Abstract
The tumour suppressor ARF (alternative reading frame), which is mutated or silenced in various tumours, has a crucial role in tumour surveillance to suppress unwarranted cell growth and proliferation. ARF has also been linked to the DNA-damage-induced response of p53 because of its ability to inhibit murine double minute 2 (MDM2). Here, however, we provide genetic evidence for a role of ARF in nucleotide excision repair (NER) that is independent of p53. Cells lacking ARF are deficient in NER. Expression of ARF restores the repair activity, which coincides with increased expression of the damaged-DNA recognition protein xeroderma pigmentosum, complementation group C (XPC). We provide evidence that, by disrupting the interaction between E2F transcription factor 4 (E2F4) and DRTF polypeptide 1 (DP1), ARF reduces the interaction of the E2F4–p130 repressor complex with the promoter of XPC to ensure high-level expression of XPC. Together, our results point to an important ‘care-taker'-type tumour-suppression function for ARF in NER through the increased expression of XPC.
Keywords: ARF, NER, XPC, E2F4, p130
Introduction
The Ink4A/ARF locus (cyclin-dependent kinase inhibitor 2A (CDKN2A) in humans) encodes two tumour-suppressor proteins, p16INK4a and p19ARF (p14ARF in humans; Quelle et al, 1995; reviewed in Sherr, 2006). ARF, derived from an alternative reading frame of the INK4A locus, is mutated, deleted or silenced in a significant number of human cancers, is coupled to the p53 pathway. In response to sustained hyperproliferative signalling, ARF is activated and, in turn, stabilizes and activates p53 by antagonizing its negative regulator, murine double minute 2 (mdm2; Kubbutat et al, 1997; Kamijo et al, 1998; Pomerantz et al, 1998; Zhang et al, 1998; Weber et al, 2000a, 2000b; Llanos et al, 2001). Activation of the ARF–p53–MDM2 pathway results in cell-cycle arrest or apoptosis and is considered to be a checkpoint that protects cells from tumorigenesis.
Interestingly, there is a growing body of evidence to suggest that ARF has additional tumour-suppressive activities that are independent of p53. The p53-independent function first became apparent when it was observed that ARF retained its ability to cause cell-cycle arrest in cells that were p53−/− or p53−/−MDM2−/− (Carnero et al, 2000; Weber et al, 2000a, 2000b; Eymin et al, 2001). Reintroduction of ARF into p53−/−MDM2−/−ARF−/− mouse embryonic fibroblasts (MEFs) caused a delayed G1-phase growth arrest (Weber et al, 2000a, 2000b). These findings imply that ARF could not be solely linked to the p53 pathway and has led to the discovery of many new targets by which it regulates the cell cycle.
ARF has been shown to inhibit ribosomal-RNA processing (Sugimoto et al, 2003), to interact with topoisomerase I (Ayrault et al, 2004) and to interact physically and re-localize several transcription factors, including E2F transcription factor 1 (E2F1; Martelli et al, 2001), forkhead box M1B transcription factor (Kalinichenko et al, 2004) and c-Myc (Datta et al, 2004; Qi et al, 2004). Consequently, it has been reported that ARF affects gene expression (Eymin et al, 2001; Martelli et al, 2001; Datta et al, 2002, 2004; Kuo et al, 2003; Rocha et al, 2003; Aslanian et al, 2004; Qi et al, 2004). We reported previously that ARF could specifically regulate E2F target gene expression by binding to DRTF polypeptide 1 (DP1), the functional partner of E2F family of factors, and inhibiting the interaction between DP1 and E2F1. Upon ARF expression there was a loss of DP1 binding to the promoter of an E2F target gene (Datta et al, 2005).
The role of ARF in DNA repair has not been investigated in depth. One study implicated ARF in processing DNA photoproducts that are generated following exposure to ultraviolet radiation (Sarkar-Agrawal et al, 2004). DNA damages incurred by ultraviolet radiation are repaired by nucleotide excision repair (NER). NER involves three basic biochemical steps consisting of damage recognition, dual incision and gap-filling DNA synthesis (Li et al, 1998). It is currently acknowledged that repair proteins act in a highly coordinated manner and assemble sequentially on the site of DNA damage to return it to its native state (Volker et al, 2001). Seven of these repair proteins belong to the xeroderma pigmentosum (XP) group (XPA-G) (de Laat et al, 1999; Masutani et al, 1999; Mullenders & Berneburg, 2001).
