Abstract
OBJECTIVE
Adipose inflammation plays a central role in obesity-related metabolic and cardiovascular complications. However, few human adipose-secreted proteins are known to mediate these processes. We hypothesized that microarray mRNA profiling of human adipose during evoked inflammation could identify novel adipocytokines.
RESEARCH DESIGN AND METHODS
Healthy human volunteers (n = 14) were treated with intravenous endotoxin (3 ng/kg lipopolysaccharide [LPS]) and underwent subcutaneous adipose biopsies before and after LPS. On Affymetrix U133Plus 2.0 arrays, adipose mRNAs modulated >1.5-fold (with P < 0.00001) were selected. SignalP 3.0 and SecretomeP 2.0 identified genes predicted to encode secreted proteins. Of these, 86 candidates were chosen for validation in adipose from an independent human endotoxemia protocol (N = 7, with 0.6 ng/kg LPS) and for exploration of cellular origin in primary human adipocytes and macrophages in vitro.
RESULTS
Microarray identified 776 adipose genes modulated by LPS; 298 were predicted to be secreted. Of detectable prioritized genes, 82 of 85 (96% [95% CI 90–99]) were upregulated (fold changes >1.0) during the lower-dose (LPS 0.6 ng/kg) validation study and 51 of 85 (59% [49–70]) were induced greater than 1.5-fold. Treatment of primary adipocytes with LPS and macrophage polarization to M1 proinflammatory phenotype increased expression by 1.5-fold for 58 and 73% of detectable genes, respectively.
CONCLUSIONS
We demonstrate that evoked inflammation of human adipose in vivo modulated expression of multiple genes likely secreted by adipocytes and monocytes. These included established adipocytokines and chemokines implicated in recruitment and activation of lymphocytes, adhesion molecules, antioxidants, and several novel genes with unknown function. Such candidates may represent biomarkers and therapeutic targets for obesity-related complications.
Activation of innate and adaptive immunity is a crucial link between adiposity and its metabolic complications (1–4). In rodents, modulation of toll-like receptor-4 (5), tumor necrosis factor (TNF) receptors (6), chemokines, and downstream kinases (7) attenuate diet-induced obesity and insulin resistance. Further, cross talk between immune cells and adipocytes promotes an inflammatory, insulin-resistant state in obesity. A key initiating event in adipose inflammation is recruitment of T-lymphocytes (8,9) and monocyte/macrophages (10,11) with elaboration of inflammatory adipocytokines that modulate metabolic signaling (12–15). Despite experimental evidence in rodent models, most evidence supporting these concepts in humans derives from observational and correlative studies (16–18). Indeed, validated adipokines that mediate, or serve as biomarkers for, complications of human adiposity remain limited.
Expression of inflammatory, insulin-signaling, and lipid genes are perturbed in adipose of obese humans (19–21). Recently, the in vitro secretome of subcutaneous and visceral primary human adipocytes was described and includes many unexplored proteins modulated during adipogenesis (1,22). Remarkably, less than half of genes found in the human subcutaneous adipocyte secretome were previously found in the murine 3T3-L1 preadipocyte secretome (22), underscoring the importance of studies in human tissue.
Experimental human endotoxemia can provide unique insights into the relationship of inflammation to metabolic disturbance in man (23,24). Others and we have shown that endotoxemia induces acute metabolic, lipoprotein, and oxidant responses that resemble the chronic changes in insulin resistance and metabolic syndrome (25,26). Notably, endotoxemia induces adipose inflammation (27) with activation of several adipose inflammatory cascades, including cytokines, chemokines, and suppressor of cytokine signaling (SOCS) molecules (26) that attenuate insulin signaling and are implicated in obesity and type 2 diabetes (28).
We applied microarray mRNA profiling of human adipose during endotoxemia to identify novel inflammation-induced adipose genes. We focused on genes predicted to be secreted and validated our findings in vivo through independent experiments of low-grade human inflammation. Finally, we identified in vitro the likely human adipose cellular source of these top candidates.
RESEARCH DESIGN AND METHODS
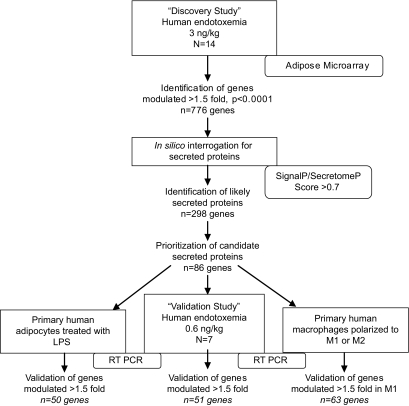
An overview of our design is provided in Fig. 1. All clinical studies were performed with approval of University of Pennsylvania Institutional Review Board after written informed consent from all subjects.
FIG. 1.
Overview of study design and results. The discovery study utilized microarray analysis of human adipose tissue isolated during in vivo endotoxemia (3 ng/kg) to identify genes modulated by inflammation (n = 776). Bioinformatics tools (SignalP 3.0 or SecretomeP 2.0 score >0.7) established which of these were likely to encode secreted proteins (n = 298). This list was prioritized to create a subset of genes (n = 86) based upon 1) novelty, 2) degree of induction, 3) sustained upregulation, 4) prior publication as secreted proteins in vitro or in vivo, 5) evidence for functional roles in inflammation, insulin signaling, or adipose biology, and 6) known association with diseases of interest. The validation study involved RT-PCR analysis of these 86 genes in human adipose isolated during in vivo endotoxemia (0.6 ng/kg) to confirm microarray findings and to identify genes modulated during lower-grade, more physiologic inflammation. Cell studies in primary human adipocytes after treatment with LPS and macrophages after polarization to inflammatory (M1) phenotype were also performed to determine likely cellular origin of these 86 genes.
Discovery human endotoxemia protocol.
