Abstract
SUMMARY
Allergic airway disease is characterized by eosinophilic inflammation, mucus hypersecretion and increased airway resistance. Fungal antigens are ubiquitous within the environment and are well know triggers of allergic disease. Bacterial products are also frequently encountered within the environment and may alter the immune response to certain antigens. The consequence of simultaneous exposure to bacterial and fungal products on the lung adaptive immune response has not been explored. Here we show that oropharyngeal aspiration of fungal lysates (Candida albicans, Aspergillus fumigatus) promotes airway eosinophilia, secretion of Th2 cytokines and mucus cell metaplasia. In contrast, oropharyngeal exposure to bacterial lysates (Pseudomonas aeruginosa) promotes airway inflammation characterized by neutrophils, Th1 cytokine secretion and no mucus production. More importantly, administration of bacterial lysates together with fungal lysates deviates the adaptive immune response to a Th1 type associated with neutrophilia and diminished mucus production. The immunomodulatory effect that bacterial lysates have on the response to fungi is TLR4-independent but MyD88 dependent. Thus, different types of microbial products within the airway can alter the host's adaptive immune response, and potentially impact the development of allergic airway disease to environmental fungal antigens.
Keywords: T helper cells, Lung inflammation, cytokines, immune responses, eosinophils
INTRODUCTION
Allergic airway disease is characterized by eosinophilic inflammation, mucus hypersecretion and increased airway resistance [1–3]. This response results from antigen-specific Th2 cell activation characterized by IL-4, IL-5 and IL-13 production [4]. IL-4 and IL-5 stimulate eosinophil maturation and activation while IL-13 is an important mediator of mucus production [4, 5]. Among inhaled allergens that can trigger this type of immune response are a number of fungal species [6]. These agents can stimulate allergic inflammation without causing active infection in the immunocompetent host [6–8]. It is unclear why airway exposure to fungal antigens leads to a Th2 immune response but may result from specific antigenic determinants of the fungi that preferentially stimulate Th2 responses. However, CD4 T cells from mice with transgenic T-cell receptors specific for a particular Aspergillus antigen differentiate ex vivo into IFNγ producing Th1 cells rather than Th2 cells [9]. Alternatively, it could be the lung environment with its costimulatory cytokine milieu that predisposes to a Th2 immune response to fungal antigens. Interestingly, not all individuals develop allergic airway inflammation in response to inhaled fungal allergens despite ubiquitous exposure [10]. The `hygiene hypothesis' proposes that lung exposure to certain bacterial components (e.g. CpG DNA) could modulate the Th2 immune response [11–13]. Thus, CD4 T cell immune response to specific protein antigens could be modulated by exposure to bacterial microbial products.
We have previously demonstrated that direct airway exposure to Aspergillus fumigatus lysates promotes a Th2 biased immune response in the lungs of mice [5]. This is similar to the immune response to A. fumigatus seen in humans with allergic bronchopulmonary aspergillosis (ABPA) [14] and is characterized by prominent lung eosinophilia and mucus production. It is also associated with antigen-specific CD4 T cell IL-4 production characteristic of a Th2 bias [5]. Using this animal model, we have now examined whether the Th2 immune response is unique to fungal antigens or whether it results from direct lung exposure to bacterial lysates as well. Here we demonstrate that, like A. fumigatus, Candida albicans antigens also promote Th2 inflammation characterized by airway eosinophilia and mucus production, whereas P. aeruginosa antigen exposure promotes a Th1 immune response characterized by neutrophilia without mucus production. Further, the presence of P. aeruginosa products are capable of modulating the adaptive immune response to fungal antigens independent of TLR4 signaling but dependent on MyD88 signaling.
