Abstract
Introduction
We examined the association between mammographic density and single-nucleotide polymorphisms (SNPs) in genes encoding CYP1A1, CYP1B1, aromatase, 17β-HSD, ESR1, and ESR2 in pre- and early perimenopausal white, African-American, Chinese, and Japanese women.
Methods
The Study of Women's Health Across the Nation is a longitudinal community-based cohort study. We analyzed data from 451 pre- and early perimenopausal participants of the ancillary SWAN Mammographic Density study for whom we had complete information regarding mammographic density, genotypes, and covariates. With multivariate linear regression, we examined the relation between percentage mammographic breast density (outcome) and each SNP (primary predictor), adjusting for age, race/ethnicity, parity, cigarette smoking, and body mass index (BMI).
Results
After multivariate adjustment, the CYP1B1 rs162555 CC genotype was associated with a 9.4% higher mammographic density than the TC/TT genotype (P = 0.04). The CYP19A1 rs936306 TT genotype was associated with 6.2% lower mammographic density than the TC/CC genotype (P = 0.02). The positive association between CYP1A1 rs2606345 and mammographic density was significantly stronger among participants with BMI greater than 30 kg/m2 than among those with BMI less than 25 kg/m2 (Pinteraction = 0.05). Among white participants, the ESR1 rs2234693 CC genotype was associated with a 7.0% higher mammographic density than the CT/TT genotype (P = 0.01).
Conclusions
SNPs in certain genes encoding sex steroid metabolism enzymes and ESRs were associated with mammographic density. Because the encoded enzymes and ESR1 are expressed in breast tissue, these SNPs may influence breast cancer risk by altering mammographic density.
Introduction
High mammographic breast density, the density of the breast on mammography, is one of the strongest known risk factors for breast cancer [1]. High breast density (dense tissue on 50% or more of the breast) could account for up to one third of breast cancer cases [2]. Factors such as body mass index, parity, age, smoking, and physical activity jointly account for only a small proportion of the variability in mammographic density [3]. In contrast, mammographic density has a strong genetic component. Twin studies have demonstrated that heritability (the proportion of variance attributable to genetic factors) accounts for 60% of the variance in mammographic density [4,5].
It is feasible that genetic variation in sex steroids or in estrogen receptors (ESRs) produced in breast tissue could lead to differing degrees of proliferation that may be manifest radiographically as interindividual differences in mammographic density. The presence of sex steroid metabolic enzymes and ESRs in breast tissue [6-24] suggests that local activation of estrogen to potentially reactive metabolites within breast tissue may play a role in initiating and promoting carcinogenesis [18]. Such enzymes include CYP1A1, CYP1B1, and 17β-hydroxysteroid dehydrogenase (17β-HSD). In addition to metabolizing environmental carcinogens (for example, polycyclic aromatic hydrocarbons), CYP1A1 has high activity with the 17β-estradiol substrate [25,26]. CYP1A1 forms mainly 2-hydroxyestrone, and to a lesser degree, some 4-hydroxyestrone, from estrone. In contrast, CYP1B1 predominantly catalyzes formation of potentially carcinogenic catechol estrogens, especially 4-hydroxyestrogens [6,26-28]. The implication of 4-hydroxy catechol estrogens in carcinogenesis suggests a key role for CYP1B1 in carcinogenesis [19,27,29,30]. CYP19A1 is the gene encoding the aromatase enzyme that catalyzes the formation of aromatic C18 estrogens from C19 androgens [6,31]. Type I 17β-HSD is the enzyme responsible for interconversion of estrone and estradiol [32]. In addition to potential local effects of these enzymes on breast tissue, ESR-estrogen interactions stimulate breast epithelial cell growth [33]. Single-nucleotide polymorphisms (SNPs) in genes encoding sex steroid-metabolizing enzymes or receptors have effects on the hormonal milieu of the breast and on levels of potential mammary carcinogens [6].
A few studies have explored associations between mammographic density and SNPs in genes encoding CYP1A1, CYP1B1, aromatase, 17β-HSD, ESR1, and ESR2 [34-39]. However, most studies were focused on postmenopausal women [36,37]. Premenopausal breast density may be more highly heritable than is postmenopausal density [40], and some genes may be associated with premenopausal but not with postmenopausal density [4]. The goal of this study was to examine the association between mammographic density and SNPs in genes encoding CYP1A1, CYP1B1, aromatase, type I 17β-HSD, ESR1, and ESR2 in a group of pre- and early perimenopausal white, African-American, Chinese, and Japanese women.
Materials and methods
To determine the association between SNPs in genes encoding sex steroid-metabolizing enzymes and ESRs and mammographic density, we used data from women who participated in the SWAN ancillary Mammographic Density Study and the SWAN Genetics Study, which are described later. All protocols were IRB approved at participating sites, and all participating women provided signed, written informed consent.
The Study of Women's Health Across the Nation (SWAN)
SWAN is a multisite longitudinal community-based cohort study of 3,302 midlife women, serving as the parent study for the Mammographic Density ancillary study. In brief, at baseline, women were aged 42 to 52 years and premenopausal (reporting no change in usual menstrual pattern) or early perimenopausal (reporting change in menstrual pattern but occurrence of menstruation in the past 3 months), had an intact uterus and one or more ovaries, were not pregnant or lactating, and were not using exogenous reproductive hormones [41]. Initiation of exogenous hormones after the baseline visit did not preclude inclusion in the longitudinal cohort study. Each of the seven study sites enrolled white women in addition to women of one other self-identified racial/ethnic group: African-American women (Boston, Detroit area, Chicago, and Pittsburgh), Japanese women (Los Angeles), Hispanic women (New Jersey), and Chinese women (Oakland, California). SWAN participants completed questionnaires and underwent fasting blood sampling annually.
The SWAN Mammographic Density Study
Three SWAN clinical sites (Los Angeles, Oakland, Pittsburgh) participated in the SWAN Mammographic Density ancillary study, which retrieved and analyzed existing participants' mammograms that had been performed by accredited mammography facilities as a part of routine medical care.
At the time of enrollment into the ancillary study, 1,248 participants were active at the three sites. Of these, 22 (2%) women were ineligible because of bilateral breast surgery, 82 (7%) were not recruited because of having an abbreviated follow-up, and 89 (7%) refused to participate. Thus, 1,055 (85%) women were eligible and agreed to participate in the mammographic density study; of these, 1,005 women had at least one mammographic density assessment.
By using previously published methods, a single expert reviewer quantified mammographic density (that is, the percentage of the breast composed of dense tissue) [42]. The reader assessed mammographic density by using the craniocaudal view of the mammogram of the right breast [43]. If a participant reported prior breast surgery involving the right breast, mammograms of the left breast were used for density assessments. A compensating polar planimeter was used to measure the total breast area (in square centimeters) and the area of dense breast tissue (in square centimeters). Percentage density was calculated as the area of dense breast tissue divided by the area of the breast. A repeated review of a 10% random subset of mammograms for intrarater reliability yielded an intraclass correlation coefficient for percentage mammographic density of 0.96 [43].
Our goal was to examine associations of SNPs with mammographic density among pre- and early perimenopausal participants. Of the 1,005 participants with at least one assessment of mammographic density, we chose one mammogram for each of the 643 pre- or early perimenopausal SWAN Mammographic Density study participants. If more than one mammogram was available for given participant, we selected the mammogram temporally closest to the preceding annual follow-up visit that was flanked by pre- or early perimenopausal status on the visits before and after the mammogram. For example, if a participant had mammographic density assessments from two mammograms during her premenopausal stage and one mammogram during her early perimenopausal stage, we chose a single mammogram for the participant by picking the mammogram that was temporally closest to its preceding annual follow-up visit. Mammograms that occurred more than 3 months before baseline and mammograms obtained during the use of current exogenous reproductive hormones were excluded.
The SWAN Genetics Study
The SWAN Genetics Study genotyped 25 SNPs relating to sex-steroid metabolism and estrogen receptors (Figure 1, Table 1). Of the 1,988 women who were eligible (that is, still participating and providing blood for the SWAN parent study at the follow-up year 5 visit), 88% agreed to participate in the genetics study. Details regarding specimen collection, specimen processing, and genotyping were previously reported [44]. Genotyping was performed by using TaqMan (Roche Molecular Systems, Inc., Pleasanton, CA) and an ABI 7900 HT sequence detection system (Applied Biosystems Inc., Foster City, CA, USA).
Figure 1.
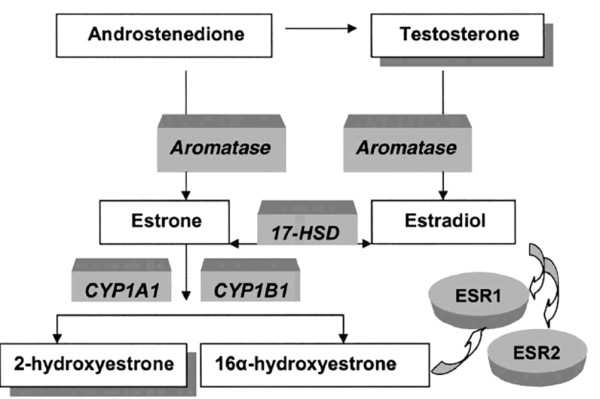
Functions of SWAN genetics sex steroid metabolism enzymes and receptors. Used with permission of Sowers and colleagues [93].
Table 1.
