Abstract
Despite the importance of epidermal growth factor receptor (EGFR) in animal development and malignant transformation, surprisingly little is known about the regulation of its expression. Here, we report a novel zinc finger and G-patch domain-containing protein, ZIP. We demonstrated that ZIP acts as a transcription repressor through the recruitment of the nucleosome remodelling and deacetylase complex. Transcriptional target analysis revealed that ZIP regulates several cellular signalling pathways including EGFR pathways that are critically involved in cell proliferation, survival, and migration. We showed that ZIP inhibits cell proliferation and suppresses breast carcinogenesis, and that ZIP depletion leads to a drastic tumour growth in vivo. We found that ZIP is downregulated in breast carcinomas and that its level of expression is negatively correlated with that of EGFR. Our data indicate that ZIP is a novel transcription repressor and a potential tumour suppressor. These findings may shed new light on the EGFR-related breast carcinogenesis and might offer a potential new target for breast cancer therapy.
Keywords: breast cancer, EGFR, gene regulation, transcription repressor
Introduction
Growth factors and their transmembrane receptor kinases have important functions in an array of cellular behaviours including cell proliferation, survival, adhesion, migration, and differentiation (Yarden and Sliwkowski, 2001). The epidermal growth factor receptor (EGFR) family consists of four transmembrane receptors, including EGFR (HER1/erbB-1), HER2 (erbB-2/neu), HER3 (erbB-3), and HER4 (erbB-4) (Yarden and Sliwkowski, 2001). These proteins are composed of an extracellular ligand-binding domain and an intracellular tyrosine kinase domain, joined by a transmembrane segment. On ligand binding, EGFR family proteins undergo conformational changes in the ectodomain, which facilitate the formation of homo/heterodimers or oligomers triggering tyrosine kinase phosphorylation (Zandi et al, 2007). As a consequence, second-messenger pathway cascades, including mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3 kinase (PI3K), are activated, ultimately leading to the alteration of cellular behaviours (Zandi et al, 2007).
Since the identification of a link between EGFR and a transforming viral oncogene v-erb-B (Downward et al, 1984), it has been well established that EGFR is involved in malignant transformation and progression of a broad variety of cancers (Holbro et al, 2003; Chan et al, 2006). Indeed, EGFR overexpression have been reported in cancers originating from bladder, brain, breast, cervical, uterine, colon, esophageal, glioma, lung, ovarian, pancreatic, and renal cell (Chan et al, 2006). This deregulation is often associated with a more aggressive phenotype and accordingly worse survival of the cancer patients (Nicholson et al, 2001). This scenario makes the EGFR family an ideal target to be exploited for cancer therapeutics. Current anti-EGFR therapies include monoclonal antibodies, such as cetuximab, panitumumab, and matuzumab, which target the extracellular domain of EGFR, and small-molecule tyrosine kinase inhibitors, such as gefitinib (Iressa) and erlotinib, which target the receptor catalytic domain (Mendelsohn and Baselga, 2006).
Despite the extensive molecular and functional characterization of EGFR and a continuing effort in pursuing anti-EGFR cancer therapies, little is known about the mechanism underlying the regulation/deregulation of EGFR expression. This issue is of particular importance as it is noted that amplifications in the EGFR gene were restricted to region of the regulatory sequence in the 5′-end of intron 1 and associated with EGFR expression in epithelial breast tumours (Brandt et al, 2006), implying the importance of transcriptional regulation of EGFR in breast carcinogenesis.
Transcriptional repression can be mediated by several mechanisms. One repression mechanism involves the recruitment of corepressor complexes (Hu and Lazar, 2000; Rosenfeld et al, 2006), many of which contain subunits that possess histone deacetylase (HDAC) activity. HDACs act to deacetylate histones and hence convert chromatin into a repressive state (Rosenfeld et al, 2006).
The Mi-2/nucleosome remodelling and deacetylase (NuRD) complex has important functions in animal development and physiology (Ahringer, 2000). This complex is a multi-subunit protein assembly with both histone deacetylation and chromatin remodelling ATPase activities and functions primarily in gene transcriptional repression (Zhang et al, 1998; Denslow and Wade, 2007). To date, the NuRD complex has been documented to mediate the transcription repression by distinct sequence-specific transcription factors, including p53, Ikaros, Hunchback, Tramtrack69, KAP-1, BCL-6, and FOG-1 (Bowen et al, 2004; Denslow and Wade, 2007). It is believed that every subunit of this complex exhibits heterogeneity at the protein and/or gene level and that the functional specialization of the NuRD complex is largely determined by incorporation of unique gene products into the complex (Bowen et al, 2004; Denslow and Wade, 2007).
In this work, we describe the identification and functional characterization of ZIP, a novel zinc finger and G-patch domain-containing protein. We demonstrated that ZIP recruits the NuRD complex to EGFR promoter and represses EGFR transcription. We show that ZIP inhibits cell proliferation and suppresses breast carcinogenesis. These data support a role for ZIP as a novel transcription repressor and a potential tumour suppressor.
Results
Cloning and characterization of ZIP
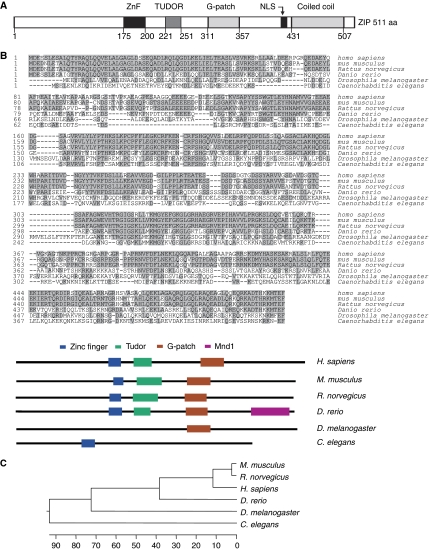
We cloned a gene, ZIP (for ZInc finger and G-Patch domain-containing of its protein product), from a mammary cDNA library. The cDNA of ZIP is 1882 bp in length (GeneBank ID BC032612) and contains an open reading frame encoding for a protein of 511 amino acids. The predicted molecular mass of this protein is ∼55.6 kDa, with a theoretical isoelectric point of 5.49. The corresponding gene is mapped to chromosome 20q13.3 and consists of seven exons and six introns. Bioinformatics analysis indicates that ZIP harbours a CCCH or C3H1 type of zinc finger, a TUDOR domain, a G-patch domain, a coiled-coil domain, and a nuclear localization signal (Figure 1A). Amino-acid sequence alignment reveals that human ZIP shares 77.9% identity with its mouse homologue and the similarity of the amino-acid sequence of ZIP with homologues in other organisms is 76.7% in Rattus norvegicus, 49.2% in Danio rerio, 19.4% in Caenorhabditis elegans, and 24.1% in Drosophila melanogaster (Figure 1B). Phylogenetic analysis also indicates that ZIP is an evolutionarily well-conserved gene (Figure 1C).
Figure 1.
Cloning and characterization of ZIP. (A) A schematic representation of the structure of ZIP. The following conserved domains are shown: ZnF (zinc finger), TUDOR, G-patch, and coiled coil. (B) Amino-acid sequence alignment of ZIP from different species. Shaded residues represent conserved regions (upper panel), and conserved domains of ZIP homologues from different species are indicated (lower panel). (C) Phylogenetic analysis of evolutionary relationships among homologues of ZIP proteins from different species.
