Abstract
Background
KRAS mutations occur in 35–45% of metastatic colorectal cancers (mCRC) and preclude responsiveness to EGFR-targeted therapy with cetuximab or panitumumab. However, less than 20% patients displaying wild-type KRAS tumors achieve objective response. Alterations in other effectors downstream of the EGFR, such as BRAF, and deregulation of the PIK3CA/PTEN pathway have independently been found to give rise to resistance. We present a comprehensive analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression in mCRC patients treated with cetuximab or panitumumab, with the aim of clarifying the relative contribution of these molecular alterations to resistance.
Methodology/Principal Findings
We retrospectively analyzed objective tumor response, progression-free (PFS) and overall survival (OS) together with the mutational status of KRAS, BRAF, PIK3CA and expression of PTEN in 132 tumors from cetuximab or panitumumab treated mCRC patients. Among the 106 non-responsive patients, 74 (70%) had tumors with at least one molecular alteration in the four markers. The probability of response was 51% (22/43) among patients with no alterations, 4% (2/47) among patients with 1 alteration, and 0% (0/24) for patients with ≥2 alterations (p<0.0001). Accordingly, PFS and OS were increasingly worse for patients with tumors harboring none, 1, or ≥2 molecular alteration(s) (p<0.001).
Conclusions/Significance
When expression of PTEN and mutations of KRAS, BRAF and PIK3CA are concomitantly ascertained, up to 70% of mCRC patients unlikely to respond to anti-EGFR therapies can be identified. We propose to define as ‘quadruple negative’, the CRCs lacking alterations in KRAS, BRAF, PTEN and PIK3CA. Comprehensive molecular dissection of the EGFR signaling pathways should be considered to select mCRC patients for cetuximab- or panitumumab-based therapies.
Introduction
Colorectal cancer (CRC) is the third cause of cancer-related death in the western world [1]. Despite improvements in the therapeutic armamentarium for metastatic CRC (mCRC), the 5-year overall survival (OS) remains poor, with a median survival of 18 to 21 months [2]. Additional drugs, as well as further insights about the mechanisms of resistance, are needed to improve clinical outcome. Treatment options for mCRC nowadays include the chimeric IgG1 monoclonal antibody (moAb) cetuximab and the human IgG2 moAb panitumumab [3], [4]. Both molecules bind to the Epidermal Growth Factor Receptor (EGFR), leading to inhibition of its downstream signaling, providing a meaningful clinical benefit. Objective response rates in unselected populations of mCRC, however, are limited to 8–12% for these agents when used as monotherapy in first [5] and subsequent lines of treatment [4], [6], [7].
We and others have previously shown that somatic KRAS mutations (a key effector of the EGFR initiated signaling) can independently impair the efficacy of panitumumab or cetuximab [8]–[10]. This led the U.S. and European health authorities to restrict the use of these agents for patients with wild-type KRAS mCRC only [11]–[13]. Although this decision is expected to ameliorate the therapeutic index in this selected population, the objective response rate is still very limited. In fact, it is restricted to 17% (vs 0% in KRAS mutated) for panitumumab monotherapy [10], to 12.8% (vs 1.2% in KRAS mutated) for cetuximab monotherapy [14] and to 59–61% (vs 36–33% in KRAS mutated) for cetuximab plus either irinotecan- or oxaliplatin-based chemotherapy, respectively [15], [16]. These findings clearly suggest that other factors, such as alterations in other EGFR effectors, including members of the RAS-MAPK or PI3K pathways could drive resistance to anti-EGFR therapy.
BRAF is the principal downstream effector of KRAS [17], [18] and its oncogenic V600E mutation is mutually exclusive with KRAS mutations in CRCs [19]. We and others have recently demonstrated that the V600E mutation can also preclude responsiveness to panitumumab or cetuximab in mCRC patients and cellular models of CRC [20]. The PIK3CA gene is mutated in approximately 20% of CRCs [21]. PIK3CA mutations occurring in the ‘hotspots’ located in exon 9 (E542K, E545K) and exon 20 (H1047R) are oncogenic in CRC cellular models [22], [23]. The PIK3CA gene encodes for a lipid kinase that regulates, alongside with KRAS, signaling pathways downstream of the EGFR. PI3K initiated signaling is normally inhibited by PTEN (phosphatase and tensin homologue deleted on chromosome ten). We and others have previously shown that loss of PTEN expression, which occurs in 30% of sporadic cases, is associated with lack of response to cetuximab [24], and that PIK3CA/PTEN deregulation may be a biomarker of resistance in KRAS wild-type patients [25], [26] and cellular models of CRC [27].