RESULTS
A. fumigatus and C. albicans induce eosinophilic airway inflammation with mucus hypersecretion
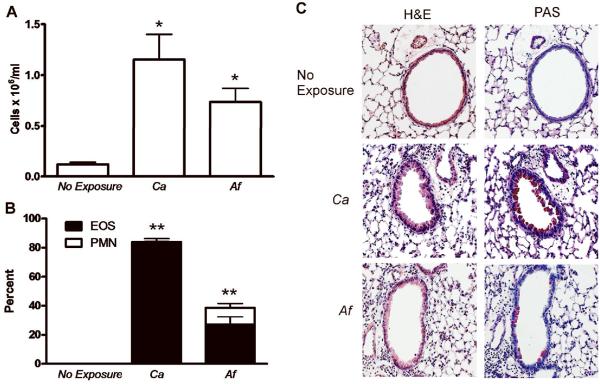
We have previously demonstrated that direct inoculation of the lung with killed A. fumigatus hyphal antigens without previous immunization and in the absence of adjuvent induces a Th2 immune response characterized by airway eosinophilia and mucus production [5]. To determine whether this response is specific to A. fumigatus or common to other fungal antigens, we examined the pulmonary immune response to Candida albicans. In certain organs, live C. albicans infections have previously been shown to induce a Th2 immune response but the immune response to killed lysates in the lung has not been determined. Using a chronic exposure model in which mice are exposed to antigens three times over 14 days, we found that similar to A. fumigatus, killed C. albicans antigens also generated significant airway inflammation as measured by BALF cell number (Figure 1A). C. albicans-induced inflammation was also characterized by airway eosinophils with minimal numbers of neutrophils (Figure 1B) and marked airway epithelial mucus production compared to nonexposed mice (Figure 1C). Further, we found similar eosinophilic inflammation with mucus production to Aspergillus niger (data not shown). We conclude that exposure of the lung to fungal lysates results in a clear Th2 immune response characterized by accumulation of eosinophils and mucus production.
FIG. 1.
A. fumigatus (Af) and C. albicans (Ca) induce eosinophilic airway inflammation with mucus hypersecretion. C57Bl/6J mice were untreated (no exposure) or exposed to 5 μg of either A. fumigatus or C. albicans on day 0, 7 and 14 with lung harvest on day 19. (A) BALF cell numbers. (B) Percentage cell type in BALF as determined by manual differential cell count of neutrophils (PMN) and eosinophils (EOS). Values represent the mean ± SEM of 4 mice per group and are representative of experiments performed twice. Statistical significance was determined by one-way ANOVA; *p<0.01 compared with non-exposed mice; **p<0.0001 eosinophil percentages compared with those in non-exposed mice. (C) Lungs were inflation fixed, sectioned and stained with H&E and PAS. Representative images are 100x magnification.
P. aeruginosa induces neutrophilic airway inflammation without mucus hypersecretion
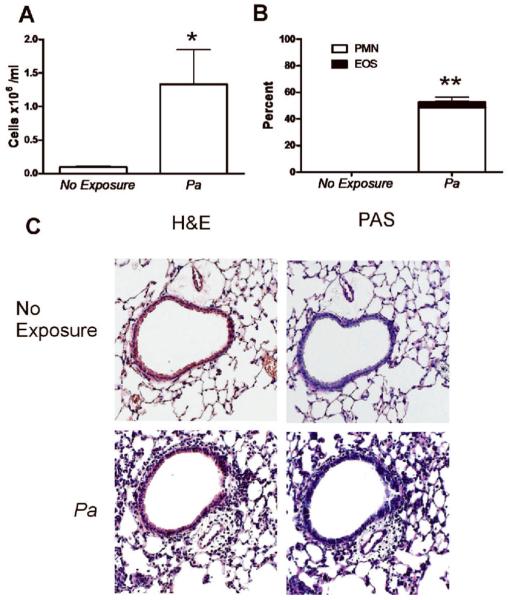
To determine whether the lung environment favors a Th2 bias to any microbial antigens or whether this type of immune response was specific to fungi, we examined the response to bacterial antigens. P. aeruginosa is a Gram negative bacterium that is frequently associated with a variety of chronic lung diseases [15–21]. We therefore exposed mice to cellular lysates of P. aeruginosa and found that lung exposure also induced airway inflammation with high numbers of cells in BALF (Figure 2A). However, unlike the airway eosinophilia elicited by fungal antigens, the cellular composition in the BALF was largely neutrophils with very few eosinophils (Figure 2B). Additionally, despite the lung tissue inflammation observed after P. aeruginosa exposure, no airway epithelial mucus staining was observed (Figure 2C). Thus, direct lung exposure to P. aeruginosa induces an immune response in the lung characterized by neutrophilia instead of eosinophilia and with no mucus production. These results demonstrate that the lung environment alone is not sufficient to dictate the type of immune response generated to microbial pathogens.