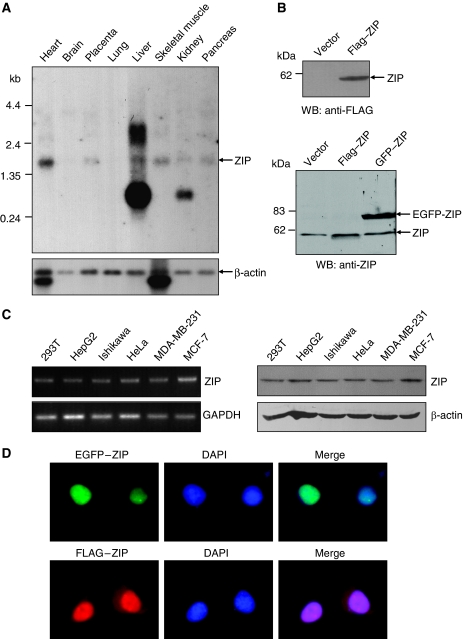
To confirm the existence of ZIP transcript(s) and to examine the expression profile of ZIP, we analysed the expression of ZIP mRNA by Northern blotting with Clontech's human multiple tissue blots. The results indicate that ZIP gene transcribes an ∼1.8 kb message in various tissues (Figure 2A). In the liver and kidneys, additional transcripts were detected (Figure 2A). We focused our research on the ∼1.8 kb transcript because it is the transcript that we initially cloned and it is the transcript that exhibits a broader tissue distribution.
Figure 2.
Expression and subcellular localization of ZIP. (A) Northern blotting analysis of ZIP mRNA expression in different tissues. (B) Western blotting analysis of ZIP protein expression. MCF-7 cells were transfected with empty vector or FLAG-ZIP or EGFP-ZIP. Cellular proteins were prepared and western blotting was performed with anti-FLAG (upper panel) or anti-ZIP (lower panel). MCF-7 (ZIP): MCF-7 cells overexpressing ZIP. (C) RT–PCR (left panel) and western blotting (right panel) analysis of ZIP expression in different cancer cell lines. GAPDH and β-actin were used as internal controls. (D) Subcellular localization of ZIP protein. MCF-7 cells were transfected with EGFP-ZIP (upper panel) or FLAG-ZIP (lower panel). Twenty-four hours after transfection, EGFP fluorescence and rhodamine staining of FLAG were visualized by fluorescence microscopy. DAPI staining was included to visualize the cell nucleus.
To examine the expression of ZIP protein, a FLAG-tagged ZIP expression construct (FLAG-ZIP) was transfected into mammary carcinoma MCF-7 cells. Twenty-four hours after transfection, cellular proteins were extracted and analysed by western blotting with a monoclonal antibody against FLAG. The results indicate that ZIP is expressed as a protein of ∼56 kDa (Figure 2B, upper panel). Western blotting analysis of endogenous ZIP along with overexpressed FLAG-ZIP or enhanced green fluorescent protein (EGFP)-tagged ZIP (EGFP-ZIP) proteins with polyclonal antibodies against ZIP, which we generated with recombinant ZIP (364–511 aa), indicate that ZIP has an apparent Mr of ∼56 kDa (Figure 2B, lower panel), confirming its predicted molecular weight. In addition, both reverse transcriptase (RT)–PCR (left panel) and western blotting (right panel) analyses detected ZIP expression in various cell lines (Figure 2C).
To gain insight into the biological function of the ZIP protein, we first examined its subcellular localization. Both fluorescent imaging of EGFP-ZIP and immunostaining of FLAG-ZIP in MCF-7 cells indicate that ZIP is primarily a nuclear protein (Figure 2D), suggesting that ZIP may function primarily in the nucleus.
ZIP binds DNA and recognizes specific DNA sequences
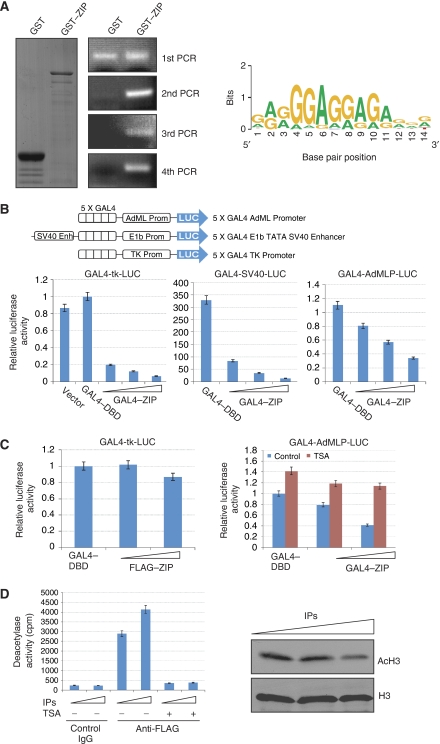
Transcriptional regulation is a primary research focus in our laboratory (Shang and Brown, 2002; Zhang et al, 2004, 2006, 2007; Wu et al, 2005, 2006; Shang, 2006; Shi et al, 2007; Liang et al, 2009). The presence of a zinc finger domain in ZIP prompted us to investigate the hypothesis that ZIP might recognize and bind to specific genomic sequences. We, therefore, performed cyclic amplification and selection of target (CASTing) assays to search for putative DNA-binding sequences for ZIP by screening double-stranded random oligonucleotides using a glutathione S-transferase fusion protein (GST–ZIP) immobilized on glutathione Sepharose 4B beads. As shown in Figure 3A, GST–ZIP fusion protein was found to bind DNA sequences specifically after the second round of binding and amplification reaction; DNA products were only detected with GST–ZIP, but not with GST, after this round. We performed a total of nine rounds of binding and amplification reactions. After that, the final PCR products were cloned and sequenced. Of 93 sequences that were cloned and sequenced, 80 contained a GA-rich DNA element GGAGG/AAG/AA (Figure 3A).
Figure 3.
ZIP is a DNA-binding protein and possesses intrinsic transcription repression activity. (A) CASTing assay. Binding and amplification reactions were done with GST or GST–ZIP fusion proteins. Coomassie blue staining of the purified GST and GST–ZIP fusion proteins and the results from PCR amplification of bound DNA up to four rounds are shown on the left. The computational result using Weblogo (http://weblogo.berkeley.edu) for conserved nucleotides within ZIP-binding sequences is shown on the right. (B) Transcription repression by ZIP. The schematic diagram shows the GAL4-luciferase reporters. For reporter assays, MCF-7 cells were transfected with different amounts of GAL4–ZIP expression construct together with the indicated GAL4-luciferase reporter. Twenty-four hours after transfection, cells were harvested and luciferase activity was measured and normalized to that of renila. Each bar represents the mean±s.d. for triplicate experiments. (C) Reporter assays with FLAG-ZIP transfection (left panel) or with TSA treatment (right panel). MCF-7 cells were transfected with indicated plasmids and were treated with TSA or left untreated. Cells were then harvested and luciferase activity was measured and normalized to that of renila. Each bar represents the mean±s.d. for triplicate experiments. (D) In vitro HDAC activity assay for the ZIP complex. Nuclear extracts from HeLa cells stably transfected with FLAG-ZIP were immunoprecipitated with anti-FLAG antibody. Increased amounts of immunoprecipitates (IPs) were incubated either with [3H]acetate-labelled HeLa histones for deacetylase activity determination by liquid scintillation counting of released [3H]acetate (left panel) or with calf thymus histones followed by immunoblotting analyses with antibodies against acetylated H3 and total H3 (right panel).