Through its crucial role in DNA-damage recognition and initiation of NER, the xeroderma pigmentosum, complementation group C (XPC) protein epitomizes a first line of defence against carcinogenesis (Sugasawa et al, 1998; Volker et al, 2001). Cells deficient in XPC are impaired in the removal of cyclobutane–pyrimidine dimers (CPDs) and 6-4 pyrimidine pyrimidones (6-4PPs) from the global genome, emphasizing the important function that XPC has in these processes (Venema et al, 1991; Emmert et al, 2000). In global genomic repair, XPC recognizes CPDs and 6-4PPs and recruits the transcription/repair factor TFIIH, which is crucial in the assembly of the NER complex that leads to excision of the damaged strand. XPC, however, is dispensable for the removal of DNA lesions from the transcribed strand of active genes (Venema et al, 1990; Emmert et al, 2000). Although the biochemical properties of XPC in repair have been characterized in some detail (Venema et al, 1991; Sugasawa et al, 1998, 2001; Emmert et al, 2000; Volker et al, 2001), much remains to be elucidated with regards to its regulation and interaction with other tumour-suppression pathways. Here, we provide evidence for a link between the tumour suppressor ARF and the repair factor XPC.
Results And Discussion
ARF is required for efficient NER
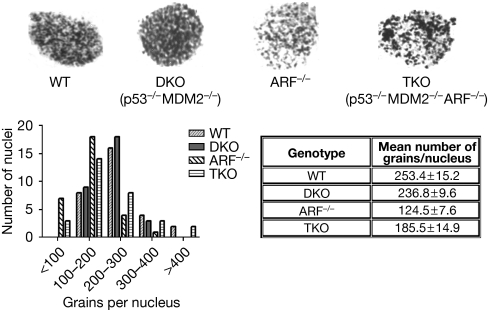
To investigate whether ARF has a role in NER, we compared the extent of DNA repair in wild-type MEFs and in ARF-null MEFs treated with ultraviolet irradiation. The repair activity was measured by unscheduled DNA synthesis (UDS), an assay that measures repair re-replication following excision of the damaged DNA strand. In UDS, the level of repair is correlated with the amount of tritium-labelled thymidine (3H-TdR) incorporated into the DNA of mammalian cells, which are not in the S phase of the cell cycle. UDS was performed using strategies described by Smith et al (2000). Briefly, the cells were pre-labelled using 3H-TdR to identify the S-phase cells. The cells were then subjected to ultraviolet irradiation (10 J/m2) and maintained in a medium containing 3H-TdR, in the absence of serum for 3 h. The cells were then incubated in a medium containing unlabelled TdR for 1 h, fixed and coated with emulsion (EM-1), and developed for UDS measurement. We found that repair synthesis—grains corresponding to 3H-TdR incorporation—in the ARF-null MEFs was significantly reduced compared with the wild-type MEFs, as can be seen in representative nuclei (Fig 1). The quantification shows that most nuclei counted for wild-type MEFs showed 200–300 grains per nucleus, whereas most nuclei in ARF-null MEFs showed 100–200 grains per nucleus (Fig 1).
Figure 1.
ARF has a role in nucleotide-excision repair that is independent of p53. The nucleotide-excision-repair activity following ultraviolet irradiation was measured in WT, ARF−/−, p53−/−MDM2−/− (DKO) and p53−/−MDM2−/−ARF−/− (TKO) MEFs by UDS (Smith et al, 2000). UDS was quantified by counting the number of 3H-thymidine grains per nucleus in 30 nuclei per cell type. Autoradiographs of the grains in nuclei are shown in the upper panel, and grains per nucleus and means±s.e.m. are shown in the lower panel. TKO compared with DKO P=0.0058 and WT compared with ARF−/− P<0.0001. ARF, alternative reading frame; DKO, double knockout; MDM2, murine double minute 2; MEF, mouse embryonic fibroblast; TKO, triple knockout; UDS, unscheduled DNA synthesis; WT, wild type.