As previously described (26), healthy volunteers, aged 18–40 years, with BMI 18–30 kg/m2 were recruited from the Delaware Valley. Exclusions included past medical history including inflammatory diseases, medication or supplement use, or pregnancy. Participants underwent three Clinical and Translational Research Center visits: visit one for screening; visit two for dietary run-in; and visit three for inpatient stay comprising an acclimatization phase and a postlipopolysaccharide (post-LPS) study phase. Serial whole blood samples were collected before and 2, 4, 6, 8, 12, 16, and 24 h after intravenous bolus of 3 ng/kg U.S. standard reference endotoxin (LPS; lot No. CC-RE-LOT-1 + 2; Clinical Center, National Institutes of Health, Bethesda, MD). Serial subcutaneous adipose samples were collected by core needle aspiration through a 4 mm gluteal incision from distinct sites, 30 min before and 4, 12, and 24 h after LPS; samples were treated with RNA Later and stored at −70°C. This report focuses on the subset that underwent microarray mRNA profiling of adipose (N = 14).
Microarray.
Adipose tissue RNA was extracted using RNeasy Lipid Tissue Mini kit (Qiagen, Valencia, CA) and underwent extensive quality control (QC) (including spectrometry, picogreen quantitation, and electrophoresis for RNA quality) before first-strand cDNA synthesis (100 ng), cRNA synthesis, and hybridization to Affymetrix GeneChip Human Genome U133Plus 2.0 arrays (Santa Clara, CA) in the Penn Microarray Facility (www.me.upenn.edu/microarr/). Microarrays were washed and then stained with amplified fluorescence markers, and signals were collected on a confocal scanner at 3 μm resolution (excitation at 570 nm). Affymetrix Microarray Suite 5.0 was used to quantitate expression levels of average signal from two sequential scans. Array signal intensities passed a two-stage QC process including affyQCReport package. All arrays had similar background values and distributions of intensity.
Adipose microarray statistical and informatic methods
Identification of candidate LPS-modulated genes.
Arrays were normalized, adjusted for background signal using robust multiarray analysis (29), and filtered by removing probesets with little evidence of expression using the PM/MM probes (63) or expression levels consistently less than threefold background. For the remaining 13,646 genes, mixed effects models using shrinkage estimates of the variance components were fit, with the goal of identifying genes with highly significant fold changes in expression between baseline and each time point (4, 12, and 24 h). Genes were selected for further study if the permutation-based P value for the null hypothesis test of a fold change of unity at any time point was less than 1 × 10−5 (30). Permutation-based false discovery rates were assessed at each time point (31). To increase the chance of identifying genes with detectable changes at the protein level, we required a minimum cutoff of at least 1.5-fold change in intensities per gene. Analyses were performed using Bioconductor (www.bioconductor.org) run under R 2.6 (R Foundation for Statistical Computing, Vienna, Austria) with libraries including affy, maanova, and panp.
Secreted protein identification.
We interrogated in silico our candidate LPS-modulated genes using the validated protein informatic software SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (32,33) and SecretomeP 2.0 (http://www.cbs.dtu.dk/services/SecretomeP/) (34) to identify the subset of human genes with highest probability of encoding either signal peptides (SignalP) or protein sequences predictive of nonclassic secretion (SecretomeP). For SignalP, we used a cutoff d-score of 0.7 as the score above which frequency of containing a signal peptide is greatly increased in known eukaryotic proteins (32). A SecretomeP cutoff of 0.7 was chosen as the cutoff above which Bendsten et al. (34) demonstrated that all positive training examples and no negative training examples were classified as nonclassically secreted. Genes encoding sequences that met the cutoff value for either classic or nonclassic secretion comprise our preliminary candidate “human adipose tissue inflammatory secretome.”
Prioritization for validation.
For initial prioritization, we focused on upregulated genes as being more likely to have detectable and increased protein levels in adiposity-related disease. Thus, candidate inflammatory, adipose-secreted proteins were prioritized as follows: 1) novelty genes with unknown functions, 2) >threefold upregulation at any time point, 3) >1.5-fold upregulation at two or more time points, 4) prior publication as secreted proteins in vitro or in vivo, 5) evidence for functional roles in inflammation, insulin signaling, or adipose biology, and 6) known association with diabetes, obesity, metabolic syndrome, or cardiovascular disease. Genes that did not meet any of the criteria were not studied further. This resulted in a list of 86 candidates (including positive controls interleukin [IL]-6, TNF, and monocyte chemotactic protein [MCP]-1) that were selected for validation and identification of adipose tissue cell of origin.
Functional classification.
Prioritized genes were uploaded to DAVID (Database for Annotation, Visualization, and Integrated Discovery) Bioinformatics Resource and the Gene Functional Classification tool was utilized to generate clusters of related and overrepresented genes (35,36). The enrichment score (geometric mean of −log P values) is presented.
Validation human endotoxemia protocol.
Healthy volunteers (n = 10) were recruited from the same population as Discovery Study, using similar criteria. Eligible participants underwent dietary counseling and were admitted for inpatient stay including acclimatization and post-LPS phases. Serial whole blood (at 0, 2, 4, 6, 12, 16, and 24 h) and subcutaneous gluteal adipose tissue samples (at 0, 4, 12, and 24 h) were collected before and after intravenous infusion of 0.6 ng/kg endotoxin and processed as above. We focused on the subset (n = 7) in whom adipose tissue samples were assayed by quantitative PCR.
Cell studies
Primary human adipocyte studies.
Fresh human adipose tissue obtained from abdominal surgery specimens via the Penn Human Adipose Resource was the source of primary human adipocytes. Briefly, the specimen was digested with collagenase (Roche Applied Science, Indianapolis, IN), filtered, and centrifuged, and the cellular layer was resuspended in OF medium (DMEM/F12 plus penicillin/streptomycin, biotin [4 mg/l] and pantothenate [8 mg/l]) with 20% FBS and plated at 30,000 cells/cm2. Once confluent, cells were differentiated in serum-free differentiation medium (OF media, insulin [20 nmol/l], hydrocortisone [1 μmol/l], dexamethasone [250 nmol/l], human transferrin [10 mg/l], T3 [0.2 nmol/l], isobutylmethylxanthine [500 μmol/l], and peroxisome proliferator–activated receptor-γ agonist GW347845 [2 μmol/l] [gift from GlaxoSmithKline, King of Prussia, PA]) until 80% mature (7–10 days). Cells were washed and treated ±LPS 100 ng/ml in DMEM/F12 with 0.1% FBS plus penicillin/streptomycin for 2, 4, 12, and 24 h for two independent experiments, in triplicate. Unless noted, reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Primary human macrophage studies.
