Abstract
OBJECTIVE
Two independent genome-wide association studies for type 2 diabetes in Japanese subjects have recently identified common variants in the KCNQ1 gene that are strongly associated with type 2 diabetes. Here we studied whether a common variant in KCNQ1 would influence BMI as well as insulin secretion and action and predict future type 2 diabetes in subjects from Sweden and Finland.
RESEARCH DESIGN AND METHODS
Risk of type 2 diabetes conferred by KCNQ1 rs2237895 was studied in 2,830 type 2 diabetic case subjects and 3,550 control subjects from Sweden (Malmö Case-Control) and prospectively in 16,061 individuals from the Malmö Preventive Project (MPP). Association between genotype and insulin secretion/action was assessed cross- sectionally in 3,298 nondiabetic subjects from the Prevalence, Prediction and Prevention of Diabetes (PPP)-Botnia Study and longitudinally in 2,328 nondiabetic subjects from the Botnia Prospective Study (BPS). KCNQ1 expression (n = 18) and glucose-stimulated insulin secretion (n = 19) were measured in human islets from nondiabetic cadaver donors.
RESULTS
The C-allele of KCNQ1 rs2237895 was associated with increased risk of type 2 diabetes in both the Malmö Case-Control (odds ratio 1.23 [95% CI 1.12–1.34]; P = 5.6 × 10−6) and the prospective (1.14 [1.06–1.22]; P = 4.8 × 10−4) studies. Furthermore, the C-allele was associated with decreased insulin secretion (corrected insulin response [CIR] P = 0.013; disposition index [DI] P = 0.013) in the PPP-Botnia Study and in the BPS at baseline (CIR P = 3.6 × 10−4; DI P = 0.0058) and after follow-up (CIR P = 0.0018; DI P = 0.0030). C-allele carriers showed reduced glucose-stimulated insulin secretion in human islets (P = 2.5 × 10−6).
CONCLUSIONS
A common variant in the KCNQ1 gene is associated with increased risk of future type 2 diabetes in Scandinavians, which partially can be explained by an effect on insulin secretion.
Recently, two independent genome-wide association studies in Japanese subjects have shown that single nucleotide polymorphisms (SNPs) in the KCNQ1 gene (rs2074196, rs2237892, rs2237895, rs2283228, and rs2237897) are associated with type 2 diabetes (1,2). We have previously replicated association of rs2074196 and rs2237892 reported by Yasuda et al. (1) in Scandinavian subjects. Here we studied rs2237895, which is the only of the replicated variants by Unoki et al. (2) in a Danish population with a minor allele frequency >5% (43%). KCNQ1 encodes for a voltage-gated potassium channel that is highly expressed in cardiac muscle, pancreas, intestine, and kidney. Mutations in the KCNQ1 gene cause the long QT syndrome and deafness (3).
Here we studied whether rs2237895 increases risk of type 2 diabetes and/or affects insulin secretion and action in several Swedish and Finnish cross-sectional and prospective cohorts including a total of 28,067 individuals.
RESEARCH DESIGN AND METHODS
Characteristics of the study participants are reported in Table 1. Malmö Case-Control consisted of 2,830 diabetic case subjects from the Malmö Diabetes Registry (4) and 3,550 nondiabetic control subjects from the Malmö Diet and Cancer Study (5) in southern Sweden. All case subjects had Scandinavian origin, age at onset >35 years, C-peptide ≥0.3 nmol/l, and no GAD antibody. Control subjects had fasting blood glucose <5.5 mmol/l and A1C <6.0% (6).
TABLE 1.
