Abstract
Sleep need is affected by developmental stage and neuronal plasticity, but the underlying mechanisms remain unclear. The fragile X mental retardation gene Fmr1, whose loss-of-function mutation causes the most common form of inherited mental retardation in humans, is involved in synaptogenesis and synaptic plasticity, and its expression depends on both developmental stage and waking experience. Fmr1 is highly conserved across species and Drosophila mutants carrying dFmr1 loss-of-function or gain-of-function mutations are well characterized: amorphs have overgrown dendritic trees with larger synaptic boutons, developmental defects in pruning, and enhanced neurotransmission, while hypermorphs show opposite defects, including dendritic and axonal underbranching and loss of synapse differentiation. We find here that dFmr1 amorphs are long sleepers and hypermorphs are short sleepers, while both show increased locomotor activity and shortened lifespan. Both amorphs and hypermorphs also show abnormal sleep homeostasis, with impaired waking performance and no sleep rebound after sleep deprivation. An impairment in the circadian regulation of sleep cannot account for the altered sleep phenotype of dFmr1 mutants, nor can an abnormal activation of glutamatergic metabotropic receptors. Moreover, overexpression of dFmr1 throughout the mushroom bodies is sufficient to reduce sleep. Finally, dFmr1 protein levels are modulated by both developmental stage and behavioral state, with increased expression immediately after eclosure and after prolonged wakefulness. Thus, dFmr1 expression dose-dependently affects both sleep and synapses, suggesting that changes in sleep time in dFmr1 mutants may derive from changes in synaptic physiology.
Keywords: Drosophila, synaptic plasticity, dFmr1, Fmr1, fragile X, FMRP, sleep, lifespan
Introduction
Sleep is present in all animal species carefully studied so far, yet its functions remain unclear (Cirelli and Tononi, 2008). It is clear, however, that there is a strong link between sleep need and synaptic plasticity. In mammals, sleep amount and intensity peak during developmental periods characterized by maximal neuronal plasticity, synaptic pruning, and high metabolic activity (Feinberg, 1982; Cowan et al., 1984; Chugani, 1998; Carskadon et al., 2004). In Drosophila, sleep is abundant immediately after eclosure (Shaw et al., 2000), when neuronal pruning takes place and flies explore the environment for the first time (Balling et al., 1987; Tessier and Broadie, 2008). In adult flies, rats, and humans, learning, enriched experience, and the occurrence of synaptic potentiation during waking increase sleep need and sleep intensity (Huber et al., 2004, 2007, 2008; Ganguly-Fitzgerald et al., 2006; Faraguna et al., 2008). Moreover, in adult rats, established molecular and/or electrophysiological markers of synaptic strength are high after wakefulness and low after sleep in cortex and hippocampus (Vyazovskiy et al., 2008). Despite this evidence the molecular mechanisms underlying the link between sleep need, experience-dependent plasticity, and developmental stage remain largely unknown.
The fragile X mental retardation gene Fmr1 is involved in synaptogenesis and synaptic plasticity and its expression depends on both developmental phase and experience. In Drosophila (Tessier and Broadie, 2008) and mammals (Lu et al., 2004; Singh et al., 2007), Fmr1 levels are high early in development and decline with age. Moreover, Fmr1 expression is reduced by sensory deprivation in flies (Tessier and Broadie, 2008), and increased by sensory stimulation and exposure to an enriched environment in mammals (Weiler et al., 1997; Todd and Mack, 2000; Todd et al., 2003a; Irwin et al., 2005). Fmr1 protein product, fragile X mental retardation protein (FMRP), is present in dendritic spines (Feng et al., 1997), and loss of FMRP in mice and humans results in abnormal spines, immature synapses (Hinton et al., 1991; Comery et al., 1997; Irwin et al., 2002; Pan et al., 2004; Restivo et al., 2005) and altered synaptic plasticity (Huber et al., 2002; Li et al., 2002; Koekkoek et al., 2005; Larson et al., 2005; Zhao et al., 2005; Desai et al., 2006). Consistent with the results in mammals, loss of FMRP in Drosophila is associated with overgrown dendritic trees, larger synaptic boutons (Pan et al., 2004), and developmental defects in pruning in the mushroom bodies (Tessier and Broadie, 2008). Importantly, dFmr1 overexpression results in the opposite phenotype, with dendritic and axonal underbranching and loss of synapse differentiation (Pan et al., 2004; Pan and Broadie, 2007).
Here we show that daily sleep amount increases when dFmr1 expression decreases, and decreases when dFmr1 expression increases. These effects are modulated by developmental stage, and overexpression of dFmr1 throughout the mushroom bodies is sufficient to reduce sleep. We also show that dFmr1 protein levels increase in waking relative to sleep. Thus, although certainly not the only one, dFmr1 may be an important molecular mechanism linking sleep need to neuronal plasticity.
Materials and Methods
Animals.
Flies were cultured and tested at 20°C, 68% humidity, on yeast, dark corn syrup and agar food. Unless otherwise stated genotypes are as described in Flybase (Drysdale, 2008). The following stocks were ordered from the Bloomington Stock Center: P (Wigren et al., 2007) Fmr1EP3517(6928; from here on EP3517) (Spradling et al., 1995, 1999), Fmr1Δ50M (6930), from here on Δ50M (Zhang et al., 2001); Fmr1Δ113M (6929; from here on Δ113M) (Zhang et al., 2001); Df(3R)Exel6265 (7732) (Parks et al., 2004). Two pan-neuronal GAL4 drivers were used: P{nSyb-GAL4}, referred herein as nSyb-GAL4, was donated by Dr. Julie Simpson [Howard Hughes Medical Institute (HHMJ) Janelia Farm, Ashburn, VA]; P{ELAV-GeneSwitch} (Osterwalder et al., 2001), referred herein as ELAV-GeneSwitch, was donated by Dr. Benjamin White (National Institute of Mental Health, Bethesda, MD). The GAL4 drivers P{GawB}30Y, P{GawB}201Y, P{GawB}238Y, and P{GawB}c309, herein referred to as 30Y, 201Y, 238Y, c309, were donated by Dr. Amita Sehgal (HHMI and University of Pennsylvania Medical School, Philadelphia, PA). To remove possible modifiers and allow comparisons in a common genetic background all alleles were outcrossed before testing into the white1118 (w1118) background over at least 3 consecutive generations. Since GAL4 drivers and EP3517 contain a white transgene (w+), inheritance of these alleles was determined by eye color. Δ50M alleles were continually outcrossed to homozygous w1118; EP3517 flies that were in turn generated by crossing heterozygous w1118; EP3517/dFmr1+ (outcrossed) flies. Heterozygous w1118; Δ50M/EP3517 flies were then selected based on eye color, and either crossed again to outcrossed w1118; EP3517 homozygotes to continue the outcrossing, or to other w1118; Δ50M/EP3517 heterozygous flies to produce amorphic homozygous w1118; Δ50M with white color eyes. Hypermorphs were generated by crossing w1118; EP3517/+ (outcrossed) flies to w1118; GAL4/+ flies (outcrossed), where the GAL4 driver could be either nSyb-GAL4 or ELAV-GeneSwitch. The w1118; GAL4/EP3517 progeny were selected by eye color and to determine whether individual insertions have an effect on sleep, were compared with progeny that inherited either EP3517, or the GAL4 driver, or neither (w1118). PCR was used to confirm the genotype as determined by eye color selection, or when the latter could not be used. Whenever possible, comparisons were made between siblings and always between flies from the same genetic background and same age.
Locomotor activity, sleep, and measures of sleep intensity.
Within 8 h after eclosure from the pupae case, individual flies were placed inside glass tubes with enough food for 1 week of recording using the Drosophila Activity Monitor System (DAMS, Trikinetics). If flies were being monitored for more than 1 week, they were transferred to fresh tubes on a weekly basis within 3 h from lights on. Unless otherwise stated, flies were kept in a 12 h light/dark (LD) cycle. DAMS monitors were housed inside environmental chambers (ThermoForma) where temperature and humidity were kept constant. Data analysis was performed using custom-designed software developed in our laboratory and written in Matlab (Mathworks) (Huber et al., 2004). Sleep and wakefulness were determined for consecutive 1-min epochs. Wakefulness was defined as any period of at least 1 min characterized by activity (≥1 count/min). Sleep was defined as any period of uninterrupted behavioral immobility (0 counts/min) lasting >5 min, because previous work (Shaw et al., 2000; Huber et al., 2004; Cirelli et al., 2005) has demonstrated that such periods of quiescence are associated with an increase in arousal threshold. Indeed, in the current experiments we confirmed that wild-type controls, amorphs, and hypermorphs were significantly less likely to respond to a complex stimulus after they had been immobile for at least 5 min, relative to when the stimulus was administered while flies were awake and moving around (n of tested flies, percentage of flies reacting when awake/percentage after 5 min of immobility, p value according to Mann–Whitney test: wild type = 39, 74/60%, p = 0.04; amorphs (Δ50M) = 39, 74/67%, p = 0.029; hypermorphs (nSyb-GAL4→EP3517) = 85, 75/50%, p = 0.04). Brief awakenings were defined as waking periods when a single activity count occurred between two sleep periods (i.e., >5 min no activity, one activity count, >5 min no activity). Mean waking activity over a given time interval was calculated by dividing the number of beam crossings by wake time (in minutes). In pilot studies, locomotor activity was also assessed by measuring distance traveled/minute inside an experimental chamber (Putz and Heisenberg, 2002). We found no difference between Δ50M amorphs and hypomorphs (EP3517 = 82 ± 10 mm/min, n = 32; Δ50M = 67 ± 7 mm/min, n = 26; p = 0.164 Mann–Whitney test).
Sleep deprivation (SD) was performed as described previously (Huber et al., 2004). Briefly, during SD flies remained in the DAMS monitor, which was placed vertically inside a framed box able to rotate along its major axis under the control of a motor. Since locomotor activity during SD was continuously recorded, the extent of sleep loss could be calculated for each individual fly. Only flies that during SD lost >90% of their baseline sleep were included in the analysis.