Human monocytes (peripheral blood mononuclear cells, >95% expression of CD14 and HLA-DR) were isolated from donor blood after apheresis and elutriation in the Penn Center for AIDS Research. As described (37), cells were plated at 3 × 105 cells/cm2 and cultured in RPMI with 20% FBS and penicillin/streptomycin then supplemented with 100 ng/ml MCSF (Sigma, St. Louis, MO) to promote differentiation to macrophages over 7 days. In two independent experiments, each in triplicate, macrophages were washed and treated for 16 h with RPMI + 5% FBS alone, 100 ng/ml LPS and 20 ng/ml Interferon Gamma (R&D Systems, Minneapolis, MN) for differentiation to the M1 phenotype, or 20 ng/ml recombinant human IL4 (R&D Systems) for differentiation to the M2 phenotype (38).
General laboratory methods
RNA extraction, cDNA synthesis, and quantitative PCR.
RNA was isolated from adipose tissue, adipocytes, and macrophages using Trizol reagent (Invitrogen, Carlsbad, CA) and was reverse transcribed (500 ng) to cDNA (High Capacity cDNA Archive Kit; ABI, Foster City, CA). Expression of genes was determined by real-time PCR (ABI7900 system) using TaqMan Universal PCR MasterMix primers and probes. To control for between-sample differences, mRNA levels were normalized to β-actin for each sample by subtracting the Ct for β-actin from the Ct for gene of interest, producing a ΔCt value. The ΔCt for each post-treatment sample was compared with the mean ΔCt for all pretreatment samples using the relative quantitation 2−ΔΔCt method to determine fold change from baseline (39).
Metabolic and inflammatory markers.
As previously described (26), plasma levels of TNF-α, IL-6 (Linco Multiplex ELISAs on Luminex IS100; Austin, TX), and C-reactive protein (high-sensitivity Latex turbidimetric immunoassay; Wako, Osaka, Japan) were measured in duplicate per manufacturers' guidelines. Intra- and interassay coefficients of variation for TNF ELISA were 8.66 and 20.4%, respectively, and for IL-6, 8.7 and 10.9%, respectively. Plasma lipids and glucose were measured enzymatically on Hitachi 912 automated chemistry (Wako Diagnostics, Richmond, VA) (26). Subject data are reported as means ± SD for continuous variables and as proportions for categorical variables. Effects of endotoxemia on clinical parameters were tested by mixed-effects modeling or repeated-measures ANOVA (26).
Statistical analysis of quantitative PCR data.
For validation, study adipose tissue fold changes at each time point were estimated using the method of generalized estimating equations to account for within-subject correlations. For adipocytes and macrophages, analysis of mRNA fold change utilized geometric means of the independent replicated samples. For the time point with peak fold change, the means and 95% CIs are reported for all studies. Correlation analyses between studies were performed by Spearman's rank correlation of the log10 peak fold change. The proportions of upregulated genes for validation, adipocyte, and macrophage studies were reported, and their 95% exact binomial CIs were calculated using Clopper and Pearson (40). Analyses were performed using the R 2.6 and Stata 10 (StataCorp, College Station, TX).
RESULTS
Baseline characteristics of participants and clinical responses to endotoxemia.
Participants in the discovery and validation studies had similar baseline characteristics with normal blood pressure, plasma lipoproteins, and BMI, although the validation study enrolled more female participants (Table 1). In the discovery study, as reported (26), there was a transient clinical and biochemical inflammatory response to LPS 3 ng/kg, whereas the lower LPS dose of 0.6 ng/kg in the validation study produced a subclinical response with modest increases in plasma cytokines but no significant symptoms or fever (Table 2).
TABLE 1.
Baseline characteristics of the discovery and validation study samples
| Discovery study 3 ng/kg endotoxin (n = 14) | Validation study 0.6 ng/kg endotoxin (n = 7) | |
|---|---|---|
| Age (years) | 25.0 ± 6.5 | 24.4 ± 3.1 |
| Female sex, n (%) | 4 (28%) | 5 (71%) |
| Race | ||
| White (%) | 12 (86%) | 6 (86%) |
| African American (%) | 2 (14%) | 1 (14%) |
| Systolic blood pressure (mg Hg) | 121 ± 9 | 115 ± 7.9 |
| BMI (kg/m2) | 24.0 ± 4.0 | 24.4 ± 2.6 |
| Fasting glucose (mmol/l) | 4.68 ± 0.28 | 3.89 ± 0.77 |
| Total cholesterol (mmol/l) | 4.53 ± 0.52 | 4.65 ± 0.60 |
| LDL cholesterol (mmol/l) | 2.46 ± 0.67 | 2.21 ± 0.56 |
| HDL cholesterol (mmol/l) | ||
| Men | 1.33 ± 0.26 | 1.24 ± 0.05 |
| Women | 1.69 ± 0.34 | 1.81 ± 0.19 |
| Triglycerides (mmol/l) | 0.86 ± 0.39 | 1.0 ± 0.3 |
Data are means ± SD unless otherwise indicated.
TABLE 2.
Clinical and inflammatory responses to endotoxemia
| Discovery study (n = 14) |
Validation study (n = 7) |
|||||
|---|---|---|---|---|---|---|
| Pre-LPS | Post-LPS | Time of peak | Pre-LPS | Post-LPS | Time of peak | |
| TNF (pg/ml) | 3.0 ± 1.1 | 507 ± 251* | 2 h | 1.0 ± 0.6 | 22 ± 13† | 2 h |
| IL-6 (pg/ml) | 3.2 ± 17.1 | 1,607 ± 650* | 2 h | 1.7 ± 1.8 | 107 ± 31* | 2 h |
| Heart rate (bpm) | 76 ± 11 | 107 ± 13.6* | 4 h | 64 ± 11 | 80 ± 15‡ | 4 h |
| Temperature (οC) | 36.5 ± 0.3 | 37.8 ± 0.9* | 4 h | 36.4 ± 0.4 | 36.7 ± 0.4 | 4 h |
| White blood cell count | 6.5 ± 1.2 | 13.8 ± 3.2* | 12 h | 6.1 ± 1.4 | 8.4 ± 1.9‡ | 4 h |
| High-sensitivity C-reactive protein (mg/l) | 0.49 ± 0.21 | 42.4 ± 8.4† | 24 h | 1.5 ± 1.5 | 16.7 ± 7.6† | 24 h |
Data are means ± SD unless otherwise indicated. For each study, within-subject pre- vs. post-LPS comparisons are reported.
