Abstract
Genomic copy number aberrations and corresponding transcriptional deregulation in the cancer genome have been suggested to have regulatory roles in cancer development and progression. However, functional evaluation of individual genes from lengthy lists of candidate genes from genomic datasets presents a significant challenge. Here we report effective gene selection strategies to identify potential driver genes based on systematic integration of genome scale data of DNA copy numbers and gene expression profiles. Using regional pattern recognition approaches, we discovered the most probable copy number-dependent regions and 50 potential driver genes. At each step of gene selection process, functional relevance of the selected genes was evaluated by estimating the prognostic significance of the selected genes. Further validation using small interference RNA (siRNA)-mediated knockdown experiments demonstrated proof-of-principle evidence for the potential driver roles of the genes in HCC progression (i.e., NCSTN and SCRIB). In addition, systemic prediction of drug responses implicated the association of the 50 genes with specific signaling molecules (mTOR, AMPK, and EGFR). In conclusion, the application of an unbiased and integrative analysis of multidimensional genomic datasets can effectively screen for potential driver genes and provides novel mechanistic and clinical insights into pathobiology of HCC.
Keywords: CGH, microarray, mTOR, AMPK, nicastrin
Introduction
Microarray technologies have been used to define the global gene expression patterns in human cancer and have successfully identified gene expression signatures or novel tumor classes as well as prognostic information (1–4). However, since the gene expression profile is only a snapshot of complex genetic interactions, it is difficult to discriminate the genes driving the neoplastic process (“driver” genes) from by-stander genes (“passenger” genes). Array-based comparative genomic hybridization (CGH) analyses have also identified DNA copy number changes in many cancers. Some of these copy number-altered loci have been linked to disease pathogenesis and clinical behavior (5–8), but many of the copy number-altered genes appear not to be expressed. Thus, it seems reasonable to suggest that the combined analysis of both datasets may reveal the causally altered genes in both copy numbers and corresponding gene expression (9–12). However, dauntingly complex genomic and epigenomic interactions in cancer and the current limitations in sensitivity and specificity of genomic data may impede the identification of the regulatory genes (13). It is therefore challenging to evaluate the biological function and clinical utility of the lengthy candidate genes obtained from genomic data.
In this study, we aimed to establish a gene selection strategy to identify potential driver genes by integrating genomic data of copy numbers and gene expression profiles. Guiding our gene selection process by estimating the prognostic significance of the selected genes, we identified 50 potential driver genes. Further computational and experimental interrogations suggested that the identified genes represent potential driver genes for HCC providing both new insights into the pathobiology of liver cancer and therapeutic opportunities.
Materials and Methods
CGH Profiling
Genomic DNA of 15 HCC samples and a normal reference liver were extracted using DNeasy extraction kit (Qiagen, Valencia, CA). After labeling tumors and a reference normal liver DNA samples with Cy5 and Cy3 fluorescent dye, respectively, hybridizations and data acquisition were performed following the manufacturer’s instructions (Nimblegen, Madison, WI). The raw data were normalized as described previously (14).
Generation of TM and TCM
The regional copy number alteration was estimated by T-statistic–based sliding window approach, T-statistic map (TM). Before constructing TM, considering the absolute copy number values exceeding 0.5 in at least three samples to be significant, 309,939 out of 3,080,000 probes were identified as having non-trivial changes. TM scores were calculated as the T-statistic values which were obtained by applying one-sample T-test on the probe values for 15 samples within a moving window. We chose 100 Kb as the best-fit window size as it most accurately reflects the genomic copy number changes when various different window sizes (100 Kb, 1 Mb, and 10 Mb) were tested (data not shown). The TM scores generated by less than 5 probes in a moving window were regarded as missing values. The threshold for significant TM score was determined as the highest TM score from 100 random datasets which were generated by randomly ordering the probe positions. Significant TM regions were determined by a segmentation algorithm. The probes above the threshold were determined as valid probes. If a valid probe was not neighbored within 100 Kb of another valid probe, it was regarded invalid. Then, if the valid probes were neighbored over 5 Mb, the probe position between them was segmented. Segments less than 100 Kb were excluded.
Regional concordance of transcriptional regulation in 139 HCC expression profiles were constructed using transcriptome correlation map (TCM) as described previously (13). Briefly, for each gene, the sum of pair-wise Pearson’s correlation coefficients of the gene expression levels in neighboring n genes was calculated. Moving window size was determined to include the largest significant genes with the smallest neighboring genes, n=24. The threshold for significant TCM values and the segments were determined by the same algorithms used in the TM analysis.