Characteristics of the study participants
| Malmö Case-Control |
MPP | PPP-Botnia Study | BPS | ||
|---|---|---|---|---|---|
| Case subjects | Control subjects | ||||
| n (men/women) | 2,830 (1,667/1,163) | 3,550 (1,340/2,210) | 16,061 (10,416/5,645) | 3,298 (1,538/1,760) | 2,328 (1,065/1,263) |
| Age (years) | 57.9 ± 11.5 | 57.5 ± 6.0 | 45.5 ± 6.9 | 48.5 ± 15.9 | 45.5 ± 13.6 |
| BMI (kg/m2) | 29.6 ± 5.5 | 25.1 ± 3.6 | 24.3 ± 3.3 | 26.2 ± 4.2 | 25.6 ± 3.9 |
| Pfasting glucose (mmol/l) | 11.89 ± 4.34 | 4.78 ± 0.36 | 5.45 ± 0.56 | 5.16 ± 0.55 | 5.52 ± 0.57 |
| HOMA-IR | 3.08 (2.38) | 0.67 (0.46) | 0.95 (1.16) | 0.60 (0.50) | 0.58 (0.40) |
| CIR | — | — | 149 (127) | 157.7 (163.3) | 112.2 (124.6) |
| ISI | — | — | 165 (170) | 144.9 (111.9) | 147.6 (111.3) |
| DI | — | — | 23,393 (27,302) | 22,535 (25,855) | 15,717 (18,334) |
| Follow-up time (years) | — | — | 24.8 (4.7) | — | 7.6 (5.2) |
| KCNQ1 rs2237895 RAF | 0.44 | 0.40 | 0.41 | 0.49 | 0.47 |
Data are means ± SD or median (interquartile range) unless otherwise indicated. Baseline characteristics are shown for MPP and BPS. RAF, risk allele frequency.
The Malmö Preventive Project (MPP) is a large population-based prospective study from the city of Malmö, Sweden, consisting of 16,061 nondiabetic subjects, 2,063 of whom developed type 2 diabetes during a 24.8-year median follow-up period (7). Diagnosis of diabetes was confirmed from patient records or fasting plasma glucose ≥7.0 mmol/l.
The Prevalence, Prediction and Prevention of Diabetes (PPP)-Botnia Study is a population-based study from the Botnia region of western Finland. The current study includes 3,298 nondiabetic subjects (fasting plasma glucose <7.0 mmol/l and 2-h plasma glucose <11.1 mmol/l).
The Botnia Study started in 1990 at the west coast of Finland, aiming at identification of genes increasing susceptibility to type 2 diabetes in members from families with type 2 diabetes (8). The prospective part included 2,770 nondiabetic family members and/or their spouses (1,263 men and 1,507 women; mean age 45 years), 138 of whom developed type 2 diabetes during a 7.7-year (median) follow-up period (9). All subjects were given information about exercise and healthy diet and subjected at 2- to 3-year intervals to a new oral glucose tolerance test (OGTT). The current analyses includes 2,328 nondiabetic individuals (fasting plasma glucose <7.0 mmol/l and 2-h plasma glucose <11.1 mmol/l) with available longitudinal measurements of insulin secretion and action.
Measurements.
In MPP, fasting blood samples were drawn at baseline and follow-up visit for measurement of plasma glucose. In the PPP-Botnia Study, blood samples were drawn at 0, 30, and 120 min of the OGTT. In the Botnia Prospective Study (BPS), blood samples were drawn at −10, 0, 30, 60, and 120 min of the OGTT both at baseline and at follow-up. Insulin sensitivity index (ISI) from the OGTT was calculated as 10,000/√(Pfasting glucose × Pfasting insulin × mean OGTTglucose × mean OGTTinsulin) (10). The basal homeostasis model assessment–insulin resistance (HOMA-IR) index was calculated from fasting insulin and glucose concentrations (http://www.dtu.ox.ac.uk). β-Cell function was assessed as corrected insulin response (CIR) during OGTT (CIR = 100 × insulin30/[glucose30 × (glucose30−3.89)]) or as disposition index (DI), i.e., insulin secretion adjusted for insulin sensitivity (DI = CIR × ISI) (11).
Genotyping.
Genotyping was performed by an allelic discrimination method with a TaqMan assay on the ABI 7900 platform (Applied Biosystems, Foster City, CA). We obtained an average genotyping success rate of 93.2%, and the average concordance rate, based on 2,944 duplicate comparisons using a KASPar competitive allele-specific PCR system (Kbioscience, Hoddesdon, U.K.), was 99.9%. Hardy-Weinberg equilibrium was fulfilled in all studied populations (P > 0.20).
Glucose-stimulated insulin secretion.