*P < 0.0001,
†P < 0.005,
‡P < 0.05.
Endotoxemia modulates expression of many human adipose tissue genes.
We identified 696 genes at 4 h, 130 genes at 12 h, and 24 genes at 24 h that met criterion for selection. The false discovery rates for the three time points were 3 × 10−5, 4 × 10−4, and 0.001, respectively, suggesting strong evidence of differential expression among these genes. Many genes were modulated at multiple time points, resulting in 776 unique genes modulated at any time point (696 at 4 h, 68 additional at 12 h, and 12 additional at 24 h). Approximately half the genes were upregulated (fold change >1.5), the remaining were downregulated (Table 3).
TABLE 3.
Summary of data for modulated genes and secreted proteins in adipose during human endotoxemia
| Time post-LPS | Modulated genes | Secreted | Classically secreted |
|---|---|---|---|
| 4 h | |||
| Total number | 696 | 266 | 198 (75) |
| Upregulated | 414 (61) | 168 (63) | |
| 12 h | |||
| Total number | 130 | 72 | 57 (80) |
| Upregulated | 75 (58) | 45 (64) | |
| 24 h | |||
| Total number | 24 | 15 | 11 (72) |
| Upregulated | 11 (46) | 8 (46) |
Data are n or n (%). Number of genes up- or downregulated >1.5 fold are shown.
A subset of modulated adipose tissue genes is predicted to encode secreted proteins.
Based on the SignalP 3.0 and SecretomeP 2.0 analysis, a total of 266 genes at 4 h, 72 at 12 h, and 15 at 24 h were predicted to encode secreted proteins (Table 3). This corresponds to 298 unique genes and represents 38% of all modulated genes, suggesting an over-representation of secreted proteins relative to the entire Affymetrix U133Plus 2.0 array (∼24%) and the human genome (15–20%). The majority of these (75%) were predicted to be classically secreted. More were upregulated (63% at 4 h and 12 h, 54% at 24 h) than downregulated.
For the current report, only upregulated genes predicted to encode secreted proteins were chosen for further validation (n = 86). These inflammatory, human adipose tissue–secreted factors included cytokines (e.g., IL6, TNF), chemokines (e.g., CXCL9, CCL4), growth factors (e.g., BMP-2), acute-phase reactants (e.g., PTX3, HP), adhesion molecules (e.g., ICAM1, VCAM), cell surface markers (e.g., CD14, CD274), antioxidants (e.g., metallothioneins), and novel genes with unknown functions (e.g., C19orf59, KIAA0247). In addition, several prioritized genes were identified that have been implicated in inflammatory processes in human or animal models but not described in adipose function (e.g., CEACAM, ISG15, CX3CL1). The heatmap in supplemental Fig. 1, available in an online appendix at http://diabetes.diabetesjournals.org/content/early/2009/06/23/db09-0256/suppl/DC1 depicts fold changes (ranked from highest to lowest fold change) in the discovery study adipose, as determined by microarray. Greatest mRNA modulation occurred at 4 h, but some genes had later induction or sustained modulation over time.
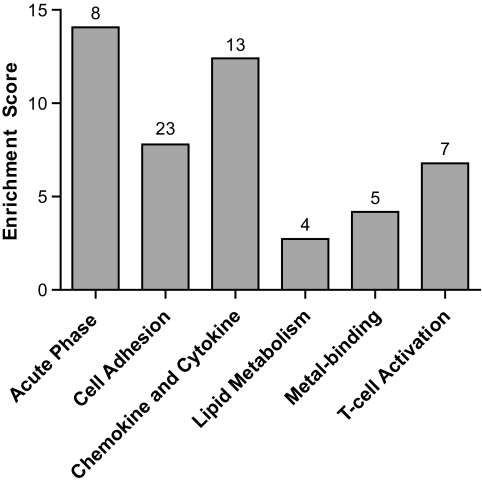
The majority of prioritized genes fall into six functional categories defined by DAVID. Figure 2 depicts the overrepresented categories that include cell adhesion, chemokine and cytokine, acute-phase response, T-cell activation, metal binding, and lipid metabolism. Remarkably, 28 genes including many unknowns remained uncategorized. Supplemental Table 1 (available in an online appendix) gives the full lists of genes in each category.
FIG. 2.
Database for DAVID functional categories with enrichment scores. Numbers of genes in each category are listed above each bar. The majority of genes are categorized into inflammatory pathways but some cluster into lipid and metal-binding categories. Many genes (28 of 86), often with unknown functions, did not classify into any known categories.
Inflammatory modulation of many prioritized genes replicates in human adipose tissue samples during an independent validation endotoxemia study.