SNPs examined in the SWAN Genetics Study
| rs Number | Other common designation of SNP | Affected allelesa | Region of affected DNA sequence | Change in amino acid |
| 17-β HSD chromosome 17q11-q21 | ||||
| rs615942 | HSD615942b | G/T | Missense, amino acid position 55, codon position 2, C → A exon 2 | Ser → Tyr |
| rs592389 | HSD592389 | G/T | 3' Near gene | N/A |
| rs2830 | HSD17B2830 | A/G | 5' Untranslated region exon 1 | N/A |
| ESR1 chromosome 6q25.1 | ||||
| rs9340799 | ESRA464, XbaI RFLP | A/G | Intron 1 | N/A |
| rs2234693 | ESRA418, PvuII RFLP | T/C | Intron 1 | N/A |
| rs728524 | ESR728524 | A/G | Intron 4 | N/A |
| rs3798577 | ESR3798577 | T/C | 3' Untranslated region exon 8 | N/A |
| ESR2 chromosome 14q23.2 | ||||
| rs1256030 | ESR1256030 | C/T | Intron 2 | N/A |
| rs1255998 | ESR1255998 | C/G | 3' Untranslated region exon 9 | N/A |
| rs1256049 | ESR1256049, V328V, RsaI | G/A | Synonymous, amino acid position 328 (valine), codon position 3 exon 6 | N/A |
| CYP1A1 chromosome 15q22-q24 | ||||
| rs2606345 | CYP2606345, -1806 | C/A | Intron 1 | N/A |
| rs4646903 | CYP1A1250, MspI, m1, T6235C | T/C | 3' Near gene | N/A |
| rs1531163 | CYP1531163, -11781 promoterc | A/G | Synonymous, amino acid position 294 (lysine), codon position 3 exon 8 | N/A |
| rs1048943 | CYP1048943, CYP1A1*2C, A2455G, m2, 6750 A>G | A/G | Missense, amino acid position 462, codon position 1 A → G exon 7 | Ile → Val |
| CYP1B1 chromosome 2p21 | ||||
| rs1800440 | CYP1800440, N453S, A4390G, CYP1B1*4 | A/G | Missense, amino acid position 453, codon position 2 A → G exon 3 | Asn → Ser |
| rs162555 | CYP162555 d | T/C | Intron 2 | N/A |
| rs1056836 | CYP1056836, CYP1B1*3, Leu432Val, 4326C>G, C1294G, m1 | C/G | Missense, amino acid position 432, codon position 1 G → C exon 3 | Val → Leu |
| CYP19A1 chromosome 15q21.1 | ||||
| rs700519 | CYP19R264 | C/T | Missense, amino acid position 264, codon position 1 C → T exon 7 | Arg → Cys |
| rs2414096 | CYP194096 | G/A | Intron 2 | N/A |
| N/A | CYP194947, hCV8234946 | A/G | Intron 1 | N/A |
| rs1008805 | CYP198805 | T/C | Intron 1 | N/A |
| rs2446405 | CYP196405a | T/A | Intron 1 | N/A |
| rs2445759 | CYP195759 | G/T | Intron 1 | N/A |
| rs936306 | CYP196306 | C/T | Intron 1 | N/A |
| rs749292 | CYP199292 | G/A | Intron 1 | N/A |
aReference (higher frequency) allele is listed first. bEncodes Coenzyme A synthase. cEncodes lectin, mannose-binding, 1-like. dEncodes LOC732 hypothetical protein. eEncodes gliomedine.
Between three and eight SNPs per gene were selected based on use in previous genetics studies, a review of the literature, and information from gene databases (National Center for Biotechnology SNP database [45] and Celera [46]). The original SNP selection process is discussed in the first SWAN Genetics Study manuscript [44]. The SWAN Genetics Study searched for published literature supporting the biologic significance of SNPs chosen. SNPs were chosen if they were thought potentially to influence circulating sex hormone levels [47,48] or disease patterns: breast cancer [49-51], ovarian cancer [52], and bone mineral density [53,54].
Of the 643 premenopausal or early perimenopausal SWAN participants with available mammographic density information, at least partial genotyping data were available for 463 (72%) women. For this analysis, we excluded one participant lacking genotyping data for 24 of the 25 SNPs and an additional 11 participants who were missing information for one or more covariates. Thus, the analytic sample for this study comprised the 451 pre- and early perimenopausal women for whom complete information was available regarding mammographic density, genotypes, and covariates.
Questionnaire-based and anthropometric measures
At baseline and at each annual follow-up visit, SWAN participants were asked to complete standardized questionnaires and underwent measurement of height and weight for calculation of body mass index (BMI, weight in kilograms divided by the square of the height in meters). We took information regarding age, race/ethnicity, reproductive history, medication use, smoking, and alcohol intake from annual questionnaires.
Statistical analysis
Allele frequencies in the SWAN Genetics Study were estimated by race/ethnicity (Mendel Version 8.0 [55]). Hardy-Weinberg equilibrium (HWE) was assessed by using Fisher's Exact tests [55]. Because of the multiple statistical tests performed, we considered a P value of < 0.01 as the criterion to reject the null hypothesis of HWE.
Creating a separate model for each of the 25 SNPs, we used multivariate linear regression to examine the relation between percentage mammographic breast density (outcome) and SNP (primary predictor). Based on previously published studies, we considered the following candidate covariates: age, race/ethnicity, number of live births, BMI, oral contraceptive use, menopausal hormone use, cigarette smoking, and alcohol intake) [3,43,56-68]. Of these candidate covariates, age, number of live births, and BMI were included in all models, based on previously well-established associations with mammographic density. The remaining candidate covariates (cigarette smoking, alcohol intake, oral contraceptive use, and menopausal hormone use) were evaluated for model inclusion by using backwards regression performed on data from the 643 pre- and early perimenopausal participants of the SWAN Mammographic Density study. We used a P value of 0.10 as the cutoff for covariate inclusion. In addition, because each site recruited a specific racial/ethnic group in addition to non-Hispanic whites, a combined variable was created for race/ethnicity and study site; this variable was included in all models. Categories for this variable were whites in Oakland, Chinese in Oakland, whites in Los Angeles, Japanese in Los Angeles, white in Pittsburgh, or African American in Pittsburgh. Age at mammogram (continuous), race/ethnicity-study site, number of live births (continuous), current cigarette smoking (yes/no), and BMI (continuous) were the covariates retained in the final models. We modeled the alleles as acting in either an additive (aa versus Aa and AA, where the effect of the Aa genotype is half the effect of the AA genotype) or recessive (aa/Aa versus AA) manner, in which A is the minor allele.
Because of prior studies showing that associations of sex steroid-related SNPs may be more evident among women with BMI greater than 25 [69], we conducted secondary analyses wherein we added an SNP*BMI interaction term to the multivariable linear regression models. Because we suspected that the sample size of certain racial/ethnic subgroups may have been too small to allow detection of SNP*race/ethnicity interactions, and because the allele frequencies for 13 of the 25 SNPs differed by more than 0.20 between the ethnic groups, we repeated all of our analyses in the subsample of white participants, the largest racial/ethnic subgroup. All regression analyses were performed with the software program R [70].
Results
Baseline characteristics of the participants
Baseline characteristics of the analytic sample (N = 451 with mammographic density, genotyping, and covariate data) are displayed in Table 2. No notable differences in characteristics were found between the overall mammographic density sample (N = 643) and the analytic sample. The median age of the participants in the analytic sample was 48.7 years (Table 2). Median BMI was 24.4 kg/m2. Mean percentage mammographic density was 43.6%. Forty-nine percent of the participants in the analytic sample were white, 24% were Chinese, 22% were Japanese, and 6% were African American. Twenty-six percent were premenopausal, and 74% were early perimenopausal at the visit immediately preceding mammography (Table 2).
Table 2.
Characteristics of the study participants: analytic sample of the current study (N = 451)
| Mean | SD | Median | Number | % | |
| Age at mammogram, years | 48.6 | 2.6 | 48.7 | ||
| BMI, kg/m2 | 25.9 | 5.9 | 24.4 | ||
| <25 | 251 | 56 | |||
| 25–30 | 111 | 25 | |||
| >30 | 89 | 20 | |||
| Percentage mammographic density | 43.6 | 19.5 | 44.7 | ||
| Age at first full-term birth | 28.1 | 5.5 | 28.0 | ||
| Ethnicity | |||||
| African American | 26 | 6 | |||
| White | 219 | 49 | |||
| Chinese | 109 | 24 | |||
| Japanese | 97 | 22 | |||
| Study site | |||||
| Oakland | 184 | 41 | |||
| Los Angeles | 169 | 37 | |||
| Pittsburgh | 98 | 22 | |||
| Smoking currently | 31 | 7 | |||
| Menopausal status at time of mammogram | |||||
| Early perimenopausal | 332 | 74 | |||
| Premenopausal | 118 | 26 | |||
| Parity | |||||
| 0 | 77 | 17 | |||
| 1 | 76 | 17 | |||
| 2 | 196 | 43 | |||
| 3 | 76 | 17 | |||
| 4 | 19 | 4 | |||
| ≥ 5 | 7 | 2 |
Baseline menopause stage information was missing for one participant whose baseline mammogram was chosen; the participant was early perimenopausal at the subsequent (first annual) visit.
Hardy-Weinberg Equilibrium assessment
We examined allele frequencies by ethnicity (Table 3) and assessed HWE (see Table 4). Within racial/ethnic subgroups, only CYP194947 showed significant deviation from HWE. Because this SNP was also the SNP with the highest frequency (8%) of missing genotypes, methodologic considerations related to genotyping of this SNP may have contributed to deviation from HWE.