ZIP possesses intrinsic transcription repression activity accompanied by histone deacetylation
The fact that ZIP harbours a zinc finger and the result of CASTing assays suggest that ZIP may indeed be a DNA-binding protein and may thus be involved in transcriptional regulation. To determine whether ZIP does in fact possess a trans-acting activity, we fused ZIP to the C-terminus of GAL4 DNA-binding domain and tested the transcription activity of the fused construct in MCF-7 cells. We used three different GAL4-driven luciferase reporter systems, which differ in basal promoter elements (Figure 3B). The results show that ZIP drastically repressed the reporter activity in a dose-dependent manner in all of the three reporter systems. In the meanwhile, overexpression of FLAG-ZIP did not affect the activity of GAL4-driven reporter (Figure 3C, left panel), suggesting that ZIP must be physically associated with DNA to exert its transcription repression activity. Similar results were also obtained in the endometrial carcinoma cell line ECC-1 and the lung carcinoma cell line A549 (data not shown).
As stated above, one common mechanism of gene transcription repression is through the recruitment of corepressor complexes that contain subunits with HDAC activities (Hu and Lazar, 2000; Rosenfeld et al, 2006). To determine whether HDAC activity is required for ZIP-mediated gene repression, we measured the reporter activity in cells treated with trichostatin A (TSA), a specific HDAC inhibitor. The results indicate that TSA treatment was able to almost completely alleviate the repression of the reporter activity by ZIP construct (Figure 3C, right panel), suggesting that ZIP-mediated repression was associated with a HDAC activity.
To further support this, nuclear extracts from HeLa cells stably expressing FLAG-ZIP were immunoprecipitated with the anti-FLAG antibody. The ZIP-containing complex was then tested for HDAC activity by incubating the immunoprecipitates with [3H]acetate-labelled HeLa histones. In vitro HDAC activity was measured by quantifying the release of radiolabelled acetyl groups from purified hyperacetylated HeLa histones. We found that FLAG-ZIP immunoprecipitates from HeLa cell extracts had HDAC activity and that treatment of the immunoprecipitates with TSA reduced HDAC activity to background levels (Figure 3D, left panel). In addition, incubating the immunoprecipitates with calf thymus bulk histones followed by immunoblotting also indicates that the acetylation level of H3 was greatly reduced (Figure 3D, right panel). All these experiments support the hypothesis that the ZIP complex is associated with a histone deacetylation activity.
ZIP is physically associated with the NuRD complex
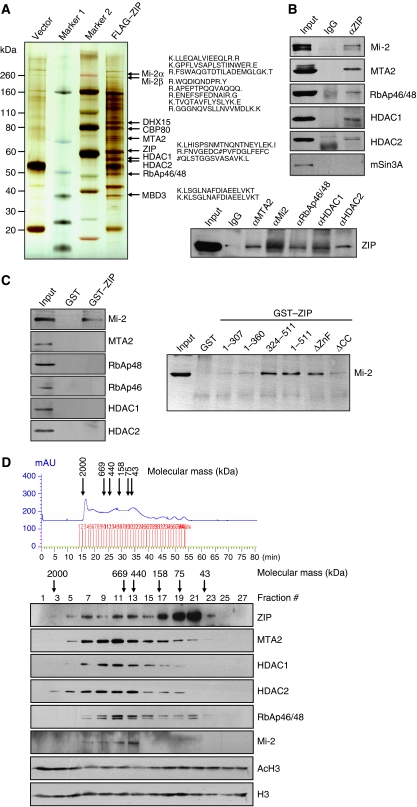
To further elucidate the molecular mechanism underlying ZIP-mediated transcription repression, ZIP-containing protein complexes were affinity purified from nuclear extracts of HeLa cells stably expressing FLAG-ZIP with the anti-FLAG antibody that was immobilized on agarose beads. The purified protein complex was resolved on SDS–PAGE and silver stained (Figure 4A). Mass spectrometry analysis identified, in addition to ZIP, DHX15 [DEAH (Asp-Glu-Ala-His) box polypeptide 15], and CBP80 (nuclear cap-binding complex subunit 1), protein components of the NuRD complex, including Mi-2α, Mi-2β, RbAp46/48, MTA2, HDAC1, HDAC2, and MBD3 (Figure 4A), suggesting that ZIP is physically associated with the NuRD complex in vivo.
Figure 4.
Physical association of ZIP with the NuRD complex. (A) Mass spectrometry analysis of ZIP-associated proteins. Nuclear extracts from HeLa cells stably expressing FLAG-ZIP were prepared and subjected to affinity-purification with anti-FLAG antibody that was immobilized on agarose beads. The purified protein complex was resolved on SDS–PAGE and silver stained, and the bands were retrieved and analysed by mass spectrometry. DHX15: DEAH (Asp-Glu-Ala-His) box polypeptide 15; CBP80: nuclear cap-binding complex subunit 1. Complete amino-acid sequences from mass spectrometry analysis are included in Supplementary data 3. (B) Co-immunoprecipitation of ZIP and the components of the NuRD complex. Whole-cell lysates from HeLa cells were prepared and immunoprecipitation was performed with anti-ZIP followed by immunoblotting with antibodies against indicated proteins (upper panel), or immunoprecipitated with antibodies against Mi-2, MTA2, RbAp46/48, HDAC1, HDAC2, or IgG followed by immunoblotting with anti-ZIP (lower panel). (C) ZIP interacts directly with Mi-2 in vitro. GST pull-down assays were performed with GST–ZIP and in vitro transcribed/translated components of the NuRD complex (left panel) or with GST–ZIP (1-511) or GST-fused ZIP deletion mutants (number represents the amino-acid position; ΔZnF: ZIP without zinc finger; ΔCC: ZIP without coiled-coil domain) and in vitro transcribed/translated Mi-2 (right panel). (D) Co-fractionation of ZIP and the NuRD complex by FPLC. Cellular extracts from HeLa cells were fractionated on Superose 6 size exclusion column. The chromatographic profile with the elution positions of calibrating proteins of known molecular mass is shown. The chromatographic fractions were analysed by western blotting with antibodies against indicated proteins or were first incubated with bulk histones and then analysed by western blotting with anti-acetylated H3 (AcH3) or anti-H3. Equal volumes from each fraction were analysed.
To confirm an in vivo interaction between ZIP and the NuRD complex, total proteins from HeLa cells were extracted and immunoprecipitated with the antibodies against ZIP. The immunoprecipitates were then immunoblotted with antibodies against the components of the NuRD complex and also against mSin3A. The results show that the components of the NuRD complex, but not mSin3A, could be efficiently co-immunoprecipitated with ZIP (Figure 4B, upper panels). Reciprocal immunoprecipitations with antibodies against the components of the NuRD complex, including Mi-2, RbAp46/48, MTA2, HDAC1, and HDAC2, and immunoblotting with anti-ZIP also revealed that ZIP is co-immunoprecipitated with the components of the NuRD complex (Figure 4B, lower panel).
GST pull-down assays were then performed to investigate the molecular details of the interaction between ZIP and the NuRD complex. Bacterially expressed GST–ZIP proteins were purified and incubated with in vitro transcribed/translated components of the NuRD complex. The results of these experiments indicate that ZIP only interacts directly with Mi-2, suggesting that the recruitment of the NuRD complex by ZIP in its transcription repression is through an interaction of ZIP with Mi-2 (Figure 4C, left panel). Further analyses by GST pull-down assays with deletion mutants of ZIP revealed that the coiled-coil domain of ZIP is responsible for the interaction of ZIP with Mi-2 (Figure 4C, right panel).