FIG. 2.
P. aeruginosa (Pa) induces neutrophilic airway inflammation without mucus hypersecretion. (A) BALF cell numbers after exposure to P. aeruginosa (5 μg) according to the protocol described in Figure 1. Statistical significance determined by the students t-test, four mice per group, *p<0.05 compared with non-exposed mice. (B) Percentage cell type in BALF by cell type as determined by manual differential cell count of neutrophils (PMN) and eosinophils (EOS). Values represent the mean ± SEM of 4 mice per group and are representative of experiments performed twice. Statistical significance was determined by one-way ANOVA; **p<0.001 compared with neutrophil percentages in non-exposed mice. (C) Lungs were inflation fixed, sectioned and stained with H&E and PAS. Representative images are 100x magnification.
C. albicans induces Th2 cytokine production while P. aeruginosa induces Th1 and Th17 cytokine production
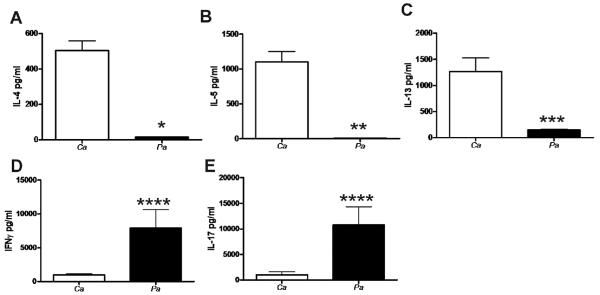
To define the in vivo CD4 T cell response to C. albicans and P. aeruginosa, we examined IFNγ, IL-4, IL-5 and IL-13 production upon CD4 T cell restimulation with either C. albicans or P. aeruginosa. CD4 T cells from C. albicans-exposed mice were restimulated with C. albicans lysates and CD4 T cells from P. aeruginosa-exposed mice were restimulated with P. aeruginosa lysates. CD4 T cells from C. albicans-exposed mice produced higher levels of IL-4, IL-5 and IL-13 when compared to CD4 T cells from P. aeruginosa-exposed mice (Figure 3A, 3B and 3C) correlating with the eosinophilic lung inflammation and mucus production observed in the mice exposed to C. albicans. Interestingly, CD4 T cells from mice exposed to P. aeruginosa produced significantly more IFNγ compared to CD4 T cells from mice exposed to C. albicans (Figure 3D), in agreement with the neutrophilic inflammation in the P. aeruginosa-exposed mice. Thus, lung exposure to fungi, but not to bacterial products, promoted CD4 T cell Th2 differentiation.
FIG. 3.
C. albicans (Ca) induces Th2 cytokine production while P. aeruginosa (Pa) induces Th1 and Th17 cytokine profile. (A) IL-4, (B) IL-5, (C) IL-13, (D) IFNγ and (E) IL-17 production by ex vivo restimulated splenic CD4 T cells after in vivo exposure to C. albicans or P. aeruginosa, as determined by ELISA. In vitro data presented are the means ± SEM of cytokine production by CD4 T cells isolated from a single mouse from a total of four individual mice per group. SEM represents the variation in cytokine production for the four individual mice. Statistical significance determined by the students t-test *, p<0.0001, **, p<0.001, ***, p<0.01, ****, p<0.05 compared with values in C. albicans exposed mice. Results are representative of two individual experiments.
An additional subset of helper T cells, Th-17, has been recently described and are characterized by their ability to produce the proinflammatory cytokine IL-17. IL-17, produced primarily by T lymphocytes early in the course of bacterial infection, promotes neutrophil recruitment and survival [22, 23], is released in response to antigen presenting cell-derived IL-23 and represents important cross-talk between the innate and adaptive immune systems [23]. We measured CD4 T cell IL-17 production in response to lung exposure to C. albicans and P. aeruginosa microbial products. Ex vivo restimulated CD4 T cells from mice exposed to P. aeruginosa produced high levels of IL-17 while CD4 T cells from C. albicans-exposed mice produced low levels (Figure 3E). These results correlate with the selective accumulation of neutrophils in P. aeruginosa-exposed mice but not in C. albicans-exposed mice. Thus, fungal products do not induce CD4 T cell responses characterized by IL-17 production.