Taken together, data from retrospective analyses show that BRAF and PIK3CA/PTEN alterations could represent additional tools for selecting mCRC patients for EGFR-targeted treatment [20], [24]–[26]. Nevertheless, a clear identification of which biomarkers should be employed together with KRAS in the clinical setting remains to be defined. This is because in these retrospective analyses: a) different determinants were evaluated in each study; and b) an overlapping among some of these biomarkers may occur in individual patients. In the present study, we performed the first comprehensive mutational analysis of KRAS, BRAF, and PIK3CA, alongside with the evaluation of PTEN expression in a cohort of 132 mCRC treated patients.
Results
Distribution and overlap of molecular alterations in individual tumors
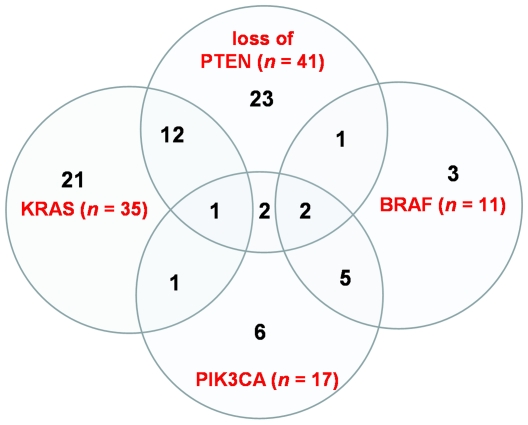
As shown in Figure 1 , analysis of tumors from a cohort of 132 patients (for clinico-pathological features, see Table 1 ) led to the identification of 104 molecular alterations. Specifically, we detected 35 KRAS mutations (26.5%), 11 BRAF mutations (8.3%), 17 PIK3CA mutations (12.3%), and 41 (out of 114 evaluable) loss of PTEN expression (36.0%). Mutations of KRAS occurred in codon 12 in 24 cases (68.6%) and in codon 13 in 10 cases (28.6%); a double point mutation involving both codons was detected in one case (2.9%). PIK3CA mutations were found either in exon 9 (4 cases) or in exon 20 (13 cases). The 11 mutations of BRAF were all V600E substitutions.
Figure 1. Representation of the distribution of molecular alterations in individual tumors of the 132 patients: mutations of KRAS and BRAF occurred in a mutually exclusive manner, while an overlapping pattern was observed between other alterations.
Table 1. Patients' baseline characteristics.
| Number of patients | 132 |
| Median age (years) [range] | 63.5 [26–85] |
| Gender (male/female) | 86/46 |
| Primary tumour site | |
| Colon | 78 |
| Sigma-rectum junction | 19 |
| Rectum | 31 |
| Other* | 4 |
| EGFR targeted therapy | |
| Cetuximab | 109 |
| Panitumumab | 23 |
| Previous chemotherapy (%) | |
| Irinotecan based | 117 (88.6%) |
| Fluoropyrimidine/capecitabine based | 115 (87.1%) |
| Oxaliplatin based | 105 (79.5%) |
| No. of previous cancer treatments for advanced disease prior anti-EGFR moAbs (%) | |
| None | 13 (9.8%) |
| One | 19 (14.4%) |
| Two | 65 (49.2%) |
| Three | 29 (22.0%) |
| More than three | 6 (4.5%) |
| Cutaneous toxicity (%) | |
| 0 | 21 (15.9%) |
| 1 | 67 (50.7%) |
| 2–3 | 37 (28.0%) |
| Unknown | 7 (5.3%) |
Other: in two cases, primary tumor site was small bowel, in one case duodenum and in one case primary tumor sites were multiple (colon and rectum).
Mutations of KRAS and BRAF occurred in a mutually exclusive manner, while an overlapping pattern was observed among other alterations. The most frequent overlapping fingerprints were PTEN loss and KRAS mutations (co-occurring in 13 patients), and BRAF and PIK3CA mutations (in 7 patients) ( Figure 1 ).