To further consolidate the in vivo association of ZIP and the NuRD complex, protein fractionation experiments were carried out through a high salt extraction and size exclusion approach by fast protein liquid chromatography (FPLC) using Superose 6 size columns. The result of the experiment revealed that native ZIP in HeLa cells could be eluted in chromatographic fractions with apparent molecular masses much greater than that of the monomeric protein; ZIP immunoreactivity could be detected in elutes with high molecular masses and with a relatively symmetrical peak centred around ∼669 kDa, and the elution pattern of ZIP in chromatographic fractions with high molecular masses was largely overlapped with that of the NuRD complex proteins, including MTA2, HDAC1, HDAC2, RbAp46/48, and Mi-2 and was accompanied by HDAC activities, as assayed by incubating these fractions with the bulk histones and then immunoblotted with anti-acetylated H3 (Figure 4D), supporting the hypothesis that ZIP is associated with the NuRD complex in vivo.
Identification of the transcriptional targets for ZIP
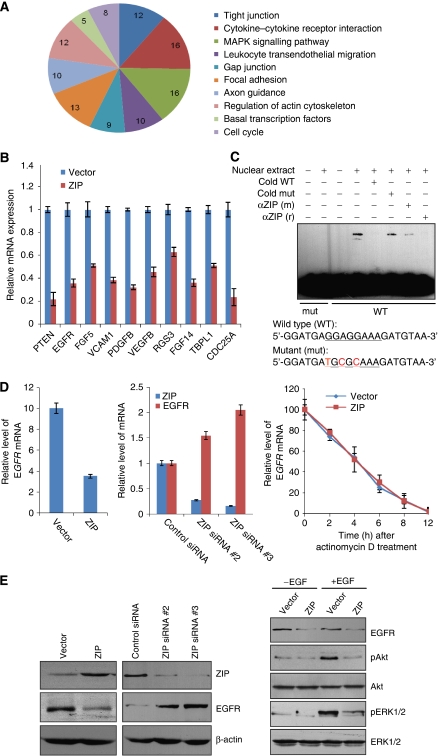
On the basis of the DNA-binding element that we identified in CASTing assays, we searched the Eukaryotic Promoter Database (EPD) (http://cmgm.stanford.edu/help/manual/databases/epd.html) for genes that might be potentially targeted by ZIP. The search yielded 383 genes containing the putative ZIP-binding sites in their 5′-upstream regulatory regions, including EGFR (Supplementary data 1).
Next, we decided to identify potential downstream targets of ZIP in the human genome using Chromatin ImmunoPrecipitation-DNA selection and ligation (ChIP-DSL). ChIP experiments were first conducted in MCF-7 cells with ZIP antibodies. After ChIP, ZIP-associated DNAs were amplified using nonbiased conditions, labelled, and hybridized to AVIVA's Hu20K arrays. Relative confidence prediction scores were generated by quantile normalization across each probe followed by an analysis using a two-state Hidden Markov model (Mukherjee and Mitra, 2005). These scores included both probe intensity and width of probe cluster. Triplicate experiments eliminated stochastic false positives, after which peaks that reproducibly appeared at least twice in the three replicates were included. The detailed results of the ChIP-DSL experiments are deposited in GEO Datasets (accession ID: GSE13905) and summarized in Supplementary data 2. These experiments identified a cohort of genes including EGFR whose promoters are targeted by ZIP. These genes were then classified into cellular signalling pathways using MAS software (http://bioinfo.capitalbio.com/mas) with a P value cutoff of less than 10−3, and these analyses identified several cellular signalling pathways, including tight junction, MAPK signalling pathway, gap junction, focal adhesion, and cell cycle, which are critically involved in cell proliferation, survival, and migration (Figure 5A). To verify the ChIP-DSL results, the mRNA expression of selected genes representing each of the pathways was measured by quantitative real-time PCR in MCF-7 cells with ZIP overexpression. The results of these experiments corroborated with the ChIP-DSL results and support ZIP as a transcription repressor (Figure 5B).
Figure 5.
Identification of EGFR as a target for ZIP. (A) Classification of the genes identified in ChIP-DSL experiments with MAS software (http://bioinfo.capitalbio.com/mas). The statistically significant (P<0.001) pathways are shown and the numbers indicate the numbers of the pathway-associated genes. (B) Verification of ChIP-DSL results by measuring the mRNA expression of the selected genes representing each of the pathways using real-time RT–PCR. (C) ZIP binds to a specific sequence within EGFR promoter. Gel shift assay was performed using MCF-7 cell nuclear extracts and radiolabelled EGFR promoter sequences containing wild type or mutated putative ZIP-binding element (underscored). Anti-ZIP (αZIP) includes antibodies from mouse (m) or from rabbit (r). (D) Regulation of EGFR expression by ZIP. MCF-7 cells were transfected with ZIP or ZIP siRNAs, total RNAs were prepared and analysed for EGFR mRNA expression by real-time RT–PCR. EGFR mRNA stability assay (right panel) was performed in MCF-7 cells that were transfected with ZIP expression construct and were treated with 5 μM of actinomycin D for different times. The data were normalized against the expression of GAPDH. Each point represents the mean±s.d. for triplicate measurements. (E) Regulation of EGFR expression by ZIP. MCF-7 cells were transfected with ZIP or ZIP siRNAs, total cellular proteins were prepared and analysed for EGFR protein expression by western blotting (left panel). MCF-7 cells were transfected with vector or ZIP. Twenty-four hours after transfection, cells were switched to conditioned media (without growth factors) for another 24 h followed by addition of EGF for 6 h. Total cellular proteins were prepared and analysed for the expression and phosphorylation status of AKT and ERK (right panel).
EGFR was identified in both the EPD interrogation and the ChIP-DSL analysis. As EGFR has an important function in malignant transformation (Holbro et al, 2003; Chan et al, 2006), and as the molecular mechanisms underlying breast carcinogenesis have been another research focus in our laboratory (Shang and Brown, 2002; Sun et al, 2004, 2009; Yin et al, 2004; Zhang et al, 2004; Wu et al, 2005; Shang, 2006; Shi et al, 2007; Chen et al, 2008), we concentrated our research on EGFR. Examination of EGFR gene promoter identified a GGAGGAAA sequence at positions −1157 to −1149 in the 5′ upstream region that closely resembles the GGAGG/AAG/AA element that we identified in CASTing assays for ZIP binding. In vitro gel shift experiments confirmed that ZIP protein can specifically bind to an EGFR promoter sequence containing this element, and mutations in this element abrogated ZIP binding (Figure 5C). These results strongly support the hypothesis that EGFR is a direct target for ZIP. Measurements of EGFR expression by quantitative real-time RT–PCR in MCF-7 cells with either ZIP overexpression or ZIP knockdown revealed that ZIP overexpression was associated with decreased EGFR expression and ZIP knockdown was associated with increased EGFR expression (Figure 5D, left and middle panels), again confirming the observation from ChIP-DSL experiments and supporting EGFR as a target for ZIP. The decreased EGFR mRNA level was not due to mRNA degradation, as mRNA stability assays for EGFR showed that ZIP did not affect the stability of EGFR mRNA (Figure 5D, right panel). Consistent with these results, protein expression measured by western blotting indicates that EGFR protein was elevated in cells with ZIP knockdown and was decreased in cells with ZIP overexpression (Figure 5E, left panel). Collectively, these data support the proposition that EGFR is a downstream target for ZIP. More importantly, the regulation of EGFR by ZIP may be physiologically significant as downregulation of EGFR by ZIP was associated with decreased levels of AKT and ERK phosphorylation, the downstream events in EGFR signalling (Figure 5E, right panel).