To determine whether the function of ARF in NER was dependent on p53, we analyzed p53−/−Mdm2−/− (double knockout (DKO)) MEFs and p53−/−Mdm2−/−ARF−/− (triple knockout (TKO)) MEFs. By comparing the four genotypes—wild-type, ARF−/−, p53−/−Mdm2−/− (DKO) and p53−/−Mdm2−/−ARF−/− (TKO) MEFs—we observed that the extent of repair was comparable between the wild-type and DKO MEFs, and that it was significantly more than the repair activity observed in the ARF−/− and TKO MEFs (Fig 1). Together, these results suggest that ARF has a function in NER independently of p53.
We hypothesized that if ARF has a function in NER then by expressing it in ARF-null MEFs we should see a reversal of the repair-deficient phenotype. We used an adenovirus expressing ARF to infect the ARF−/− MEFs and, for comparison, we infected the ARF−/− MEFs with a control adenovirus. As expected, re-introduction of ARF restored the degree of repair to a level similar to that seen in the wild-type MEFs (supplementary Fig S1 online).
Involvement of ARF in the removal of 6-4PPs and CPDs
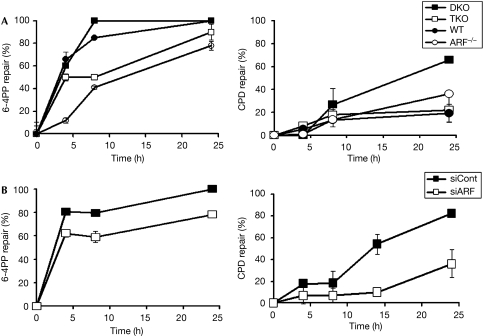
As UDS measures DNA synthesis after the excision of DNA fragments containing 6-4PPs and CPDs, we compared the rates of removal of CPD and 6-4PP in MEFs lacking ARF (TKO and ARF−/−) with wild-type MEFs and with DKO MEFs using a previously described procedure (Eveno et al, 1995; Li et al, 2006). Genomic DNA was isolated from the MEFs at various time points after ultraviolet irradiation. The DNA was denatured and subjected to dot-blot assays. The membrane was assayed using antibodies against 6-4PPs (clone 64M2) and CPDs (clone TDM2) in each cell type. Quantification was obtained by using the Image J program and calculated by comparing the intensities of the signals produced at the indicated times to that of the corresponding signal at time zero. We found that the TKO MEFs and the ARF−/− MEFs were considerably impaired compared with the wild-type or DKO MEFs in removing 6-4PPs (Fig 2). CPD removal, however, was much less efficient in the MEFs, especially at an early time point (4 h).
Figure 2.
ARF increases 6-4PP and CPD removal. (A) 6-4PP and CPD removal rates were compared between DKO, TKO, WT and ARF−/− MEFs, and (B) in DKO cells in which ARF was knocked down by the expression of an ARF shRNA (see supplementary information online for methods). Percentages of 6-4PP and CPD removed in three experiments were quantified and plotted. 6-4PP, 6-4 pyrimidine pyrimidone; ARF, alternative reading frame; Cont, control; CPD, cyclobutane pyrimidine dimer; DKO, double knockout; MEF, mouse embryonic fibroblast; shRNA, short-hairpin RNA; TKO, triple knockout; WT, wild type.
Surprisingly, we did not see any significant difference in CPD removal between wild-type MEFs and ARF-null MEFs (Fig 2). However, the DKO MEFs showed high levels of CPD repair. The extent of CPD removal in DKO MEFs was much higher than that reported for rodent cells (Tang et al, 2000). For example, at 24 h post-ultraviolet irradiation, close to 70% of the CPDs were removed in DKO MEFs, whereas in the other MEFs only about 20% were removed (Fig 2). We hypothesize that the efficient removal of CPD in the DKO MEFs results from the high expression of ARF in those cells (Datta et al, 2005; supplementary Fig S2 online), as similar efficient removal was detected by overexpressing ARF in ARF−/− cells (data not shown).
To confirm further the role of ARF in the repair of 6-4PPs and CPDs, we used DKO cells in which the level of ARF had been knocked down by the expression of an ARF short-hairpin RNA. Knockdown of ARF was confirmed by using Western blot (supplementary Fig S2 online). The DKO cells, with or without ARF knockdown, were compared for the removal of CPDs and 6-4PPs. Clearly, depletion of ARF caused a significant impairment in the removal levels of CPD and 6-4PP (Fig 2, lower panels), confirming the idea that ARF has a significant function in the repair of ultraviolet-damaged DNA.