Islets from 27 human cadaver donors were provided by the Nordic network for clinical islets transplantation by the courtesy of Olle Korsgren, Uppsala University, Uppsala, Sweden. The experimental protocol for isolation of islets was approved by the ethics committee of Uppsala University and performed in accordance with local institutional and Swedish national regulations.
Purified islets were collected under a stereomicroscope at room temperature. The islets were either directly subjected to Affymetrix analysis (see below) or preincubated for 30 min at 37°C in Krebs Ringer bicarbonate (KRB) buffer, pH 7.4, supplemented with 10 mmol/l N-2 hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 0.1% BSA, and 1 mmol/l glucose. Each incubation vial contained 12 islets in 1.0 ml KRB buffer solution and treated with 95% O2/5% CO2 to obtain constant pH and oxygenation. After preincubation, the buffer was changed to a medium containing either 1 or 20 mmol/l glucose. The islets were then incubated for 1 h at 37°C in a metabolic shaker (30 cycles per min). Immediately after incubation an aliquot of the medium was removed for measurement of insulin using a radioimmunoassay (Linco Research, Saint Charles, MO). Glucose-stimulated insulin secretion is expressed as fold change of insulin release from the islets by comparing release at 20 mmol/l with release using the 1 mmol/l glucose medium.
Expression of the KCNQ1 gene in human pancreatic islets.
Total RNA was isolated with the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany). RNA quality and concentration were measured using an Agilent 2100 bioanalyzer and Nanodrop ND-1000 equipment, respectively.
The microarrays were performed following the Affymetrix standard protocol. Briefly, total RNA 100–300 ng was processed following the GeneChip Expression 3′-Amplification Reagents One-cycle cDNA Synthesis Kit instructions (Affymetrix, Santa Clara, CA) to produce double-stranded cDNA. This was used as a template to generate biotin-targeted cRNA following manufacturer's specifications. Additionally, 15 μg of the biotin-labeled cRNA was fragmented to strands between 35 and 200 bases in length, 10 μg of which was hybridized onto the GeneChip Human Gene 1.0 ST whole transcript based–assay overnight in the GeneChip Hybridization Oven 6400 using standard procedures. The arrays were washed and stained in a GeneChip Fluidics Station 450. Scanning was carried out with the GeneChip Scanner 3000, and image analysis was performed using GeneChip Operating Software. The array data were summarized and normalized with robust multiarray analysis method using the software Expression Console (Affymetrix).
Statistical analyses.
Data are presented as means ± SD and, if not normally distributed, as median (interquartile range). The risk of developing type 2 diabetes is expressed as odds ratio (OR) using logistic regression analyses adjusted for age, sex, and BMI. Because men and women in MPP were included at different time periods, we adjusted for this using inclusion period and an interaction term inclusion period × sex. Genotype-phenotype correlations were studied using linear regression analyses adjusted for age, sex, and BMI (where appropriate). A robust variance estimate was used to adjust for within-pedigree dependence in BPS, treating each pedigree as an independent entity when calculating the variance of the estimates. Nonnormally distributed variables (insulin, HOMA-IR, ISI, CIR, and DI) were logarithmically (natural) transformed for analyses. For analysis of data from human pancreatic islets, one-way ANOVA was used to assess association between genotype and phenotype (expression and insulin secretion) and Spearman's rank correlation test was used to assess association of KCNQ1 mRNA level with insulin release. All statistical analyses were performed under an additive model with the Statistical Package for the Social Sciences v. 16.0 (SPSS, Chicago, IL) and STATA SE v. 10.1 (StataCorp, College Station, TX). Meta-analyses were performed with Metan using fixed-effect models using the inverse variance method. Inter-study heterogeneity was tested by Cochran Q and the I2 measure as implemented in STATA. Two-sided P values <0.05 were considered statistically significant.
RESULTS
Effect of KCNQ1 rs2237895 on risk of type 2 diabetes and glucose levels.