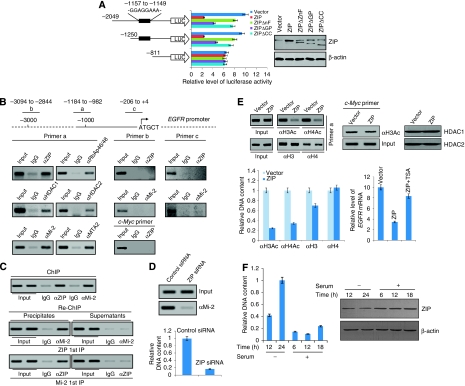
We next cloned a 2049 bp fragment of the 5′ regulatory region of EGFR gene and constructed luciferase reporters to test whether ZIP might be capable of repressing EGFR promoter-driven luciferase reporter. In these experiments, MCF-7 cells were co-transfected with the EGFR promoter-driven luciferase constructs and the expression constructs for ZIP or ZIP domain deletion mutants. Twenty-four hours after transfection, cellular lysates were prepared and analysed for luciferase activity. As shown in Figure 6A, ZIP was able to repress the EGFR promoter activity, but only when the sequence GGAGGAAA was present. EGFR promoter that lacked this sequence did not respond to ZIP. In addition, both the zinc finger and the coiled-coil domains appeared to be essential for ZIP's repression activity, whereas the G-patch domain was somewhat less important. These data further support the hypothesis that ZIP specifically targets EGFR and represses its transcription, and that the zinc finger and the coiled-coil domains are central to that process.
Figure 6.
Transcriptional regulation of EGFR by ZIP. (A) ZIP represses EGFR promoter-driven luciferase activity. MCF-7 cells were co-transfected with EGFR promoter-driven luciferase constructs and the expression constructs for wild type ZIP or ZIP deletion mutants (ZIPΔZnF: ZIP without zinc finger; ZIPΔGP: ZIP without G-patch; ZIPΔCC: ZIP without coiled coil). Twenty-four hours after the transfection, cells were collected and luciferase activity was measured and normalized to that of renila. Each bar represents the mean±s.d. for triplicate experiments. The expression of ZIP and its mutants was examined by western blotting. (B) The recruitment of ZIP and the NuRD complex on the EGFR promoter. ChIP assays were performed in MCF-7 cells using indicated antibodies and primer pairs and also on c-Myc promoter. (C) ZIP and the NuRD complex exist in the same protein complex on the EGFR promoter. ChIP and Re-ChIP experiments were performed with indicated antibodies and primer pair a. (D) The requirement of ZIP for the recruitment of the NuRD complex on the EGFR promoter. MCF-7 cells were transfected with control siRNA or ZIP-specific siRNA. Twenty-four hours after the transfection, cells were collected for ChIP experiments with antibodies against Mi-2. Both conventional semi-quantitative PCR (left panel) and quantitative PCR (right panel) were performed for the measurement. (E) ZIP recruitment on the EGFR promoter is associated with changes of the histone acetylation status in the region. MCF-7 cells were transfected with vector or ZIP expression construct. H3 and H4 acetylation levels were analysed by both conventional semi-quantitative ChIP and quantitative ChIP, and the protein expression of HDAC1 and HDAC2 was measured by western blotting. The H3 acetylation status of c-Myc promoter was also tested. Right lower panel: MCF-7 cells were transfected with vector or ZIP expression construct, treated with TSA, and analysed for EGFR mRNA expressions by real-time RT–PCR. Each bar represents the mean±s.d. for triplicate measurements. (F) ZIP recruitment to the EGFR promoter is associated with serum-deprived states. MCF-7 cells were grown in normal media to a 60% confluence and switched to serum-free media for different times followed by growth in media replenished with serum for different times. Cells were then collected for quantitative ChIP experiments with antibodies against ZIP. Each bar represents the mean±s.d. for triplicate measurements. The expression of ZIP in corresponding cells was examined by western blotting.
To test whether ZIP represses the expression of EGFR through recruitment of the NuRD complex to the EGFR promoter, ChIP assays were performed in MCF-7 cells with antibodies against ZIP, RbAp46/48, HDAC1, HDAC2, Mi-2, MTA2, or control IgG. These experiments revealed that both ZIP and the NuRD components occupied the promoter of EGFR gene spanning the putative ZIP-binding element but not in unrelated regions (Figure 6B). To further support the hypothesis that ZIP and the NuRD complex interact and exist in the same protein complex on EGFR promoter, sequential ChIP or ChIP/Re-ChIP experiments (Shang et al, 2000; Zhang et al, 2004) were performed. In these experiments, soluble chromatins were first immunoprecipitated with antibodies against ZIP. Both the supernatants and immunoprecipitates were subsequently re-immunoprecipitated with antibodies against Mi-2, and vice versa. The results of the experiments indicate that in precipitates, the EGFR promoter that was immunoprecipitated with antibodies against ZIP could be re-immunoprecipitated with antibodies against Mi-2, whereas in the supernatants, antibodies against Mi-2 found no detectable EGFR promoter (Figure 6C). The same results held when the initial ChIP was done with antibodies against Mi-2; EGFR promoter could only be detected in precipitates, but not in supernatants after Re-ChIP with antibodies against ZIP. Taken together, these experiments support the idea that ZIP recruits and is physically associated with the NuRD complex on the EGFR promoter.
We then investigated the requirement for ZIP in the recruitment of the NuRD complex to the EGFR promoter. For this purpose, MCF-7 cells were transfected with ZIP-specific or mock siRNAs. The recruitment of Mi-2 subunit of the NuRD complex on the EGFR promoter was then measured by both semi-quantitative ChIP and quantitative ChIP. As shown in Figure 6D, the recruitment of Mi-2 was almost abrogated in cells transfected with ZIP-specific siRNA, supporting the idea that ZIP is required for the recruitment of the NuRD complex to the EGFR promoter.
To verify a functional interaction between ZIP and the NuRD complex on the EGFR promoter, we investigated histone acetylation status in the promoter region of EGFR in MCF-7 cells. To this end, MCF-7 cells were transfected with FLAG-ZIP, and both semi-quantitative ChIP and quantitative ChIP assays were performed using antibodies against acetylated H3 or H4. The results of these experiments indicate that overexpression of ZIP was associated with a significant decrease of the acetylation level of histone H3 and H4, whereas the H3 acetylation status of c-Myc promoter was not affected (Figure 6E). The reduced levels of acetylated histone H3 and H4 were not due to the loss of H3 and H4 nor the upregulation of HDACs by ZIP (Figure 6E). The reduction in acetylated histone H3 and H4 at EGFR promoter strongly suggests that EGFR promoter is actively deacetylated in the presence of ZIP, supporting a functional connection between ZIP and the NuRD complex. Corroborating this idea, TSA treatment led to an alleviated ZIP repression of EGFR mRNA expression as quantified by real-time RT–PCR in MCF-7 cells (Figure 6E, right lower panel). Furthermore, the action of ZIP appeared to be associated with a cellular environment that is unfavourable to cell proliferation, as ZIP recruitment to the EGFR promoter appeared to be linked to serum-deprived states when measured by quantitative ChIP in MCF-7 cells (Figure 6F).