Statistical differences in sleep parameters were assessed by Kruskal–Wallis test (p < 0.05), followed by Mann–Whitney rank sum test for post hoc analysis. All charts show mean ± SE.
Complementation analysis.
The Δ113M allele was outcrossed into the w1118 background using the same crossing strategy that generated outcrossed Δ50M. When testing Δ50M with Δ113M, heterozygous flies (w1118; Δ50M/EP3517 × w1118; Δ113M/EP3517) were crossed together to produce white (Δ50M/Δ113M), medium red (w1118; Δ50M/EP3517 or w1118;Δ113M/EP3517), and dark red (w1118; EP3517) eyed progeny.
To test Df(3R)6265 in combination with Δ50M, females heterozygous for deficiency (w1118; Df(3R)Exel6265/+) were crossed to heterozygous Δ50M males (w1118; EP3517/Δ50M). This cross produced dark red (w1118; EP3517/Df(3R)Exel6265), medium red (Df(3R)Exel6265/Δ50M or EP3517/+), and white (+/Δ50M) eye progeny. After testing sleep, flies were harvested and genotype was determined by PCR. Primer pairs 157F (caacactttttgcggctgta) to 185R (gttcggttcgtcgacttcat) were used to specifically amplify the wild-type template without the EP3517 insertion; 160F (cggtgagctgtcacttagca) to Sp1(acacaacctttcctctcaacaa) were used to amplify the template that had EP3517; 81F (atactataccgctgcgaagagc) to 82R (caggaacacagagcttgagaaa) were used to amplify sequences outside the dFmr1 region to confirm that the template was present.
Controversy exists over the semilethal phenotype of the dFmr1 null alleles, which was described for Δ50M and Δ113M but not for other amorphic dFmr1 alleles (Dockendorff et al., 2002; Morales et al., 2002). Our crosses between Δ50M/EP3517 heterozygous parents confirmed the semilethal phenotype, because only 30% (n = 1250, p < 0.05 χ2 test) of the expected Δ50M amorph males and 19% (n = 897, p < 0.05 χ2 test) of the expected Δ50M amorph females eclosed. The number of eclosed flies is higher than previously reported, because eclosure rate is compared relative to progeny that inherited EP3517, which also has a semilethal phenotype in males (89% eclosed, n = 599, p < 0.05 χ2 test) and females (63% eclosed, n = 667, p < 0.05 χ2 test). Although the semilethal phenotype could result from a second site mutation (Gao, 2002), this is unlikely because (1) this mutation would have to be present in all the described null alleles with a semilethal phenotype; (2) hypomorphs such as EP3517 flies also have reduced semilethality, suggesting that the strength of this phenotype depends on the reduced expression of dFmr1. The Δ113M/Δ50M combination was also recessive lethal, with only 22% (n = 195, p < 0.05 χ2 test) of the expected progeny eclosing.
Western blots.
Flies were harvested at the indicated time and frozen immediately at −70°C. Flies were then selected and pooled in groups of 4 based on their behavioral state before harvesting. Proteins were extracted from fly heads (4 flies/group) after homogenization in 20 μl Tris-Triton (10 mm Tris, pH 7.4, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate). After adding 20 μl Lammeli's buffer the protein extract was boiled at 95°C for 5 min and the sample was loaded (13 μl) into two separate lanes (technical duplicates) on a 4–20% polyacrylamide gel (Bio-Rad), separated by Tris-HCl gel electrophoresis (Bio-Rad) in 1× tris/Glycine/SDS Buffer (Bio-Rad), and transferred to 0.45 μm pore size nitrocellulose membrane (Bio-Rad) in 1× Tris base/Glycine/Methanol blotting buffer. Blots were blocked for 1 h at room temperature (RT) in freshly made 5% nonfat dry milk (Bio-Rad) in 1× TBS with 0.01% Tween 20 (Sigma). After washing (1× TBS, 0.01% Tween 20), blots were incubated with primary antibody (1× TBS, 0.01% Tween 20, 5% BSA) overnight at 4°C, washed again (1× TBS, 0.01% Tween 20), and incubated at RT with secondary antibody (horseradish peroxidase conjugated anti-mouse IgG antibodies, Millipore) in 1× TBS, 0.01% Tween 20, 5% nonfat dry milk. Immunoreactive bands were detected using autoradiographic enhanced chemiluminescence (ECL-Plus, Amersham) and captured using the Typhoon 9410 Variable Mode Imager (Amersham). Band intensities were quantified using ImageQuant software (Amersham). All time points were tested at least in two independent experiments on two different blots. All blots were loaded with a common protein extract from Drosophila heads to allow normalization and comparison across blots. Primary antibodies (Developmental Studies Hybridoma Bank, University of Iowa) included 5A11 (Okamura et al., 2004) and used a 1:1000 dilution to detect Drosophila FMRP (DFMRP) and E7, and a 1:2000 dilution to detect β-tubulin as a loading control. Statistical analysis was based on Student's unpaired t test between samples (p < 0.05).
Lifespan.
Lifespan was measured by collecting newly eclosed flies, placing them in single DAMS tubes and then recording their sleep/waking cycle at 20°C until death. Flies were transferred to new tubes with fresh food every week. Lifespan curves were generated by plotting the daily percentage of survivorship as a function of time in days.
Circadian analysis.
Parents and progeny were reared in 12 h light/dark conditions. Flies were harvested within 8 h of eclosure, separated into monitor tubes, kept in 12 h light/dark conditions for 6 d, and then kept in constant darkness (DD). Autocorrelations were done using standard software (Levine et al., 2002).
RU486 and MPEP treatment.
Flies treated with RU486 were housed in monitor tubes with the regular cornmeal molasses food containing 500 μm RU486. Flies were kept in the conditions previously described and transferred to tubes with fresh media each week during the first 3 h of the light phase. The RU486 was dissolved in 80% ethanol before adding it to the melted (60°C) cornmeal molasses food. Control food was made by adding the same amount of 80% ethanol. 2-Methyl-6-(phenylethynyl)pyridine (MPEP)-treated flies were housed in monitor tubes containing media at one end consisting of 86 μm MPEP, 5% sucrose and 2% agar. Vehicle-treated flies were housed in monitor tubes containing 5% sucrose and 2% agar.
Results
As dFmr1 expression increases, daily sleep amount decreases
Different sleep abnormalities have been described in patients with fragile X syndrome, including reduced time spent in REM sleep and increased sleep fragmentation (Musumeci et al., 1995; Miano et al., 2008). Studies in mammals, however, are complicated by the presence of two paralogues to Fmr1, FXR1 and FXR2, which could compensate for the loss of Fmr1. We therefore focused on Drosophila, where only one gene, dFmr1, is present. We compared sleep throughout the entire lifespan in a full spectrum of mutants ranging from flies that had complete loss of dFmr1 expression (amorphs) to flies with increased dFmr1 expression (hypermorphs). All alleles were outcrossed into a w1118 background before testing (see Materials and Methods). Flies were monitored from 0 to 8 h after eclosure until they died, but results are shown for the first 42 d of life, after which not enough flies survived in all mutant classes to allow comparisons. Almost every day after eclosure both males and females with the loss of function mutation Δ50M (amorphs) slept significantly more than wild-type flies (Fig. 1A). On average, throughout the 42 d period, daily sleep amount increased relative to wild-type flies by 4 h in males and 3 h in females (Fig. 1B). In males, the increase in daily sleep amount peaked on day 12 after eclosure (58%), while in females the largest increase was observed at day 7 (32%) and at day 41 (45%) (Fig. 1C).
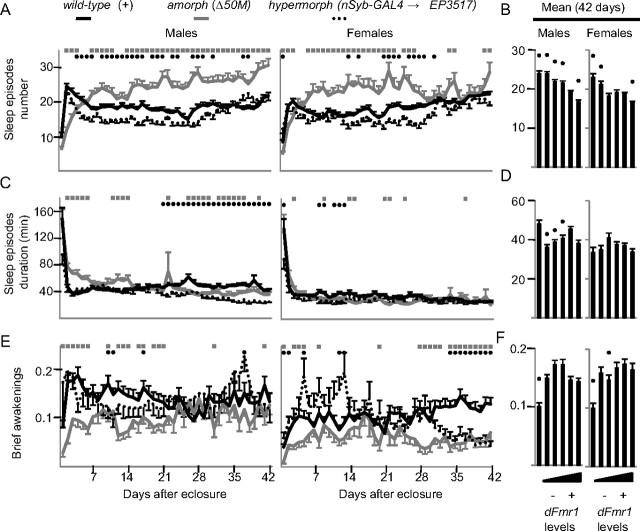
Figure 1.