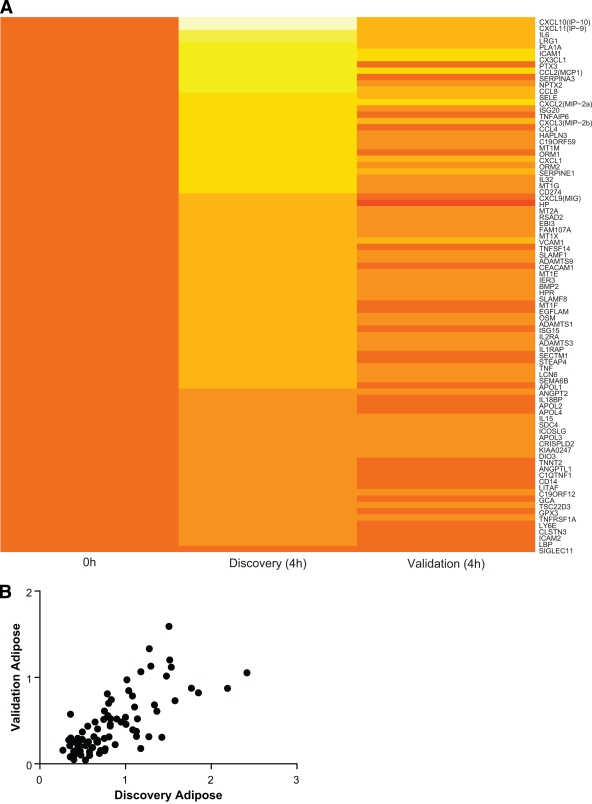
We replicated endotoxemia-induced adipose tissue genes in an independent study of lower-grade endotoxemia (0.6 ng/kg). This dose resulted in modest cytokine induction without clinically significant responses and therefore may more closely reflect the modest inflammation observed in human obesity. Of the 86 prioritized genes, 85 (98% [95% CI 94–100]) were confirmed to be expressed (Ct value <37). During 0.6 ng/kg endotoxemia, 82 of 85 (96% [90–99]) were increased (fold changes >1.0) and 51 of 85 (60% [49–70]) were upregulated by greater than 1.5-fold. The heatmap in supplemental Fig. 2 depicts mRNA expression changes over time. Compared with the discovery study, the lower-dose LPS validation study induced a more modest but similar pattern of expression (Fig. 3A). Remarkably, there was a relatively high correlation (r = 0.64, P < 0.0001) of peak mRNA fold change between the discovery and validation studies (Fig. 3B).
FIG. 3.
Comparison between endotoxin-induced adipose mRNA fold change in discovery versus validation study. A: Heatmap depicting direct comparison of 4 h log10 expression changes of 86 high-priority genes in discovery study versus validation study, ranked in order of log10 fold change in discovery study. The National Center for Biotechnology Information gene identification names are listed on the right. Time 0 values are set to one (log10 changes = 0). Color coding: 0 is set to orange as seen at baseline, yellow indicates upregulation, red indicates downregulation. B: Correlation (Spearman) between log10 peak fold change of 85 detectable genes in discovery versus validation study. r = 0.64; P < 0.0001.
The majority of the putative adipose tissue inflammatory secretome is expressed in human adipocytes and induced by LPS in vitro.
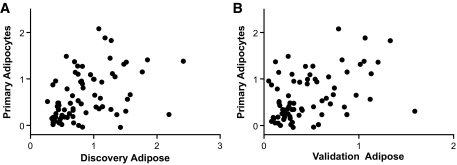
Analysis of primary human adipocytes revealed that 77 of 86 (90%) prioritized genes had detectable expression (Ct value <37) in resting adipocytes, 68 of 77 (88% [95% CI 79–94]) were increased (>1.0-fold), and 50 of 77 (65% [53–75]) were upregulated by greater than 1.5-fold after 100 ng/ml LPS. There was a remarkable correlation of peak mRNA fold change between discovery human adipose and adipocytes (Fig. 4A) as well as between validation adipose and adipocytes (Fig. 4B), suggesting that adipocytes are a major source of inflammatory modulated genes in human adipose tissue in vivo.
FIG. 4.
Comparison between endotoxin-induced mRNA fold change in human adipose versus primary adipocytes. A: Spearman correlation between log10 peak fold change of 77 detectable genes in primary human adipocytes (after 100 ng/ml LPS) and human adipose in discovery study (3 ng/kg endotoxin). r = 0.49; P < 0.0001. B: Correlation between log10 peak fold change of 77 detectable genes in primary human adipocytes (after 100 ng/ml LPS) and human adipose in validation study (0.6 ng/kg endotoxin). r = 0.42; P < 0.0001.
Many candidate genes are expressed in human macrophages and upregulated during polarization to the M1-phenotype.
Next, expression was tested in classically activated proinflammatory (M1) and alternatively activated anti-inflammatory (M2) macrophages (41,42) relative to undifferentiated macrophages. In resting macrophages, 72 of 86 mRNAs (84%) were detectable (Ct <37) whereas 66 of 72 (92% [95% CI 83–97]) were increased (>1.0-fold) and 63 of 72 (88% [78–94]) were upregulated by >1.5-fold during polarization to the M1 phenotype, supporting their potential role in inflammatory macrophage signaling in human adipose tissue. In contrast, genes upregulated in adipose tissue tended to be unchanged or downregulated in M2 macrophages; only 22 of 72 (31% [21–43]) were increased >1.0-fold and 14 of 72 (20% [11–31]) were upregulated >1.5-fold.
A summary of findings in the discovery and validation studies as well as in primary human adipocytes and human macrophages is presented in supplementary Table 2, available in an online appendix. Overall, of the 51 genes confirmed to be upregulated >1.5-fold in human adipose tissue in our validation study, the majority (46 of 51, 90%) was induced by LPS in adipocytes and/or M1 macrophages (Table 4).
TABLE 4.
Induction of validated adipose genes (n = 51) by LPS in human adipocytes and during polarization to M1 macrophages
| Up in adipocytes | Not up in adipocytes | |
|---|---|---|
| Up in M1 | 32 (63%) | 9 (18%) |
| Macrophages | CCL8, CXCL10, MT2A | CXCL9, IL2RA, SLAMF1 |
| Not up in M1 | 5 (10%) | 5 (10%) |
| Macrophages | TNFSF14, ICOSLG, ANGPT2 | LCN6, DIO3, CEACAM1 |
Data are n (%). Number of genes upregulated >1.5 fold in respective cell-type are shown, along with examples of genes in each category.
DISCUSSION
This mRNA microarray analysis of human adipose tissue during experimental endotoxemia is the first attempt to identify novel adipocytokines using an in vivo evoked adipose inflammatory model. Our strategy identified genes encoding established adipokines implicated in obesity-related insulin resistance as well as emerging adipose tissue–secreted factors (1) and multiple secreted proteins with unknown adipose functions. These experimental human studies are internally consistent, provide independent validation, and define the likely cellular origin. Such candidates may represent new biomarkers and therapeutic targets for the treatment of obesity-related complications.
