kiselev NA, Bottcher B, Berriman JA, Borisova GP, Ose V, Pumpens P. Cell. 1994;77:943. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 7.Dryden KA, Wieland SF, Whitten-Bauer C, Gerin JL, Chisari FV, Yeager M. Mol. Cell. 2006;22:843. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 8.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. Proc. Natl. Acad. Sci. USA. 2008;105:9216. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingfield PT, Stahl SJ, Williams RW, Steven AC. Biochemistry. 1995;34:4919. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 10.Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT. Biochemistry. 1996;35:7412. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 11.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Nature. 1997;386:91. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 12.Böttcher B, Wynne SA, Crowther RA. Nature. 1997;386:88. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 13.Wynne SA, Crowther RA, Leslie AG. Mol. Cell. 1999;3:771. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 14.Steven AC, Conway JF, Cheng N, Watts NR, Belnap DM, Harris A, Stahl SJ, Wingfield PT. Adv. Virus Res. 2005;64:125. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- 15.Ceres P, Zlotnick A. Biochemistry. 2002;41:11525. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 16.Kegel WK, van der Schoot P. Biophys. J. 2004;86:3905. doi: 10.1529/biophysj.104.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heck AJ, Van Den Heuvel RH. Mass Spectrom. Rev. 2004;23:368. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 18.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 19.Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Chem. Rev. 2007;107:3544. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 20.Benjamin DR, Robinson CV, Hendrick JP, Hartl FU, Dobson CM. Proc. Natl. Acad. Sci. USA. 1998;95:7391. doi: 10.1073/pnas.95.13.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenzen K, Vannini A, Cramer P, Heck AJ. Structure. 2007;15:1237. doi: 10.1016/j.str.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Fuerstenau SD, Benner WH, Thomas JJ, Brugidou C, Bothner B, Siuzdak G. Angew. Chem. 2001;113:559. doi: 10.1002/1521-3773(20010316)40:6<9822::aid-anie9822>3.3.co;2-i. Angew. Chem. Int. Ed.2001, 40, 541. [DOI] [PubMed] [Google Scholar]
- 23.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Anal. Chem. 1998;70:2236. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 24.Böttcher B, Vogel M, Ploss M, Nassal M. J. Mol. Biol. 2006;356:812. doi: 10.1016/j.jmb.2005.11.053. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzen K, Versluis C, van Duijn E, van den Heuvel RHH, Heck AJR. Int. J. Mass Spectrom. 2007;268:198. [Google Scholar]
- 26.van Duijn E, Simmons DA, van den Heuvel RH, Bakkes PJ, van Heerikhuizen H, Heeren RM, Robinson CV, van der Vies SM, Heck AJ. J. Am. Chem. Soc. 2006;128:4694. doi: 10.1021/ja056756l. [DOI] [PubMed] [Google Scholar]
- 27.Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA. Proc. Natl. Acad. Sci. USA. 2005;102:15821. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belnap DM, Grochulski WD, Olson NH, Baker TS. Ultramicroscopy. 1993;48:347. doi: 10.1016/0304-3991(93)90110-j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer S, Verma CS. Proc. Natl. Acad. Sci. USA. 1999;96:9613. doi: 10.1073/pnas.96.17.9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingfield PT. unpublished results. [Google Scholar]
- 31.Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJ. J. Mol. Biol. 2008;379:385. doi: 10.1016/j.jmb.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alverdi V, Mazon H, Versluis C, Hemrika W, Esposito G, van den Heuvel R, Scholten A, Heck AJ. J. Mol. Biol. 2008;375:1380. doi: 10.1016/j.jmb.2007.11.053. [DOI] [PubMed] [Google Scholar]
- 33.Nie Z, Tzeng YK, Chang HC, Chiu CC, Chang CY, Chang CM, Tao MH. Angew. Chem. 2006;118:8311. Angew. Chem. Int. Ed.2006, 45, 8131. [Google Scholar]
- 34.van den Heuvel RH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJ, Langridge D, van der Oost J, Hoyes J, Hec AJ. Anal. Chem. 2006;78:7473. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 35.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int. J. Mass Spectrom. 2007;261:1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.