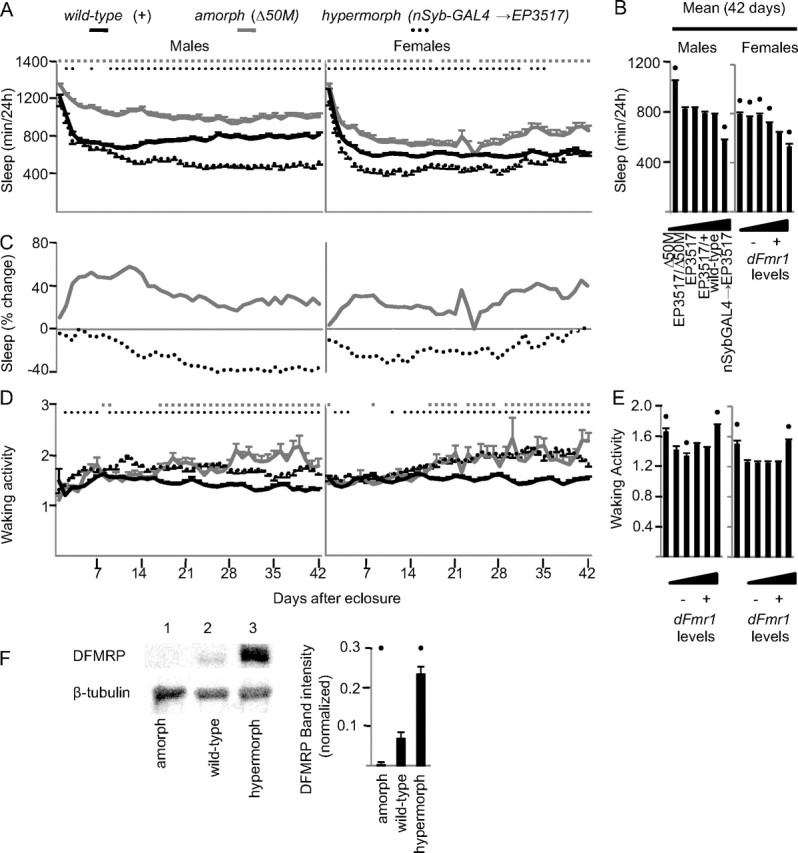
Inverse relationship between dFmr1 levels and daily sleep amount. A, B, Daily sleep amount from eclosure to day 42 and average values for the entire 42 d period. Allele combinations are arranged from left to right based on increasing levels of dFmr1 expression. C, Percentage change in sleep amount compared with wild-type flies over the first 42 d after eclosure. D, E, Waking activity (number of beam crossing/min) for the first 42 d after eclosure and average values for the entire 42 d period. Statistics were based in all cases on Kruskal–Wallis test for multiple comparisons, followed by post hoc analysis using Mann–Whitney test to compare mutant genotype to wild type. In A and D, significant differences (p < 0.05) relative to wild type are indicated by gray squares (amorphs) and black circles (hypermorphs). In B and E, black dots indicate significant differences relative to wild type. Number of flies for each genotype for day 1 and day 42 were as follows: males: Δ50M, n = 60/34; EP3517/Δ50M, n = 73/49; EP3517, n = 112/81; EP3517/+, n = 68/56; dFmr1+, n = 112/92; nSyb-GAL4->EP3517, n = 47/39; females: Δ50M, n = 45/21; EP3517/Δ50M, n = 51/25; EP3517, n = 85/56; EP3517/+, n = 62/56; dFmr1+, n = 117/102; nSyb-GAL4→EP3517, n = 38/32. F, DFMRP levels in amorphs, hypermorphs, and wild-type flies as measured by Western blot (β-tubulin was used as loading control). A black dot indicates a significant difference (p < 0.05) from wild type as determined by Student's t test.
dFmr1 overexpression was driven from the EP element EP3517, which is inserted in the dFmr1 promoter region, using the pan-neuronal driver nSyb-GAL4. On most days, both male and female dFmr1 hypermorphs had reduced daily sleep amount relative to wild-type flies, but the effect was no longer present in females after day 35 (Fig. 1A). In males, the decrease in daily sleep amount became progressively larger after eclosure and peaked after ∼4 weeks of age (−40%), while in females the effect was present immediately after eclosure and peaked at day 10 (−31%) (Fig. 1C). On average, throughout the 42 d period, daily sleep amount decreased relative to wild-type flies by >3 h in males and 2 h in females (Fig. 1B). Western blots confirmed the loss of DFMRP in amorphs, and its increased levels in hypermorphs (Fig. 1F).
We also tested different combinations of hypomorphic (partial loss of function) dFmr1 mutations, as shown in Figure 1B, where the dFmr1 allele combinations are arranged left to right from the lowest to the highest dFmr1 expression. As dFmr1 expression increased, the average daily sleep amount over the 42 d period decreased in both sexes. However, females appeared to be more sensitive than males to even small changes in dFmr1 levels. Specifically, the amorph mutation (Δ50M) similarly increased the average daily sleep amount in males (34%) and females (29%) compared to wild type. In males, however, the next strongest hypomorph (EP3517/Δ50M) showed only a 6% (p = 0.5) increase in sleep amount, while in females, the EP3517/+ hypomorph, which has a small loss in dFmr1 expression, still showed an 11% increase in daily sleep amount. Moreover, the sleep phenotype of EP3517/+ females was intermediate between wild-type and amorph after eclosure, but became closer to the amorph phenotype after 2 weeks of age (data not shown).
In contrast to sleep, waking locomotor activity increased relative to wild-type in both amorphs and hypermorphs, and did not consistently change in the hypomorphs (Fig. 1D,E). Thus, the effects of dFmr1 mutations on the sleep phenotype are dissociable from the effects on locomotor activity. Moreover, the increased waking activity in amorphs is consistent with previous reports (Dockendorff et al., 2002) and indicates that their long sleep is unlikely to be a sign of sickness.
dFmr1 levels consistently affect the number but not the duration of sleep episodes
To further characterize the changes in sleep behavior, we analyzed the number and duration of sleep episodes in dFmr1 mutants. In wild-type flies, the number of sleep episodes rose sharply during the first 2 d after eclosure, declined within the first week, and then stabilized (Fig. 2A). The number of sleep episodes in amorphs, instead, increased after eclosure and remained higher than in wild-type flies for most of the 42 d period (Fig. 2A). Hypermorphs followed the same trend as in wild-type flies, but with a more pronounced decrease after the first week that continued until approximately day 36. Averaged across 42 d after eclosure, the inverse relationship between sleep episode number and dFmr1 expression was significant for both sexes (Fig. 2B). Sleep episode duration instead did not show consistent changes (Fig. 2C,D). Of note, male amorphs had longer sleep episodes than wild-type flies during the first week after eclosure, which accounts for their increased daily sleep amount during this period (Fig. 1A) despite a decrease in the number of sleep episodes (Fig. 2A). Thus, changes in sleep amount in dFmr1 mutants are primarily due to changes in the number of sleep episodes.
Figure 2.
Effects of changes in dFmr1 expression on sleep parameters. Daily values from eclosure until day 42 (A, C, E) and means across the entire 42 d period (B, D, F). The number of brief awakenings is per hour of sleep. Number of flies and statistics are as in Figure 1.
Deep and consolidated sleep, such as sleep occurring after sleep deprivation, is characterized by a reduction in the number of brief awakenings (Huber et al., 2004). Throughout their lifespan, amorphs often had fewer brief awakenings per day than wild-type flies (Fig. 2E), an effect that over the entire 42 d period resulted in a significant decrease (−30% in males, −42% females) (Fig. 2F). dFmr1 overexpression instead did not consistently affect the number of brief awakenings.
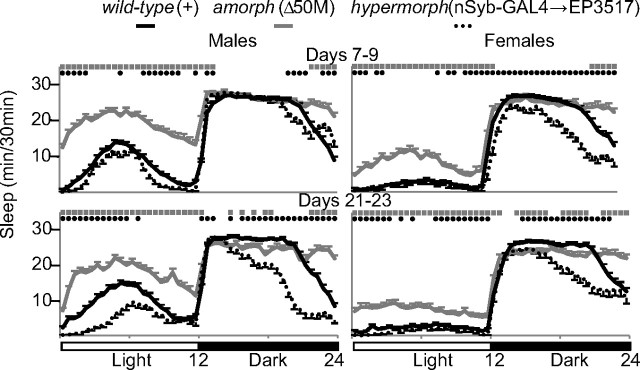
Drosophila sleep mainly at night, although males also tend to have a “siesta” during the day (Huber et al., 2004). Loss of dFmr1 expression increased the daily sleep amount both by increasing sleep during the day, and by prolonging sleep at the end of the dark phase (Fig. 3). Moreover, while wild-type flies wake up in anticipation of light onset, amorphs continue to sleep during this period. Opposite effects are seen with dFmr1 overexpression, which decreased sleep during the latter half of the night and, in males, also during the day (Fig. 3).
Figure 3.
Effects of changes in dFmr1 expression on the diurnal sleep pattern. Hypnograms illustrating sleep amount (min/30 min) throughout the 24 h period. Sleep amounts were averaged over days 7–9 (top) and days 21–23 (bottom). Number of flies and statistics are the same as in Figure 1.
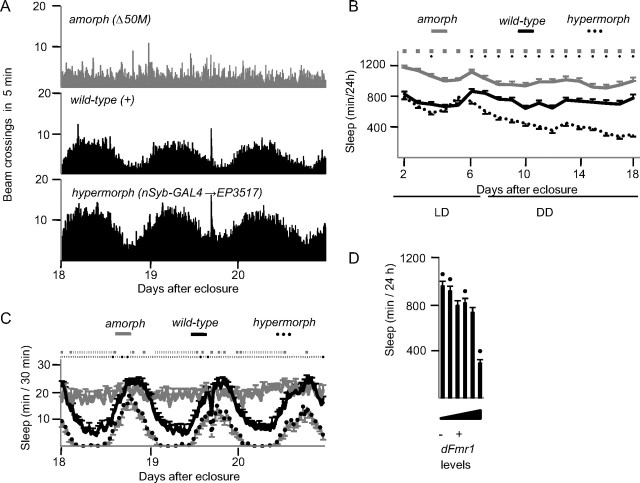
The effects of dFmr1 mutations on sleep persist in constant darkness
Previous reports found that dFmr1 amorphs are arrhythmic in constant darkness, while flies overexpressing dFmr1 using a tim-GAL4 driver had a lengthened period (Dockendorff et al., 2002; Morales et al., 2002). We maintained mutant flies in 12 h LD conditions for 6 d after eclosure, and then switched them to DD for 2 weeks. Figure 4A shows locomotor activity plots during the last 3 d in DD, and confirms the lack of rhythmicity in amorphs [rhythmicity index (RI) = 0.17] and the persistence of a strong rest/activity rhythm in both wild-type flies (RI = 0.6) and hypermorphs (RI = 0.63). The inverse relationship between dFmr1 expression and daily sleep amount persisted in DD (Fig. 4B,C). Specifically, amorphs showed a similar increase in daily sleep amount relative to wild-type in both LD (+32%; mean of days 18–20) and DD (+31%), suggesting perhaps a ceiling effect. As expected due to the arrhythmic phenotype, amorphs slept at all times of day and night (Fig. 4C). Hypomorphs had a stronger increase in sleep amount relative to wild-type in DD than LD (EP3517/Δ50M, +25% vs + 2%; EP3517, +8% vs + 4%; EP3517/+, +11% vs + 1%; mean of days 18–20) (Fig. 4D). Hypermorphs also showed a stronger sleep phenotype in DD than LD (−59% vs −26%, mean of days 18–20) (Fig. 4D). Thus, the effects of dFmr1 on sleep continue and in some cases are even stronger in constant darkness, and persist when circadian rhythmicity is lost.