Table 3.
Allele frequencies of SWAN Genetics Study participants by race/ethnicity
| Estimated frequencies | ||||||
| Locus | Allele | All | African | White | Chinese | Japanese |
| 17β-HSD | ||||||
| rs615942 | G | 0.52 | 0.64 | 0.47 | 0.55 | 0.56 |
| T | 0.48 | 0.36 | 0.53 | 0.45 | 0.44 | |
| rs592389 | A | 0.50 | 0.41 | 0.475 | 0.56 | 0.56 |
| C | 0.50 | 0.59 | 0.53 | 0.44 | 0.44 | |
| rs2830 | A | 0.50 | 0.59 | 0.53 | 0.44 | 0.44 |
| G | 0.50 | 0.41 | 0.47 | 0.56 | 0.56 | |
| ESR1 | ||||||
| rs9340799 | A | 0.72 | 0.73 | 0.65 | 0.77 | 0.82 |
| G | 0.28 | 0.27 | 0.35 | 0.23 | 0.18 | |
| rs2234693 | C | 0.46 | 0.46 | 0.46 | 0.41 | 0.48 |
| T | 0.54 | 0.54 | 0.54 | 0.59 | 0.52 | |
| rs728524 | A | 0.89 | 0.76 | 0.98 | 0.81 | 0.81 |
| G | 0.11 | 0.24 | 0.02 | 0.19 | 0.188 | |
| rs3798577 | C | 0.46 | 0.41 | 0.48 | 0.45 | 0.45 |
| T | 0.54 | 0.59 | 0.52 | 0.55 | 0.55 | |
| ESR2 | ||||||
| rs1256030 | C | 0.61 | 0.75 | 0.57 | 0.70 | 0.58 |
| T | 0.39 | 0.25 | 0.43 | 0.30 | 0.42 | |
| rs1255998 | C | 0.68 | 0.50 | 0.89 | 0.37 | 0.54 |
| G | 0.32 | 0.50 | 0.11 | 0.63 | 0.46 | |
| rs1256049 | A | 0.18 | 0.11 | 0.02 | 0.45 | 0.30 |
| G | 0.82 | 0.89 | 0.98 | 0.55 | 0.70 | |
| CYP1A1 | ||||||
| rs2606345 | A | 0.37 | 0.18 | 0.66 | 0.06 | 0.05 |
| C | 0.63 | 0.82 | 0.34 | 0.94 | 0.95 | |
| rs4646903 | A | 0.75 | 0.71 | 0.89 | 0.54 | 0.61 |
| G | 0.25 | 0.29 | 0.11 | 0.46 | 0.39 | |
| rs1531163 | A | 0.88 | 0.66 | 0.95 | 0.84 | 0.80 |
| G | 0.12 | 0.34 | 0.05 | 0.16 | 0.20 | |
| rs1048943 | A | 0.88 | 0.98 | 0.96 | 0.75 | 0.76 |
| G | 0.12 | 0.02 | 0.04 | 0.25 | 0.24 | |
| CYP1B1 | ||||||
| rs1800440 | A | 0.89 | 0.90 | 0.80 | 1.00 | 1.00 |
| G | 0.11 | 0.10 | 0.20 | 0.004 | 0.006 | |
| rs162555 | C | 0.12 | 0.16 | 0.19 | 0.04 | 0.01 |
| T | 0.88 | 0.84 | 0.81 | 0.96 | 0.99 | |
| rs1056836 | C | 0.69 | 0.33 | 0.59 | 0.88 | 0.86 |
| G | 0.31 | 0.67 | 0.41 | 0.12 | 0.14 | |
| CYP19 | ||||||
| rs700519 | A | 0.12 | 0.13 | 0.04 | 0.14 | 0.29 |
| G | 0.88 | 0.88 | 0.96 | 0.86 | 0.71 | |
| rs2414096 | A | 0.43 | 0.25 | 0.50 | 0.46 | 0.32 |
| G | 0.57 | 0.75 | 0.50 | 0.54 | 0.68 | |
| 4947a | A | 0.52 | 0.71 | 0.55 | 0.39 | 0.49 |
| G | 0.48 | 0.29 | 0.45 | 0.61 | 0.51 | |
| rs1008805 | C | 0.36 | 0.16 | 0.42 | 0.28 | 0.37 |
| T | 0.64 | 0.84 | 0.58 | 0.72 | 0.63 | |
| rs2446405 | A | 0.32 | 0.52 | 0.18 | 0.47 | 0.45 |
| T | 0.68 | 0.48 | 0.82 | 0.53 | 0.55 | |
| rs936306 | C | 0.73 | 0.40 | 0.85 | 0.70 | 0.60 |
| T | 0.27 | 0.60 | 0.15 | 0.30 | 0.40 | |
| rs2445759 | G | 0.96 | 0.92 | 0.93 | 1.00 | 1.00 |
| T | 0.04 | 0.08 | 0.07 | 0.004 | 0.003 | |
| rs749292 | A | 0.44 | 0.51 | 0.44 | 0.50 | 0.36 |
| G | 0.56 | 0.49 | 0.56 | 0.50 | 0.64 | |
aNo corresponding dbsnp rs number; commonly referred to as CYP194947.
Table 4.
Hardy-Weinberg equilibrium evaluation by race/ethnicity
| All | African | White | Chinese | Japanese | ||||||
| Locus | P value | Number | P value | Number | P value | Number | P value | Number | P value | Number |
| HSD | ||||||||||
| rs615942 | 0.0741 | 716 | 1 | 56 | 0.0691 | 364 | 0.2962 | 139 | 0.632 | 157 |
| rs592389 | 0.0462 | 714 | 0.4159 | 56 | 0.075 | 361 | 0.3061 | 138 | 0.7462 | 156 |
| rs2830 | 0.0356 | 715 | 0.4074 | 56 | 0.1147 | 362 | 0.1189 | 139 | 0.7504 | 156 |
| ESR1 | ||||||||||
| rs9340799 | 0.5221 | 720 | 0.1818 | 56 | 0.1726 | 365 | 0.6308 | 140 | 0.5958 | 157 |
| rs2234693 | 0.8786 | 718 | 0.5964 | 56 | 0.5327 | 364 | 1 | 139 | 0.4255 | 157 |
| rs728524 | 0.0724 | 719 | 0.4825 | 56 | 1 | 364 | 1 | 140 | 0.7874 | 157 |
| rs3798577 | 0.9362 | 715 | 0.7783 | 55 | 0.8299 | 363 | 1 | 139 | 1 | 156 |
| ESR2 | ||||||||||
| rs1256030 | 0.6411 | 715 | 0.0811 | 56 | 1 | 363 | 0.1591 | 140 | 0.7379 | 154 |
| rs1255998 | <0.00005 | 712 | 0.2985 | 56 | 0.41 | 361 | 0.199 | 138 | 1 | 155 |
| rs1256049 | <0.00005 | 679 | 1 | 46 | 0.1804 | 344 | 0.3854 | 132 | 0.5638 | 155 |
| CYP1A1 | ||||||||||
| rs2606345 | <0.00005 | 715 | 0.6693 | 55 | 0.9088 | 360 | 1 | 141 | 1 | 157 |
| rs4646903 | <0.00005 | 706 | 0.7502 | 55 | 0.4137 | 356 | 1 | 138 | 0.1252 | 155 |
| rs1531163 | 0.0092 | 712 | 0.5642 | 56 | 0.5508 | 359 | 0.3384 | 139 | 0.3173 | 156 |
| rs1048943 | 0.0001 | 714 | 1 | 56 | 1 | 362 | 0.6582 | 138 | 0.0452 | 156 |
| CYP1B1 | ||||||||||
| rs1800440 | <0.00005 | 685 | 0.347 | 47 | 0.0629 | 345 | 1 | 135 | 1 | 156 |
| rs162555 | 0.0033 | 716 | 1 | 56 | 0.2346 | 364 | 0.1518 | 138 | 1 | 156 |
| rs1056836 | <0.00005 | 704 | 0.0591 | 53 | 0.2778 | 355 | 1 | 139 | 0.3159 | 155 |
| CYP19 | ||||||||||
| rs700519 | 0.0756 | 712 | 0.1854 | 56 | 1 | 360 | 0.0694 | 138 | 0.7032 | 156 |
| rs2414096 | 0.9384 | 719 | 0.7184 | 56 | 0.1193 | 365 | 0.2394 | 141 | 0.3603 | 155 |
| 4947a¶ | <0.00005 | 654 | 0.0195 | 41 | 0.5857 | 344 | <0.00005 | 129 | <0.00005 | 138 |
| rs1008805 | 0.7465 | 717 | 0.6143 | 56 | 0.2444 | 364 | 0.2942 | 140 | 0.7366 | 155 |
| rs2446405 | 0.0376 | 716 | 0.7845 | 55 | 0.7187 | 364 | 0.6048 | 139 | 0.7456 | 156 |
| rs936306 | 0.0031 | 717 | 1 | 56 | 0.3074 | 364 | 0.552 | 140 | 0.7366 | 155 |
| rs2445759 | 1 | 682 | 1 | 45 | 1 | 344 | 1 | 135 | 1 | 156 |
| rs749292 | 0.6463 | 715 | 0.4183 | 55 | 0.2371 | 361 | 0.6075 | 141 | 1 | 156 |
aNo corresponding dbsnp rs number; commonly referred to as CYP194947.