The biological effect of the transcription repression of EGFR by ZIP
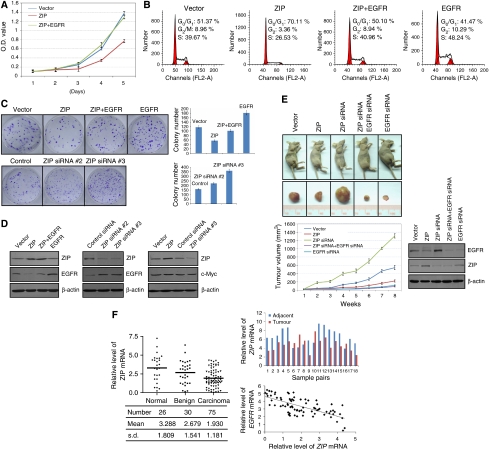
To determine whether the transcription repression of EGFR by ZIP could extend to a physiologically relevant response in breast cancer cells, we first examined the effect of ZIP on cell proliferation and colony formation. For these experiments, we created MCF-7 cell clones that stably express ZIP. These clones were expanded and their growth was measured by MTT assays. The results indicate that ZIP has a significant inhibitory effect on MCF-7 cell proliferation, which could be rescued by overexpressing EGFR (Figure 7A). Moreover, flow cytometry analyses revealed that EGF-promoted cell cycle progression is significantly delayed when ZIP is overexpressed; there was a significant accumulation of cells in G0/G1 phase under this condition. Conversely, ectopic expression of EGFR was able to overcome the inhibitory effect of ZIP (Figure 7B). Furthermore, the colony formation assays indicate that ZIP overexpression is associated with a decreased colony number, which could be rescued by EGFR overexpression, whereas ZIP knockdown is associated with an increased colony number (Figure 7C). All these experiments support a role for ZIP in the inhibition of cell proliferation and indicate that ZIP does so, at least in part, by downregulation of EGFR expression.
Figure 7.
ZIP inhibits cell proliferation and suppresses tumorigenesis. (A) Inhibition of cell proliferation by ZIP. MCF-7 cells were stably transfected with vector, ZIP expression construct, or ZIP plus EGFR expression constructs. The growth curve of the cells was measured by MTT assays. (B) Inhibition of cell cycle progression by ZIP. MCF-7 cells were transfected with vector or indicated expression constructs. Twenty-four hours after the transfection, the cells were switched to conditioned medium without serum for another 24 h. The cells were then cultured in medium containing EGF for 12 h and were collected for cell cycle analysis by flow cytometry. Experiments were repeated three times and the data from a representative experiment are shown. (C) Colony formation assay. MCF-7 cells stably expressing corresponding plasmids were maintained in culture media for 14 days under the presence of 1 mg/ml G418 and stained with crystal violet. The number of colonies in each condition was counted and expressed as mean±s.d from triplicate experiments. (D) The protein expression of ZIP, EGFR, and c-Myc under indicated experimental conditions was examined by western blotting analysis. (E) ZIP suppresses breast tumorigenesis. MCF-7 cells with lentivirus-delivered ZIP overexpression, ZIP knockdown, EGFR knockdown or ZIP and EGFR double-knockdown were transplanted into ovariectomized athymic mice. Tumours were measured weekly using a vernier calliper and the volume was calculated according to the formula: π/6 × length × width2. Each point represents the mean±s.d. for different animal measurements (n=6). The levels of EGFR and ZIP proteins in these tumours were examined by western blotting. (F) The expression of ZIP mRNA is downregulated in breast carcinomas and its level is negatively correlated with the level of EGFR mRNA. Total RNAs were extracted and the expression of ZIP and EGFR mRNA was measured by real-time RT–PCR with GAPDH as the reference. ZIP mRNA levels in normal, benign, and carcinoma breast samples were showed in left. ZIP mRNA levels in paired samples of breast carcinomas versus adjacent normal mammary tissues were showed in right upper panel. The relative level of ZIP expression was plotted against the relative level of EGFR expression and shown in right lower panel.
ZIP suppressed breast carcinogenesis
To further support the anti-proliferative effect of ZIP and to investigate its possible role in breast carcinogenesis, we transplanted five types of breast tumours developed from MCF-7 cells into ovariectomized athymic mice (BALB/c, Charles River, Beijing, China). The transplanted tumours either had ZIP expression unchanged (infected with lentiviruses carrying an empty vector), ZIP overexpression (infected with lentiviruses carrying the ZIP gene), ZIP specific knockdown (infected with lentiviruses carrying a specific siRNA for ZIP), ZIP and EGFR double-knockdown (infected with lentiviruses carrying specific siRNA for ZIP and EGFR) or EGFR-specific knockdown (infected with lentiviruses carrying specific siRNA for EGFR). Growth of the implanted tumours was measured in mice (n=6 for each group) over a period of 8 weeks. The results indicate that, in athymic mice receiving tumour with ZIP overexpression, tumour growth was significantly suppressed, and in athymic mice receiving tumour with ZIP knockdown, tumour growth was dramatically enhanced (Figure 7E). As expected, in athymic mice receiving tumour with ZIP and EGFR double-knockdown or with EGFR knockdown, tumour growth was significantly inhibited (Figure 7E). These observations strongly support a role of ZIP in suppressing tumorigenesis and also support the targeting of EGFR by ZIP.
As stated before, the expression of EGFR is deregulated in a variety of human epithelial tumours, including breast cancer, and this deregulation is often associated with a more aggressive phenotype and worse survival of the cancer patients (Nicholson et al, 2001). To substantiate the functional link between ZIP and EGFR and to extend the physiological relevance of this link, we collected normal mammary tissue from 26 mammary reductions, 30 lumps from benign breast lesions, and 75 carcinoma samples from breast cancer patients. The expression of ZIP mRNA was analysed by real-time RT–PCR in these samples. We found that ZIP expression is downregulated in breast carcinomas (Figure 7F). Intriguingly, there appeared to be a progressive decrease in ZIP mRNA levels from normal to benign to malignant samples; compared with that in normal tissue, the mean ZIP mRNA level was lower in benign lesions and was even lower in carcinoma samples. Decreased ZIP mRNA expression was also evident in majority of breast carcinomas compared with adjacent normal tissues (Figure 7F, right upper panel). EGFR mRNA levels in carcinoma samples were also analysed and were plotted by the levels of ZIP mRNA (Figure 7F, right lower panel). Statistical analysis found a Pearson correlation coefficient of −0.6942 (P<0.0001) and a Spearman correlation coefficient of −0.6614 (P<0.0001), indicating a strong negative correlation between the expression of these two genes in breast carcinomas. These data further support a role for ZIP in EGFR regulation and in breast carcinogenesis.
Discussion
ZIP as a novel transcription repressor
The ZIP protein appears to be a gene-specific repressor that acts to actively repress its target genes. It is a modular protein with several important functional domains: a CCCH zinc finger, a TUDOR domain, a G-patch, and a coiled-coil domain. Our experiments demonstrated that the coiled-coil domain of ZIP is responsible for its interaction with Mi-2 thus the recruitment of the NuRD complex. Although additional activities such as the recognition of RNA and other proteins have been described (Gamsjaeger et al, 2007), one of the hallmark features of zinc finger structure is its DNA-binding capability. Indeed, our experiments show that ZIP is capable of binding DNA. CASTing and gel shift assays indicate that ZIP recognizes specific sequences, and ChIP experiments demonstrate that ZIP is recruited to target gene promoters. In addition, ZIP is able to repress the transcription of EGFR promoter and the zinc finger domain is essential for this activity.