ARF stimulates the expression of XPC protein
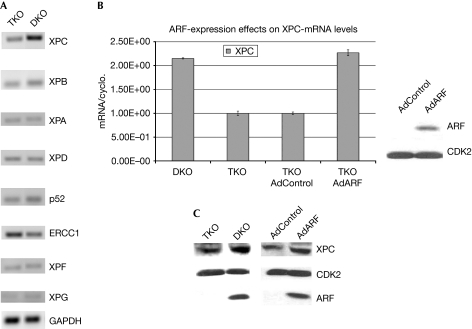
To determine the mechanism by which ARF participates in NER, we investigated whether the genes that are essential for NER are regulated by ARF. We compared the RNA isolated from ARF+ cells (DKO MEFs) with those from ARF− cells (TKO MEFs) using semi-quantitative reverse transcription PCR (RT–PCR). The messenger-RNA (mRNA) levels of XPC, XPB, XPA, XPD, p52 (one of the subunits of TFIIH), excision repair cross-complementing rodent repair deficiency, complementation group 1 (ERCC1), XPF and XPG were analysed (Fig 3A). We observed that the transcript level of XPC in ARF+ cells was substantially higher than in ARF− cells. This observation was also seen by using quantitative RT–PCR (Fig 3B). Furthermore, we found that infecting ARF− (TKO MEFs) cells with ARF-expressing adenovirus brought the transcript levels of XPC to that observed in the ARF+ cells (DKO MEFs; Fig 3B). Moreover, expression of ARF increased the protein level of XPC (Fig 3C). The observation that ARF stimulates the expression of XPC is significant because XPC has an invaluable role in DNA-damage recognition and the initiation step of NER (Sugasawa et al, 1998; Volker et al, 2001). We did not see any significant difference in the level of the XPE gene product damage-specific DNA binding protein 2 (DDB2) in the ARF-null MEFs in response to ARF expression (data not shown).
Figure 3.
ARF stimulates the expression of XPC. (A) Semi-quantitative RT–PCR and (B) quantitative RT–PCR assays were used to measure the expression of genes that are essential for nucleotide-excision repair in TKO and DKO MEFs. (C) Protein levels of XPC in the extracts (0.15 mg) of DKO and TKO MEFs were compared using Western-blot assays. A comparison of XPC level in TKO cells infected with control or ARF-expressing adenovirus is shown on the right lanes. Ad, adenovirus; ARF, alternative reading frame; CDK2, cyclin-dependent kinase 2; DKO, double knockout; ERCC1, excision repair cross-complementing group 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MEF, mouse embryonic fibroblast; mRNA, messenger RNA; RT–PCR, reverse transcription PCR; TKO, triple knockout; XPC, xeroderma pigmentosum group C.
ARF disrupts E2F4 repressor complexes to stimulate XPC
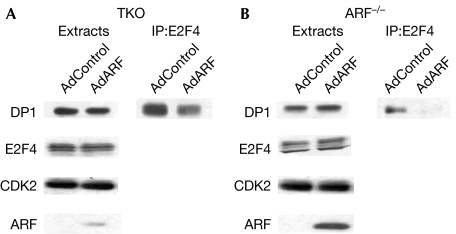
As there was a substantial increase in XPC transcript level on ARF expression, we directed our attention to the promoter of XPC. A previous report (Cam et al, 2004) indicated that the XPC gene is repressed by the E2F4–p130 repressor complex. Previous studies have shown that the tumour suppressor ARF binds to DP1 to inhibit the interaction between DP1 and E2F1 (Datta et al, 2005). We considered the possibility that ARF would disrupt the interaction between the E2F4–p130 repressor complex and the XPC promoter by dissociating the interaction between E2F4 and DP1, which would result in the loss of DNA binding. To investigate whether ARF reduces the interaction between E2F4 and DP1, we infected TKO MEFs and ARF-null MEFs with either control adenovirus or ARF-expressing adenovirus. Extracts from the infected cells were immunoprecipitated with the E2F4 antibody and the immunoprecipitates were assayed for the presence of DP1 by Western blot using an DP1 antibody. Consistent with our prediction, there was indeed a reduced association between E2F4 and DP1 in the presence of ARF expression in both cell types (Figs 4A,B).
Figure 4.