Associations between SNPs and percentage mammographic density
We examined the association between percentage mammographic breast density and each of the SNPs in models adjusted for age, race/ethnicity-study site, parity, smoking, and BMI (Tables 5 and 6).
Table 5.
Percentage mammographic density as a function of single-nucleotide polymorphism: recessive modelsa
| Entire analytic sample (N = 451) | Whites only (N = 219) | |||||
| Age, race/ethnicity-study site,b parity, smoking, body mass index | Age, study site, parity, smoking, body mass index | |||||
| Locus | β | SD | P value | β | SD | P value |
| 17-β HSD | ||||||
| rs615942 G | -0.18 | 1.73 | 0.92 | 0.78 | 2.55 | 0.76 |
| rs592389 A | -0.81 | 1.71 | 0.64 | 0.97 | 2.55 | 0.70 |
| rs2830 A | 1.75 | 1.65 | 0.29 | 2.69 | 2.59 | 0.30 |
| ESR1 | ||||||
| rs9340799 A | 4.40 | 2.79 | 0.12 | 6.67 | 3.50 | 0.06 |
| rs2234693 T | 3.37 | 1.81 | 0.06 | 7.04 | 2.67 | 0.01 |
| rs728524 A | 1.43 | 5.33 | 0.79 | N/A | N/A | N/A |
| rs3798577 T | 3.04 | 1.79 | 0.09 | 1.14 | 2.75 | 0.68 |
| ESR2 | ||||||
| rs1256030 C | 0.72 | 2.10 | 0.73 | 0.80 | 2.94 | 0.79 |
| rs1255998 C | -0.14 | 2.25 | 0.95 | -11.59 | 11.92 | 0.33 |
| rs1256049 G | -5.58 | 3.21 | 0.08 | -6.98 | 16.85 | 0.68 |
| CYP1A1 | ||||||
| rs2606345 C | -0.21 | 2.15 | 0.92 | -0.09 | 2.32 | 0.97 |
| rs4646903 A | 0.83 | 2.45 | 0.73 | -1.11 | 6.45 | 0.99 |
| rs1531163 A | 0.09 | 4.47 | 0.98 | -5.50 | 16.87 | 0.74 |
| rs1048943 A | 1.14 | 3.99 | 0.77 | N/A | N/A | N/A |
| CYP1B1 | ||||||
| rs1800440 A | -2.29 | 3.92 | 0.56 | -1.35 | 4.50 | 0.76 |
| rs162555 T | 9.36 | 4.62 | 0.04 | 8.23 | 5.45 | 0.13 |
| rs1056836 C | 2.48 | 2.51 | 0.33 | 1.37 | 3.07 | 0.66 |
| CYP19A1 | ||||||
| rs700519 G | -4.59 | 6.12 | 0.45 | N/A | N/A | N/A |
| rs2414096 G | -0.83 | 1.94 | 0.67 | 1.33 | 2.81 | 0.64 |
| CYP194947¶ A | 1.30 | 1.68 | 0.44 | 3.93 | 2.83 | 0.17 |
| rs1008805 T | -3.56 | 2.24 | 0.11 | -2.36 | 3.07 | 0.44 |
| rs2446405 T | 1.99 | 2.31 | 0.39 | -4.96 | 5.78 | 0.39 |
| rs936306 C | -6.17 | 2.82 | 0.03 | -16.36 | 5.96 | 0.01 |
| rs2445759 G | -7.30 | 9.18 | 0.43 | -10.36 | 11.98 | 0.39 |
| rs749292 G | 0.40 | 1.93 | 0.84 | 3.53 | 2.98 | 0.24 |
aReference allele was the allele with the higher frequency in the overall analytic sample and is indicated in the first column for each SNP.
bBecause each site recruited a specific racial/ethnic group in addition to non-Hispanic whites, a combined variable was created for race/ethnicity and study site.
Table 6.
Percentage mammographic density as a function of single-nucleotide polymorphism: additive modelsa
| Entire analytic sample (N = 451) | Whites only (N = 219) | |||||
| Age, race/ethnicity-study siteb, parity, smoking, body mass index | Age, study site, parity, smoking, body mass index | |||||
| LOCUS | β | SD | P value | β | SD | P value |
| 17-β HSD | ||||||
| rs615942 G | -0.68 | 1.02 | 0.50 | -0.68 | 1.55 | 0.66 |
| rs592389 G | -1.01 | 1.02 | 0.32 | -0.59 | 1.56 | 0.71 |
| rs2830 A | 1.05 | 1.01 | 0.30 | 0.63 | 1.56 | 0.69 |
| ESR1 | ||||||
| rs9340799 A | 1.58 | 1.22 | 0.20 | 3.11 | 1.77 | 0.08 |
| rs2234693 T | 1.44 | 1.06 | 0.18 | 4.08 | 1.61 | 0.01 |
| rs728524 A | 3.35 | 1.77 | 0.06 | -9.14 | 6.50 | 0.16 |
| rs3798577 T | 1.01 | 1.05 | 0.34 | -0.36 | 1.65 | 0.83 |
| ESR2 | ||||||
| rs1256030 C | 0.79 | 1.10 | 0.47 | 0.76 | 1.64 | 0.64 |
| rs1255998 C | -0.42 | 1.30 | 0.75 | -2.38 | 2.61 | 0.36 |
| rs1256049 G | -1.02 | 1.53 | 0.51 | -2.28 | 4.34 | 0.60 |
| CYP1A1 | ||||||
| rs2606345 C | -1.32 | 1.40 | 0.34 | -1.00 | 1.69 | 0.56 |
| rs4646903 T | 1.49 | 1.19 | 0.21 | 1.34 | 2.25 | 0.55 |
| rs1531163 A | -0.33 | 1.60 | 0.84 | -4.51 | 3.70 | 0.23 |
| rs1048943 A | 0.20 | 1.55 | 0.90 | 1.19 | 4.05 | 0.77 |
| CYP1B1 | ||||||
| rs1800440 A | 0.72 | 1.58 | 0.65 | 1.73 | 1.84 | 0.35 |
| rs162555 T | 0.52 | 1.64 | 0.75 | 0.45 | 2.01 | 0.82 |
| rs1056836 C | -0.37 | 1.22 | 0.78 | -1.39 | 1.62 | 0.39 |
| CYP19A1 | ||||||
| rs700519 C | 1.59 | 1.72 | 0.36 | -3.46 | 4.00 | 0.39 |
| rs2414096 G | 0.77 | 1.11 | 0.49 | 2.75 | 1.74 | 0.12 |
| CYP194947c A | 0.99 | 0.96 | 0.30 | 2.67 | 1.62 | 0.10 |
| rs1008805 T | -0.02 | 1.12 | 0.99 | -0.65 | 1.66 | 0.70 |
| rs2446405 T | 0.81 | 1.20 | 0.50 | -3.02 | 2.05 | 0.14 |
| rs936306 C | -0.92 | 1.25 | 0.46 | -3.67 | 2.12 | 0.09 |
| rs2445759 G | -3.47 | 2.36 | 0.14 | -5.19 | 2.86 | 0.07 |
| rs749292 G | 0.67 | 1.08 | 0.53 | 2.83 | 1.67 | 0.09 |
aReference allele was the allele with the higher frequency in the overall analytic sample and is indicated in the first column for each SNP. bBecause each site recruited a specific racial/ethnic group in addition to non-Hispanic whites, a combined variable was created for race/ethnicity and study site. cNo corresponding dbsnp rs number; commonly referred to as CYP194947.
In the fully adjusted recessive models (adjusted for age, race/ethnicity-study site, parity, smoking, and BMI), the CYP1B1 rs162555 CC genotype was associated with 9.4% higher percentage mammographic density than the TC/TT genotype (P = 0.04). The CYP19A1 rs936306 TT genotype was associated with 6.2% lower percentage mammographic density than TC/CC genotype (P = 0.03) (Table 5). In contrast to analyses restricted to white participants, ESR1 rs2234693 was not significantly associated with mammographic density in either recessive or additive models that included the entire analytic sample (Tables 5 and 6).
Interaction by BMI
In additive models, CYP1A1 rs2606345 was significantly associated with BMI (1.1 kg/m2 higher for each A allele; P = 0.03) and CYP19A1 rs2414096 (1.1 kg/m2 lower for each A allele; P = 0.01; data not shown). Similarly, in recessive models restricted to white participants, the CYP194947 GG genotype was associated with a 2.1 kg/m2 lower BMI compared with the GA/AA genotype (P = 0.05), and the CYP19A1 rs749292 AA genotype was associated with a 2.3 kg/m2 lower BMI than the GA/GG genotype (P = 0.05; data not shown).
To determine whether associations between SNPs and mammographic density varied according to BMI, we added BMI*SNP interaction terms to multiple linear regression models that included age, race/ethnicity-study site, smoking, parity, and BMI as covariates and percentage mammographic density as the outcome (data not shown). In additive models, the CYP1A1 rs2606345-mammographic density association was significantly different (stronger) among participants with BMIs greater than 30 kg/m2 compared with participants with BMIs less than 25 kg/m2 (Pinteraction = 0.05). Specifically, among participants with BMIs less than 25 kg/m2, percentage mammographic density was 0.57% higher for each CYP1A1 rs2606345 C allele; in contrast, among participants with BMIs greater than 30 kg/m2, percentage mammographic density was 6.1% higher for each additional C allele. The associations of SNPs with mammographic density did not significantly differ by BMI category for CYP19A1 rs2414096, CYP19A1 rs749292, or CYP194947. However, we may not have had adequate statistical power to detect an SNP*BMI interaction when BMI was categorized into tertiles.