Figure 4.
Inverse relationship between dFmr1 levels and daily sleep amount in constant darkness. Flies were kept in a 12 h LD cycle until day 6 after eclosure, then kept in DD until day 20. A, Number of beam crossings in 5 min intervals for the last 3 d in DD (days 18–20). B, Daily sleep amount in LD and DD. C, Sleep pattern during the last 3 d in DD. In B and C, significant differences (p < 0.05) relative to wild type are indicated by gray squares (amorphs) and black circles (hypermorphs). D, Daily sleep amount averaged over the last 3 d in DD for all genotypes tested (arranged from left to right as in Fig. 1). A black circle indicates significant difference from wild type. Number of flies: Δ50M, n = 29; EP3517/Δ50M, n = 34; EP3517, n = 59; EP3517/+, n = 39; dFmr1+, n = 35; nSyb-GAL4→EP3517, n = 29.
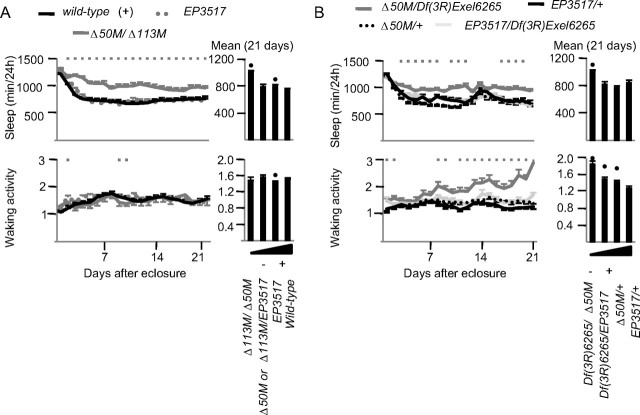
Complementation analysis confirms the link between decreased dFmr1 expression and increased sleep amount
Our crossing scheme, designed to place dFmr1 alleles in a common genetic background, permits the independent segregation of loci that are not closely linked to dFmr1. However, to completely rule out the possibility that a closely linked mutation next to the dFmr1 locus was responsible for the sleep phenotype we also tested the null alleles Δ113M and Df(3R)6265 in combination with Δ50M. If the loss of dFmr1 is responsible for the increase in daily sleep amount, other amorphic combinations that result in no wild-type dFmr1+ allele being present should also produce a long sleep phenotype. Indeed, Δ113M failed to complement Δ50M, since Δ113M/Δ50M heterozygous flies showed a long sleep phenotype relative to wild-type flies (Fig. 5A). Waking activity in Δ113M/Δ50M heterozygous flies was instead similar to that in wild-type flies (Fig. 5A).
Figure 5.
Complementation analysis. A, Daily sleep amount and waking activity for the first 21 d after eclosure in Δ50M/Δ113M recombinants (gray line), EP3517 homozygotes (gray dots), and wild-type flies (black line). Average values over the 21 d period are shown on the right bar graphs. Numbers of flies: Δ50M/Δ113M, n = 20; Δ50M/EP3517 or Δ113M/EP3517 combined, n = 34; EP3517, n = 33; dFmr1+, n = 112. Kruskal–Wallis test followed by Mann–Whitney test; square boxes indicate days when Δ50M/Δ113M is significantly different from wild type. Black dots in the bar graphs indicate significant difference (p < 0.05 Mann–Whitney) from wild type. B, Daily sleep amount and waking activity for the first 21 d after eclosure after combining Δ50M with Df(3R)Exel6265. After recording sleep data, flies were genotyped by PCR: EP3517/Df(3R)Exel6265, n = 28; Df(3R)Exel6265/Δ50M, n = 28; EP3517/+, n = 28; +/Δ50M, n = 32. Kruskal–Wallis test followed by Mann–Whitney test. Gray boxes indicate significant difference between Df(3R)Exel6265/Δ50M and EP3517/+ (no other differences between genotypes were found). Black dots in the bar graphs indicate significant difference (p < 0.05 Mann-Whitney) from EP3517/+.
Both Δ50M and Δ113M were produced by mobilizing the EP3517 insertion, and thus may share common genetic variations from the original chromosome. To test a dFmr1 null allele from a completely independent source, we crossed Δ50M with Df(3R)Exel6265, a small deletion (85F10–85F16) that includes the dFmr1 locus (Wilson et al., 2008). Consistent with the previous results, Df(3R)Exel6265/Δ50M flies showed increased daily sleep amount, as well as higher waking activity levels, relative to control flies (EP3517/+ heterozygotes) (Fig. 5B). Thus, the complementation results support the conclusion that the loss of dFmr1 expression is responsible for the long sleep phenotype. The alternative possibility, such as a second-site mutation causing the increased sleep amount, is very unlikely, because the amorphic alleles tested here were derived from independent sources and should not have second-site mutations in common.
Both loss and gain in dFmr1 expression result in loss of sleep homeostasis
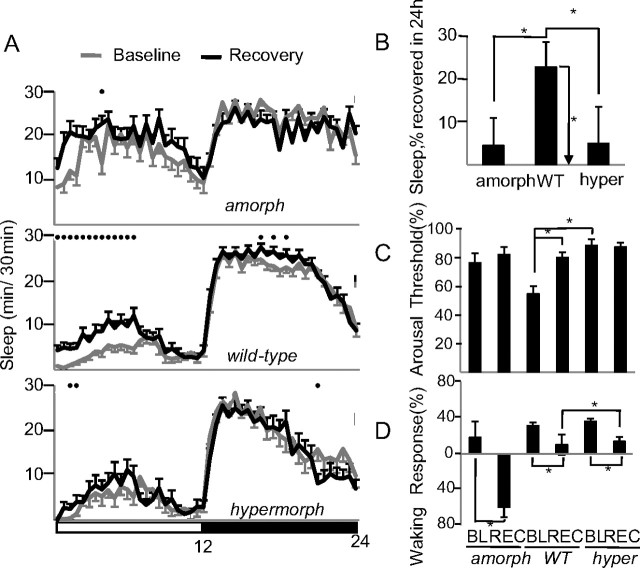
In flies, as in mammals, sleep duration and sleep intensity increase after sleep deprivation to compensate for sleep loss (Huber et al., 2004). As expected, after 24 h of sleep deprivation wild-type males slept longer during the first 6 h of recovery sleep [54 min (recovery − baseline), p < 0.05], while time asleep did not increase significantly in amorphs (47 min, p = 0.1) and hypermorphs (15 min, p = 0.5) (Fig. 6A). Across the entire 24 h period following sleep deprivation wild-type flies recovered 23% of the sleep lost, while dFmr1 mutants did not recover a significant amount of sleep (Fig. 6B). The duration of sleep episodes, a measure of sleep intensity, also increased during the first 6 h of recovery sleep in wild-type flies, but not in amorphs and hypermorphs (data not shown).
Figure 6.
Effects of changes in dFmr1 expression on the response to sleep deprivation. A, Hypnograms displaying sleep patterns over a 24 h period the day before sleep deprivation (baseline) and the day after 24 h of sleep deprivation (recovery). Only flies (all males) that lost >90% of their sleep during sleep deprivation were included in this analysis. Amorphs (n = 18) are Δ50M homozygotes, wild-type (n = 12) are dFmr1+ homozygotes, and hypermorphs (n = 30) have nSyb-GAL4 driving EP3517. Black circles indicate significant differences between baseline and recovery (p < 0.05 Mann–Whitney). B, Amount of sleep recovered during the entire 24 h of recovery expressed as percentage of sleep lost during sleep deprivation (same flies as in A). Horizontal bars indicate differences between mutants and wild type (*p < 0.05, Mann–Whitney). Only wild-type flies showed a significant increase in sleep duration during recovery relative to baseline (vertical bar, p < 0.05 signed rank test). C, Arousal thresholds measured as the percentage of sleeping flies that react to a complex stimulus of low intensity (Huber et al., 2004; Cirelli et al., 2005) (*p < 0.05, Mann–Whitney). D, Waking performance measured as the percentage increase in the number of beam crossings during the minute following the delivery of the stimulus relative to the minute before the stimulation. All flies had been active (i.e., awake) during the minute before the delivery of the stimulus (*p < 0.05, Mann–Whitney). In C and D, the number of flies (all males) is amorphs = 27, wild-type = 62, hypermorphs = 74. Flies were tested during the first 6 h of the light period in baseline (BL) and recovery (REC).
It was previously found that dFmr1 amorphs respond to odors and light similarly to flies with a wild-type dFmr1+ transgene (McBride et al., 2005). We measured the escape response to a complex stimulus, which included noise, vibration, and shadow, during the first 6 h of the light period before (baseline) and after sleep deprivation (recovery). In agreement with previous studies (Huber et al., 2004; Cirelli et al., 2005), when the stimulus was applied to sleeping wild-type flies, ∼55% of them failed to respond during baseline, and the number of nonresponders increased to ∼80% after sleep deprivation, an indication that postdeprivation sleep is deeper (with higher arousal thresholds) (Fig. 6C). In contrast, most amorphs (77%) and hypermorphs (89%) already failed to respond to the same stimulus before sleep deprivation, and the number of responders did not change afterward. This suggests that dFmr1 mutants have deeper sleep than wild-type flies in baseline conditions, and a further increase in arousal threshold may be difficult to measure due to a ceiling effect.