ARF dissociates the E2F4–DP1 interaction. (A) TKO MEFs or (B) ARF-null MEFs were infected with control or ARF-expressing adenovirus for 16 h. Cell extracts (1 mg) were immunoprecipitated (IP) using the E2F4 antibody, and the immunoprecipitates were subjected to Western blot analysis using the DP1 antibody. The levels of E2F4, DP1 and ARF in the extracts (0.1 mg) of the infected cells are also shown. Ad, adenovirus; ARF, alternative reading frame; CDK2, cyclin-dependent kinase 2; DKO, double knockout; DP1, DRTF polypeptide 1; E2F4, E2F transcription factor 4; MEF, mouse embryonic fibroblast; TKO, triple knockout.
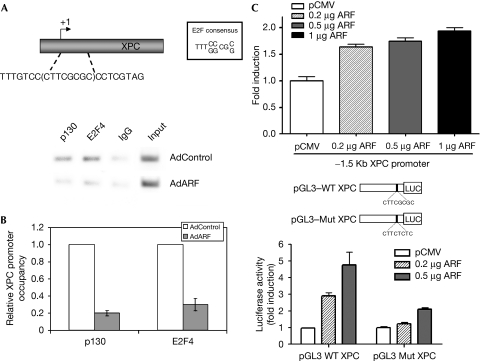
To determine whether the expression of ARF has a regulatory effect on the E2F4–p130 repressor complex and its interaction with the promoter of XPC, we carried out chromatin-immunoprecipitation (ChIP) experiments. The mouse XPC promoter contains a canonical E2F binding site between −3 and +5 positions relative to the transcription start site. We designed primers to detect E2F4 and p130 binding to this region of the XPC promoter. TKO MEFs were either infected with control adenovirus or ARF-expressing adenovirus. At 16 h post infection, the cells were cross-linked and the chromatin was sonicated and processed for ChIP using E2F4 and p130 antibodies. The amounts of XPC promoter DNA associated with the immunoprecipitated chromatin were quantified by using real-time PCR. As shown in Fig 5A,B, the expression of ARF resulted in a considerable decrease of E2F4 and p130 bound to the XPC promoter. To confirm that the XPC promoter is indeed a target of ARF-mediated activation, we generated a luciferase reporter construct with a −1.5 kb fragment of the XPC promoter. ARF-stimulated expression from the native promoter is shown in Fig 5C, upper panel. To assess further the role of the E2F site in the XPC promoter, sequences between −51 and +7 positions of the mouse XPC promoter, with intact or mutated E2F sites, were used to generate luciferase reporter constructs. Expression of the reporter gene was measured in serum-starved cells, a condition in which the repressor complex of E2F4 is abundant. The expression of ARF clearly stimulated transcription from the promoter with an intact E2F site, whereas little stimulation was observed from the promoter construct with a mutated E2F site (Fig 5C, lower panel). Among the NER genes, the ERCC1 and XPC promoters contain E2F binding sites; however, we detected a decrease in ERCC1 expression in DKO MEFs that overexpress ARF. It is possible that ERCC1 is a target of the activator E2Fs, which are inhibited by ARF (Martelli et al, 2001). Taken together, these observations show that the expression of ARF stimulates XPC transcription by downregulating the repressor complex of E2F4.
Figure 5.
ARF stimulates the expression of XPC by disrupting the repressor complex of E2F4. TKO MEFs were infected with either control or ARF-expressing adenovirus and subjected to ChIP assay. XPC promoter DNA associated with immunoprecipitated chromatin was quantified by using real-time PCR. An aliquot of the RT–PCR sample was analysed by agarose gel electrophoresis (A), and the quantitative RT–PCR results are shown in panel (B). In panel (C), NIH-3T3 cells were serum starved for 12 h followed by transfection with a 1.5 kB fragment of the XPC and a smaller fragment of the promoter (−51 to +7) containing the E2F site or a mutant promoter construct along with the indicated amounts of ARF. Luciferase activity was measured. Results from three experiments were plotted. ARF, alternative reading frame; ChIP, chromatin immunoprecipitation; E2F4, E2F transcription factor 4; MEF, mouse embryonic fibroblast; Mut, mutant; RT–PCR, reverse transcription PCR; TKO, triple knockout; WT, wild type; XPC, xeroderma pigmentosum group C.