Microarray analysis of gene expression is established for discovery of disease-related genes (43). Indeed, mRNA profiling of adipose has identified novel genes involved in obesity and type 2 diabetes (44). Several reports identified genes involved in inflammation, insulin signaling, and lipid metabolism as differentially expressed in adipose tissue of obese humans (19–21). We also identified many of these genes suggesting that our model may be useful in human adipokine discovery. For example, 9 of 89 genes differentially expressed in omental adipose of obese subjects (20) were also upregulated in our study (ADRB2, IFIT2, IL10RA, IL6, ISG20, JAK2, LDL4, MAP3K4, and NGFR). In adipocytes cultured ex vivo from subcutaneous adipose of obese versus nonobese Pima Indians, 38 upregulated genes were also increased in our study (21), including 13 likely secreted proteins that we prioritized for validation (APOL1, APOL3, CCL2, CCL4, CD14, CXCL1, CXCL2, CXCL3, ICAM2, IER3, MT2A, SERPINE1, and TNFRSF1A).
The dose of LPS (3 ng/kg) used in the discovery study induced an acute and more marked inflammatory state than that found in obesity. This superphysiologic activation of innate immunity allowed us to identify in vivo a broad range of adipose inflammatory genes, with the caveat that the degree of modulation may be greater than that in human adiposity. To explore our findings in a setting more relevant to the low-grade inflammation in central obesity, we validated adipose tissue upregulation in an independent human study of very-low-dose endotoxemia (0.6 ng/kg) that results in modest cytokine induction without significant clinical inflammatory responses. Notably, the majority of adipose tissue–upregulated genes identified in the discovery study were also induced in the validation study, suggesting that they may be modulated during the more modest adipose inflammation of obesity.
Our in silico–predicted adipose inflammatory secretome is remarkable because it included 1) many established adipocytokines including TNFα, IL-6, MCP-1, plasminogen activator inhibitor (PAI)-1, and selectin E; 2) secreted inflammatory molecules with known immune functions but no known role in adipose or metabolic signaling (e.g., CX3CL1, ISG15, and EBI3); 3) a number of proteins recently identified in the human visceral adipose secretome (1) (e.g., CSF-1, PAI-1, ADAMSTS, and PTX3); and 4) multiple genes encoding secreted proteins without known functions. Functional classification of the 86 genes revealed several expected inflammatory categories including acute-phase response, chemokines and cytokines, as well as cell adhesion molecules. The overrepresentation of T-cell–activating genes is of particular interest, given the recent implication of T-cell recruitment in obesity-related adipose tissue inflammation and insulin resistance (2,8,45). An inflammatory impact on adipocyte metabolic and redox activities is suggested by excess representation of lipid and metal-binding gene categories. Notably, 28 genes, many with completely unknown functions, remained uncategorized, highlighting the potential for this model to provide novel insights into human adipose biology.
As macrophage infiltration is a key event in adipose tissue inflammation and insulin resistance (10,11), we expected that the evoked inflammatory state would induce factors involved in recruitment and differentiation of monocytes. Chemokines such as MCP1, CSF1, CXCL3, and CX3CL1 were upregulated at early time points. Macrophage cell markers, such as CD14 and CD64, and macrophage-secreted proteins such as MIP-1 (CCL3), IL8, and IL18BP were also induced by LPS. In fact, many of the overexpressed genes are known to be macrophage secreted, and our validation studies in human M1 macrophages confirmed that ∼75% of genes induced in adipose tissue by LPS were also upregulated during in vitro polarization to inflammatory macrophages.
Macrophages are polarized from a resting state to “classically activated” (M1) or “alternatively activated” (M2) phenotypes with proinflammatory or anti-inflammatory activities, respectively (41,42). In mouse models, obesity induces a phenotypic switch in adipose tissue macrophages (ATMs) from a predominantly M2 phenotype that protects against insulin resistance to an M1 inflammatory state that confers sensitivity to TNFα-induced insulin resistance (46). In humans, ATMs possess a complex phenotype with some M2 markers but M1-like proinflammatory cytokine production (47). Of note, our predicted adipose inflammatory secretome contained 22 genes found to be upregulated in the M1 phenotype compared with resting macrophages (48), and our human macrophage studies confirmed M1-macrophages as a potential source of many inflammatory molecules.
T-cell recruitment to adipose tissue has been recently identified in obesity-induced adipose inflammation. In fact, some findings suggest T-cells are found in adipose and can induce insulin resistance before macrophage recruitment (2,8,45). These inflammatory T-cells secrete cytokines/chemokines that switch resident ATMs to the M1 phenotype while recruiting additional circulating monocytes. Several potent T-cell chemokines, including CCL4, CXCL9, CXCL10, CXCL11, and IL15, were highly induced by endotoxin. Additionally, overexpression of T-cell–induced genes (ISGs, ICOSLG, and IRFs) was detected, suggesting increased T-cell activity.
We confirmed in vitro that both human adipocytes and M1-proinflammatory macrophages are likely cellular sources for the vast majority of our adipose inflammatory secretome. By focusing on these cells, we have excluded other adipose components (e.g., stromal cells). Of genes validated as upregulated in human adipose inflammation, however, only five were not induced by LPS in either adipocytes or macrophages. This finding suggests these cells are the principle source of inflammatory modulation.
Our study used subcutaneous gluteal adipose tissue from healthy humans. Although visceral adipose tissue is commonly held to be more active in obesity-related inflammation, it appears that the evoked inflammatory response in subcutaneous adipose tissue approaches a similar phenotype. Many genes previously found to be overexpressed in visceral adipose were upregulated in subcutaneous adipose during in vivo endotoxemia. Several of the proteins identified in a recently published human visceral adipose secretome (1) were also upregulated in our discovery microarray (NETO1, RBP, PAI-1, THSD1, CD14, PTX3, LUM, and SPON2). These findings support our use of subcutaneous adipose tissue. In fact, a recent analysis revealed that both subcutaneous and visceral depots displayed a proinflammatory phenotype with obesity (49).