The immune response to C. albicans is shifted from Th2 to Th1 by the presence of P. aeruginosa
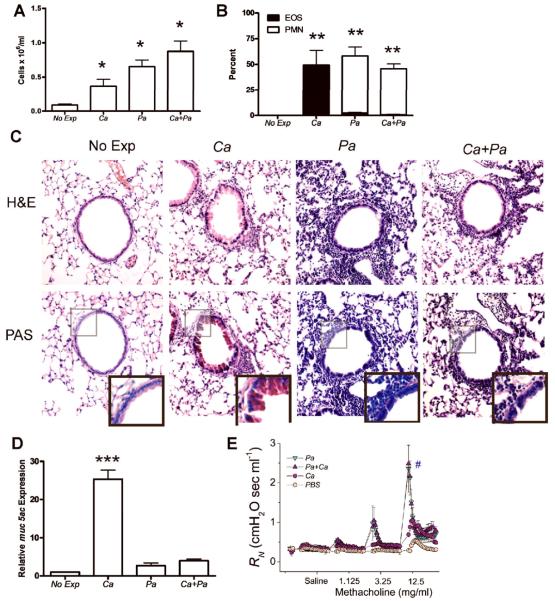
In chronic bacterial infections of the lung, such as those associated with CF, fungi, such as A. fumigatus and C. albicans are often also detected [18, 24–27]. While bacterial colonization is clearly detrimental and often treated aggressively [16, 17, 26], the effects of fungal airway infections on lung function, bacterial virulence or host response are almost completely unknown. To determine how the presence of bacteria can affect the host immune response, we examined the immune response in the lung to the combined exposure of directly administered C. albicans and P. aeruginosa microbial products. As expected, C. albicans, P. aeruginosa and the co-exposure to C. albicans and P. aeruginosa all resulted in airway inflammation as measured by BALF cell number when compared to nonexposed mice (Figure 4A). However, while C. albicans exposure resulted in predominantly eosinophils accumulating in the airway, exposure to C. albicans and P. aeruginosa resulted in the immune response changing from eosinophilic to neutrophilic (Figure 4B). Both the percent and total number of eosinophils were greatly reduced upon co-exposure. Mice exposed to C. albicans, P. aeruginosa or both developed peribronchial airway inflammation on H&E staining when compared to nonexposed mice (Figure 4C). However, while PAS staining of the lung tissue demonstrated epithelial mucus production in C. albicans exposed mice (Figure 4C), exposure to both C. albicans and P. aeruginosa together resulted in an inflammatory response characterized by an absence of mucus production (Figure 4C). Lung mucus staining was further quantified by determining the expression of the muc5ac gene (a common mucin gene in the lung) [28] in the lung tissue using real time RT-PCR. C. albicans exposure induced a 25-fold increase in levels of muc5ac gene expression when compared to nonexposed control mice while exposure to P. aeruginosa or both C. albicans and P. aeruginosa resulted in less than an 4-fold increase in muc5ac gene expression (Figure 4D). These results demonstrate that the presence of bacteria does not significantly alter the magnitude of inflammation but rather dictates the type of inflammation elicited.
FIG. 4.
The adaptive immune response to C. albicans (Ca) is shifted from a Th2 toward Th1 by the presence of P. aeruginosa (Pa). (A) BALF cell numbers after exposure to either C. albicans or P. aeruginosa (5 μg) or both using the protocol described in Figure 1. (B) BALF cell percentage by cell type as determined by manual differential cell count of neutrophils (PMN) and eosinophils (EOS). Values represent the mean ± SEM of 4 mice per group and are representative of experiments performed twice. Statistical significance determined by one-way ANOVA; *, p<0.05 compared with nonexposed mice, **, p< 0.0001 compared with EOS or PMN in non-exposed mice. (C) C57BL/6J mice were exposed to either C. albicans, P. aeruginosa or both as in Figure 1. Lungs were inflation fixed, sectioned and stained with H&E and PAS. Representative images are 100x magnification and PAS inlay are 200x magnification. (D) Real time RT-PCR using total lung tissue to detect muc5ac after oropharyngeal exposure to either C. albicans or P. aeruginosa or both. Expression of muc5ac is presented relative to expression in non-exposed C57BL/6J mice. Statistical significance determined by the students t-test, 4 mice per group and are representative of experiments performed twice; ***, p<0.0001 compared with either non-exposed, P. aeruginosa exposed or P. aeruginosa and C. albicans exposed mice. (E) Airway resistance in response to methacholine after oropharyngeal exposure to either phosphate buffered saline (PBS), C. albicans or P. aeruginosa or both. Data presented are the means + SEM of four mice per group. #p<0.05 between P. aeruginosa or combined P. aeruginosa and C. albicans compared to PBS or C. albicans alone (two-way ANOVA).