In a different experiment, the same complex stimulus was applied during waking. As expected, in wild-type flies, the stimulation triggered a smaller locomotor response after sleep deprivation relative to baseline (Fig. 6D), an indication that sleep deprivation impaired performance. Both amorphs and hypermorphs also showed smaller responses after sleep loss relative to baseline, but the response in sleep deprived hypermorphs was significantly bigger than in sleep deprived wild-type flies (Fig. 6D), suggesting that the former were less affected by sleep loss. In summary, despite the significant difference in daily sleep amounts amorphs and hypermorphs responded similarly to sleep deprivation: they both failed to show a homeostatic sleep response (no changes in sleep duration or sleep intensity), and they both showed deficits in waking performance, although amorphs may be more affected than hypermorphs.
dFmr1 expression levels depend on both age and experience
The effects that dFmr1 loss or gain have on sleep depend on developmental stage (Figs. 1 and 2), consistent with the fact that dFmr1 expression is regulated in an age-dependent manner (Tessier and Broadie, 2008). We assayed DFMRP protein levels in fly heads at 1, 3, 7, 14, and 21 d after eclosure to confirm that dFmr1+ expression is developmentally regulated. As expected, DFMRP was never detected in amorphs (Fig. 7A). In both wild-type and hypermorphs, DFMRP levels were highest immediately after eclosure, declined steeply on day 3, reached the lowest levels 7 d after eclosure (>10-fold decrease relative to eclosure, ∼2-fold decrease relative to day 3), and then remained at very low levels afterward (Fig. 7A). Hypermorphs had increased expression compared with wild-type flies on all tested days, with the smallest change on day 1 (+275%, p = 0.06), and the biggest difference on day 21 (+312%, p < 0.05).
Figure 7.
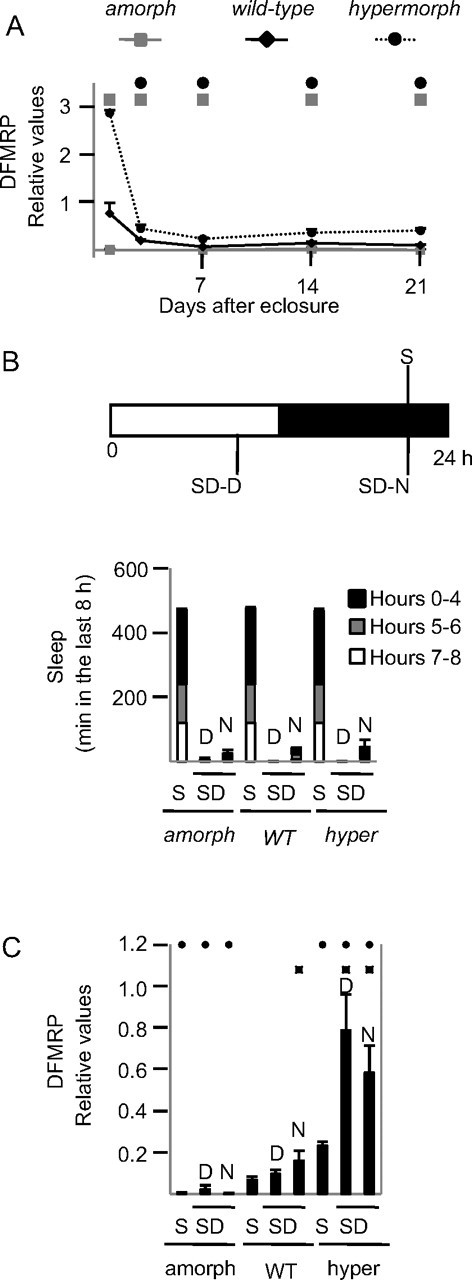
Changes in DFMRP levels due to developmental stage and behavioral state. A, Relative dFmr1 levels in sleeping males 1, 3, 7, 14, and 21 d after eclosure. Males were harvested 8 h after lights off and selected for maximal sleep (asleep >90% of the previous 8 h). To exclude potentially sick flies, mean waking activity (crossings/min of waking) the day before harvesting was required to be >0.7. B, The three experimental groups. C, Average sleep amounts during the last 8 h before harvesting. S flies were required to spend >90% of the previous 8 h asleep. SD-D flies were required to stay awake >90% of the last 8 h, with no sleep in the last 4 h before harvesting. Since flies mainly sleep at night SD-N is more difficult to enforce than SD-D. Criteria for SD-N flies were therefore less stringent: > 80% awake over the last 8 h, with <13% of sleep in the last 2 h before harvesting. Mean waking activity the day before harvesting was >0.7 in all flies. C, Relative DFMRP expression levels as measured in Western blots. Black squares indicate, within the same genotype, significant differences relative to S flies (p < 0.05, Student's t test). Black circles indicate, within the same behavioral state, significant differences relative to wild-type (p < 0.05, Student's t test).
A previous study found that DFMRP expression does not change over the 24 h cycle (Inoue et al., 2002), but these experiments did not control for behavioral state. Experiments in mammals, however, have found that enriched experience and sensory stimulation increase Fmr1 expression (Todd and Mack, 2000; Todd et al., 2003a). We therefore measured DFMRP levels in three groups of 7-d-old male flies. One group of sleeping flies (S) was collected at night (4 A.M.) after 8 h of sleep, and two groups of sleep deprived flies were collected after being kept awake for 8 h either at night (SD-N, 4 A.M.) or during the day (SD-D, 4 P.M.). As expected, S flies spent most of the 8 h before harvesting asleep, whereas SD flies were awake at least 90% of the time (Fig. 7B). DFMRP levels increased in both wild-type and hypermorph SD flies relative to S flies. The increase was significant after sleep loss during the night in wild-type flies (percentage increase relative to S, SD-D = 41%, p = 0.2; SD-N = 132%, p < 0.05), and after sleep loss during both day and night in hypermorphs (SD-D = 237%, p < 0.05; SD-N = 150% p < 0.05). Thus, DFMRP expression levels increase with waking experience independent of time of day or light. Moreover, the increase appears to depend on dFmr1 expression levels, since the increase was higher in hypermorphs than in wild-type flies.
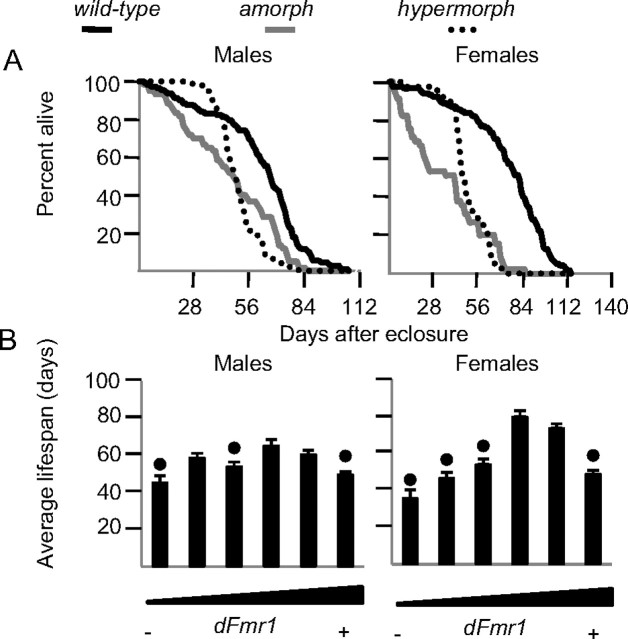
Either increasing or decreasing dFmr1 expression reduces lifespan
Amorph and hypermorph dFmr1 flies of both sexes had decreased lifespan, but with a different time course (Fig. 8A). Specifically, male and female amorphs lacked the initial lag phase that occurs before wild-type flies start dying at a logarithmic rate. In contrast, the lag phase in hypermorphs was short, followed by a steep logarithmic death rate. Interestingly, similar to the changes in sleep, the effects on lifespan were more pronounced in females than males (Fig. 8B). Except for EP3517/+ heterozygotes, other hypomorph flies also tended to have reduced lifespan, and more so in females than males (Fig. 8B). Thus, an intermediate level of dFmr1 expression seems to be optimal for longevity. Moreover, at least in hypermorphs, dFmr1 must affect lifespan through the brain, because its overexpression was neuron-specific. In humans decreased (<7 h) and increased (>8 h) sleep is associated with an increased risk of mortality (Hublin et al., 2007), but whether dFmr1 effects on lifespan are mediated by its effects on sleep is unknown.
Figure 8.
Effects of changes in dFmr1 expression on lifespan. A, Longevity curves for amorph (Δ50M), wild-type (dFmr1+), and hypermorphic (nSyb-GAL4 driving EP3517) flies. B, Average lifespan for all male and female dFmr1 mutants tested. Genotypes are arranged from left to right in order of increasing dFmr1+ expression as in Fig. 1 (same number of flies). Black dots indicate significant differences from wild type (p < 0.05, Mann–Whitney test).
Overexpression of dFmr1 in adult flies also decreases daily sleep amount
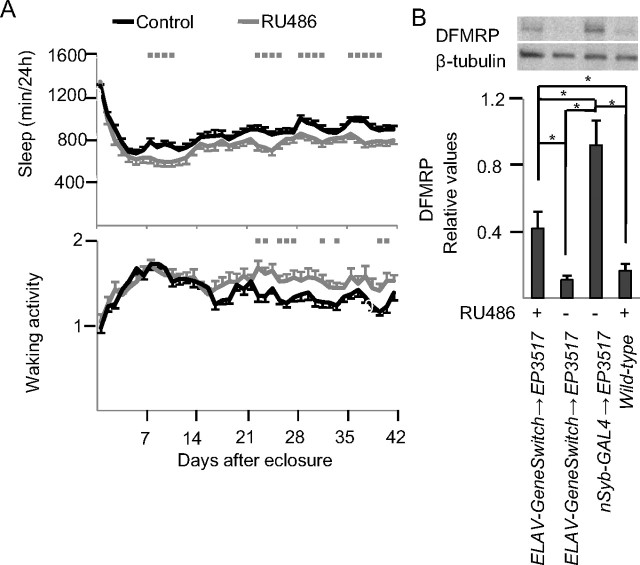
The effects on sleep described so far were due to constitutive changes in dFmr1 levels. To determine whether dFmr1 overexpression could decrease daily sleep amount when induced after eclosure we used the ELAV-GeneSwitch driver to drive expression pan-neuronally only when the progesterone analog RU486 is added to the media (Osterwalder et al., 2001). Relative to flies of same genotype that were fed control media, flies fed RU486 media slept significantly less (RU486 = 776 ± 23 min; control = 883 ± 19 min, p < 0.05 Mann–Whitney) and tended to have higher waking activity (RU486 = 1.50 ± 0.047 beam crossings per waking min; control = 1.39 ± 0.027 beam crossing per waking min, Mann–Whitney p = 0.14) (Fig. 9A). The decrease in daily sleep amount, however, was not as pronounced as that previously observed using the constitutive nSyb-GAL4 driver (Fig. 1). Western blots analysis showed that DFMRP levels were increased using both drivers, but the difference relative to wild-type levels was >2-fold larger when using the nSyb-GAL4 system (Fig. 9B). Thus, the greater effects on sleep of constitutive, relative to inducible, dFmr1 overexpression may be due to the higher levels of DFMRP reached in the first condition, although other effects due to dFmr1 miserexpression during early development cannot be excluded. In any case, overexpression of dFmr1 can decrease daily sleep amount independent of developmental changes before eclosure, since it is also effective when induced in adult flies.