Analyses restricted to white participants
In analyses restricted to whites (n = 219), we detected two associations that were similar to those seen in the overall analytic sample (for example, CYP19A1 rs936306 in recessive models, CYP19A1 rs2414096 in additive models). In white participants, the ESR1 rs2234693 CC genotype was associated with a 7.0% higher percentage mammographic density than the CT/TT genotype (P = 0.01; Table 5); this finding was also apparent in additive models (P = 0.01; Table 6). The association between ESR1 rs2234693 and mammographic density varied by ethnicity; the association was stronger among whites than among Japanese (interaction P value, 0.09) or Chinese (interaction P value, 0.03) participants.
Discussion
In pre- and early perimenopausal women, SNPs involving CYP1B1 (rs162555 CC genotype), CYP19A1 (rs936306 TT/CC genotype), and ESR1 (rs2234693 CC genotype) were each significantly positively associated with mammographic density. Associations between several SNPs (CYP1A1 rs2606345, CYP194947, CYP19A1 rs749292, CYP19A1 rs2414096) and mammographic density were attenuated after adjustment for BMI. Percentage mammographic density varied at least 3% per allele for the statistically significant associations. These differences in mammographic density according to genotype are of a clinically relevant magnitude, given that each 1% increment in mammographic density is associated with a 2% higher relative risk of breast cancer [2]. Several SNP-mammographic density associations varied significantly by ethnicity.
Several of our findings are novel. As far as we know, other publications have not reported information regarding associations between mammographic density and the following SNPs: 17β-HSD rs615942, 17β-HSD rs592389, 17β-HSD rs2830, ESR1 rs728524, ESR1 rs3798577, ESR2 rs1256030, ESR2 rs1255998, ESR2 rs1256049, CYP1B1 rs162555, CYP1B1 rs1800440, CYP19A1 rs700519, CYP19A1 rs2446405, CYP19A1 rs2445759, CYP19A1 rs1008805, CYP19A1 rs936306, CYP19A1 rs2414096, CYP19A1 rs749292, CYP194947, CYP1A1 rs1531163, or CYP1A1 rs2606345.
Our finding of an association between ESR1 rs2234693 and mammographic density among white women conflicts with some prior studies. The association between ESR1 rs2234693 and mammographic density was described in three reports from the EPIC study. In the first EPIC report, the T allele was associated with higher mammographic density [39], whereas in this study, the CC genotype is associated with higher mammographic density.
The second EPIC analysis found a statistically significant difference in mammographic density between hormone therapy users and never-users of hormone therapy among women the CT or TT genotype, but not among those with the CC genotype [71].
The third EPIC analysis reported no association between ESR1 rs2234693 and mammographic density [36]; the discrepancy among studies may be because the previous study used a different mammographic density measurement technique, had a less heterogenous study population, and focused on postmenopausal women.
We found an association between CYP1B1 rs1056836 and mammographic density that neared statistical significance only before adjustment, but not after adjustment, for BMI. These results may be consistent with three previously published studies [35,36,38].
A cross-sectional observational European study of white women found statistically significantly higher mammographic density in carriers of at least one ESR1 rs9340799 A allele [39]. Although we had similar results, our findings were not statistically significant, possibly because of the smaller number of participants in our study or the younger age of our participants.
The other SNPs involved in sex steroid metabolism or estrogen receptors were not significantly associated with mammographic density in the present study. As with our study, past studies reported absence of an association between mammographic density and CYP1A1 rs1048943 and CYP1A1 rs4646903 [35,38].
Although previously published studies have not included a systematic examination of sex steroid metabolism SNPs and mammographic density, some previously studied SNPs may be linked with the SNPs that we examined. We searched Haploview version 4.1 (Daly Lab, Cambridge, MA) with Hapmap genotype data to search for information regarding linkage disequilibrium for each of the three SNPs that we found to be associated with mammographic density and other SNPs previously studied in relation to mammographic density. Linkage diseqilibrium R2 values for ESR1A1 rs2234693 (which we found to be associated with mammographic density) and rs9340799 (which prior studies found to be associated with mammographic density) range from 0.234 to 0.55, depending on the ethnic group. For CYP19A1 rs936306 (which we found to be associated with mammographic density) and rs10046 (which prior studies found not to be associated with mammographic density), R2 values range from 0.017 to 0.193. Linkage-disequilibrium information is not currently available for CYP1B1 rs162555 on Hapmap. Although LD information was not available for rs162555, we note that its chromosomal location is not close to the other two previously studied CYP1B1 SNPs.
Our findings have a biologic rationale. A local influence of sex steroid metabolism SNPs on breast tissue is suggested by prior breast cancer studies. For example, ESR1 rs2234693 has been associated with duration of breast cancer survival [72], degree of breast cancer differentiation [73], age at breast cancer diagnosis [74], and receptor status of breast cancer tumors [75,76]. Likewise, CYP19A1 rs936306 may be associated with breast cancer disease-free survival [77]. Case-control studies of breast cancer risk related to ESR1 rs2234693 [73,76,78-87] and in relation to CYP19A1 rs936306 [77,88] are conflicting. Inconsistent results of breast cancer case-control studies are likely due to differences in ethnicity and menopausal status of participants across studies. Reasons exist to suspect that associations of SNPs with mammographic density may vary by BMI, as we found for CYP1A1 rs2606345. Sex steroid metabolism (for example, peripheral aromatization of androstenedione) varies by BMI, so that effects of sex steroid SNPs on breast tissue may be more pronounced among obese women. Although prior studies have not examined whether associations of CYP1A1 rs2606345 with mammographic density vary by BMI, a prior study reported that the association of an ESR1 SNP with increased breast cancer risk was apparent only among women with BMI greater than 25 kg/m2 [69].
Strengths of our study included its multiethnic study population, use of validated and reproducible mammographic density-assessment techniques, rigorous attention to genotyping methods, and collection of detailed information regarding key covariates related to mammographic density. However, this study did not directly assess sex steroid activity in breast tissue samples. Furthermore, although our sample size was relatively large, its heterogeneity may have precluded detection of statistically significant race-specific associations or interactions of SNPs with mammographic density. Finally, the observational study design precluded coordination of mammographic density with menstrual-cycle phase. Relations between SNPs and mammographic density may have been diluted because we analyzed mammograms taken during varying menstrual phases. Breasts are more radiographically dense during the luteal phase [89-91], although a recent study found that variation in mammographic density over the menstrual cycle may be subtle (that is, may not be statistically significant) [92].
Conclusions
In conclusion, SNPs involving sex steroid metabolism enzymes and ESR1 may be associated with mammographic density in pre- and early perimenopausal women. Future studies relating these SNPs to mammographic density not only should adjust for BMI but also should consider interactions by BMI. The mechanisms underlying the association (for example, increased proliferation of epithelial and stromal cells) require elucidation. Because these enzymes and ESR1 are expressed in target tissues, these SNPs (or genetic factors with which they are in linkage disequilibrium) may alter breast cancer risk by altering mammographic density. These findings inform the understanding of biologic influences on mammographic density, a strong risk factor for breast cancer.
Abbreviations
BMI: body mass index; CYP: cytochrome P450; ESR: estrogen receptor; HSD: hydroxysteroid dehydrogenase; HWE: Hardy Weinberg equilibrium; SNP: single-nucleotide polymorphism; SWAN: Study of Women's Health Across the Nation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CC contributed to study conception, study design, analysis and interpretation of data, manuscript drafting, and revision. JS and MS contributed to study design. MS, SC, EG, LH, LB, MS, GG, and JS contributed to analysis and interpretation of data and manuscript revision.
Acknowledgments
Acknowledgements
The Study of Women's Health Across the Nation (SWAN) has grant support from the National Institutes of Health (NIH), DHHS, through the National Institute on Aging (NIA), the National Institute of Nursing Research (NINR), and the NIH Office of Research on Women's Health (ORWH) (grants NR004061; AG012505, AG012535, AG012531, AG012539, AG012546, AG012553, AG012554, AG012495, AG017719). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIA, NINR, ORWH, or the NIH. Dr. Crandall's work was supported by National Institutes of Health research grant 5K12 AG01004-08 from the National Institute on Aging and by the Michael Parr Fund for Women's Health Research and Education at the Iris Cantor-UCLA Women's Health Center.
Clinical Centers: University of Michigan, Ann Arbor, MaryFran Sowers, PI; Massachusetts General Hospital, Boston, MA, Robert Neer, PI 1994–1999; Joel Finkelstein, PI 1999 to present; Rush University, Rush University Medical Center, Chicago, IL, Lynda Powell, PI; University of California, Davis/Kaiser, Ellen Gold, PI; University of California, Los Angeles, Gail Greendale, PI; University of Medicine and Dentistry, New Jersey Medical School, Newark, Gerson Weiss, PI 1994 to 2004; Nanette Santoro, PI 2004 to present; and the University of Pittsburgh, Pittsburgh, PA, Karen Matthews, PI.
NIH Program Office: National Institute on Aging, Bethesda, MD, Marcia Ory 1994 to 2001; Sherry Sherman, 1994 to present; National Institute of Nursing Research, Bethesda, MD, Program Officers.
Central Laboratory: University of Michigan, Ann Arbor, Daniel McConnell (Central Ligand Assay Satellite Services).