Estimation of the Prognostic Values of Gene Sets
The prognostic value (P0) of a selected gene set was evaluated by using a clustering algorithm and log-rank test as described previously (4). Then, a permutation test was performed to compare the prognostic value of a selected region with those from other regions. 2,000 random datasets were generated by randomly selecting the same number of genes from outside the selected regions (or genes) to be tested. For each random dataset, hierarchical clustering was performed to stratify the HCCs into two groups, and the log-rank test P-value (Pm) between the groups was calculated. Iterative hierarchical clustering for the 2,000 random datasets was performed by a command-line version of Cluster 3.0 (15). The significance of the permutation test was taken as the frequency of events Pm < P0 from the 2,000 trials. Prognostic impact score (PIS) was calculated as − log10 (permutation P-value). A permutation P < 0.05 (PIS > 1.3) was regarded as significant.
siRNA-mediated Gene Knockdown
HepG2 and HuH-7 cells were transfected with siGENOME SMARTpool siRNAs (Dharmacon, Lafayette, CO) for the selected target genes using lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Non-targeting siRNA (siGENOME Non-Targeting Pool #1 siRNA) was used as control. After 96 hrs incubation with siRNAs, the cell viability was assessed by MTT assay.
Screening of Therapeutic Drugs using in trans correlated gene signatures
Raw data of Connectivity Map (build01) with two different Affymetrix platforms (i.e., hgu133a and HT-hgu133a) were downloaded from authors’ website (www.broad.mit.edu/cmap/) and normalized independently using the RMA method implemented in Bioconductor (http://www.bioconductor.org). For each of the in trans gene signatures, positive or negative signatures were assigned according to the algebraic sign of the correlation coefficient of each gene. For each treatment instance, the average connectivity score for the in trans gene signatures was calculated as the representative connectivity score (Savg). The connectivity scores (Si) for each in trans gene signature to drug treatment instance (i) were calculated based on Kolmogorov-Smirnov test and the significance of the connectivity of a perturbagen was evaluated by a permutation test for 10,000 times as described previously (16).
Additional procedures are described in Supplementary Materials and Methods.
Results
Comparison of Gene Copy Number and Transcription Levels in HCC
We generated fine-scale whole genome copy number profiles of 15 HCC samples using CGH microarrays containing 3,080,000 isothermal oligonucleotide probes. To represent the HCC heterogeneity, we selected 5 tumor samples from each of the subgroups of A, B, and HB which were previously defined as having homogeneous expression patterns with prognostic differences [Supplementary Table S1] (1, 4). First, we screened for the copy number altered regions in HCC by applying the T-statistic map (TM) method. TM was optimized to identify chromosomal regions with low stringent but high regional concordance of copy number changes, which yielded 57 TM regions. These regions were highly concordant with previous findings including the recurrent gains at 1q, 3q, 6p, 7q, 8q, 17q, 20q, 22q and the losses at 1p, 6q, 8p, 9p, 14q, 16q, and 17p (Fig. 1A and SI Table 2A)(7, 10, 13, 17, 18).
Figure 1.
Identification of genomic regional patterns of DNA copy numbers and mRNA expression in HCC. A, T-statistic map (TM ) was constructed based on one-sample T-statistic values of the copy numbers in 15 HCCs with moving window size 100 Kb. B, Gene expression profiles of 139 HCC samples in the order of chromosomal location. C, Transcriptome correlation map (TCM) for the gene expression profiles of 139 HCC. Thresholds for TM (29.9) and TCM (28.7) were determined by 100 random permutation tests.
To assess the chromosomal regional influence of the gene expression, we next rearranged the previous 139 HCC gene expression data (4) according to a sequence map of the human genome (hg17), which revealed a strong regional concordance of gene expression patterns across the heterogeneous tumor samples (Fig. 1B). This regional pattern can be shown more clearly with a transcriptome correlation map (TCM). TCM was previously used to identify concordant regional coexpression which was thought to be largely due to the copy number alteration (Fig. 1C)(19). With a TCM, we identified 48 regions in which gene expression levels were likely to be copy number-dependent (Supplementary Table S2B). Remarkably high TCM scores were observed in chromosome 8, implying strong influence of the copy numbers on gene expression levels. This agrees well with the previous studies indicating marked copy number aberration of chromosome 8 and its regulatory role in cancer development and progression (20,21). We then cross-compared the TM and TCM regions, which yielded 25 overlapped common regions (CR) spanning 112.8 Mb and containing 580 genes (Supplementary Table S2C). These regions may represent the most plausible regions in which copy numbers and expressions are concomitantly deregulated.
Prognostic Impact of the Copy Number-Dependent Regions
We hypothesized that if genomic alterations of the genes in CR are critical events during HCC development, the gene expression patterns in CR would be well associated with patient prognosis. We evaluated the prognostic relevance of the selected regions by calculating prognostic impact score (PIS) which may represent the relative prognostic significance compared to the non-selected regions. The TM and TCM regions showed significant log-rank test P-values, but their PIS were not significant compared to the other regions (Fig. 2A). However, the genes residing in the common regions (CR) showed a significant prognostic value (PIS=1.62), which may suggest that the genes in CR play regulatory roles in the HCC progression (Fig. 2A). In particular, 4 out of the 25 CR segments (CR1:1q21.3-23.2, CR3: 1q42.11-42.12, CR13: 7q36.1, and CR20: 8q24.11-24.22) were highly predictive for patient survival (P<0.001, Fig. 2B). CR13 was significant in predicting tumor recurrence as well as survival (Fig. 2C). Deregulated genes in these regions (1q21, 1q42, 7q36, and 8q24) may therefore have a high probability of functioning as potential driver genes.