Figure 9.
Sleep effects of dFmr1 overexpression in the adult. A, Daily sleep amount and waking activity from eclosure to day 42 and average values for the entire 42 d period. All flies shared the same genotype (w1118; ELAV-GeneSwitch/EP3517), but their food contained either vehicle (control) or RU486. Gray squares (in A) indicate significant difference from controls (p < 0.05, Mann–Whitney test). Number of flies for day 1 and day 42 were as follows: RU486 = 26/24, Control = 36/29. B, Western blot analysis using an antibody that binds DFMRP (β-tubulin was used as loading control).
The mGluR antagonist MPEP does not rescue the long sleeping phenotype in adult amorphs
It has been demonstrated that several phenotypes associated with the loss of dFmr1 are due to abnormally increased activity of the metabotropic glutamate receptors (mGluRs) (Bear et al., 2004). Consistent with this hypothesis, it was shown that treating adult dFmr1 amorphs with mGluR antagonists rescues defective courtship memory (McBride et al., 2005). In the same flies, however, the treatment failed to rescue the defect in circadian rest/activity rhythms and the morphological defects in the mushroom bodies (McBride et al., 2005). As shown above, we found that inducing dFmr1 in adult flies is sufficient to reduce sleep amount. Thus, we wanted to determine whether MPEP, a specific inhibitor of mGluRs, can reduce sleep in adult flies and rescue the long-sleeping phenotype in amorphs. After eclosure, flies were tested for 2 d on cornmeal molasses media and then transferred to sucrose media containing either vehicle or 86 μm MPEP, a concentration previously demonstrated to rescue behavioral defects (McBride et al., 2005). Treatment with MPEP had no effect on the long sleeping phenotype of delta50M amorphs (Fig. 10A). MPEP also did not affect sleep in the hypomorphs (EP3517), which sleep normally compared with wild-type flies (Fig. 10A). MPEP treatment instead increased waking activity levels in EP3517 flies over the first 4 d of treatment, and motor activity also tended to increase in the amorphs (Fig. 10B). Thus, blocking mGluR signaling in adult flies cannot rescue the delta50M long sleeping phenotype.
Figure 10.
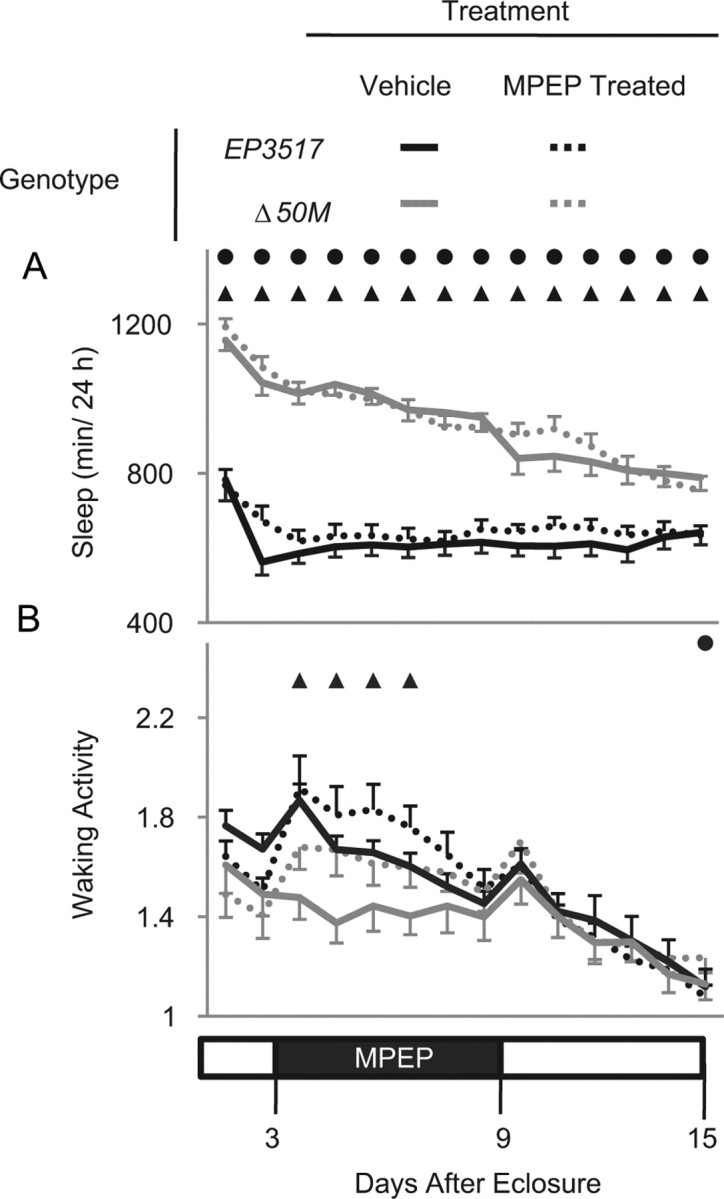
MPEP, an mGluR antagonist, does not rescue the long sleeping phenotype due to loss of dFMR1. Age-matched flies, harvested within 24 h of eclosure, were tested on cornmeal molasses media for 2 d. On the third day (at lights on), flies were transferred to tubes with sucrose media containing vehicle or 86 μm MPEP, tested for 6 d, and then transferred to fresh sucrose media and monitored for seven additional days. A, Daily sleep amount (min/24 h) before, during, and after MPEP treatment. B, Waking activity (number of beam crossings/min) before, during, and after MPEP treatment. Solid circles indicated significant difference between MPEP-treated Δ50M amorphs and MPEP-treated EP3517 hypomorphs; triangles represent significant difference between untreated Δ50M amorphs and untreated EP3517 hypomorphs (p < 0.05, Mann–Whitney test). Number of flies were as follows: MPEP-treated EP3517 = 48, MPEP-treated Δ50M = 39, untreated EP3517 = 41, untreated Δ50M = 32.
Overexpression of dFmr1 in the mushroom bodies is sufficient to decrease daily sleep amount
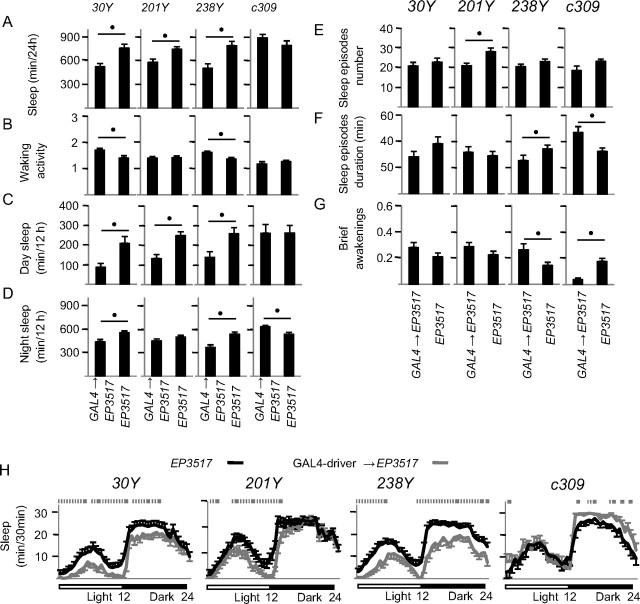
To start investigating whether the sleep effects of dFmr1 overexpression are region-specific we focused on the mushroom bodies, a large area of the fly brain where dFmr1 is necessary for normal synaptic pruning (Tessier and Broadie, 2008), and where mutations that affect neuronal activity are also able to affect sleep (Joiner et al., 2006; Pitman et al., 2006). We crossed EP3517 with 4 GAL4 lines (30Y, 201Y, 238Y, c309) known to drive expression in the mushroom bodies, as well as in other discrete brain regions (Yang et al., 1995; Joiner and Griffith, 1999; Kurusu et al., 2002; Joiner et al., 2006). Sleep parameters were averaged across days 15–19 after eclosure, because by this time the pan-neuronal overexpression of dFmr1 produces a robust short-sleeping phenotype (Fig. 1A,B). In combination with EP3517, 201Y, 30Y and 238Y reduced sleep (−22%, −31%, −36%, respectively; p < 0.05), while c309 tended to increase sleep (Fig. 11A). By comparison, during the same time period, the pan-neuronal overexpression of dFmr1 produced a 37% decrease in sleep time relative to EP3517 flies. Importantly, inheriting the GAL4-drivers alone did not alter the sleep phenotype, as measured by comparing siblings that inherited the driver relative to w1118 from the same crosses (Mann–Whitney test; data not shown). Similar results were obtained when sleep was averaged across the first 19 d after eclosure (rather than days 15–19 only), except that the increase in daily sleep time with c309 became significant (13%, p < 0.05). 30Y, 201Y, 238Y reduced sleep primarily during the day, while c309 increased sleep only at night (Fig. 11C,D). Further analysis of sleep episodes and brief awakenings (Fig. 11E–G) showed that c309 consolidated sleep into longer bouts, while the effects of 30Y, 201Y, 238Y were neither consistent nor profound. As before, the effects on sleep and waking activity could be dissociated, because the latter did not change with 201Y or c309 but increased with 30Y and 238Y (Fig. 11B).