SWAN Repository: University of Michigan, Ann Arbor, MaryFran Sowers.
Coordinating Center: New England Research Institutes, Watertown, MA, Sonja McKinlay, PI 1995 to 2001; University of Pittsburgh, Pittsburgh, PA, Kim Sutton-Tyrrell, PI 2001 to present.
Steering Committee: Chris Gallagher, Chair
Susan Johnson, Chair
We thank the study staff at each site and all the women who participated in SWAN.
Contributor Information
Carolyn J Crandall, Email: ccrandall@mednet.ucla.edu.
Mary E Sehl, Email: MSehl@mednet.ucla.edu.
Sybil L Crawford, Email: Sybil.Crawford@umassmed.edu.
Ellen B Gold, Email: ebgold@ucdavis.edu.
Laurel A Habel, Email: Laurel.Habel@kp.org.
Lesley M Butler, Email: Lesley.Butler@colostate.edu.
MaryFran R Sowers, Email: mfsowers@umich.edu.
Gail A Greendale, Email: GGreenda@mednet.ucla.edu.
Janet S Sinsheimer, Email: Janet@mednet.ucla.edu.
References
- American Cancer Society . Breast Cancer Facts & Figures 2007–2008. Atlanta, Georgia, American Cancer Society Inc; 2007. [Google Scholar]
- Boyd NF, Byng JW, Jong RA, Fishell EK, Little LE, Miller AB, Lockwood GA, Tritchler DL, Yaffe MJ. Quantitative classification of mammographic densities and breast cancer risk: Results from the Canadian National Breast Screening Study. J Natl Cancer Inst. 1995;87:670–675. doi: 10.1093/jnci/87.9.670. [DOI] [PubMed] [Google Scholar]
- Vachon CM, Kuni CC, Anderson K, Anderson VE, Sellers TA. Association of mammographically defined percent breast density with epidemiologic risk factors for breast cancer (United States) Cancer Causes Control. 2000;11:653–662. doi: 10.1023/a:1008926607428. [DOI] [PubMed] [Google Scholar]
- Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MR, Giles GG, Tritchler D, Chiarelli A, Yaffe MJ, Hopper JL. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med. 2002;347:886–894. doi: 10.1056/NEJMoa013390. [DOI] [PubMed] [Google Scholar]
- Ursin G, Lillie EO, Lee E, Cockburn M, Schork NJ, Cozen W, Parisky YR, Hamilton AS, Astrahan MA, Mack T. The relative importance of genetics and environment on mammographic density. Cancer Epidemiol Biomarkers Prev. 2009;18:102–112. doi: 10.1158/1055-9965.EPI-07-2857. [DOI] [PubMed] [Google Scholar]
- Mitrunen K, Hirvonen A. Molecular epidemiology of sporadic breast cancer: the role of polymorphic genes involved in oestrogen biosynthesis and metabolism. Mutat Res. 2003;544:9–41. doi: 10.1016/s1383-5742(03)00016-4. [DOI] [PubMed] [Google Scholar]
- Hellmold H, Rylander T, Magnusson M, Reihner E, Warner M, Gustafsson JA. Characterization of cytochrome P450 enzymes in human breast tissue from reduction mammoplasties. J Clin Endocrinol Metab. 1998;83:886–895. doi: 10.1210/jcem.83.3.4647. [DOI] [PubMed] [Google Scholar]
- Honma N, Takubo K, Sawabe M, Arai T, Akiyama F, Sakamoto G, Utsumi T, Yoshimura N, Harada N. Estrogen-metabolizing enzymes in breast cancers from women over the age of 80 years. J Clin Endocrinol Metab. 2006;91:607–613. doi: 10.1210/jc.2005-1967. [DOI] [PubMed] [Google Scholar]
- Sasano H, Frost AR, Saitoh R, Harada N, Poutanen M, Vihko R, Bulun SE, Silverberg SG, Nagura H. Aromatase and 17 beta-hydroxysteroid dehydrogenase type 1 in human breast carcinoma. J Clin Endocrinol Metab. 1996;81:4042–4046. doi: 10.1210/jcem.81.11.8923858. [DOI] [PubMed] [Google Scholar]
- Sasano H, Nagura H, Harada N, Goukon Y, Kimura M. Immunolocalization of aromatase and other steroidogenic enzymes in human breast disorders. Hum Pathol. 1994;25:530–535. doi: 10.1016/0046-8177(94)90127-9. [DOI] [PubMed] [Google Scholar]
- Shibuya R, Suzuki T, Miki Y, Yoshida K, Moriya T, Ono K, Akahira J, Ishida T, Hirakawa H, Evans DB, Sasano H. Intratumoral concentration of sex steroids and expression of sex steroid-producing enzymes in ductal carcinoma in situ of human breast. Endocr Relat Cancer. 2008;15:113–124. doi: 10.1677/ERC-07-0092. [DOI] [PubMed] [Google Scholar]
- Singh N, Mitra AK, Garg VK, Agarwal A, Sharma M, Chaturvedi R, Rath SK. Association of CYP1A1 polymorphisms with breast cancer in North Indian women. Oncol Res. 2007;16:587–597. doi: 10.3727/000000007783629972. [DOI] [PubMed] [Google Scholar]
- Song D, Liu G, Luu-The V, Zhao D, Wang L, Zhang H, Xueling G, Li S, Desy L, Labrie F, Pelletier G. Expression of aromatase and 17beta-hydroxysteroid dehydrogenase types 1, 7 and 12 in breast cancer: an immunocytochemical study. J Steroid Biochem Mol Biol. 2006;101:136–144. doi: 10.1016/j.jsbmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- McKay JA, Melvin WT, Ah-See AK, Ewen SW, Greenlee WF, Marcus CB, Burke MD, Murray GI. Expression of cytochrome P450 CYP1B1 in breast cancer. FEBS Lett. 1995;374:270–272. doi: 10.1016/0014-5793(95)01126-y. [DOI] [PubMed] [Google Scholar]
- Haas S, Pierl C, Harth V, Pesch B, Rabstein S, Bruning T, Ko Y, Hamann U, Justenhoven C, Brauch H, Fischer HP. Expression of xenobiotic and steroid hormone metabolizing enzymes in human breast carcinomas. Int J Cancer. 2006;119:1785–1791. doi: 10.1002/ijc.21915. [DOI] [PubMed] [Google Scholar]
- Singh S, Chakravarti D, Edney JA, Hollins RR, Johnson PJ, West WW, Higginbotham SM, Cavalieri EL, Rogan EG. Relative imbalances in the expression of estrogen-metabolizing enzymes in the breast tissue of women with breast carcinoma. Oncol Rep. 2005;14:1091–1096. [PubMed] [Google Scholar]
- Licznerska BE, Wegman PP, Nordenskjold B, Wingren S. In situ levels of oestrogen producing enzymes and its prognostic significance in postmenopausal breast cancer patients. Breast Cancer Res Treat. 2007;112:15–23. doi: 10.1007/s10549-007-9819-x. [DOI] [PubMed] [Google Scholar]
- Modugno F, Knoll C, Kanbour-Shakir A, Romkes M. A potential role for the estrogen-metabolizing cytochrome P450 enzymes in human breast carcinogenesis. Breast Cancer Res Treat. 2003;82:191–197. doi: 10.1023/B:BREA.0000004376.21491.44. [DOI] [PubMed] [Google Scholar]
- Sasano H, Suzuki T, Nakata T, Moriya T. New development in intracrinology of breast carcinoma. Breast Cancer. 2006;13:129–136. doi: 10.2325/jbcs.13.129. [DOI] [PubMed] [Google Scholar]
- Wen W, Ren Z, Shu XO, Cai Q, Ye C, Gao YT, Zheng W. Expression of cytochrome P450 1B1 and catechol-O-methyltransferase in breast tissue and their associations with breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:917–920. doi: 10.1158/1055-9965.EPI-06-1032. [DOI] [PubMed] [Google Scholar]
- Iscan M, Klaavuniemi T, Coban T, Kapucuoglu N, Pelkonen O, Raunio H. The expression of cytochrome P450 enzymes in human breast tumours and normal breast tissue. Breast Cancer Res Treat. 2001;70:47–54. doi: 10.1023/a:1012526406741. [DOI] [PubMed] [Google Scholar]
- Huang Z, Fasco MJ, Figge HL, Keyomarsi K, Kaminsky LS. Expression of cytochromes P450 in human breast tissue and tumors. Drug Metab Dispos. 1996;24:899–905. [PubMed] [Google Scholar]
- Liehr JG, Ricci MJ. 4-Hydroxylation of estrogens as marker of human mammary tumors. Proc Natl Acad Sci USA. 1996;93:3294–3296. doi: 10.1073/pnas.93.8.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Fuqua SA. Estrogen receptor and breast cancer. Semin Cancer Biol. 2001;11:339–352. doi: 10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- Perera FP. Environment and cancer: who are susceptible? Science. 1997;278:1068–1073. doi: 10.1126/science.278.5340.1068. [DOI] [PubMed] [Google Scholar]
- Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology. 2003;144:3382–3398. doi: 10.1210/en.2003-0192. [DOI] [PubMed] [Google Scholar]
- Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17 beta-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci USA. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager JD, Liehr JG. Molecular mechanisms of estrogen carcinogenesis. Annu Rev Pharmacol Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res. 2000;60:3440–3444. [PubMed] [Google Scholar]
- Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H, Williams RH. Williams Textbook of Endocrinology. 11. Philadelphia: Saunders/Elsevier; 2008. [Google Scholar]