Figure 2.
Prognostic impacts of T-statistic map (TM) or transcriptome correlation map (TCM) regions. A, Kaplan-Meir plot analyses (left) and prognostic impact scores (PIS)(right) of the genes located in the TM, TCM, and CR regions are shown, respectively. To show the significance of the PIS, the distribution of log-rank test P-values generated from 2,000 random datasets (Pm) were plotted with the log-rank test P-values calculated from the selected gene sets (P0, red line). B, Kaplan-Meir plot analyses and log-rank tests for overall survival and recurrence free survival (C) in the HCC subgroups based on expression similarities of the genes located in the CR1, CR3, CR13, and CR20.
Screening of the Potential Driver Genes
Next, we tested whether the previous correlation-based gene selection method (9, 11) can identify the potential driver genes which have prognostic relevance. Before gene selection, we evaluated the genome-wide influence of all copy number changes on gene expression. The distribution correlation coefficients showed significant global influence of the copy number changes on the gene expression levels (Supplementary Fig. S1A). Moreover, the copy number profile of each sample was best correlated with its corresponding expression levels, demonstrating that the overall copy number changes substantially reflected the gene expression profiles in each individual genome (Supplementary Fig. S1B).
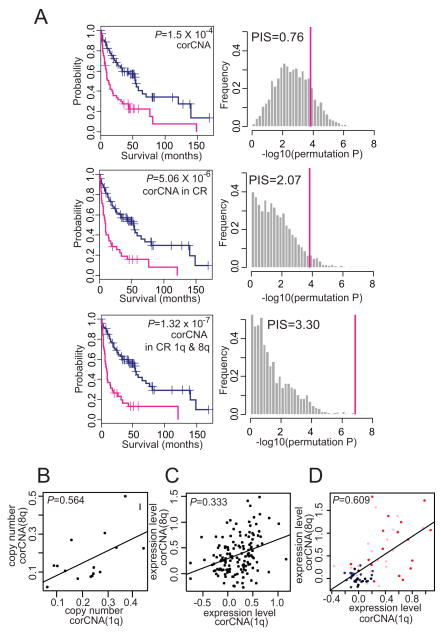
Confident that the copy number changes were strongly reflected in the corresponding gene expression levels, we selected 379 correlated copy number alteration (corCNA) genes based on the correlation between the expression levels and the copy numbers (P<0.01, r>0.649, Pearson’s correlation test, false discovery rate=0.0019). To validate the correlation by an independent method, we measured the copy numbers of two corCNA genes (POLR2K and C1orf43) in 52 HCC samples using qPCR (Supplementary Fig. S1C,D). However, the prognostic impact of the 379 genes was not significantly different from the non-correlated genes (PIS=0.76) even though the log-rank test revealed a significant difference (Fig. 3A, top). We next selected the corCNA genes residing in CR, which yielded 50 genes. Strikingly, these 50 corCNA genes showed a strong prognostic impact (P=5.06 × 10−6, PIS=2.03, Fig. 3A, middle). This result suggests that our gene selection strategy using the regional pattern recognition approach can effectively select the functionally relevant genes. Thus, we reasoned that these 50 genes were the most probable candidate driver genes (Table 1).
Figure 3.
Correlation of gene copy numbers and transcriptional levels. A, Kaplan-Meir plot analyses (left) and prognostic impact scores (PIS) (right) of 379 (top), 50 (middle), and 30 (bottom) Correlated copy number alteration (corCNA) genes are shown, respectively. B, scatter plot for the average copy numbers of the corCNA genes at 1q vs. 8q in 15 HCC dataset. C-D, Scatter plots for the average gene expression levels of corCNA genes at 1q vs. 8q in 139 HCC (C) and GSE6764 dataset (D). Pathological phenotypes of cirrhotic liver (n = 13, black), dysplastic nodules (n = 17, blue), early HCC (n = 18, pink), and advanced HCC (n = 17, red) are indicated with different colors in D. The significance of the correlation coefficients was evaluated by 10,000 random permutation test (P = 0.042, P = 0.005, respectively).
Table 1.