Association of clinical variables and objective tumor response
Among clinical variables (see Table 1 ), only cutaneous toxicity was associated with objective response (Wald's test: p = 0.002; direction of response = 0-1-≥2). Other clinical variables including gender, site of primary tumor (colon, sigmoid-rectum junction, rectum, other sites), and age were not significantly associated with objective tumor response (p = 0.491, 0.490 and 0.904, respectively; p values were obtained by Fisher's exact test for site and gender and by Wald's test for age). Since patients selected in this cohort were treated with mixed lines of previous chemotherapy regimens (although the vast majority received 2–3 previous lines of treatment, see Table 1 ), we also studied the association between the number of previous chemotherapy lines and objective tumor response, showing that this variable did not exert any effect (Wald's test: p = 0.536).
Multivariate analyses of molecular alterations and objective response
Multivariate analysis including all four molecular alterations (adjusted by cutaneous toxicity and number of previous chemotherapy lines) showed that only KRAS mutations and loss of PTEN expression were independently associated with lack of objective response (p = 0.001 and <0.001, respectively) ( Table 2 ). Importantly, the Bayesian informative criterion according to Schwarz suggested that a model including KRAS mutations and loss of PTEN expression was overall the best strategy in identifying non-responsive patients in our cohort.
Table 2. Multivariate analysis of objective response done with exact logistic regression in the cohort of 132 patients evaluated in the study.
| Molecular alteration | Odds Ratio of response | CI 95% | p value |
| KRAS Mutant versus wild type | 0.06 | 0.001–0.469 | 0.001 |
| BRAF Mutant versus wild type | 0.32 | 0.000–4.175 | 0.379 |
| PIK3CA Mutant versus wild type | 0.19 | 0.000–1.701 | 0.146 |
| PTEN normal versus loss | 23.89 | 3.136–997.754 | <0.001 |
Odds ratio values are adjusted by score of cutaneous toxicity and number of previous chemotherapy lines.
Multivariate analyses of molecular alterations and survival
The multivariate Cox analysis for OS confirmed that the role of KRAS mutations and loss of PTEN was significant in conferring worse clinical outcome (HR = 1.72, p = 0.043 and HR = 0.54, p = 0.012, respectively), with also BRAF mutations exerting a detrimental borderline effect (HR = 2.31, p = 0.093). As for PFS, only KRAS mutations were associated with decreased survival (HR = 1.65, p = 0.033), whereas none of the other molecular alterations was demonstrated to independently affect clinical outcome ( Table 3 ).
Table 3. Multivariate analysis of survival done with exact logistic regression in the cohort of 132 patients evaluated in the study.
| Molecular alteration | PFS Hazard Ratio (CI 95%) | p value | OS Hazard Ratio (CI 95%) | p value |
| KRAS (mutant versus wild type) | 1.65 (1.041–2.601) | 0.033 | 1.72 (1.017–2.903) | 0.043 |
| BRAF (mutant versus wild type) | 1.39 (0.521–3.685) | 0.513 | 2.31 (0.867–6.131) | 0.093 |
| PIK3CA (mutant versus wild type) | 1.79 (0.801–4.017) | 0.156 | 1.63 (0.815–3.269) | 0.166 |
| PTEN (normal versus loss) | 0.77 (0.501–1.167) | 0.213 | 0.54 (0.332–0.874) | 0.012 |
Hazard ratio values are adjusted by score of cutaneous toxicity and number of previous chemotherapy lines.
PFS: progression-free survival; OS: overall survival.
Multivariate analyses of molecular alterations and clinical outcome in patients with wild-type KRAS mCRC
Our cohort consists of retrospective cases and, for this reason, also KRAS mutated patients were included in the analysis. After the decision of health authorities to restrict the use of cetuximab and panitumumab to wild-type KRAS mCRC [11]–[13], [28], this subgroup has achieved utmost relevance. Accordingly, we focused our analysis on the effect of BRAF and PIK3CA mutations and loss of PTEN in the remaining 96 wild-type KRAS patients. At multivariate analysis, loss of PTEN confirmed a significant association with lack of response (p<0.001), while BRAF and PIK3CA were not significant (p = 0.265, 0.075, respectively) ( Table 4 ).
Table 4. Multivariate analysis of objective response done with exact logistic regression among KRAS wild-type patients.
| Molecular alteration | Odds Ratio of response | CI 95% | p value |
| BRAF mutant versus wild type | 0.24 | 0.000–3.093 | 0.265 |
| PIK3CA mutant versus wild type | 0.14 | 0.000–1.203 | 0.075 |
| PTEN normal versus loss | 30.46 | 3.831–1436.461 | <0.001 |
Odds ratio values are adjusted by score of cutaneous toxicity and number of previous chemotherapy lines.