The current functional characterization of the CCCH zinc finger and the G-patch domains, together with their presence in the ZIP protein, raise a distinct possibility that ZIP may function in mRNA processing. Although a role for ZIP in pre-mRNA splicing cannot be ruled out, our experiments demonstrate that ZIP had no apparent effect on EGFR mRNA degradation. Rather, we found that it could bind to specific DNA sequences and be recruited to target gene promoters; it interacts with the NuRD complex, both physically and functionally; and it possesses an intrinsic transcription repression activity. These observations, together with the fact that ZIP contains a TUDOR domain, which is believed to be a chromatin-presenting module reading the methylated histone marks (Kim et al, 2006), and the fact that we saw a comparatively lower importance for the G-patch domain in ZIP's repression of EGFR transcription, all argue against ZIP's primary function being the regulation of mRNA turnover, at least in this case.
The physical and functional connection between ZIP and the NuRD complex
Our experiments show that in the course of repressing transcription, ZIP recruits the NuRD complex. We show that ZIP's repression of transcription activity is associated with changes in histone acetylation status and that it is sensitive to HDAC inhibitors. This indicates that ZIP and the NuRD complex are functionally connected. As stated earlier, the NuRD complex has been found to mediate the function of several transcription repressors. It is believed that subunit composition of this complex is highly heterogeneous, and this heterogeneity may contribute to the functional specialization of the NuRD complex (Bowen et al, 2004; Denslow and Wade, 2007). In this context, it is interesting to note that the ZIP–NuRD complex was also co-purified with DHX15 or PrPp43p, a member of RNA helicases (Tanner and Linder, 2001) that are implicated in all processes involving RNA molecules, including transcription, editing, splicing, ribosome biogenesis, RNA export, translation, RNA turnover, and organelle gene expression, and CBP80, a component of the m7GpppG-binding complex (Calero et al, 2002). In addition to the possibility that these proteins may have yet unidentified functions outside of mRNA processing, it is intriguing to speculate that ZIP may also have a function in pre-mRNA splicing, especially considering that ZIP contains a CCCH-type of zinc finger and a G-patch domain, both of which have been featured in proteins functioning in mRNA processing. Alternatively, ZIP may coordinate an active coupling between transcription regulation and pre-mRNA splicing. Coupling between transcription and RNA processing has now been recognized as a key regulatory mechanism of gene expression (Bentley, 2005). However, this mechanism is reasonably easy to understand in transcription activation, in which mRNA production is promoted, whereas in transcriptional repression, in which mRNA synthesis is inhibited, the logic of this mechanism is not clear. Nevertheless, the coupling concept in transcriptional repression cannot be totally ruled out because it is probably a safe assumption that the transcription process is not yet fully understood.
Transcription regulation of EGFR by ZIP
It has been well established that EGFR is involved in malignant transformation and cancer progression (Holbro et al, 2003; Chan et al, 2006). Indeed, overexpression of this cellular receptor has been demonstrated in a host of human tumour types, and this overexpression often signifies a more aggressive phenotype and accordingly worse survival (Chan et al, 2006). Therefore, understanding the transcription-regulating mechanisms that control EGFR proto-oncogene expression is of great importance. However, surprisingly little is known about the transcription regulation of the EGFR gene. Except for several enhancer elements: one in direct proximity to the promoter and two others in intron 1 (+1788 to +2318) and upstream of the promoter (−1409 to −1109), the 5′-regulatory sequence of the EGFR gene contains a GC-rich promoter without any consensus sequences, such as TATA or CAAT boxes (Brandt et al, 2006). As for trans-acting factors, the basal transcription of the EGFR gene is believed to be regulated by Sp1 and the EGFR gene has also been identified as a target for c-Jun (Brandt et al, 2006).
Amplification of oncogenes is a common mechanism in the initiation and progression of malignant tumours that can circumvent basic transcription mechanisms. It is interesting to note that microsatellite analysis showed that amplifications in the EGFR gene were restricted to region of the regulatory sequence in the 5′-end of intron 1 and were associated with EGFR expression in epithelial breast tumours (Brandt et al, 2006). Also intriguingly, amplifications involving the above-described sequences of intron 1 of EGFR have been noted in normal-appearing epithelial and stromal breast tissue, next to the respective benign and malignant tumours, leading to a suggestion that this genetic alteration may represent the first initial hits in breast carcinogenesis (Brandt et al, 2006).
In this report, we showed that ZIP acts to negatively regulate the transcription of the EGFR gene. We identified a sequence, GGAGGAAA, in the 5′ upstream region of EGFR that closely resembles the ZIP-binding sequence as isolated in CASTing assays. ChIP assays revealed that ZIP is recruited to the EGFR promoter region that contains this sequence, and gel shift experiments demonstrated that mutations in this sequence abrogated ZIP binding. Reporter assays indicate that the transcription repression activity of ZIP on EGFR promoter is dependent on the presence of this sequence. It is expected that more trans-acting factors and cis-acting elements for the EGFR gene will be identified in the future. Such efforts will benefit for the better understanding of the patho-physiological functions of EGFR.
Physiological significance of EGFR regulation by ZIP
In most cell types, EGFR is found in amounts from 2 × 104 to 2 × 105 receptors per cell, whereas in cancer cells, EGFR is overexpressed and often can reach >106 receptors per cell for many cancer types (Brandt et al, 2006). High concentration of EGFR would result in amplified/sustained EGFR signalling, and eventually uncontrolled cellular effects, malignant transformation, and tumour progression. Indeed, positive correlations have been reported between increased amounts of this receptor and worse survival of cancer patients, poor response to chemotherapy, and even failure of endocrine therapy in breast cancer (Barrett-Lee, 2005).
Therefore, keeping an appropriate concentration of EGFR is critical for normal physiological cell behaviours. The expression and the activity of EGFR inside cells must be kept in balance at all times. Once this balance is skewed, regardless genetic or epigenetic including transcription regulatory causes, aberrant EGFR signalling would lead to abnormal cellular behaviours and malignant transformation. Our identification of the negative regulation of EGFR expression by ZIP positions ZIP as an important denominator in the balance equation of the expression/activity of EGFR. By downregulating EGFR expression, ZIP would inhibit the EGFR activity and dampen EGFR signalling. It would thus suppress the tumorigenic potential of EGFR. In this sense, ZIP could be viewed as a potential tumour suppressor gene. We show that the expression of ZIP is downregulated in breast carcinoma samples and there is a negative correlation of the expression between ZIP and EGFR. In support of ZIP's tumour suppressing role, it is interesting to note that one of the recently identified methylated loci in brain tumours corresponds to the ZIP gene (Ordway et al, 2006).
Another interesting feature of ZIP is its pattern of tissue distribution. In addition to the transcript described in this paper, at least one larger and another smaller transcript exist in a tissue-specific and highly expressed manner. Specifically, these two additional transcripts were highly expressed in liver, and the smaller one appears to be restricted to the kidneys. The molecular mechanisms underlying breast carcinogenesis have been a primary research interest in our laboratory, but other potential roles for ZIP and its isoforms, particularly in these other organs, need to be investigated. Gene ablation experiments are currently underway to study these issues. Future investigations will need to also address the scope and variety of the cellular functions of the ZIP protein.
Materials and methods
CASTing assay
A library of single-stranded oligonucleotides containing the sequences 5′-GACTCGAGACTCCTAGGATGCGCA(N)20CGTCTATGTCAGTGAAGCTTCGAT-3′ was generated and double-stranded oligonucleotides were produced. The double-stranded oligonucleotides were purified and incubated with GST-fused ZIP protein bound to glutathione beads in a binding buffer containing poly (dI-dC) and BSA. After a 30 min rotating incubation at room temperature, the beads were washed for eight times with cold binding buffer without poly (dI-dC) and then boiled for 5 min in sterilized H2O. The eluted oligonucleotides were used for PCR amplification. The amplified products were subsequently used for a second round of selection. After nine rounds of amplification, PCR products were cloned into pGEM-Teasy vector, transformed into DH5α competent host cells, and sequenced.