List of 50 potential driver genes
| Chr. | cytoband | Gene symbol | Correlation Coefficient | Correlation P-value | Copy numbers | mRNA (mean) | Chr. | cytoband | Gene symbol | Correlation Coefficient | Correlation P-value | Copy numbers | mRNA (mean) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1 | q21.3 | C1orf43 | 0.693 | 4.17E-03 | 0.366 | 0.324 | chr8 | q22.2 | POLR2K | 0.706 | 3.25E-03 | 0.291 | 0.805 |
| chr1 | q21.3 | HAX1 | 0.668 | 6.47E-03 | 0.314 | 0.083 | chr8 | q22.3 | NCALD | 0.712 | 9.42E-03 | 0.086 | 0.204 |
| chr1 | q21.3 | ADAR | 0.803 | 3.14E-04 | 0.187 | 0.546 | chr8 | q22.3 | RRM2B | 0.788 | 8.23E-04 | 0.182 | 0.198 |
| chr1 | q22 | CCT3 | 0.724 | 2.28E-03 | 0.313 | 1.084 | chr8 | q22.3 | AZIN1 | 0.805 | 2.94E-04 | 0.205 | 0.271 |
| chr1 | q23.1 | CD1C | 0.716 | 2.70E-03 | 0.164 | −0.432 | chr8 | q24.12 | TAF2 | 0.659 | 7.56E-03 | 0.159 | 0.556 |
| chr1 | q23.2 | NCSTN | 0.668 | 6.48E-03 | 0.147 | 0.486 | chr8 | q24.13 | DERL1 | 0.754 | 1.15E-03 | 0.209 | 0.224 |
| chr1 | q31.3 | CFHR2 | 0.733 | 1.88E-03 | 0.106 | −1.513 | chr8 | q24.13 | ZHX1 | 0.672 | 8.49E-03 | 0.193 | −0.08 |
| chr1 | q42.12 | PYCR2 | 0.795 | 1.17E-03 | 0.201 | 0.751 | chr8 | q24.13 | TATDN1 | 0.814 | 3.92E-04 | 0.229 | 0.654 |
| chr6 | p21.31 | C6orf107 | 0.655 | 8.04E-03 | 0.198 | 0.075 | chr8 | q24.13 | NDUFB9 | 0.754 | 1.15E-03 | 0.152 | 0.466 |
| chr6 | q16.3 | CCNC | 0.737 | 2.61E-03 | −0.046 | −0.315 | chr8 | q24.13 | KIAA0196 | 0.678 | 5.46E-03 | 0.136 | 0.414 |
| chr6 | q22.31 | MAN1A1 | 0.668 | 6.49E-03 | −0.146 | −1.293 | chr8 | q24.3 | TSTA3 | 0.68 | 5.25E-03 | 0.153 | 0.401 |
| chr6 | q22.31 | SERINC1 | 0.678 | 5.48E-03 | −0.156 | −0.825 | chr8 | q24.3 | SCRIB | 0.717 | 2.64E-03 | 0.163 | 1.102 |
| chr7 | q22.1 | EPHB4 | 0.743 | 5.65E-03 | 0.286 | 0.189 | chr8 | q24.3 | HSF1 | 0.654 | 8.12E-03 | 0.129 | 0.736 |
| chr7 | q22.1 | CUTL1 | 0.73 | 1.99E-03 | 0.275 | 0.081 | chr8 | q24.3 | KIFC2 | 0.677 | 7.76E-03 | 0.218 | 0.141 |
| chr8 | p22 | PCM1 | 0.649 | 8.82E-03 | −0.23 | −0.225 | chr16 | q23.1 | KARS | 0.899 | 5.16E-06 | 0.012 | 0.072 |
| chr8 | p21.1 | ELP3 | 0.745 | 2.23E-03 | −0.204 | −0.122 | chr19 | p13.12 | ILVBL | 0.914 | 1.86E-06 | 0.167 | −0.253 |
| chr8 | p21.1 | HMBOX1 | 0.771 | 7.57E-04 | −0.204 | −0.055 | chr19 | p13.12 | BRD4 | 0.769 | 2.13E-03 | 0.163 | 0.209 |
| chr8 | q21.13 | MRPS28 | 0.771 | 7.75E-04 | 0.087 | −0.072 | chr19 | p13.12 | WIZ | 0.739 | 1.64E-03 | 0.163 | 0.463 |
| chr8 | q22.1 | KIAA1429 | 0.699 | 3.76E-03 | 0.206 | 0.332 | chr19 | p13.12 | CYP4F11 | 0.68 | 5.24E-03 | 0.109 | −0.891 |
| chr8 | q22.1 | UQCRB | 0.676 | 5.67E-03 | 0.096 | 0.628 | chr19 | p13.12 | RAB8A | 0.722 | 2.38E-03 | 0.289 | −0.081 |
| chr8 | q22.1 | PTDSS1 | 0.818 | 1.96E-04 | 0.154 | 0.365 | chr19 | p13.11 | FAM32A | 0.769 | 8.09E-04 | 0.346 | 0.15 |
| chr8 | q22.1 | PGCP | 0.666 | 6.74E-03 | 0.041 | 0.388 | chr19 | p13.11 | C19orf42 | 0.735 | 6.49E-03 | 0.3 | 0.366 |
| chr8 | q22.1 | MTDH | 0.669 | 6.37E-03 | 0.245 | 0.282 | chr19 | p13.11 | MYO9B | 0.825 | 1.75E-03 | 0.291 | 0.096 |
| chr8 | q22.2 | STK3 | 0.686 | 4.78E-03 | 0.096 | 0.184 | chr19 | q12 | CCNE1 | 0.76 | 1.62E-03 | 0.363 | 0.174 |
| chr8 | q22.2 | COX6C | 0.799 | 3.54E-04 | 0.172 | 0.508 | chr19 | q12 | C19orf2 | 0.791 | 4.45E-04 | 0.069 | 0.437 |