To determine whether the repair defects in the ARF-deficient cells resulted from reduced expression of XPC, we added back XPC in ARF-deficient TKO cells by infecting them with XPC-expressing lentivirus. Cells infected with either control virus (pLVx) or XPC-expressing virus (pLVx-XPC) were selected for three days with puromycin. XPC expression was confirmed using Western blot (supplementary Fig S2 online). These cells, along with DKO cells, were subjected to the UDS assay. Expression of XPC in ARF-deficient TKO cells caused a significant stimulation of UDS (supplementary Fig S3 online). The expression of XPC stimulated UDS in TKO cells to a level comparable with the DKO cells. These results further confirm the idea that NER deficiency in cells lacking ARF results from the reduced expression of XPC.
Here, we have established that ARF functions as a ‘care-taker' type of tumour suppressor and has a important function in the maintenance of genomic integrity. We have shown that ARF is required for high-level expression of XPC, a factor involved in damage recognition and a crucial component in the first step of NER. Previous studies (Adimoolam & Ford, 2002) have indicated a role for p53 in stimulating the expression of XPC after DNA damage. Consistent with that, we observed an increase in XPC mRNA in ultraviolet-irradiated wild-type MEFs (1.6-fold increase after correcting for loading; supplementary Fig S1 online). However, the DKO MEFs, which lack both p53 and MDM2, express XPC at a much higher level (supplementary Fig S1 online). The DKO cells, unlike other MEFs, also overexpress ARF (supplementary Fig S1 online). Our observations made in DKO cells suggest that ARF is able to stimulate XPC expression independently of p53. ARF activates XPC expression by de-repressing the promoter through disruption of the E2F4 repressor complex. De-regulation of the E2F4 repressor complex is a new mechanism, and it is likely that ARF uses this mechanism to stimulate other genes that are repressed by the E2F4–p130 repressor complex.
Speculation
The E2F4–p130 repressor complex regulates the expression of several DNA-repair genes, including RAD54, BARD1, MLH1 and MSH2 (Ren et al, 2002); E2F4 also regulates the expression of the checkpoint genes CHK1 and MAD2 (Ren et al, 2002). Therefore, the observation that ARF disrupts the interaction of the E2F4–p130 complex with a target DNA-repair gene provides support to the idea that ARF would stimulate pathways that are significant in maintaining genomic integrity. For example, RAD54 participates in homologous-recombination-mediated double-strand break repair, whereas MLH1 and MSH2 are crucial for mismatch repair. It will be interesting to determine whether ARF stimulates these pathways. It is noteworthy that ARF−/− cells show defective DNA-damage response after ionizing-radiation treatment, which is partly due to impaired activation of p53 in these cells (Khan et al, 2004). There is some evidence that nucleolar ARF re-localizes to the nucleoplasm after DNA damage (Lee et al, 2005), which might be important in stimulating the DNA-damage-response genes regulated by the E2F4–p130 complex. Furthermore, ARF expression is stimulated by supra-physiological mitogenic signals. We speculate that the ‘care-taker' function of ARF would be beneficial for a cell responding to strong mitogenic signals.
Methods
Cells. Wild-type, ARF−/− (Kamijo et al, 1999), p53−/−Mdm2−/−ARF−/− (TKO; Weber et al, 2000a, 2000b), p53−/−Mdm2−/− (DKO) and NIH-3T3 MEFs were maintained in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Hyclone, Logan, UT, USA), supplemented with 1% L-glutamine (Invitrogen) and 1% non-essential amino acids (Invitrogen).
UDS assay. NER was measured using UDS as described previously (Smith et al, 2000). Cells were plated overnight on coverslips. The next morning, cells were pre-labelled in serum-free media for 1 h with 3H-TdR1 > (10 μCi/ml) to identify the S-phase cells. Cells were then irradiated using ultraviolet subtype C radiation at a total dose of 10 J/m2 in the absence of culture medium. After this irradiation, the cells were again placed in 3H-TdR in serum-free media for 3 h. The cells were then incubated with non-radioactive thymidine for an additional hour, fixed, treated with EM-1 (Amersham, Pisctaway, NJ, USA) and developed for UDS measurement.