In the Malmö Case-Control study, the C-allele of KCNQ1 rs2237895 was more frequent in case subjects than in control subjects (0.44 and 0.40, respectively; χ2 P = 9.5 × 10−6), yielding an age-, sex-, and BMI-adjusted OR of 1.23 (95% CI 1.12–1.34; P = 5.6 × 10−6). The same SNP (C-allele) predicted future type 2 diabetes in the MPP (1.14 [1.06–1.22]; P = 4.7 × 10−4). In the MPP, nondiabetic carriers of the C-allele showed higher fasting plasma glucose levels both at baseline (P = 0.033) and after the 25-year follow-up period (P = 1.2 × 10−6) (Table 2). Also in the PPP-Botnia study, the C-allele carriers showed elevated fasting plasma glucose concentrations (P = 0.0067) (Table 2).
TABLE 2.
Metabolic effects of KCNQ1 rs2237895 in nondiabetic individuals from the studied populations
| n | Genotype |
Additive model |
|||||
|---|---|---|---|---|---|---|---|
| AA | AC | CC | β | SE | P | ||
| MPP | |||||||
| Baseline | |||||||
| BMI (kg/m2) | 12,326 | 24.0 ± 3.1 | 24.0 ± 3.1 | 24.0 ± 3.0 | −0.035 | 0.040 | 0.37 |
| Fasting plasma glucose (mmol/l) | 12,328 | 5.41 ± 0.55 | 5.42 ± 0.54 | 5.42 ± 0.55 | 0.015 | 0.007 | 0.033 |
| 2-h plasma glucose (mmol/l) | 6,718 | 6.28 ± 1.58 | 6.36 ± 1.65 | 6.36 ± 1.67 | 0.050 | 0.026 | 0.052 |
| Follow-up | |||||||
| BMI (kg/m2) | 12,271 | 26.9 ± 3.9 | 26.8 ± 3.9 | 26.8 ± 3.9 | 0.023 | 0.051 | 0.65 |
| Fasting plasma glucose (mmol/l) | 12,327 | 5.44 ± 0.54 | 5.46 ± 0.55 | 5.50 ± 0.55 | 0.033 | 0.007 | 1.2 × 10−6 |
| PPP-Botnia Study | |||||||
| BMI (kg/m2) | 2,991 | 26.1 ± 4.0 | 26.2 ± 4.2 | 26.1 ± 4.3 | 0.033 | 0.102 | 0.75 |
| Fasting plasma glucose (mmol/l) | 2,994 | 5.13 ± 0.53 | 5.17 ± 0.56 | 5.20 ± 0.56 | 0.038 | 0.014 | 0.0067 |
| 2-h plasma glucose (mmol/l) | 2,976 | 5.08 ± 1.53 | 5.17 ± 1.56 | 5.23 ± 1.65 | 0.074 | 0.038 | 0.053 |
| ISI | 2,838 | 145 (109) | 144 (111) | 144 (112) | 0.001 | 0.013 | 0.95 |
| HOMA-IR | 2,939 | 0.60 (0.50) | 0.60 (0.50) | 0.60 (0.50) | 0.001 | 0.014 | 0.93 |
| CIR | 2,849 | 166 (180) | 155 (158) | 157 (161) | −0.048 | 0.019 | 0.013 |
| CIR adjusted for ISI | −0.051 | 0.019 | 0.0073 | ||||
| DI | 2,814 | 23,464 (27,419) | 22,366 (25,013) | 21,866 (24,716) | −0.052 | 0.021 | 0.013 |
| BPS | |||||||
| Baseline | |||||||
| BMI (kg/m2) | 2,123 | 25.8 ± 3.9 | 25.4 ± 3.9 | 25.7 ± 3.9 | −0.065 | 0.119 | 0.59 |
| Fasting plasma glucose (mmol/l) | 2,128 | 5.59 ± 0.58 | 5.49 ± 0.56 | 5.53 ± 0.58 | −0.029 | 0.022 | 0.19 |