Interestingly, of the 50 corCNA genes, 30 genes resided in 1q (n=8) and 8q (n=22) which were previously shown to have copy number gains at the early stage of HCC development (7). The prevalence of 1q and 8q genes among 50 corCNA genes is unlikely to be due to the gene abundance in those regions (P=9.42 × 10−5, hypergeometric test). These 30 genes in the “early event” regions (1q21.3-42.12 and 8q21.13-24.13) showed the most significant prognostic relevance (P=1.32 × 10−7; PIS=3.30; Fig. 3A, bottom), indicating the importance of the early dysregulation of these genes in HCC development. The prognostic values of the 50 or 30 1q/8q corCNA genes were further validated by two independent HCC datasets (SNU (22) and GSE6764 (2)) using class prediction algorithms (for details see SI Methods). The tumor classes defined by the expression similarity of the 50 or 30 genes could successfully predict the prognostic outcome of HCC in the independent datasets (Supplementary Fig. S2 & Table S3), supporting the robustness and consistency of our finding.
The enrichment of the 1q and 8q genes among the 50 corCNA genes raised the possibility that the gene expression alterations at 1q and 8q might concomitantly occur during the early stage of HCC development. We observed that the average copy numbers of 1q and 8q corCNA genes were correlated to each other (r=0.564, P=0.028; Fig. 3B). In parallel, the gene expression levels of 1q and 8q corCNA genes were also correlated in our 139 HCC dataset (r=0.333, P=6.17 × 10−5, permutation P=0.042; Fig. 3C), and this was validated in an independent 35 HCC dataset (GSE6764, r=0.398, P=0.017; Fig. 3D). In addition, the result from GSE6764 also showed that the coexpression levels of the 1q and 8q genes corresponded with the pathological staging of the liver (cirrhosis, dysplasia, and early HCC), and advanced HCC (n=65, r=0.609, P=7.11 × 10−8, permutation P=0.005; Fig. 3D). Permutation tests from 10,000 random trials indicated that our findings are not likely to be observed by chance. Taken together, the 30 corCNA genes might be co-amplified and coexpressed during the early stage and play critical roles in HCC development and progression.
Functional Assessment and Identification of Therapeutic Targets for the 50 Genes
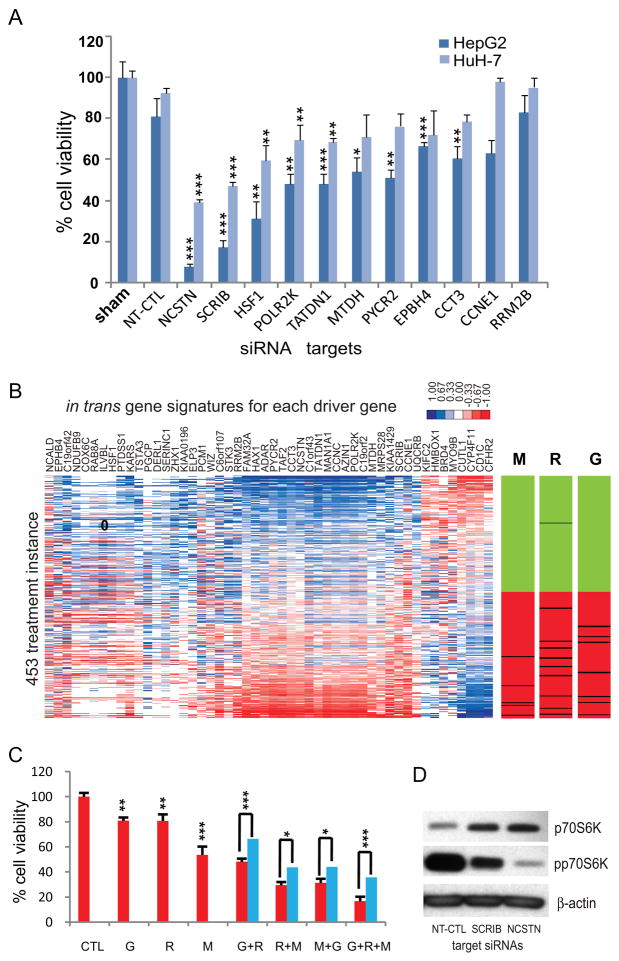
Since the 50 genes were selected by unbiased screening, we next examined their potential driver roles. We randomly chose 11 genes from the 50 genes to include the copy number-gained genes in the distinct chromosomes (CCT3, SCRIB, PYCR2, HSF1, NCSTN, EPHB4, MTDH, CCNE1, RRM2B, TATDN1, and POLR2K) and tested the siRNA-mediated knockdown effects on the cancer cell viability. Of the 11 genes, 5 siRNAs for SCRIB, NCSTN, HSF1, TATDN1, and POLR2K significantly reduced the viability of both HepG2 and HuH-7 liver cancer cell lines (P < 0.01, Fig. 4A). In particular, the loss of NCSTN and SCRIB showed the most potent growth inhibition (P < 0.001) supporting the potential driver role of these genes in HCC development and making them attractive therapeutic targets. The knockdown of each 5 effective target genes was confirmed by qRT-PCR, and its target selectivity was validated by two independent siRNAs (Supplementary Fig. S3). These results show that our screening strategy can successfully identify novel therapeutic targets for HCC. The 50 potential driver genes were identified by computational and experimental methods. However, the global influence of the 50 genes on cell function might be mediated by their target effector genes. We therefore sought to identify the putative effector genes whose expression levels were correlated in trans with expression of each driver gene (P<0.001, Pearson’s correlation test). Overall, the in trans correlated gene sets (ranged from 15 to 1,606) were enriched with the cancer-dominant functions such as protein transport/translation, oxidative stress, RNA processing/DNA replication, and metabolism/immune related functions; this may support their representative roles for the effector genes (Supplementary Fig. S4). Considering the in