CPD- and 6-4PP-removal assay. Removal rates of ultraviolet-induced photoproducts were measured as reported previously (Eveno et al, 1995; Li et al, 2006). After ultraviolet irradiation, cells were allowed to repair for various times in fresh culture medium. Cells were collected and genomic DNA was extracted using Easy-DNA (Invitrogen). For CPD removal, 1 μg of genomic DNA was denatured by adding NaOH at a final concentration of 0.4 M. EDTA was added at a final concentration of 10 mM. Samples were boiled for 10 min at 100°C. DNA was then neutralized by adding 2 M ammonium acetate (pH 7). For the removal of 6-4 PP, 1 μg of DNA was boiled at 100°C for 5 min followed by cooling for 5 min on ice. The DNA was bound to membrane using the Bio-Dot Microfilitration Apparatus (Bio-Rad, Hercules, CA, USA) according to the manufacturer's protocol for DNA blotting. The membrane was then probed with an antibody specific for CPD or 6-4PP, the chief photoproducts produced by ultraviolet subtype C treatment (anti-thymidine dimer, clone TDM2; or anti-6-4PP antibody clone 64M2) and used at a dilution of 1:3,000 and 1:100, respectively. The quantification was obtained by using Image J and was calculated by comparing the intensities of the dots produced at the indicated times to that of the corresponding dot at time zero when there was no opportunity of repair and all 100% CPDs were present.
ChIP assay. TKO (p53−/−Mdm2−/−ARF−/−) cells were either infected with control adenovirus or ARF-expressing adenovirus and processed 16 h post infection for ChIP assays. Cells were first cross-linked by the addition of formaldehyde to a final concentration of 1%, the chromatin was sonicated and immunoprecipitation was carried out using 2 μg of E2F4 antibody (C20; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and 2 μg of p130 antibody (clone C20; Santa Cruz Biotechnology). ChIP was carried out using a ChIP assay kit (Upstate, Lake Placid, NY, USA) according to the manufacturer's protocol. DNA released from the precipitated complexes was amplified by PCR alongside 0.1% of the input chromatin used to carry out the immunoprecipitation. Mouse XPC promoter (−68 to +63 bp)-specific primers (XPC forward 5′-CGGGAACAGGAACTCAGAAA-3′ and XPC reverse 5′-CAGCCCAGGGTAGGACCAC-3′) were used to carry out PCR. The PCR products were separated on agarose gels and visualized by ethidium-bromide staining.
The details for immunoprecipitation, Western blot and luciferase assays can be found in the supplementary information online. The primers used for the RNA assays are also described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank A. Wani from The Ohio State University for kindly providing us with the XPC antibody. The double-knockout and triple-knockout mouse embryonic fibroblasts were a kind gift from C. Sherr from St Jude Children's Research Hospital, TN, USA. This work was supported by National Institute of Health grant CA100035, AG 024138 and CA 77637 to P.R. and a minority supplemental grant CA100035S to C.D.-B.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adimoolam S, Ford JM (2002) p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci USA 99: 12985–12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanian A, Iaquinta PJ, Verona R, Lees JA (2004) Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev 18: 1413–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrault O, Andrique L, Larsen CJ, Seite P (2004) Human Arf tumor suppressor specifically interacts with chromatin containing the promoter of rRNA genes. Oncogene 23: 8097–8104 [DOI] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD (2004) A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 16: 399–411 [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Hannon GJ, Beach DH (2000) Loss-of-function genetics in mammalian cells: the p53 tumor suppressor model. Nucleic Acids Res 28: 2234–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Nag A, Raychaudhuri P (2002) Differential regulation of E2F1, DP1, and the E2F1/DP1 complex by ARF. Mol Cell Biol 22: 8398–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Nag A, Pan W, Hay N, Gartel AL, Colamonici O, Mori Y, Raychaudhuri P (2004) Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J Biol Chem 279: 36698–36707 [DOI] [PubMed] [Google Scholar]
- Datta A, Sen J, Hagen J, Korgaonkar CK, Caffrey M, Quelle DE, Hughes DE, Ackerson TJ, Costa RH, Raychaudhuri P (2005) ARF directly binds DP1: interaction with DP1 coincides with the G1 arrest function of ARF. Mol Cell Biol 25: 8024–8036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NG, Hoeijmakers JH (1999) Molecular mechanism of nucleotide excision repair. Genes Dev 13: 768–785 [DOI] [PubMed] [Google Scholar]
- Emmert S, Kobayashi N, Khan SG, Kraemer KH (2000) The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6-4 photoproducts. Proc Natl Acad Sci USA 97: 2151–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveno E et al. (1995) Different removal of ultraviolet photoproducts in genetically related xeroderma pigmentosum and trichothiodystrophy diseases. Cancer Res 55: 4325–4332 [PubMed] [Google Scholar]