Survival analyses shown in Table 5 demonstrated that BRAF mutations (HR = 3.75, p = 0.015) and loss of PTEN (HR = 0.43, p = 0.009), but not PIK3CA mutations (HR = 1.20, p = 0.672), were significantly associated with decreased OS, whereas none of these alterations was significantly associated with PFS.
Table 5. Multivariate analysis of survival done with exact logistic regression among KRAS wild-type patients.
| Molecular alteration | PFS Hazard Ratio (CI 95%) | p value | OS Hazard Ratio (CI 95%) | p value |
| BRAF (mutant versus wild type) | 2.03 (0.66–6.28) | 0.218 | 3.75 (1.29–10.90) | 0.015 |
| PIK3CA (mutant versus wild type) | 1.45 (0.51–4.14) | 0.492 | 1.20 (0.52–2.78) | 0.672 |
| PTEN (normal versus loss) | 0.81 (0.47–1.39) | 0.439 | 0.43 (0.22–0.81) | 0.009 |
Hazard ratio values are adjusted by score of cutaneous toxicity and number of previous chemotherapy lines.
PFS: progression-free survival; OS: overall survival.
Effect of the number of molecular alterations on clinical outcome
In light of the occurrence of multiple molecular alterations within the same tumor, we investigated our cohort by separating patients according to the actual number of molecular abnormalities in the same tumor, i.e., none vs 1 vs ≥2 alterations. Because the molecular status of one marker among PTEN, PIK3CA and BRAF was undetermined in 18 tumors, we decided to perform this analysis in the remaining 114 patients.
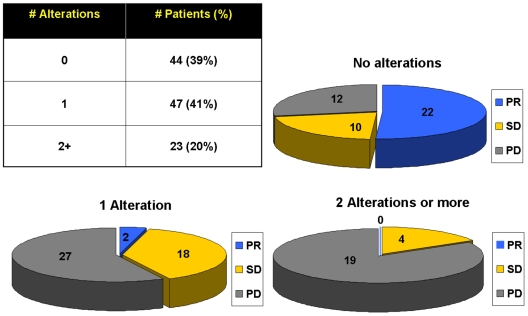
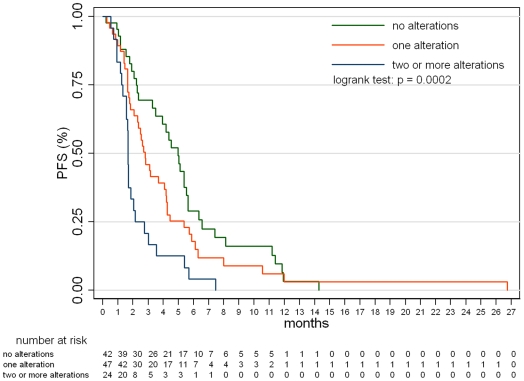
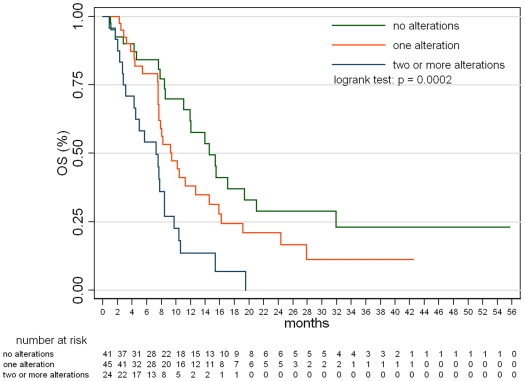
The probability of response was 51.1% (22/43) among patients with no alterations, 4.2% (2/47) with 1 alteration, and 0% (0/24) with ≥2 alterations, and these difference were statistically significant (p<0.0001 by Fisher's exact test). Figure 2 shows distribution of the number of mutations in the cohort and response to EGFR-targeted therapy according to the number of molecular abnormalities within individual tumor samples. The detrimental effect of accumulating alterations was also confirmed by the logistic model, showing that, on average, an unitary increase in the number of alteration(s), would mean a decrease in the odds of response by 96% (odds ratio = 0.04; p<0.00001). Taken together, these findings mean that a necessary but not sufficient condition to reach objective response is to have no more than one of the four molecular alterations. Similarly, survival analyses showed that patients displayed different PFS and OS depending on the number of molecular alterations in their tumors. Figures 3 and 4 show that PFS and OS were increasingly worse for none, 1 or ≥2 molecular alterations (p = 0.0002 for both PFS and OS; logrank test). The pairwise tests adjusted for multiple comparisons documented that a worse clinical outcome was observed for patients with tumors bearing ≥2 alterations vs 1 (p = 0.0198 and 0.0213 for PFS and OS, respectively) and ≥2 vs none (p = 0.0002 for both PFS and OS), but not for patients with 1 alteration vs none (p = 0.3852 and 0.3807 for PFS and OS, respectively). Median PFS was 2.8 months (5.0, 2.8 and 1.7 for patients harboring none, one or ≥2 alterations, respectively); median OS was 9.4 months (14.6, 9.3 and 7.3 for patients harboring none, one or ≥2 alterations, respectively).