| 2-h plasma glucose (mmol/l) | 2,128 | 6.10 ± 1.45 | 6.08 ± 1.45 | 6.27 ± 1.50 | 0.072 | 0.048 | 0.13 |
| ISI | 2,128 | 144 (106) | 154 (114) | 146 (109) | 0.014 | 0.018 | 0.42 |
| HOMA-IR | 2,128 | 0.61 (0.42) | 0.57 (0.37) | 0.58 (0.43) | −0.015 | 0.016 | 0.34 |
| CIR | 2,128 | 118 (131) | 115 (126) | 96 (117) | −0.089 | 0.025 | 3.6 × 10−4 |
| CIR adjusted for ISI | −0.083 | 0.024 | 7.2 × 10−4 | ||||
| DI | 2,128 | 15,934 (19,260) | 16,663 (19,049) | 14,102 (16,413) | −0.075 | 0.027 | 0.0058 |
| Follow-up | |||||||
| BMI (kg/m2) | 2,068 | 26.8 ± 4.2 | 26.4 ± 4.0 | 26.8 ± 4.3 | −0.051 | 0.136 | 0.71 |
| Fasting plasma glucose (mmol/l) | 2,128 | 5.29 ± 0.57 | 5.30 ± 0.55 | 5.33 ± 0.60 | 0.032 | 0.018 | 0.077 |
| 2-h plasma glucose (mmol/l) | 2,128 | 5.77 ± 1.76 | 5.85 ± 1.67 | 6.00 ± 1.77 | 0.127 | 0.050 | 0.011 |
| ISI | 2,127 | 101 (94) | 102 (85) | 96 (93) | −0.003 | 0.019 | 0.89 |
| HOMA-IR | 2,125 | 0.97 (0.87) | 0.93 (0.76) | 0.96 (0.90) | −0.010 | 0.018 | 0.58 |
| CIR | 2,128 | 160 (184) | 142 (160) | 130 (140) | −0.086 | 0.027 | 0.0018 |
| CIR adjusted for ISI | −0.087 | 0.027 | 0.0013 | ||||
| DI | 2,127 | 15,563 (21,424) | 14,322 (18,304) | 13,299 (17,404) | −0.089 | 0.030 | 0.0030 |
| Meta analysis | |||||||
| BMI (kg/m2) | 0.41 | ||||||
| Fasting plasma glucose (mmol/l) | 0.0078 | ||||||
| 2-h plasma glucose (mmol/l) | 0.0021 | ||||||
| ISI | 0.58 | ||||||
| HOMA-IR | 0.57 | ||||||
| CIR | 3.0 × 10−5 | ||||||
| CIR adjusted for ISI | 2.7 × 10−5 | ||||||
| DI | 2.5 × 10−4 | ||||||
Data are means ± SD or median (interquartile range) unless otherwise indicated. β and SE from linear regression analysis adjusted for age, sex, BMI, and within-pedigree dependence (BPS) denote the effect size by each C-allele (additive model) on phenotype. Meta-analysis includes the PPP-Botnia Study and the BPS at baseline. P values < 0.05 are in boldface.
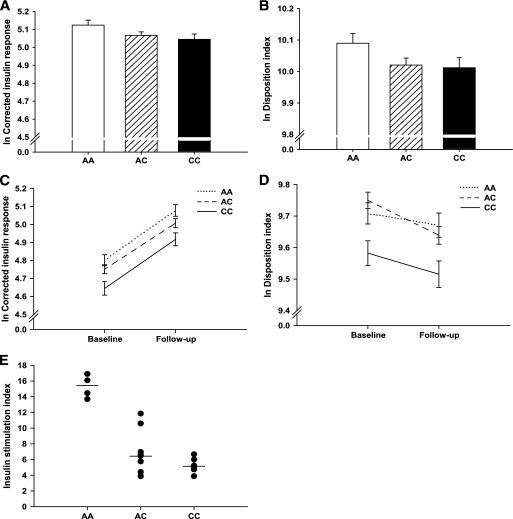
Effect of KCNQ1 rs2237895 on BMI, insulin secretion, and action.
There was no effect of the SNP on BMI in any of the cohorts. The C-allele carriers of the PPP-Botnia Study showed lower insulin response to glucose at 30 min during OGTT (CIR P = 0.013, DI P = 0.013; Table 2, Fig. 1A and B). Also in BPS, the C-allele was associated with reduced β-cell function at baseline (CIR P = 3.6 × 10−4; DI P = 0.0058) and at follow-up (CIR P = 0.0018; DI P = 0.0030) (Table 2, Fig. 1C and D). The KCNQ1 rs2237895 had no effect on insulin sensitivity estimated by HOMA-IR or ISI during OGTT.
FIG. 1.