Figure 11.
Regional specificity of the effects of dFMR1 on sleep duration. Males flies were generated by crossing w: GAL4-driver/+ to w: EP3517/+. Progeny were selected based on eye color and genotype confirmed using PCR after testing. A, Sleep time (min/24 h) for each of the 5 d tested (days 15–19 posteclosure). B, Waking activity represents the number of beam crossings per minute. C, D, Sleep time (min/12 h). E, F, Number and duration of sleep episodes throughout the 24 h period. G, Number of brief awakenings per hour of sleep. H, Hypnograms illustrating sleep amount (min/30 min) throughout the 24 h period, averaged over days 15–19. Number of flies tested were as follows (GAL4-driver driving EP3517, EP3517): 30Y = 16, 19; 201Y = 25, 20; 238Y = 17, 20; c309 = 7, 10. Significant differences (p < 0.05, Mann–Whitney test) are indicated by black circles (A–G) and gray squares (H).
Discussion
We have shown here that dFmr1 expression is inversely related to daily sleep amount: overexpression is associated with short sleep, and loss of expression with long sleep. The increase in sleep duration in dFmr1 amorphs was found with three independent alleles (Δ50M, Δ113M, Df(3R)Exel6265), while the short sleep phenotype was seen by overexpressing dFmr1 with two different drivers, nSyb-GAL4 and ELAV-GeneSwitch. The constitutive nSyb-Gal4 driver, which resulted in higher DFMRP levels, also produced a stronger sleep phenotype. Similarly, amorphic mutations produced a more severe long sleep phenotype than hypomorphic mutations. Loss of dFmr1 occurs ubiquitously in amorphs, while overexpression of dFmr1 in hypermorphs is neuron-specific. Moreover, overexpression of dFmr1 throughout the mushroom bodies was sufficient to reduce sleep. Together, these results provide strong evidence that dFmr1 expression plays an important role in determining daily sleep amount, and does so by acting in the brain.
In flies, loss of dFmr1 is associated with overgrown dendritic trees and axonal processes, larger synaptic boutons containing more synaptic vesicles (Pan et al., 2004; Pan and Broadie, 2007), increased levels of dopamine and serotonin (Zhang et al., 2005), and enhanced excitatory neurotransmission, as indicated by the fact that defects in synaptic morphology and courtship behavior in amorphs can be rescued by GABA (Chang et al., 2008). In contrast, dFmr1 overexpression is associated with undergrowth, underbranching, and loss of synapse differentiation (Pan et al., 2004; Pan and Broadie, 2007). The specific targets through which dFmr1 affects synaptic physiology remain elusive, because FMRP binds to up to 4% of all brain mRNAs (Hinton et al., 1991), and controls the translation of several synaptic and cytoskeleton-associated proteins (Li et al., 2001; Zhang et al., 2001; Todd et al., 2003b; Lu et al., 2004; Castets et al., 2005; Reeve et al., 2005; Muddashetty et al., 2007; Zalfa et al., 2007). Recently, it was suggested that many symptoms in fragile X patients may derive from abnormal activation of metabotropic glutamate receptors (Bear et al., 2004). Indeed, abnormalities in ocular dominance plasticity and dendritic spine density can be corrected when Fmr1 knock-out mice are crossed with mGluR5 deficient mice (Dölen et al., 2007). However, hyperactivity, exploration deficits and abnormal spine morphology in Fmr1 knock-out mice are rescued by prolonged exposure to an enriched environment (Restivo et al., 2005), and some defects in synaptic plasticity are corrected by infusion of BDNF (Lauterborn et al., 2007), and by enhancing Ras signaling (Hu et al., 2008). Abnormalities in multiple pathways, therefore, most likely underlie the complex syndrome associated with loss of FMRP. Consistent with this conclusion, blocking metabotropic glutamatergic signaling in dFmr1 amorph flies rescues courtship behavior, but not the defects in circadian rest/activity rhythms and mushroom bodies morphology (McBride et al., 2005), nor, as shown in the current study, the long sleeping phenotype. Instead, defects in synaptic morphology and courtship behavior in dFmr1 amorphs are rescued by GABA (Chang et al., 2008). Future experiments should therefore assess whether sleep abnormalities can also be corrected by enhancing GABAergic transmission.
Previous studies found that dFmr1 amorphs are arrhythmic when kept in constant darkness, but not when entrained to a light/dark cycle (Dockendorff et al., 2002; Inoue et al., 2002). Fmr1 and Fxr2 single knock-out mice have shorter circadian periods in constant darkness but are normally entrained by light (Zhang et al., 2008). Double Fmr1/Fxr2 knock-out mice, however, which may be closer to the Drosophila dFmr1 amorph model, are completely arrhythmic even under a light/dark cycle (Zhang et al., 2008). We found that dFmr1 amorphs were long sleepers under both light/dark and constant dark conditions. Moreover, short sleeping flies with constitutive dFmr1 overexpression had normal circadian rest/activity rhythms even in constant darkness. Thus, at least in flies, the effects of changes in dFmr1 expression on sleep duration are unlikely to be mediated by the circadian clock.
After sleep deprivation both amorphs and hypermorphs failed to consistently increase sleep duration and sleep intensity (measured by changes in sleep episode duration and arousal threshold). This lack of sleep rebound did not reflect high resistance to sleep loss, because waking performance was impaired in both amorphs and hypermorphs. The lack of a sustained sleep rebound in amorphs may reflect a ceiling effect, because these flies already sleep ∼18 h/d during baseline, and their sleep is deep based on our assessment of arousal threshold. In dFmr1 hypermorphs, instead, which have undergrown synapses, prolonged waking may not affect synaptic activity enough to produce a sustained sleep rebound. This hypothesis is, admittedly, highly speculative, but is suggested by increasing evidence in flies, rats, and humans that learning, enriched experience, and the occurrence of synaptic potentiation during waking increase sleep need and sleep intensity (Huber et al., 2004, 2007, 2008; Ganguly-Fitzgerald et al., 2006; Faraguna et al., 2008). Similarly, brain infusion of molecules that can enhance synaptic function, such as the glutamatergic agonist NMDA (Wigren et al., 2007), BDNF (Faraguna et al., 2008), and TNF (Churchill et al., 2008), increase sleep. Thus, dFmr1 amorphs may sleep longer because of higher synaptic activity due to increased synaptic density and/or enhanced neurotransmission, while dFmr1 hypermorphs with undergrown synapses may need less sleep, both during baseline and after sleep deprivation. Indeed, we found that hypermorphs performed better after sleep loss than wild-type flies, even if their performance had deteriorated from baseline levels. Consistent with this hypothesis is also our observation that in wild-type flies DFMRP levels and sleep amounts peak the first day after eclosure, when almost all experience-driven axon pruning, which depends on dFmr1, also occurs (Tessier and Broadie, 2008). Moreover, sleep amounts decrease steeply during the first 3–4 d after eclosure, when there is no further pruning. In dFmr1 amorphs, instead, pruning continues for at least 3–4 d after eclosure (Tessier and Broadie, 2008), and during the same time period sleep amounts are high and show a less significant decline.
A full rescue of the amorphs' defects in synaptic morphology occurs when dFmr1 is induced during early larval development (Gatto and Broadie, 2008). However, a partial rescue is still possible when dFmr1 is expressed at maturity, suggesting that its induction can affect synaptic plasticity after development (Gatto and Broadie, 2008). Consistent with these results is our finding that dFmr1 overexpression was still able to decrease sleep in adult flies, albeit less significantly than when it was constitutively induced. Also consistent is our finding that dFmr1 expression is sensitive to changes in behavioral state in adult flies, where we found a >2-fold increase in DFMRP levels after 8 h of forced waking relative to 8 h of sleep. While increased brain FMRP levels have been described in adult mice after acute whisker stimulation (Todd and Mack, 2000; Todd et al., 2003a), chronic exposure to an enriched environment (Irwin et al., 2005) and glutamatergic stimulation (Weiler et al., 1997), this is the first indication that dFmr1 expression in adult Drosophila increases with prolonged activity, and is consistent with the fact that sensory deprivation decreases dFmr1 mRNA and protein levels (Tessier and Broadie, 2008).
Driving dFmr1 with GAL4 lines 30Y, 201Y, and 238Y produced a decrease in daily sleep similar to that observed after pan-neuronal dFmr1 overexpression. These lines show expression in all major components of the mushroom bodies, suggesting that the latter play an important, if not unique, role in regulating sleep need. The strongest decrease in sleep duration occurred with line 30Y, which has a very extensive pattern of expression within the mushroom bodies (Yang et al., 1995), but also with line 238Y, which has more restricted expression, similar to that of line 201Y (Yang et al., 1995; Kurusu et al., 2002). In contrast, driving dFmr1 with line c309, which lacks expression in the core region of the mushroom bodies (Joiner et al., 2006), increased sleep. Thus, it is likely that different neuronal populations within the mushroom bodies have different, and perhaps even opposite, effects on sleep regulation, as previously suggested (Joiner et al., 2006). However, these GAL4 lines also show limited and not overlapping expression outside the mushroom bodies, and thus the role of other neuronal groups in sleep regulation cannot be completely ruled out.