Figure 2. Distribution of the number of mutations (table) and response to EGFR-targeted therapy (pie-charts) according to the number of molecular abnormalities within individual tumor samples.
Figure 3. Progression-free survival according to the number of molecular abnormalities within individual tumor samples.
Data from the cohort of patients with a known molecular status of all four markers.
Figure 4. Overall survival according to the number of molecular abnormalities within individual tumor samples.
Data from the cohort of patients with a known molecular status of all four markers.
Discussion
Despite the recent recommendation by ASCO [28] and by health authorities in Europe [11], [12] and US [13] of KRAS testing as a diagnostic prerequisite for EGFR-targeted cetuximab- or panitumumab-based therapies for mCRC, the response rate to either of these drugs is limited to less than 20% in wild-type KRAS patients [10], [14]. Recent data indicate that BRAF or PIK3CA mutations may contribute for additional 20–30% of resistance [20], [25], [26]. In addition, also PTEN has been proposed as an independent predictive factor of cetuximab efficacy [24], [25], [27]. However, the relative and overall contribution of each of these molecular alterations to clinical decision making remains unclear. Furthermore, whether and to what extent the occurrence of multiple molecular alterations affects clinical response and patients' survival is presently unknown. EGFR copy number assessed by FISH has also been suggested to be predictive of clinical outcome to EGFR-targeted therapies [29]–[33]. However, EGFR FISH for mCRC is undergoing inter-laboratory standardization [30] and to avoid the introduction of confounding elements we elected not to carry out this analysis. Here, we exploited the comprehensive molecular analysis of EGFR downstream effectors to ascertain their role in predicting response/resistance to cetuximab or panitumumab in mCRC. By the concomitant assessment of four molecular alterations, we were able to identify up to 70% of non-responder patients, a result that has never been achieved before. Notably, only three patients with tumors carrying a single alteration were in the subgroup of responders, (two patients with KRAS mutations and one patient with loss of PTEN expression), whereas no others showed any alteration (“quadruple negative” tumors). This suggests that previously reported outliers, i.e., very uncommon cases of mCRC with KRAS mutations responding to therapy [9], [14], [29], [34] may be patients harboring only one of these molecular alterations, thus not concurrently deregulating both MAPK and PI3K pathways.