Effect of KCNQ1 rs2237895 on β-cell function. Data are represented as the mean of the unadjusted logarithmic (natural) values for corrected insulin response and DI. Error bars denote SE of the mean. A: Decline in CIR with each C-allele (P = 0.013) in the PPP-Botnia Study. B: Decrease in DI with each C-allele (P = 0.013) in the PPP-Botnia Study. C: CIR in various genotype carriers at baseline and at follow-up in the BPS. The C-allele carriers had lower CIR at baseline (P = 3.6 × 10−4) that remained lower at follow-up (P = 0.0018). D: DI in various genotype carriers at baseline and at follow-up in the BPS. The C-allele carriers had lower DI at baseline (P = 0.0058) that remained lower at follow-up (P = 0.0030). E: Glucose-stimulated insulin secretion as fold change in insulin release at high (20 mmol/l) compared with low (1 mmol/l) glucose concentration from human islets in various genotype carriers. C-allele carriers showed lower glucose-stimulated insulin secretion (P = 2.5 × 10−6). KCNQ1 rs2237895 genotypes: AA, homozygous major allele carriers; AC, heterozygous; CC, homozygous minor (risk) allele carriers.
Expression of KCNQ1 and glucose-stimulated insulin secretion in human pancreatic islets.
We also analyzed KCNQ1 mRNA from microarray data on pancreatic islets from 18 nondiabetic human cadaver organ donors in relation to genotype. There was no significant difference in KCNQ1 expression in human islets among carriers of different genotypes (P = 0.65). Information on glucose-stimulated insulin secretion at 1 and 20 mmol/l of glucose was available for islets from 19 donors. Risk allele carriers showed lower glucose-stimulated insulin secretion measured as fold change of insulin release from the islets at 1 and 20 mmol/l glucose, i.e., stimulation index (P = 2.5 × 10−6) (Fig. 1E). These results remained unchanged when stimulation index was adjusted for the basal insulin secretion at 1 mmol/l of glucose (P = 4.7 × 10−5; supplementary Fig. 1, available online at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0246/DC1). We could not observe any correlation between KCNQ1 expression and glucose-stimulated insulin secretion (n = 10, r = 0.115, P = 0.75).
DISCUSSION
The key finding of the present study was that a common variant rs2237895 in the KCNQ1 gene was associated with increased risk of future type 2 diabetes because of impairment of β-cell function. Variants in KCNQ1 have been associated with type 2 diabetes predominantly in Asian subjects. Most of these studies had a case subject–control subject design that tends to overestimate the risk of a SNP because case subjects and control subjects usually represent two extremes of the distribution of glucose tolerance. This is to our knowledge the first population-based study using a prospective design showing that the SNP indeed increases risk of future type 2 diabetes and that this is due to failing β-cell function.
Although common variants in KCNQ1 increase susceptibility to type 2 diabetes in both Asians and Europeans, the frequency of the risk allele of most SNPs is much higher in Europeans than in Asians (92–96% in Europeans compared with 59–69% in Japanese), which most likely explains why SNPs in this gene were not significantly associated with type 2 diabetes in the initial European genome-wide association studies. In this regard, SNP rs2237895 represents an exception because the frequency of the risk allele was similar in Scandinavians (43%) and in Asians (36%) (1). The ORs for type 2 diabetes were also quite similar across studies: 1.23 in the current study, 1.31 in Asians (1), and 1.24 in Danes (2).
We also provide compelling evidence that the risk C-allele is associated with deterioration of β-cell function over time in the BPS, which thereby confirms and extends our previous observation of an association between the risk allele of another SNP (rs2237892) in KCNQ1 and impaired insulin secretion (P = 0.024) (1).
In analogy with Kir 6.2 (KCNJ11), KCNQ1 is an ATP-dependent potassium channel that is also expressed in human islets. It has been ascribed a role in insulin secretion, most likely through alterations in the membrane repolarization potential of the pancreatic β-cells. Indeed, we found that risk allele carriers had lower glucose-stimulated insulin secretion in human islets. We could not demonstrate a significant effect of the genotype on expression of the KCNQ1 gene in human islets, suggesting that the effect could be mediated through effects on splicing, translation, or posttranslational modifications. However, we cannot rule out that this lack of effect is due to low power because of limited number of human islets. In conclusion, we provide conclusive evidence that common variants in the KCNQ1 gene increase risk of future type 2 diabetes by causing impaired β-cell function.