Like dFmr1 expression, cAMP levels and cAMP-dependent protein kinase (PKA) activity in flies are inversely related to sleep duration (Hendricks et al., 2001), and inducing PKA expression in the mushroom bodies is sufficient to reduce sleep (Joiner et al., 2006). CREB is a major target of PKA, and CREB activity in flies is also inversely related to sleep duration (Hendricks et al., 2001). Like DFMRP levels, CREB activity increases during waking relative to sleep (Hendricks et al., 2001). In contrast to dFmr1, however, blocking CREB activity actually increases the response to sleep deprivation, and increasing it has no effect on sleep homeostasis (Hendricks et al., 2001). Moreover, driving a PKA transgene with the same GAL4 lines used in this study increased sleep with 201Y, decreased sleep with c309, and caused opposite effects on day and night sleep with 30Y and 238Y (Joiner et al., 2006). Thus, although cAMP and CREB are strongly involved in synaptic plasticity, it is unlikely that dFmr1 expression affects sleep by acting directly through this pathway. It remains likely, however, that the ability of dFmr1 to modulate sleep need depends on its effects on synaptic physiology.
Footnotes
This work was supported by National Institute of General Medical Sciences Grant R01 GM075315 (C.C.) and by the Canadian Institutes of Health Research (D.B.). We thank Drs. Amita Sehgal, Julie Simpson, and Benjamin White for providing fly stocks, and Dr. Reto Huber for helpful discussions.
References
- Balling A, Technau GM, Heisenberg M. Are the structural changes in adult Drosophila mushroom bodies memory traces? Studies on biochemical learning mutants. J Neurogenet. 1987;4:65–73. [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Carskadon MA, Acebo C, Jenni OG. Regulation of adolescent sleep: implications for behavior. Ann N Y Acad Sci. 2004;1021:276–291. doi: 10.1196/annals.1308.032. [DOI] [PubMed] [Google Scholar]
- Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- Chang S, Bray SM, Li Z, Zarnescu DC, He C, Jin P, Warren ST. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- Chugani HT. A critical period of brain development: studies of cerebral glucose utilization with PET. Prev Med. 1998;27:184–188. doi: 10.1006/pmed.1998.0274. [DOI] [PubMed] [Google Scholar]
- Churchill L, Rector DM, Yasuda K, Fix C, Rojas MJ, Yasuda T, Krueger JM. Tumor necrosis factor alpha: activity dependent expression and promotion of cortical column sleep in rats. Neuroscience. 2008;156:71–80. doi: 10.1016/j.neuroscience.2008.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Is sleep essential? Plos Biol. 2008;6:e216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci U S A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan WM, Fawcett JW, O'Leary DD, Stanfield BB. Regressive events in neurogenesis. Science. 1984;225:1258–1265. doi: 10.1126/science.6474175. [DOI] [PubMed] [Google Scholar]
- Desai NS, Casimiro TM, Gruber SM, Vanderklish PW. Early postnatal plasticity in neocortex of Fmr1 knockout mice. J Neurophysiol. 2006;96:1734–1745. doi: 10.1152/jn.00221.2006. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale R. FlyBase: a database for the Drosophila research community. Methods Mol Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- Faraguna U, Vyazovskiy VV, Nelson AB, Tononi G, Cirelli C. A causal role for brain-derived neurotrophic factor in the homeostatic regulation of sleep. J Neurosci. 2008;28:4088–4095. doi: 10.1523/JNEUROSCI.5510-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg I. Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res. 1982;17:319–334. doi: 10.1016/0022-3956(82)90038-3. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Waking experience affects sleep need in Drosophila. Science. 2006;313:1775–1781. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- Gao FB. Understanding fragile X syndrome: insights from retarded flies. Neuron. 2002;34:859–862. doi: 10.1016/s0896-6273(02)00740-7. [DOI] [PubMed] [Google Scholar]
- Gatto CL, Broadie K. Temporal requirements of the fragile X mental retardation protein in the regulation of synaptic structure. Development. 2008;135:2637–2648. doi: 10.1242/dev.022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- Hu H, Qin Y, Bochorishvili G, Zhu Y, van Aelst L, Zhu JJ. Ras signaling mechanisms underlying impaired GluR1-dependent plasticity associated with fragile X syndrome. J Neurosci. 2008;28:7847–7862. doi: 10.1523/JNEUROSCI.1496-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- Huber R, Tononi G, Cirelli C. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep. 2007;30:129–139. doi: 10.1093/sleep/30.2.129. [DOI] [PubMed] [Google Scholar]
- Huber R, Määttä S, Esser SK, Sarasso S, Ferrarelli F, Watson A, Ferreri F, Peterson MJ, Tononi G. Measures of cortical plasticity after transcranial paired associative stimulation predict changes in electroencephalogram slow-wave activity during subsequent sleep. J Neurosci. 2008;28:7911–7918. doi: 10.1523/JNEUROSCI.1636-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hublin C, Partinen M, Koskenvuo M, Kaprio J. Sleep and mortality: a population-based 22-year follow-up study. Sleep. 2007;30:1245–1253. doi: 10.1093/sleep/30.10.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, Nakamura A, Kobayashi S, Ishida N, Siomi H. A role for the Drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–1335. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Idupulapati M, Gilbert ME, Harris JB, Chakravarti AB, Rogers EJ, Crisostomo RA, Larsen BP, Mehta A, Alcantara CJ, Patel B, Swain RA, Weiler IJ, Oostra BA, Greenough WT. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am J Med Genet. 2002;111:140–146. doi: 10.1002/ajmg.10500. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Christmon CA, Grossman AW, Galvez R, Kim SH, DeGrush BJ, Weiler IJ, Greenough WT. Fragile X mental retardation protein levels increase following complex environment exposure in rat brain regions undergoing active synaptogenesis. Neurobiol Learn Mem. 2005;83:180–187. doi: 10.1016/j.nlm.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Joiner MA, Griffith LC. Mapping of the anatomical circuit of CaM kinase-dependent courtship conditioning in Drosophila. Learn Mem. 1999;6:177–192. [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Koekkoek SK, Yamaguchi K, Milojkovic BA, Dortland BR, Ruigrok TJ, Maex R, De Graaf W, Smit AE, VanderWerf F, Bakker CE, Willemsen R, Ikeda T, Kakizawa S, Onodera K, Nelson DL, Mientjes E, Joosten M, De Schutter E, Oostra BA, Ito M, De Zeeuw CI. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129:409–419. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- Larson J, Jessen RE, Kim D, Fine AK, du Hoffmann J. Age-dependent and selective impairment of long-term potentiation in the anterior piriform cortex of mice lacking the fragile X mental retardation protein. J Neurosci. 2005;25:9460–9469. doi: 10.1523/JNEUROSCI.2638-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterborn JC, Rex CS, Kramár E, Chen LY, Pandyarajan V, Lynch G, Gall CM. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J Neurosci. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pelletier MR, Perez Velazquez JL, Carlen PL. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol Cell Neurosci. 2002;19:138–151. doi: 10.1006/mcne.2001.1085. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O'donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, McDonald TV, Jongens TA. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Miano S, Bruni O, Elia M, Scifo L, Smerieri A, Trovato A, Verrillo E, Terzano MG, Ferri R. Sleep phenotypes of intellectual disability: a polysomnographic evaluation in subjects with Down syndrome and Fragile-X syndrome. Clin Neurophysiol. 2008;119:1242–1247. doi: 10.1016/j.clinph.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Keliă S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci SA, Ferri R, Elia M, Dal Gracco S, Scuderi C, Stefanini MC, Castano A, Azan G. Sleep neurophysiology in Fragile X patients. Dev Brain Dysfunct. 1995;8:218–222. [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Broadie KS. Drosophila fragile X mental retardation protein and metabotropic glutamate receptor A convergently regulate the synaptic ratio of ionotropic glutamate receptor subclasses. J Neurosci. 2007;27:12378–12389. doi: 10.1523/JNEUROSCI.2970-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol. 2004;14:1863–1870. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Putz G, Heisenberg M. Memories in Drosophila heat-box learning. Learn Mem. 2002;9:349–359. doi: 10.1101/lm.50402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve SP, Bassetto L, Genova GK, Kleyner Y, Leyssen M, Jackson FR, Hassan BA. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- Restivo L, Ferrari F, Passino E, Sgobio C, Bock J, Oostra BA, Bagni C, Ammassari-Teule M. Enriched environment promotes behavioral and morphological recovery in a mouse model for the fragile X syndrome. Proc Natl Acad Sci U S A. 2005;102:11557–11562. doi: 10.1073/pnas.0504984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Singh K, Gaur P, Prasad S. Fragile x mental retardation (Fmr-1) gene expression is down regulated in brain of mice during aging. Mol Biol Rep. 2007;34:173–181. doi: 10.1007/s11033-006-9032-8. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci U S A. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ. Sensory stimulation increases cortical expression of the fragile X mental retardation protein in vivo. Brain Res Mol Brain Res. 2000;80:17–25. doi: 10.1016/s0169-328x(00)00098-x. [DOI] [PubMed] [Google Scholar]
- Todd PK, Malter JS, Mack KJ. Whisker stimulation-dependent translation of FMRP in the barrel cortex requires activation of type I metabotropic glutamate receptors. Brain Res Mol Brain Res. 2003a;110:267–278. doi: 10.1016/s0169-328x(02)00657-5. [DOI] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003b;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci. 2008;11:200–208. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigren HK, Schepens M, Matto V, Stenberg D, Porkka-Heiskanen T. Glutamatergic stimulation of the basal forebrain elevates extracellular adenosine and increases the subsequent sleep. Neuroscience. 2007;147:811–823. doi: 10.1016/j.neuroscience.2007.04.046. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, Bagni C. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, Albrecht U, Tamanini F, Meijer JH, Oostra BA, Nelson DL. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Friedman DB, Wang Z, Woodruff E, 3rd, Pan L, O'Donnell J, Broadie K. Protein expression profiling of the drosophila fragile X mutant brain reveals up-regulation of monoamine synthesis. Mol Cell Proteomics. 2005;4:278–290. doi: 10.1074/mcp.M400174-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhao MG, Toyoda H, Ko SW, Ding HK, Wu LJ, Zhuo M. Deficits in trace fear memory and long-term potentiation in a mouse model for fragile X syndrome. J Neurosci. 2005;25:7385–7392. doi: 10.1523/JNEUROSCI.1520-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]