Our data indicate that single or multiple mutations of KRAS, BRAF, or PIK3CA unfavorably affect clinical outcome to cetuximab- or panitumumab-based therapies; however, the possibility that these molecular alterations could be negative prognostic biomarkers independently from targeted therapies should be taken into account. The RASCAL retrospective study conducted on 2721 CRC patients indicated that the presence of KRAS mutations is associated with a 26% increased risk of fatal outcome [35]. However, conflicting data on the same topic have been recently published. In a phase III trial reported by Karapetis et al. [14], clinical benefit in patients with wild-type KRAS mCRC was found in cetuximab treated patients but not in control patients treated with best supportive care only, thus indicating that the benefit was not due to a prognostic effect of KRAS. Moreover, Roth et al. [Proc Am Soc Clin Oncol Gastrointestinal Cancers Symposium 206, 2009; Abstr 288] tested the prognostic value per stage of KRAS and BRAF mutations using CRC tumor samples from the adjuvant PETACC3 trial, and they found no significant effects on relapse-free survival for both mutations, neither in stage II nor in stage III. Studies assessing the impact of other molecular alterations rather than KRAS mutations are limited. As for the prognostic role of PIK3CA and BRAF, in a study including 586 patients by Barault et al., decreased rates of 3-year survival were associated with mutations of at least one gene among KRAS, BRAF and PIK3CA [36]. A recent report by Tol et al. found that the presence of the BRAF V600E mutation was a negative prognostic marker in 516 patients with metastatic colorectal cancer treated with capecitabine, oxaliplatin, and bevacizumab based regimens [37]. Finally, Ogino et al. reported that, in a series of 450 patients with stage I–III CRC who underwent curative surgery, tumor PIK3CA mutation was associated with shorter cancer-specific survival. Such adverse effect of PIK3CA mutation on prognosis was consistent across most strata of clinical and tumoral predictors of patient outcome. Interestingly, this adverse effect was mainly limited to patients with KRAS wild-type tumors [38]
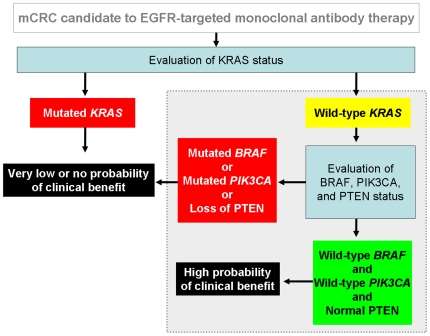
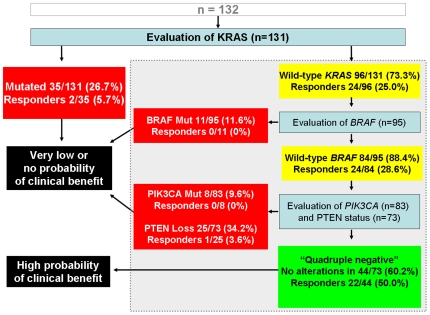
In conclusion, we document that concomitant detection of KRAS, BRAF and PIK3CA mutations and evaluation of loss of PTEN expression in mCRC patients has remarkable clinical implications by increasing the ability to predict the outcome to EGFR-targeted therapies. In light of the nature of our patient series, the most reliable indicator of the predictive value of biomarker(s) is objective tumor response. Interpretation of survival analyses should indeed take into account a possible limitation due to patients treated with mixed previous line(s) of chemotherapy including a 10% (13/132) of patients treated with first-line cetuximab monotherapy. On the other hand, the study of such patients represents a unique opportunity to ascertain the predictive value of a given biomarker without the influence of chemotherapy, either concurrent or previous, as well as of selection exerted by other treatments. In light of these considerations, we propose here a new algorithm for deciding the clinical use of EGFR-targeted monoclonal antibodies that is based on objective response rates ( Figures 5 and 6 ). This novel approach deserves validation in prospective studies with cetuximab- or panitumumab-based therapies in mCRC prior to have an impact as clinical practice-changing. Importantly, we found that approximately 20% of mCRC non-responders do not harbor mutations of KRAS, BRAF, PIK3CA nor loss of PTEN expression and we propose to define these tumors as “quadruple negative”. The lack of response in quadruple negative patients may be due to multiple reasons including but not restricted to: a) the limited sensitivity of current sequencing methods in detecting point mutations in DNA extracted from FFPE tumors [39]; b) the oncogenic deregulation of the same four genes by mechanism other then mutations (such as amplification as reported for PIK3CA); c) the occurrence of alterations in other key elements of the EGFR-dependent signal cascade (such as for example AKT or MAPK); and d) the presence of genetic alterations in tyrosine kinase receptors other than EGFR, providing an alternate pathway of survival and/or proliferation. Further molecular dissections of the EGFR-initiated oncogenic signaling cascade are likely to be helpful in improving the tailoring of EGFR targeted therapies. Overall, our results underscore the relevance of using molecular-based algorithms to shift the treatment of solid tumors into the era of personalized cancer medicine.
Figure 5. Algorithm of molecular diagnostics based on data discussed in this study for patients with mCRC candidates to cetuximab- or panitumumab-based therapies.
The area marked in grey within the dotted line box describes the hypothesis generated in this study.
Figure 6. Following evaluation of KRAS status in individual tumors, enhancement of predictability of clinical benefit may derive from assessment of the status of BRAF, PIK3CA and PTEN, as simulated here based on analyses of subgroups from the present cohort (n = 131).
We propose to define as “quadruple negative” the mCRCs lacking alterations in KRAS, BRAF, PTEN and PIK3CA.