Acknowledgments
These studies were supported by grants from the Swedish Research Council including a Linné Grant (31475113580), the Heart and Lung Foundation, the Swedish Diabetes Research Society, the European Community's Seventh Framework Programme (FP7/2007-2013), the ENGAGE project, Grant HEALTH-F4-2007-201413, a Nordic Centre of Excellence Grant in Disease Genetics, the Diabetes Programme at Lund University, the Påhlsson Foundation, the Crafoord Foundation, the Knut and Alice Wallenberg Foundation, the European Foundation for the Study of Diabetes (EFSD), the Finnish Diabetes Research Society, the Sigrid Juselius Foundation, the Folkhälsan Research Foundation, the Signe and Ane Gyllenberg Foundation, the Swedish Cultural Foundation in Finland, the Ollqvist Foundation, the Foundation for Life and Health in Finland, Jakobstad Hospital, the Medical Society of Finland, the Närpes Research Foundation, and the Vasa and Närpes Health Centers.
These studies were also supported by the Novo Nordisk Foundation. No other potential conflicts of interest relevant to this article were reported.
Human islets were provided by the Nordic network for clinical islets transplantation by the courtesy of Dr. Olle Korsgren, Uppsala, Sweden. We thank the patients for their participation and the Botnia Study Group for clinically studying the patients.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, Yamagata K, Hinokio Y, Wang HY, Tanahashi T, Nakamura N, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Takeda J, Maeda E, Shin HD, Cho YM, Park KS, Lee HK, Ng MC, Ma RC, So WY, Chan JC, Lyssenko V, Tuomi T, Nilsson P, Groop L, Kamatani N, Sekine A, Nakamura Y, Yamamoto K, Yoshida T, Tokunaga K, Itakura M, Makino H, Nanjo K, Kadowaki T, Kasuga M: Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 2.Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jorgensen T, Sandbaek A, Lauritzen T, Hansen T, Nurbaya S, Tsunoda T, Kubo M, Babazono T, Hirose H, Hayashi M, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Tai ES, Pedersen O, Kamatani N, Kadowaki T, Kikkawa R, Nakamura Y, Maeda S: SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008;40:1098–1102 [DOI] [PubMed] [Google Scholar]
- 3.Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, Keating MT: Spectrum of mutations in long-QT syndrome genes: KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation 2000;102:1178–1185 [DOI] [PubMed] [Google Scholar]
- 4.Lindholm E, Agardh E, Tuomi T, Groop L, Agardh CD: Classifying diabetes according to the new WHO clinical stages. Eur J Epidemiol 2001;17:983–989 [DOI] [PubMed] [Google Scholar]
- 5.Hedblad B, Nilsson P, Engstrom G, Berglund G, Janzon L: Insulin resistance in non-diabetic subjects is associated with increased incidence of myocardial infarction and death. Diabet Med 2002;19:470–475 [DOI] [PubMed] [Google Scholar]
- 6.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 7.Berglund G, Nilsson P, Eriksson KF, Nilsson JA, Hedblad B, Kristenson H, Lindgarde F: Long-term outcome of the Malmo preventive project: mortality and cardiovascular morbidity. J Intern Med 2000;247:19–29 [DOI] [PubMed] [Google Scholar]
- 8.Groop L, Forsblom C, Lehtovirta M, Tuomi T, Karanko S, Nissen M, Ehrnstrom BO, Forsen B, Isomaa B, Snickars B, Taskinen MR: Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes 1996;45:1585–1593 [DOI] [PubMed] [Google Scholar]
- 9.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L: Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008;359:2220–2232 [DOI] [PubMed] [Google Scholar]
- 10.Matsuda M, DeFronzo RA: Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 1999;22:1462–1470 [DOI] [PubMed] [Google Scholar]
- 11.Hanson RL, Pratley RE, Bogardus C, Narayan KM, Roumain JM, Imperatore G, Fagot-Campagna A, Pettitt DJ, Bennett PH, Knowler WC: Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am J Epidemiol 2000;151:190–198 [DOI] [PubMed] [Google Scholar]