- Kendall A, Folkerd EJ, Dowsett M. Influences on circulating oestrogens in postmenopausal women: relationship with breast cancer. J Steroid Biochem Mol Biol. 2007;103:99–109. doi: 10.1016/j.jsbmb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Rayter Z. Steroid receptors in breast cancer. Br J Surg. 1991;78:528–535. doi: 10.1002/bjs.1800780506. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Bernstein L, Berg D, Ingles SA, Salane M, Ursin G. Genetic determinants of mammographic density. Breast Cancer Res. 2002;4:R5. doi: 10.1186/bcr434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Hankinson SE, De Vivo I, Guillemette C, Ishibe N, Hunter DJ, Byrne C. Polymorphisms in steroid hormone pathway genes and mammographic density. Breast Cancer Res Treat. 2003;77:27–36. doi: 10.1023/a:1021112121782. [DOI] [PubMed] [Google Scholar]
- Warren R, Skinner J, Sala E, Denton E, Dowsett M, Folkerd E, Healey CS, Dunning A, Doody D, Ponder B, Luben RN, Day NE, Easton D. Associations among mammographic density, circulating sex hormones, and polymorphisms in sex hormone metabolism genes in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2006;15:1502–1508. doi: 10.1158/1055-9965.EPI-05-0828. [DOI] [PubMed] [Google Scholar]
- Lord SJ, Mack WJ, Berg D Van Den, Pike MC, Ingles SA, Haiman CA, Wang W, Parisky YR, Hodis HN, Ursin G. Polymorphisms in genes involved in estrogen and progesterone metabolism and mammographic density changes in women randomized to postmenopausal hormone therapy: Results from a pilot study. Breast Cancer Res. 2005;7:R336–R344. doi: 10.1186/bcr999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskarinec G, Lurie G, Williams AE, Le Marchand L. An investigation of mammographic density and gene variants in healthy women. Int J Cancer. 2004;112:683–688. doi: 10.1002/ijc.20448. [DOI] [PubMed] [Google Scholar]
- van Duijnhoven FJ, Bezemer ID, Peeters PH, Roest M, Uitterlinden AG, Grobbee DE, van Gils CH. Polymorphisms in the estrogen receptor alpha gene and mammographic density. Cancer Epidemiol Biomarkers Prev. 2005;14:2655–2660. doi: 10.1158/1055-9965.EPI-05-0398. [DOI] [PubMed] [Google Scholar]
- Vachon CM, Sellers TA, Pankratz VS. Mammographic density of the breast. N Engl J Med. 2003;348:174–175. doi: 10.1056/NEJM200301093480215. author reply 174–175. [DOI] [PubMed] [Google Scholar]
- Sowers M, Crawford S, Morgenstein D, Gold E, Greendale GA, Evans D, Neer R, Matthews K, Sherman S, Lo A, Weiss G, Kelsey J. Design, survey sampling and recruitment Methods of SWAN: a multi-center, multi-ethnic, community-based cohort study of women and the menopausal transition. In: Lobos R, Marcus R, Kelsey JL, editor. Menopause. New York: Academic Press; 2000. pp. 175–188. [Google Scholar]
- Habel LA, Dignam JJ, Land SR, Salane M, Capra AM, Julian TB. Mammographic density and breast cancer after ductal carcinoma in situ. J Natl Cancer Inst. 2004;96:1467–1472. doi: 10.1093/jnci/djh260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habel LA, Capra AM, Oestreicher N, Greendale GA, Cauley JA, Bromberger J, Crandall CJ, Gold EB, Modugno F, Salane M, Quesenberry C, Sternfeld B. Mammographic density in a multiethnic cohort. Menopause. 2007;14:891–899. doi: 10.1097/gme.0b013e318032569c. [DOI] [PubMed] [Google Scholar]
- Kardia SR, Chu J, Sowers MR. Characterizing variation in sex steroid hormone pathway genes in women of 4 races/ethnicities: the Study of Women's Health Across the Nation (SWAN) Am J Med. 2006;119:S3–S15. doi: 10.1016/j.amjmed.2006.07.001. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology SNP database http://www.ncbi.nlm.nih.gov/SNP
- Celera http://www.celera.com
- Tworoger SS, Chubak J, Aiello EJ, Ulrich CM, Atkinson C, Potter JD, Yasui Y, Stapleton PL, Lampe JW, Farin FM, Stanczyk FZ, McTiernan A. Association of CYP17, CYP19, CYP1B1, and COMT polymorphisms with serum and urinary sex hormone concentrations in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2004;13:94–101. doi: 10.1158/1055-9965.epi-03-0026. [DOI] [PubMed] [Google Scholar]
- Baghaei F, Rosmond R, Westberg L, Hellstrand M, Eriksson E, Holm G, Bjorntorp P. The CYP19 gene and associations with androgens and abdominal obesity in premenopausal women. Obes Res. 2003;11:578–585. doi: 10.1038/oby.2003.81. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Stram DO, Pike MC, Kolonel LN, Burtt NP, Altshuler D, Hirschhorn J, Henderson BE. A comprehensive haplotype analysis of CYP19 and breast cancer risk: the Multiethnic Cohort. Hum Mol Genet. 2003;12:2679–2692. doi: 10.1093/hmg/ddg294. [DOI] [PubMed] [Google Scholar]
- Gold B, Kalush F, Bergeron J, Scott K, Mitra N, Wilson K, Ellis N, Huang H, Chen M, Lippert R, Halldorsson BV, Woodworth B, White T, Clark AG, Parl FF, Broder S, Dean M, Offit K. Estrogen receptor genotypes and haplotypes associated with breast cancer risk. Cancer Res. 2004;64:8891–8900. doi: 10.1158/0008-5472.CAN-04-1256. [DOI] [PubMed] [Google Scholar]
- Bailey LR, Roodi N, Dupont WD, Parl FF. Association of cytochrome P450 1B1 (CYP1B1) polymorphism with steroid receptor status in breast cancer. Cancer Res. 1998;58:5038–5041. [PubMed] [Google Scholar]
- Aktas D, Guney I, Alikasifoglu M, Yuce K, Tuncbilek E, Ayhan A. CYP1A1 gene polymorphism and risk of epithelial ovarian neoplasm. Gynecol Oncol. 2002;86:124–128. doi: 10.1006/gyno.2002.6720. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, Orimo H. Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res. 1996;11:306–311. doi: 10.1002/jbmr.5650110304. [DOI] [PubMed] [Google Scholar]
- Willing M, Sowers M, Aron D, Clark MK, Burns T, Bunten C, Crutchfield M, D'Agostino D, Jannausch M. Bone mineral density and its change in white women: estrogen and vitamin D receptor genotypes and their interaction. J Bone Miner Res. 1998;13:695–705. doi: 10.1359/jbmr.1998.13.4.695. [DOI] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69:A1886. [Google Scholar]
- Gapstur SM, Lopez P, Colangelo LA, Wolfman J, Van Horn L, Hendrick RE. Associations of breast cancer risk factors with breast density in Hispanic women. Cancer Epidemiol Biomarkers Prev. 2003;12:1074–1080. [PubMed] [Google Scholar]
- Maskarinec G, Meng L, Ursin G. Ethnic differences in mammographic densities. Int J Epidemiol. 2001;30:959–965. doi: 10.1093/ije/30.5.959. [DOI] [PubMed] [Google Scholar]
- Maskarinec G, Pagano I, Lurie G, Wilkens LR, Kolonel LN. Mammographic density and breast cancer risk: the multiethnic cohort study. Am J Epidemiol. 2005;162:743–752. doi: 10.1093/aje/kwi270. [DOI] [PubMed] [Google Scholar]
- Martin C, Peck J, Pisano E, Davis C, McTiernan A, Hulka B. Prior HRT use, ethnicity and mammographic density. Ann Epidemiol. 2000;10:461–462. doi: 10.1016/s1047-2797(00)00157-5. [DOI] [PubMed] [Google Scholar]
- Kerlikowske K, Shepherd J, Creasman J, Tice JA, Ziv E, Cummings SR. Are breast density and bone mineral density independent risk factors for breast cancer? J Natl Cancer Inst. 2005;97:368–374. doi: 10.1093/jnci/dji056. [DOI] [PubMed] [Google Scholar]
- Riza E, Dos Santos Silva I, De Stavola B, Perry N, Karadedou-Zafiriadou E, Linos D, Remoundos DD, Linos A. Correlates of high-density mammographic parenchymal patterns by menopausal status in a rural population in Northern Greece. Eur J Cancer. 2005;41:590–600. doi: 10.1016/j.ejca.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Hofling M, Carlstrom K, Svane G, Azavedo E, Kloosterboer H, Von Schoultz B. Different effects of tibolone and continuous combined estrogen plus progestogen hormone therapy on sex hormone binding globulin and free testosterone levels: an association with mammographic density. Gynecol Endocrinol. 2005;20:110–115. doi: 10.1080/09513590400021151. [DOI] [PubMed] [Google Scholar]
- Heng D, Gao F, Jong R, Fishell E, Yaffe M, Martin L, Li T, Stone J, Sun L, Hopper J, Boyd NF. Risk factors for breast cancer associated with mammographic features in Singaporean Chinese women. Cancer Epidemiol Biomarkers Prev. 2004;13:1751–1758. [PubMed] [Google Scholar]