FPLC chromatography
HeLa nuclear extracts were prepared and dialyzed against buffer D (20 mM HEPES, pH 8.0, 10% glycerol, 0.1 mM EDTA, 300 mM NaCl) (Applygen Technologies Inc). Approximately 6 mg of nuclear protein was concentrated to 1 ml using a Millipore Ultrafree centrifugal filter apparatus (10 kDa nominal molecular mass limit), and then applied to an 850 × 20 mm Superose 6 size exclusion column (Amersham Biosciences) that had been equilibrated with buffer D containing 1 mM dithiothreitol and calibrated with protein standards (blue dextran, 2000 kDa; thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; bovine serum albumin, 67 kDa; RNase A, 13.7 kDa, all from Amersham Biosciences). The column was eluted at a flow rate of 0.5 ml/min and fractions were collected.
ChIP-DSL
ChIP samples were amplified by ligation-mediated PCR as described (Kwon et al, 2007). DNA fragmentation, biotin labelling, and hybridization were performed according to the Aviva Systems Biology protocol (http://www.avivasysbio.com) and using the Aviva's Hu20K arrays. An average ratio was calculated for each DNA probe on the array from at least three replicates. The gene was considered to be regulated if the median ratio was >2.5 and P value was <0.05.
Tissue specimens
Breast carcinoma tissues were obtained from Peking University Oncology Hospital. Samples were frozen in liquid nitrogen immediately after surgical removal and maintained at −80°C until use. All human tissue was collected using protocols approved by the Ethics Committee of the Peking University Health Science Center.
Tumour xenografts
MCF-7 breast cancer cells were plated and infected in vitro with mock or lentiviruses carrying ZIP or ZIP RNAi at MOI of 100. Forty-eight hours after infection, 5 × 106 viable MCF-7 cells in 200 μl PBS were injected into the mammary fat pads of 6- to 8-week-old female BALB/c mice (Charles River, Beijing, China). Six animals per group were used in each experiment. Seventeen β-estradiol pellets (0.72 mg/pellet, 60 day release; Innovative Research of America, Sarasota, FL) were implanted one day before the tumour cell injection. Tumours were measured weekly using a vernier calliper and the volume was calculated according to the formula: π/6 × length × width2. All studies were approved by the Animal Care Committee of Peking University Health Science Center.
Supplementary Material
Supplementary Methods
Supplementary File 1
Supplementary File 2
Supplementary File 3
Review Process File
Acknowledgments
We thank Joanne Balmer Green and James Balmer (Penn State University) for editorial assistance. This work was supported by grants (30830032, 30621002, and 30470912 to YS, 30600319 to WY, and 30500263 to JL) from National Natural Science Foundation of China and grants (863 Program: 2006AA02Z466 and 973 Program: 2005CB522404 and 2007CB914503 to YS) from the Ministry of Science and Technology of China.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahringer J (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356 [DOI] [PubMed] [Google Scholar]
- Barrett-Lee PJ (2005) Growth factor signalling in clinical breast cancer and its impact on response to conventional therapies: a review of chemotherapy. Endocr Relat Cancer 12(Suppl 1): S125–S133 [DOI] [PubMed] [Google Scholar]
- Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Brandt B, Meyer-Staeckling S, Schmidt H, Agelopoulos K, Buerger H (2006) Mechanisms of EGFR gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res 12: 7252–7260 [DOI] [PubMed] [Google Scholar]
- Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA (2002) Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol 9: 912–917 [DOI] [PubMed] [Google Scholar]
- Chan SK, Hill ME, Gullick WJ (2006) The role of the epidermal growth factor receptor in breast cancer. J Mammary Gland Biol Neoplasia 11: 3–11 [DOI] [PubMed] [Google Scholar]
- Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y (2008) The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem 283: 17969–17978 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438 [DOI] [PubMed] [Google Scholar]
- Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield MD (1984) Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature 307: 521–527 [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE (2003) The ErbB receptors and their role in cancer progression. Exp Cell Res 284: 99–110 [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA (2000) Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab 11: 6–10 [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD (2007) Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci 104: 4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Zhang H, Zhang Y, Shang Y (2009) GAS, a new glutamate-rich protein, interacts differentially with SRCs and is involved in oestrogen receptor function. EMBO Rep 10: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn J, Baselga J (2006) Epidermal growth factor receptor targeting in cancer. Semin Oncol 33: 369–385 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Mitra S (2005) Hidden Markov models, grammars, and biology: a tutorial. J Bioinform Comput Biol 3: 491–526 [DOI] [PubMed] [Google Scholar]
- Nicholson RI, Gee JM, Harper ME (2001) EGFR and cancer prognosis. Eur J Cancer 37(Suppl 4): S9–15 [DOI] [PubMed] [Google Scholar]
- Ordway JM, Bedell JA, Citek RW, Nunberg A, Garrido A, Kendall R, Stevens JR, Cao D, Doerge RW, Korshunova Y, Holemon H, McPherson JD, Lakey N, Leon J, Martienssen RA, Jeddeloh JA (2006) Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis 27: 2409–2423 [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- Shang Y (2006) Molecular mechanisms of oestrogen and SERMs in endometrial carcinogenesis. Nat Rev Cancer 6: 360–368 [DOI] [PubMed] [Google Scholar]
- Shang Y, Brown M (2002) Molecular determinants for the tissue specificity of SERMs. Science 295: 2465–2468 [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843–852 [DOI] [PubMed] [Google Scholar]
- Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, Hong M, Shang Y (2007) Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol 27: 5105–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Shi L, Li W, Yu W, Liang J, Zhang H, Yang X, Wang Y, Li R, Yao X, Yi X, Shang Y (2009) JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc Natl Acad Sci 106: 10195–10200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhang H, Wang D, Ma D, Shen Y, Shang Y (2004) DLP, a novel Dim1 family protein implicated in pre-mRNA splicing and cell cycle progression. J Biol Chem 279: 32839–32847 [DOI] [PubMed] [Google Scholar]
- Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8: 251–262 [DOI] [PubMed] [Google Scholar]
- Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, Wang D, Li R, Yi X, Zhang H, Sun L, Shang Y (2005) Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 438: 981–987 [DOI] [PubMed] [Google Scholar]
- Wu H, Sun L, Zhang Y, Chen Y, Shi B, Li R, Wang Y, Liang J, Fan D, Wu G, Wang D, Li S, Shang Y (2006) Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. J Biol Chem 281: 21848–21856 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Yin N, Wang D, Zhang H, Yi X, Sun X, Shi B, Wu H, Wu G, Wang X, Shang Y (2004) Molecular mechanisms involved in the growth stimulation of breast cancer cells by leptin. Cancer Res 64: 5870–5875 [DOI] [PubMed] [Google Scholar]
- Zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS (2007) Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal 19: 2013–2023 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sun L, Liang J, Yu W, Zhang Y, Wang Y, Chen Y, Li R, Sun X, Shang Y (2006) The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J 25: 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yi X, Sun X, Yin N, Shi B, Wu H, Wang D, Wu G, Shang Y (2004) Differential gene regulation by the SRC family of coactivators. Genes Dev 18: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang H, Liang J, Yu W, Shang Y (2007) SIP, a novel ankyrin repeat containing protein, sequesters steroid receptor coactivators in the cytoplasm. EMBO J 26: 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Supplementary File 1
Supplementary File 2
Supplementary File 3
Review Process File