We acknowledge that our acute evoked inflammatory model does not reproduce the chronic low-grade changes in diet and obesity-related adipose inflammation. However, our prioritized inflammatory adipose molecules are cogent biomarkers for obesity-related diseases. First, the transient inflammatory and metabolic responses to endotoxemia resemble those observed in insulin resistance and metabolic syndrome (23,24). Second, rodent models have proven that adipose inflammatory pathways play important roles in diet and obesity-related metabolic disturbance (5–7). Finally, mRNA microarray and secretome profiling of human adipose tissue and adipocytes identify inflammatory pathways in obesity similar to those classically activated by endotoxin (1,20,21). Additional studies are required to confirm changes at the protein level, and mechanistic studies are required to determine whether our prioritized genes are truly involved in adipose metabolic functions.
In conclusion, human endotoxemia induces adipose inflammation and upregulates many adipose genes and proteins implicated in the complications of obesity. Microarray analysis of human adipose tissue during endotoxemia also identified novel genes that may serve as potential biomarkers of and therapeutic targets for obesity-related diseases.
Acknowledgments
We acknowledge the support of the Genomic Core and Human Adipose Resource of Diabetes and Endocrine Research Center (P20-DK 019525) at the University of Pennsylvania. This work was supported by a Clinical and Translational Science Award (RFA-RM-06-002) from the National Center for Research Resources (NCRR), by RO1 HL-073278 (M.R.), P50 HL-083799-SCCOR (M.R.), and W.W. Smith Charitable Trust (#H0204) (M.R.). S.H. is supported by National Institutes of Health Grant GM-085226.
No potential conflicts of interest relevant to this article were reported.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Alvarez-Llamas G, Szalowska E, de Vries MP, Weening D, Landman K, Hoek A, Wolffenbuttel BH, Roelofsen H, Vonk RJ: Characterization of the human visceral adipose tissue secretome. Mol Cell Proteomics 2007;6:589–600 [DOI] [PubMed] [Google Scholar]
- 2.Huber J, Kiefer FW, Zeyda M, Ludvik B, Silberhumer GR, Prager G, Zlabinger GJ, Stulnig TM: CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab 2008;93:3215–3221 [DOI] [PubMed] [Google Scholar]
- 3.Kim F, Pham M, Luttrell I, Bannerman DD, Tupper J, Thaler J, Hawn TR, Raines EW, Schwartz MW: Toll-like receptor-4 mediates vascular inflammation and insulin resistance in diet-induced obesity. Circ Res 2007;100:1589–1596 [DOI] [PubMed] [Google Scholar]
- 4.Nguyen MT, Favelyukis S, Nguyen AK, Reichart D, Scott PA, Jenn A, Liu-Bryan R, Glass CK, Neels JG, Olefsky JM: A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem 2007;282:35279–35292 [DOI] [PubMed] [Google Scholar]
- 5.McCowen KC, Ling PR, Ciccarone A, Mao Y, Chow JC, Bistrian BR, Smith RJ: Sustained endotoxemia leads to marked down-regulation of early steps in the insulin-signaling cascade. Crit Care Med 2001;29:839–846 [DOI] [PubMed] [Google Scholar]
- 6.Hotamisligil GS: Inflammatory pathways and insulin action. Int J Obes Relat Metab Disord 2003;27(Suppl.):S53–S55 [DOI] [PubMed] [Google Scholar]
- 7.Shoelson SE, Lee J, Goldfine AB: Inflammation and insulin resistance. J Clin Invest 2006;116:1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kintscher U, Hartge M, Hess K, Foryst-Ludwig A, Clemenz M, Wabitsch M, Fischer-Posovszky P, Barth TF, Dragun D, Skurk T, Hauner H, Bluher M, Unger T, Wolf AM, Knippschild U, Hombach V, Marx N: T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol 2008;28:1304–1310 [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Ghosh S, Perrard XD, Feng L, Garcia GE, Perrard JL, Sweeney JF, Peterson LE, Chan L, Smith CW, Ballantyne CM: T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation 2007;115:1029–1038 [DOI] [PubMed] [Google Scholar]
- 10.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW, Jr: Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 2003;112:1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clement K, Viguerie N, Poitou C, Carette C, Pelloux V, Curat CA, Sicard A, Rome S, Benis A, Zucker JD, Vidal H, Laville M, Barsh GS, Basdevant A, Stich V, Cancello R, Langin D: Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. Faseb J 2004;18:1657–1669 [DOI] [PubMed] [Google Scholar]
- 12.Plomgaard P, Nielsen AR, Fischer CP, Mortensen OH, Broholm C, Penkowa M, Krogh-Madsen R, Erikstrup C, Lindegaard B, Petersen AM, Taudorf S, Pedersen BK: Associations between insulin resistance and TNF-α in plasma, skeletal muscle and adipose tissue in humans with and without type 2 diabetes. Diabetologia 2007;50:2562–2571 [DOI] [PubMed] [Google Scholar]
- 13.Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K, Kitazawa R, Kitazawa S, Miyachi H, Maeda S, Egashira K, Kasuga M: MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest 2006;116:1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H: Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fain JN, Bahouth SW, Madan AK: TNFα release by the nonfat cells of human adipose tissue. Int J Obes Relat Metab Disord 2004;28:616–622 [DOI] [PubMed] [Google Scholar]
- 16.Visser M, Bouter LM, McQuillan GM, Wener MH, Harris TB: Elevated C-reactive protein levels in overweight and obese adults. JAMA 1999;282:2131–2135 [DOI] [PubMed] [Google Scholar]
- 17.Poitou C, Viguerie N, Cancello R, De Matteis R, Cinti S, Stich V, Coussieu C, Gauthier E, Courtine M, Zucker JD, Barsh GS, Saris W, Bruneval P, Basdevant A, Langin D, Clement K: Serum amyloid A: production by human white adipocyte and regulation by obesity and nutrition. Diabetologia 2005;48:519–528 [DOI] [PubMed] [Google Scholar]
- 18.Olszanecka-Glinianowicz M, Zahorska-Markiewicz B, Janowska J, Zurakowski A: Serum concentrations of nitric oxide, tumor necrosis factor (TNF)-α and TNF soluble receptors in women with overweight and obesity. Metabolism 2004;53:1268–1273 [DOI] [PubMed] [Google Scholar]