trans correlated genes as putative effectors for the 50 genes, we hypothesized that the disruption of the expression of the in trans correlated genes would interrupt HCC development. To test this hypothesis, we employed Connectivity Map, an analytical resource of gene expression profiles for drug responses (16). For each of the 50 in trans gene signatures, we calculated the connectivities to the drug instances in the Connectivity Map. The connectivity score profiles of the 50 in trans gene sets showed overall similar connectivities against each treatment instance, suggesting the commonality of their connectivities to the drug responses (Fig. 4B). Therefore, we used the averaged connectivity score of the 50 in trans gene sets (Savg) as a representing connectivity to each treatment instance. We found only two perturbagens, metformin and sirolimus (rapamycin), to be significantly connected to the Savg (permutation P=0.0025 and 0.0082, respectively; Fig. 4B). In addition, gefitinib, an EGFR selective tyrosine kinase inhibitor, was also identified as having significant connectivity (Savg=−0.335 and ranked to 7th out of 453 instances). Since only one instance for gefitinib was available in Connectivity Map (build01), we performed permutation test by treating all the EGFR/IR selective tyrosine kinase inhibitors as a same perturbagen (n=7). This revealed a strong connectivity of the EGFR tyrosine kinase inhibitors to Savg (permutation P=0.0022; Fig. 4B). These results may suggest that the dysregulated potential driver genes can be interrupted by rapamycin, metformin, or gefitinib treatment.
Figure 4.
Evaluation of functional and clinical utility of the 50 in trans correlated genes. A, The siRNAs (15 nM) targeting 11 driver genes were transfected to HepG2 and HuH-7 cells for 96 hrs, and the cell viabilities were assessed by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Non-targeting control siRNA(NT-CTL) was used as a control. Each bar indicates mean percentage cell viability of three replicates compared to NT-CTL. Error bars represent mean ± SD. Significance of growth inhibition compared to NT-CTL for each cell line was evaluated by two-sample T-test (* P <0.05, ** P < 0.01, and *** P < 0.001). B, Connectivity scores for each of the in trans correlated gene signatures to Connectivity Map are shown in a heatmap ordered by the average connectivity scores for individual instances (Savg). Bar-views with the instances of metformin (M, n = 5), rapamycin (R, n = 10), and EGFR/IR selective tyrosine kinase inhibitors (i.e., gefitinib, 4,5-dianilinophthalimide, butein, tyrophostin AG-1478, and HNMPA-(AM)3) (G, n = 7). Ranked distribution of Savg for the interesting perturbagen is indicated as a black line in the ordered 453 instances. Other instances with positive and negative connectivity scores are indicated with green and red color, respectively. C, HuH-7 cells were treated with rapamycin (R, 10 nM), metformin (M, 1 mM), or gefitinib (G, 1 μM) for 48 hrs in serum free media, and the cell viability was assessed by MTT ([3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. The % cell viability of three replicates compared to the control group (red bar) and the expected viability of combination treatment (blue bar) are indicated. Error bars indicate mean ± SD. Significance of the growth inhibition effects compared to the control or expected value E were evaluated by two-sample T-test (*P<0.05, **P<0.01, and ***P<0.001). D, HuH-7 cells were transfected with siRNAs (15 nM) targeting NCSTN or SCRIB for 48 hrs, and Western blot analysis was performed using antibodies for p70 S6 kinase (p70S6K), phospho-p70 S6 kinase (pp70S6K), and β-actin.