- Eymin B, Karayan L, Seite P, Brambilla C, Brambilla E, Larsen CJ, Gazzeri S (2001) Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 20: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV et al. (2004) Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev 18: 830–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ (1998) Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA 95: 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, van de Kamp E, Chong MJ, Zindy F, Diehl JA, Sherr CJ, McKinnon PJ (1999) Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled atm function. Cancer Res 59: 2464–2469 [PubMed] [Google Scholar]
- Khan S, Guevara C, Fujii G, Parry D (2004) p14ARF is a component of the p53 response following ionizing irradiation of normal human fibroblasts. Oncogene 23: 6040–6046 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Kuo ML, Duncavage EJ, Mathew R, den Besten W, Pei D, Naeve D, Yamamoto T, Cheng C, Sherr CJ, Roussel MF (2003) Arf induces p53-dependent and -independent antiproliferative genes. Cancer Res 63: 1046–1053 [PubMed] [Google Scholar]
- Lee C, Smith BA, Bandyopadhyay K, Gjerset RA (2005) DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res 65: 9834–9842 [DOI] [PubMed] [Google Scholar]
- Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, Wani AA (2006) DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res 66: 8590–8597 [DOI] [PubMed] [Google Scholar]
- Li RY, Calsou P, Jones CJ, Salles B (1998) Interactions of the transcription/DNA repair factor TFIIH and XP repair proteins with DNA lesions in a cell-free repair assay. J Mol Biol 281: 211–218 [DOI] [PubMed] [Google Scholar]
- Llanos S, Clark PA, Rowe J, Peters G (2001) Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol 3: 445–452 [DOI] [PubMed] [Google Scholar]
- Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, DePinho RA, Livingston DM, Grossman SR (2001) p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci USA 98: 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- Mullenders LH, Berneburg M (2001) Photoimmunology and nucleotide excision repair: impact of transcription coupled and global genome excision repair. J Photochem Photobiol B 65: 97–100 [DOI] [PubMed] [Google Scholar]
- Pomerantz J et al. (1998) The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723 [DOI] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR (2004) p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431: 712–717 [DOI] [PubMed] [Google Scholar]
- Quelle DE, Ashmun RA, Hannon GJ, Rehberger PA, Trono D, Richter KH, Walker C, Beach D, Sherr CJ, Serrano M (1995) Cloning and characterization of murine p16INK4a and p15INK4b genes. Oncogene 11: 635–645 [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD (2002) E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S, Campbell KJ, Perkins ND (2003) p53- and Mdm2-independent repression of NF-kappa B transactivation by the ARF tumor suppressor. Mol Cell 12: 15–25 [DOI] [PubMed] [Google Scholar]
- Sarkar-Agrawal P, Vergilis I, Sharpless NE, DePinho RA, Runger TM (2004) Impaired processing of DNA photoproducts and ultraviolet hypermutability with loss of p16INK4a or p19ARF. J Natl Cancer Inst 96: 1790–1793 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (2006) Divorcing ARF and p53: an unsettled case. Nat Rev Cancer 6: 663–673 [DOI] [PubMed] [Google Scholar]
- Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ Jr (2000) p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol 20: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 2: 223–232 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F (2001) A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev 15: 507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Kuo ML, Roussel MF, Sherr CJ (2003) Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell 11: 415–424 [DOI] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G (2000) Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell 5: 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA 87: 4707–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, van Hoffen A, Karcagi V, Natarajan AT, van Zeeland AA, Mullenders LH (1991) Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol Cell Biol 11: 4128–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 8: 213–224 [DOI] [PubMed] [Google Scholar]
- Weber JD, Jeffers JR, Rehg JE, Randle DH, Lozano G, Roussel MF, Sherr CJ, Zambetti GP (2000a) p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev 14: 2358–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Kuo ML, Bothner B, DiGiammarino EL, Kriwacki RW, Roussel MF, Sherr CJ (2000b) Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol 20: 2517–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information