- 19.Baranova A, Collantes R, Gowder SJ, Elariny H, Schlauch K, Younoszai A, King S, Randhawa M, Pusulury S, Alsheddi T, Ong JP, Martin LM, Chandhoke V, Younossi ZM: Obesity-related differential gene expression in the visceral adipose tissue. Obes Surg 2005;15:758–765 [DOI] [PubMed] [Google Scholar]
- 20.Gomez-Ambrosi J, Catalan V, Diez-Caballero A, Martinez-Cruz LA, Gil MJ, Garcia-Foncillas J, Cienfuegos JA, Salvador J, Mato JM, Fruhbeck G: Gene expression profile of omental adipose tissue in human obesity. Faseb J 2004;18:215–217 [DOI] [PubMed] [Google Scholar]
- 21.Nair S, Lee YH, Rousseau E, Cam M, Tataranni PA, Baier LJ, Bogardus C, Permana PA: Increased expression of inflammation-related genes in cultured preadipocytes/stromal vascular cells from obese compared with non-obese Pima Indians. Diabetologia 2005;48:1784–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zvonic S, Lefevre M, Kilroy G, Floyd ZE, DeLany JP, Kheterpal I, Gravois A, Dow R, White A, Wu X, Gimble JM: Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol Cell Proteomics 2007;6:18–28 [DOI] [PubMed] [Google Scholar]
- 23.Agwunobi AO, Reid C, Maycock P, Little RA, Carlson GL: Insulin resistance and substrate utilization in human endotoxemia. J Clin Endocrinol Metab 2000;85:3770–3778 [DOI] [PubMed] [Google Scholar]
- 24.Hudgins LC, Parker TS, Levine DM, Gordon BR, Saal SD, Jiang XC, Seidman CE, Tremaroli JD, Lai J, Rubin AL: A single intravenous dose of endotoxin rapidly alters serum lipoproteins and lipid transfer proteins in normal volunteers. J Lipid Res 2003;44:1489–1498 [DOI] [PubMed] [Google Scholar]
- 25.Suffredini AF, Fromm RE, Parker MM, Brenner M, Kovacs JA, Wesley RA, Parrillo JE: The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med 1989;321:280–287 [DOI] [PubMed] [Google Scholar]
- 26.Anderson PD, Mehta NN, Wolfe ML, Hinkle CC, Pruscino L, Comiskey LL, Tabita-Martinez J, Sellers KF, Rickels MR, Ahima RS, Reilly MP: Innate immunity modulates adipokines in humans. J Clin Endocrinol Metab 2007;92:2272–2279 [DOI] [PubMed] [Google Scholar]
- 27.Vitseva OI, Tanriverdi K, Tchkonia TT, Kirkland JL, McDonnell ME, Apovian CM, Freedman J, Gokce N: Inducible Toll-like receptor and NF-κB regulatory pathway expression in human adipose tissue. Obesity 2008;16:932–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuelli B, Peraldi P, Filloux C, Chavey C, Freidinger K, Hilton DJ, Hotamisligil GS, Van Obberghen E: SOCS-3 inhibits insulin signaling and is up-regulated in response to tumor necrosis factor-α in the adipose tissue of obese mice. J Biol Chem 2001;276:47944–47949 [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP: A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–193 [DOI] [PubMed] [Google Scholar]
- 30.Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA: Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 2005;6:59–75 [DOI] [PubMed] [Google Scholar]
- 31.Ge YDS, Speed TP: Resampling-based mutliple testing for microarray data analysis. Test 2003;12:1–77 [Google Scholar]
- 32.Bendtsen JD, Nielsen H, von Heijne G, Brunak S: Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004;340:783–795 [DOI] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G: Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 1997;10:1–6 [DOI] [PubMed] [Google Scholar]
- 34.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S: Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 2004;17:349–356 [DOI] [PubMed] [Google Scholar]
- 35.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA: DAVID: database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:P3. [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 37.Lehrke M, Reilly MP, Millington SC, Iqbal N, Rader DJ, Lazar MA: An inflammatory cascade leading to hyperresistinemia in humans. PLoS Med 2004;1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez FO, Sica A, Mantovani A, Locati M: Macrophage activation and polarization. Front Biosci 2008;13:453–461 [DOI] [PubMed] [Google Scholar]
- 39.Schmittgen TD, Livak KJ: Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 40.Clopper C, Pearson S: The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 1934;404–413 [Google Scholar]
- 41.Goerdt S, Politz O, Schledzewski K, Birk R, Gratchev A, Guillot P, Hakiy N, Klemke CD, Dippel E, Kodelja V, Orfanos CE: Alternative versus classical activation of macrophages. Pathobiology 1999;67:222–226 [DOI] [PubMed] [Google Scholar]
- 42.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M: The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004;25:677–686 [DOI] [PubMed] [Google Scholar]
- 43.Sun G: Application of DNA microarrays in the study of human obesity and type 2 diabetes. Omics 2007;11:25–40 [DOI] [PubMed] [Google Scholar]
- 44.Trevino V, Falciani F, Barrera-Saldana HA: DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol Med 2007;13:527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocha VZ, Folco EJ, Sukhova G, Shimizu K, Gotsman I, Vernon AH, Libby P: Interferon-gamma, a Th1 cytokine, regulates fat inflammation: a role for adaptive immunity in obesity. Circ Res 2008;103:467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lumeng CN, Bodzin JL, Saltiel AR: Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 2007;117:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeyda M, Farmer D, Todoric J, Aszmann O, Speiser M, Gyori G, Zlabinger GJ, Stulnig TM: Human adipose tissue macrophages are of an anti-inflammatory phenotype but capable of excessive pro-inflammatory mediator production. Int J Obes 2007;31:1420–1428 [DOI] [PubMed] [Google Scholar]
- 48.Martinez FO, Gordon S, Locati M, Mantovani A: Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 2006;177:7303–7311 [DOI] [PubMed] [Google Scholar]
- 49.Dolinkova M, Dostalova I, Lacinova Z, Michalsky D, Haluzikova D, Mraz M, Kasalicky M, Haluzik M: The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol 2008;291:63–70 [DOI] [PubMed] [Google Scholar]