Interestingly, the cross-talk among the target molecules of rapamycin, metformin, and gefitinib has been reported (i.e., mTOR, AMPK, and EGFR )(23). This supports the idea that a combination of these drugs may provide clinical benefit for HCC patients (24). We found that the combined treatment with these drugs could potentiate the growth inhibition of cancer cells as compared to the expected potency calculated by Bliss independence model (for review see (25)) (Fig. 4C).
In addition, our data suggest a novel link between the 50 genes and the EGFR, AMPK, and mTOR signaling pathways. To verify this notion, we examined the knockdown effects of NCSTN and SCRIB on the mTOR signaling which is thought to be a common target for the three signaling pathways. Western blot analysis demonstrated that the knockdown of NCSTN and SCRIB could indeed inhibit the phosphorylation of p70S6 kinase, one of mTOR targets (Fig. 4D).
Discussion
In this study, we conducted combined analysis of copy numbers and gene expression with the following unbiased strategies. By applying regional pattern recognition approach, TM and TCM, we could identify the most probable copy number-dependent genes. In each step of our gene selection strategy, the functional relevance of the selected genes was evaluated by estimating the prognostic impact from the expression patterns and clinical information. Although our study was not intended to classify prognostic groups using the copy numbers or the gene expression patterns, we adopted clustering algorithm and log-rank test to calculate PIS which allowed evaluation of the functional relevance of our gene selection strategy. Our analysis revealed 50 potential driver genes which have significant prognostic relevance. Indeed, the 50 genes included many cancer related genes. For example, RRM2B, HAX1, CUTL1, HSF1, EPHB4, and MTDH were known to regulate tumorigenesis, tumor migration or adhesion. The genes related to RNA processing/transcription (e.g., ADAR, ELP3, HMBOX1, POLR2K, ZHX1, and KARS) and cell cycle regulation (e.g., BRD4, CCNC, and CCNE1) were also identified.
The functional significance of the 50 genes was further evaluated in part by siRNA-mediated knockdown experiments. Particularly, NCSTN, encoding a gamma-secretase component, showed the most potent effect on cancer cell growth in our screening. Supporting our finding, NCSTN was previously identified as one of the copy number-dependent genes in HCC (11). Moreover, inhibition of gamma-secretase has been reported to reduce leukemia cell growth via Notch and mTOR signaling pathways (26). SCRIB was also known to associate with human cancers (27), but its oncogenic role is not yet fully understood. Loss of HSF1 inhibited cancer cell growth (Fig. 4) in an agreement with the recent identification of HSF1 as an oncogene (28). Taken together, our data successfully demonstrate the potential driver roles of the 50 genes, and suggest that our strategy can efficiently identify novel therapeutic targets.
The application of the in trans correlated genes to mine the Connectivity Map allowed us to identify the therapeutic targets for the 50 genes. In the past, the Connectivity Map has been used to predict the therapeutic targets or diseases from gene expression signatures (16,29,30). Differing from these studies, we utilized in trans correlated gene signatures to predict the molecular targets which were functionally associated with each of the 50 genes. In support of our approach, the coexpressed genes in microarray data have been previously shown to be functionally related (31). Accordingly, our in silico approach could predict relevant effector targets without generating experimental signatures for each of the driver genes. Identification of common connectivities (Savg) from 50 different signatures might be also helpful in reducing by-chance findings which can be generated by applying a single signature. Indeed, our approach was able not only to predict well-known anticancer drugs with recognized therapeutic potential for HCC, such as rapamycin and gefitinib (8, 32–36), but also identified metformin as a therapeutic drug for targeting the 50 in trans gene signatures. Metformin is a widely used hypoglycemic drug for diabetes patients. Recently, several studies have shown its anticancer effect, suggesting a novel utility of the anti-metabolic drugs for cancer treatment (15,37,38). Supporting these studies, we observed for the first time the therapeutic efficacy of metformin for HCC using single or combination regimens for treating HCC cell lines in vitro (Fig. 4C).
Our analysis also predicted novel links between the 50 genes and mTOR, AMPK, and EGFR pathways. Supporting this notion, the oncogenic function of HSF1 has been reported to be mediated by mTOR (22) and EGFR (39) pathways. Likewise, we demonstrated that the knockdown effects of NCSTN and SCRIB are possibly mediated through these pathways. Although further stringent validations for the mechanism(s) might be required, our findings open new opportunities for studying the multifaceted association of the 50 genes with the three identified pathways.
In conclusion, we suggest that our unbiased gene selection strategy of integrating multidimensional genomic data can effectively select the potential driver genes and provide new biological and clinical insights into the copy number-dependent genes in HCC. Undoubtedly, our strategy can be applied to other cancer types.