Materials and Methods
Ethics statement
Samples were collected according to the ethical requirements and regulations and obtaining written consent as approved by the review board of the Ospedale Niguarda Ca' Granda, Milano, Italy and of the Ospedale San Giovanni, Bellinzona, Switzerland.
Patient population and treatment regimens
We retrospectively analyzed 132 patients with EGFR-positive mCRC at Ospedale Niguarda Ca' Granda (Milan, Italy), at the Oncogenomics Center, Institute for Cancer Research and Treatment, (Candiolo, Italy), or at the Institute of Pathology (Locarno, Switzerland). Patients gave informed consent and were treated with panitumumab- or cetuximab-based regimens at Ospedale Niguarda Ca' Granda or at the Oncology Institute of Southern Switzerland (Bellinzona, Switzerland). Patients were selected based on evidence that treatment outcome could be attributable only to administration of panitumumab or cetuximab. Patients' clinical characteristics are reported in Table 1 . With the exception of 13 patients who received cetuximab as frontline monotherapy [5], the others had failed at least one prior chemotherapy regimen. Twenty-three (17.4%) received panitumumab monotherapy, fifteen (11.4%) patients cetuximab monotherapy, and ninety-four (71.2%) cetuximab plus irinotecan-based chemotherapy. For patients who progressed on irinotecan-based chemotherapy, cetuximab was administered with irinotecan at the same dose and schedule to which they were resistant. Treatment was continued until progressive disease (PD) or toxicity occurred, as per standard criteria [40].
Clinical evaluation and tumor response criteria
Clinical response was assessed every 6–8 weeks with radiological examination (CT or MR) according to RECIST criteria. Objective tumor responses were classified into partial response (PR), stable disease (SD), and PD. Patients with SD or PD were also defined as non-responders. Response to therapy was also evaluated retrospectively by independent radiologists.
Molecular analyses
Formalin-fixed paraffin-embedded (FFPE) tumor blocks were reviewed for quality and tumor content. A single representative block, from either the primary tumor or the liver metastasis, depending on availability, containing at least 70% of malignant cells, was selected for each case. Genomic DNA was extracted using the QIAamp Mini kit (Qiagen, Chatsworth, CA, USA) according to the manufacturer's instructions. Molecular analyses were performed on tissue sample from primary tumor used for initial diagnosis in 130 out of 132 cases. In two cases only the analysis was performed on metastatic sites (liver).
PTEN expression
PTEN protein expression was evaluated by immunohistochemistry on 3 µm FFPE tissue sections as reported [24] with some modifications. Briefly, anti-PTEN Ab4 (Thermo Fisher Scientific, CA, USA) with 1∶200 dilution and PTEN Ab2 (Neomarkers, Fremont, CA) with 1∶50 dilution were used at the Niguarda Hospital and at the Institute of Pathology of Locarno, respectively. PTEN protein expression was mainly detected at cytoplasmic level, while very few cases showed also nuclear positivity. Tumors were considered negative, i.e. with loss of PTEN expression, when absence or reduction of immunostaining was seen in more than 50% of cells as compared with internal controls (i.e. vascular endothelial cells and nerves). Normal endometrium was used as external positive control. The evaluations were performed by two independent pathologists without knowledge of clinical data or results of molecular analyses.
Mutational analysis of KRAS, BRAF and PIK3CA in tumor samples
We searched for KRAS (exon 2), for BRAF (exon 15) and for PIK3CA (exons 9 and 20) mutations. KRAS exon 2 includes codons 12 and 13, BRAF exon 15 includes codon 600, PIK3CA exon 9 includes codons 542 and 545 and PIK3CA exon 20 includes codon 1047, where the large majority of mutations occur in these genes [29]. The list of primers used for mutational analysis is available from the authors upon request. All samples were subjected to automated sequencing by ABI PRISM 3730 (Applied Biosystems, Foster City, CA, USA). All mutated cases were confirmed twice with independent PCR reactions.
Statistical analyses
All data were analyzed with the suitable descriptive statistical methods, after checking their distributions by means of the Shapiro-Wilk test. Cross-tabulations of qualitative variables were analyzed with the Fisher's exact test, while comparisons between continuous variables were carried out with Student's t or Mann-Whitney U tests. Logistic regression with Wald's test, and exact logistic regression (dealing with one-way causation, such as the case where all patients in a group show a positive or negative outcome), with exact p-value (as the probability of observing a more extreme value with respect to sufficient statistics for a given regression parameter) were used to assess univariate and multivariate analysis with binary endpoint. The survival analysis was performed with the Kaplan-Meier survivor function followed by logrank test, and with the Cox model; proportional hazard assumption was checked using the Schoenfeld residuals. Statistical significance was assumed for p<0.05. All the statistical analyses were done using Stata/SE 10.1 (the StataCorp, College Station,TX-USA).