- Masala G, Ambrogetti D, Assedi M, Giorgi D, Del Turco MR, Palli D. Dietary and lifestyle determinants of mammographic breast density: a longitudinal study in a Mediterranean population. Int J Cancer. 2006;118:1782–1789. doi: 10.1002/ijc.21558. [DOI] [PubMed] [Google Scholar]
- Jeffreys M, Warren R, Gunnell D, McCarron P, Smith GD. Life course breast cancer risk factors and adult breast density (United Kingdom) Cancer Causes Control. 2004;15:947–955. doi: 10.1007/s10522-004-2473-3. [DOI] [PubMed] [Google Scholar]
- McCormack VA, dos Santos Silva I, De Stavola BL, Perry N, Vinnicombe S, Swerdlow AJ, Hardy R, Kuh D. Life-course body size and perimenopausal mammographic parenchymal patterns in the MRC 1946 British birth cohort. Br J Cancer. 2003;89:852–859. doi: 10.1038/sj.bjc.6601207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LM, Gold EB, Greendale GA, Crandall CJ, Modugno F, Oestreicher N, Quesenberry CP, Jr, Habel LA. Menstrual and reproductive factors in relation to mammographic density: the Study of Women's Health Across the Nation (SWAN) Breast Cancer Res Treat. 2008;112:165–174. doi: 10.1007/s10549-007-9840-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greendale GA, Reboussin BA, Slone S, Wasilauskas C, Pike MC, Ursin G. Postmenopausal hormone therapy and change in mammographic density. J Natl Cancer Inst. 2003;95:30–37. doi: 10.1093/jnci/95.1.30. [DOI] [PubMed] [Google Scholar]
- Kocabas NA, Sardas S, Cholerton S, Daly AK, Karakaya AE. Cytochrome P450 CYP1B1 and catechol O-methyltransferase (COMT) genetic polymorphisms and breast cancer susceptibility in a Turkish population. Arch Toxicol. 2002;76:643–649. doi: 10.1007/s00204-002-0387-x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2008. [Google Scholar]
- van Duijnhoven FJ, Peeters PH, Warren RM, Bingham SA, Uitterlinden AG, van Noord PA, Monninkhof EM, Grobbee DE, van Gils CH. Influence of estrogen receptor alpha and progesterone receptor polymorphisms on the effects of hormone therapy on mammographic density. Cancer Epidemiol Biomarkers Prev. 2006;15:462–467. doi: 10.1158/1055-9965.EPI-05-0754. [DOI] [PubMed] [Google Scholar]
- Boyapati SM, Shu XO, Ruan ZX, Cai Q, Smith JR, Wen W, Gao YT, Zheng W. Polymorphisms in ER-alpha gene interact with estrogen receptor status in breast cancer survival. Clin Cancer Res. 2005;11:1093–1098. [PubMed] [Google Scholar]
- Gonzalez-Mancha R, Galan JJ, Crespo C, Iglesias Perez L, Gonzalez-Perez A, Moron FJ, Moreno Nogueira JA, Real LM, Pascual MH, Ruiz A, Royo JL. Analysis of the ERalpha germline PvuII marker in breast cancer risk. Med Sci Monit. 2008;14:CR136–CR143. [PubMed] [Google Scholar]
- Parl FF, Cavener DR, Dupont WD. Genomic DNA analysis of the estrogen receptor gene in breast cancer. Breast Cancer Res Treat. 1989;14:57–64. doi: 10.1007/BF01805976. [DOI] [PubMed] [Google Scholar]
- Hill SM, Fuqua SA, Chamness GC, Greene GL, McGuire WL. Estrogen receptor expression in human breast cancer associated with an estrogen receptor gene restriction fragment length polymorphism. Cancer Res. 1989;49:145–148. [PubMed] [Google Scholar]
- Andersen TI, Heimdal KR, Skrede M, Tveit K, Berg K, Borresen AL. Oestrogen receptor (ESR) polymorphisms and breast cancer susceptibility. Hum Genet. 1994;94:665–670. doi: 10.1007/BF00206961. [DOI] [PubMed] [Google Scholar]
- Long JR, Kataoka N, Shu XO, Wen W, Gao YT, Cai Q, Zheng W. Genetic polymorphisms of the CYP19A1 gene and breast cancer survival. Cancer Epidemiol Biomarkers Prev. 2006;15:2115–2122. doi: 10.1158/1055-9965.EPI-06-0464. [DOI] [PubMed] [Google Scholar]
- Cai Q, Shu XO, Jin F, Dai Q, Wen W, Cheng JR, Gao YT, Zheng W. Genetic polymorphisms in the estrogen receptor alpha gene and risk of breast cancer: Results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2003;12:853–859. [PubMed] [Google Scholar]
- Hu Z, Song CG, Lu JS, Luo JM, Shen ZZ, Huang W, Shao ZM. A multigenic study on breast cancer risk associated with genetic polymorphisms of ER Alpha, COMT and CYP19 gene in BRCA1/BRCA2 negative Shanghai women with early onset breast cancer or affected relatives. J Cancer Res Clin Oncol. 2007;133:969–978. doi: 10.1007/s00432-007-0244-7. [DOI] [PubMed] [Google Scholar]
- Onland-Moret NC, van Gils CH, Roest M, Grobbee DE, Peeters PH. The estrogen receptor alpha gene and breast cancer risk (The Netherlands) Cancer Causes Control. 2005;16:1195–1202. doi: 10.1007/s10552-005-0307-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Zuloeta Ladd AM, Vasquez AA, Rivadeneira F, Siemes C, Hofman A, Stricker BH, Pols HA, Uitterlinden AG, van Duijn CM. Estrogen receptor alpha polymorphisms and postmenopausal breast cancer risk. Breast Cancer Res Treat. 2008;107:415–419. doi: 10.1007/s10549-007-9562-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Li DK, Wu J, Zhang Z, Gao E. Joint effects of the CYP1A1 MspI, ERalpha PvuII, and ERalpha XbaI polymorphisms on the risk of breast cancer: Results from a population-based case-control study in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2006;15:342–347. doi: 10.1158/1055-9965.EPI-05-0485. [DOI] [PubMed] [Google Scholar]
- Shin A, Kang D, Nishio H, Lee MJ, Park SK, Kim SU, Noh DY, Choe KJ, Ahn SH, Hirvonen A, Kim JH, Yoo KY. Estrogen receptor alpha gene polymorphisms and breast cancer risk. Breast Cancer Res Treat. 2003;80:127–131. doi: 10.1023/a:1024439202528. [DOI] [PubMed] [Google Scholar]
- Vord PJ Vande, Wooley PH, Darga LL, Severson RK, Wu B, Nelson DA. Genetic determinants of bone mass do not relate with breast cancer risk in US white and African-American women. Breast Cancer Res Treat. 2006;100:103–107. doi: 10.1007/s10549-006-9228-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Higuchi R, Modugno F, Li J, Umblas N, Lee J, Lui LY, Ziv E, Tice JA, Cummings SR, Rhees B. Estrogen receptor alpha haplotypes and breast cancer risk in older Caucasian women. Breast Cancer Res Treat. 2007;106:273–280. doi: 10.1007/s10549-007-9497-8. [DOI] [PubMed] [Google Scholar]
- Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, Persson I, Baron J, Weiderpass E. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437–R449. doi: 10.1186/bcr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir K, Darabi H, Li Y, Low YL, Li YQ, Bonnard C, Sjolander A, Czene K, Wedren S, Liu ET, Hall P, Humphreys K, Liu J. ESR1 and EGF genetic variation in relation to breast cancer risk and survival. Breast Cancer Res. 2008;10:R15. doi: 10.1186/bcr1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Kataoka N, Li C, Wen W, Smith JR, Gao YT, Shu XO, Zheng W. Haplotype analyses of CYP19A1 gene variants and breast cancer risk: Results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2008;17:27–32. doi: 10.1158/1055-9965.EPI-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist DS, Aiello EJ, Miglioretti DL, White E. Mammographic breast density, dense area, and breast area differences by phase in the menstrual cycle. Cancer Epidemiol Biomarkers Prev. 2006;15:2303–2306. doi: 10.1158/1055-9965.EPI-06-0475. [DOI] [PubMed] [Google Scholar]
- Ursin G, Parisky YR, Pike MC, Spicer DV. Mammographic density changes during the menstrual cycle. Cancer Epidemiol Biomarkers Prev. 2001;10:141–142. [PubMed] [Google Scholar]
- White E, Velentgas P, Mandelson MT, Lehman CD, Elmore JG, Porter P, Yasui Y, Taplin SH. Variation in mammographic breast density by time in menstrual cycle among women aged 40–49 years. J Natl Cancer Inst. 1998;90:906–910. doi: 10.1093/jnci/90.12.906. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan G, Chow L, Schlosser A, Yaffe MJ, Boyd NF, Martin LJ. Differences in measured mammographic density in the menstrual cycle. Cancer Epidemiol Biomarkers Prev. 2009;18:1993–1999. doi: 10.1158/1055-9965.EPI-09-0074. [DOI] [PubMed] [Google Scholar]
- Sowers MR, Wilson AL, Karvonen-Gutierrez CA, Kardia SR. Sex steroid hormone pathway genes and health-related measures in women of 4 races/ethnicities: the Study of Women's Health Across the Nation (SWAN) Am J Med. 2006;119:S103–S110. doi: 10.1016/j.amjmed.2006.07.012. [DOI] [PubMed] [Google Scholar]