Supplementary Material
Acknowledgments
We thank the staff of Laboratory Experimental Carcinogenesis for critical reading of the manuscript. This project was supported by the Intramural Research Program of the Center for Cancer Research/NCI, and, in part, by FG-4-2 of the 21C Frontier Functional Human Genome Project from the Ministry of Science & Technology in Korea.
Footnotes
The authors have no potential conflicts.
References
- 1.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–76. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 2.Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–47. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 3.Ye QH, Qin LX, Forgues M, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–23. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 4.Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–6. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 5.Paris PL, Andaya A, Fridlyand J, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13:1303–13. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco DR, Tonon G, Huang Y, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–25. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 7.Poon TC, Wong N, Lai PB, Rattray M, Johnson PJ, Sung JJ. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology. 2006;131:1262–70. doi: 10.1053/j.gastro.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Katoh H, Ojima H, Kokubu A, et al. Genetically distinct and clinically relevant classification of hepatocellular carcinoma: putative therapeutic targets. Gastroenterology. 2007;133:1475–86. doi: 10.1053/j.gastro.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 9.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patil MA, Chua MS, Pan KH, et al. An integrated data analysis approach to characterize genes highly expressed in hepatocellular carcinoma. Oncogene. 2005;24:3737–47. doi: 10.1038/sj.onc.1208479. [DOI] [PubMed] [Google Scholar]
- 11.Lee SA, Ho C, Roy R, et al. Integration of genomic analysis and in vivo transfection to identify sprouty 2 as a candidate tumor suppressor in liver cancer. Hepatology. 2008;47:1200–10. doi: 10.1002/hep.22169. [DOI] [PubMed] [Google Scholar]
- 12.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38:1386–96. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 14.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 15.Buzzai M, Jones RG, Amaravadi RK, et al. Systemic Treatment with the Antidiabetic Drug Metformin Selectively Impairs p53-Deficient Tumor Cell Growth. Cancer Res. 2007;67:6745–52. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 16.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 17.Su WH, Chao CC, Yeh SH, et al. HCC: an integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 2007;35:D727–31. doi: 10.1093/nar/gkl845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–46. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 19.Moinzadeh P, Breuhahn K, Stutzer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade--results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–41. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–30. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo HG, Park ES, Cheon JH, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–64. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 23.Guertin DA, Sabatini DM. Defining the Role of mTOR in Cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Bianco R, Garofalo S, Rosa R, et al. Inhibition of mTOR pathway by everolimus cooperates with EGFR inhibitors in human tumours sensitive and resistant to anti-EGFR drugs. Br J Cancer. 2008;98:923–30. doi: 10.1038/sj.bjc.6604269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–66. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 26.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–86. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamei Y, Kito K, Takeuchi T, et al. Human scribble accumulates in colorectal neoplasia in association with an altered distribution of beta-catenin. Hum Pathol. 2007;38:1273–81. doi: 10.1016/j.humpath.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 28.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hieronymus H, Lamb J, Ross KN, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–30. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Wei G, Twomey D, Lamb J, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–42. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–94. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahin F, Kannangai R, Adegbola O, Wang J, Su G, Torbenson M. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–5. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 33.Parent R, Kolippakkam D, Booth G, Beretta L. Mammalian target of rapamycin activation impairs hepatocytic differentiation and targets genes moderating lipid homeostasis and hepatocellular growth. Cancer Res. 2007;67:4337–45. doi: 10.1158/0008-5472.CAN-06-3640. [DOI] [PubMed] [Google Scholar]
- 34.Villanueva A, Chiang DY, Newell P, et al. Pivotal Role of mTOR Signaling in Hepatocellular Carcinoma. Gastroenterology. 2008;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philip PA, Mahoney MR, Allmer C, et al. Phase II study of Erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J Clin Oncol. 2005;23:6657–63. doi: 10.1200/JCO.2005.14.696. [DOI] [PubMed] [Google Scholar]
- 36.Thomas MB, Chadha R, Glover K, et al. Phase 2 study of erlotinib in patients with unresectable hepatocellular carcinoma. Cancer. 2007;110:1059–67. doi: 10.1002/cncr.22886. [DOI] [PubMed] [Google Scholar]
- 37.Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–73. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 38.Ben Sahra I, Laurent K, Loubat A, et al. The antidiabetic drug metformin exerts an antitumoral effect in vitro and in vivo through a decrease of cyclin D1 level. Oncogene. 2008;27:3576–86. doi: 10.1038/sj.onc.1211024. [DOI] [PubMed] [Google Scholar]
- 39.O’Callaghan-Sunol C, Sherman MY. Heat shock transcription factor (HSF1) plays a critical role in cell migration via maintaining MAP kinase signaling. Cell Cycle. 2006;5:1431–7. doi: 10.4161/cc.5.13.2915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.