Acknowledgments
We thank Salvatore Artale for clinical care, Federico Pozzi for data management, Carlo Zanon and Silvia Benvenuti for sequence analysis and data management, and Roberta Schiavo for supervision of tumor samples procurement.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from Italian Association for Cancer Research (AIRC), Italian Ministry of Health, Regione Piemonte, Italian Ministry of University and Research, Association for International Cancer Research (AICR-UK), EU FP6 MCSCs contract 037297, CRT Progetto Alfieri, Oncosuisse grants OCS-01921-08-2006 and OCS-02301-08-2008, Fondazione Ticinese per la Ricerca sul Cancro (Tessin Foundation for Cancer Research), and OCGO (Oncologia Ca Granda Onlus) Fondazione. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Sargent DJ, Wieand HS, Haller DG, Gray R, Benedetti JK, et al. Disease-free survival versus overall survival as a primary endpoint for adjuvant colon cancer studies: individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2005;23:8664–8670. doi: 10.1200/JCO.2005.01.6071. [DOI] [PubMed] [Google Scholar]
- 3.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 5.Pessino A, Artale S, Sciallero S, Guglielmi A, Fornarini G, et al. First-line single-agent cetuximab in patients with advanced colorectal cancer. Ann Oncol. 2008;19:711–716. doi: 10.1093/annonc/mdm516. [DOI] [PubMed] [Google Scholar]
- 6.Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 8.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 9.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 10.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 11.European Medicines Agency website, European Public Assessment Report. http://www.emea.europa.eu/pdfs/human/opinion/40511307en.pdf. Downloaded on March 31, 2009.
- 12.European Medicines Agency website, European Public Assessment Report. http://www.emea.europa.eu/pdfs/human/opinion/Erbitux_28040208en.pdf. Downloaded on March 31, 2009.
- 13. http://www.fda.gov/AboutFDA/CentersOffices/CDER/ucm172905.htm.
- 14.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 16.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 17.Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 18.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. Embo J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, et al. Tumorigenesis: RAF/RAS oncogenes and mismatchrepair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 20.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 21.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuels Y, Diaz J, Schmidt-Kittler O, Cummins JM, Delong L, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 24.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 26.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 27.Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68:1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, et al. American Society of Clinical Oncology Provisional Clinical Opinion: Testing for KRAS Gene Mutations in Patients With Metastatic Colorectal Carcinoma to Predict Response to Anti-Epidermal Growth Factor Receptor Monoclonal Antibody Therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 29.Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 30.Moroni M, Sartore-Bianchi A, Veronese S, Siena S. EGFR FISH in colorectal cancer: what is the current reality? Lancet Oncol. 2008;9:402–403. doi: 10.1016/S1470-2045(08)70109-8. [DOI] [PubMed] [Google Scholar]
- 31.Sartore-Bianchi A, Moroni M, Veronese S, Carnaghi C, Bajetta E, et al. Epidermal growth factor receptor gene copy number and clinical outcome of metastatic colorectal cancer treated with panitumumab. J Clin Oncol. 2007;25:3238–3245. doi: 10.1200/JCO.2007.11.5956. [DOI] [PubMed] [Google Scholar]
- 32.Cappuzzo F, Finocchiaro G, Rossi E, Jänne PA, Carnaghi C, et al. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19:717–723. doi: 10.1093/annonc/mdm492. [DOI] [PubMed] [Google Scholar]
- 33.Personeni N, Fieuws S, Piessevaux H, De Hertogh G, De Schutter J, et al. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: a fluorescent in situ hybridization study. Clin Cancer Res. 2008;14:5869–5876. doi: 10.1158/1078-0432.CCR-08-0449. [DOI] [PubMed] [Google Scholar]
- 34.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 35.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 36.Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–2259. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 37.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 38.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchetti A, Gasparini G. K-ras mutations and cetuximab in colorectal cancer. N Engl J Med. 2009;360:833–834. doi: 10.1056/NEJMc082346. [DOI] [PubMed] [Google Scholar]
- 40.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]