Recent studies using models of combine ovalbumin (OVA) and LPS sensitization and challenge have suggested that a Th1 dominant immune response to OVA, characterized by neutrophilia and INFγ production, results in a greater increase in airway hyperresponsiveness (AHR, PenH) in response to methacholine [29]. To determine if immunodeviating the immune response to C. albicans from eosinophilic to neutrophilic airway inflammation with P. aeruginosa had an impact on AHR, we measured airway resistance (RN) following methacholine challenge in mice exposed to PBS, C. albicans, P. aeruginosa or a combination of C. albicans and P. aeruginosa antigens. Airway exposure to C. albicans alone showed increased airway resistance (Rn) when compared to PBS exposed controls (Figure 4E). Interestingly however, exposure to P. aeruginosa, either alone or in combination with C. albicans resulted in a significantly increased degree of AHR when compared to exposures to PBS or C. albicans alone. These results suggest that shifting of the immune response to C. albicans by P. aeruginosa leads to a more severe asthma phenotype, which may be the result of greater airway injury and more direct access of nebulized methacholine to underlying smooth muscle.
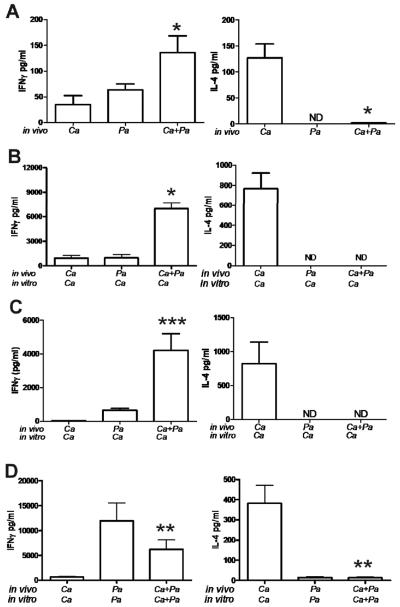
To determine whether the airway neutrophilia observed after co-exposure to C. albicans and P. aeruginosa is associated with differential airway cytokines, we measured IL-4 and IFNγ in BALF after exposure to C. albicans and P. aeruginosa. The co-exposure to C. albicans and P. aeruginosa led to high levels of IFNγ and low levels of IL-4 (Figure 5A), suggesting that P. aeruginosa may alter the differentiation of antigen specific CD4 T cells from Th2 to Th1. To test this hypothesis we then isolated splenic CD4 T cells from mice co-exposed to C. albicans and P. aeruginosa and restimulated them in vitro with C. albicans and found that antigen co-exposure resulted in high levels of IFNγ while IL-4 was undetectable (Figure 5B). Similar results were found when CD4 T cells were isolated directly from the lung of mice co-exposed to C. albicans and P. aeruginosa (Figure 5C). Additionally, when splenic CD4 T cells from mice co-exposed to C. albicans and P. aeruginosa in vivo and restimulated with both microbial products in vitro, there was still preferential production of IFNγ over IL-4 (Figure 5D). Taken together, these results support the conclusion that the Th2 immune response induced by fungal microbial products is redirected to a Th1 immune response in the presence of P. aeruginosa products.
FIG. 5.
BAL and CD4 T cell cytokine production after exposure to C. albicans (Ca), P. aeruginosa (Pa) or both. (A) Production in BALF from mice exposed to C. albicans (Ca), P. aeruginosa (Pa) or both. Data presented are the means + SEM of four mice per group. **, p<0.05 compared with values in C. albicans exposed mice. Production from (B) splenic CD4 T cells or (C) pulmonary T cells restimulated in vitro with C. albicans antigens after in vivo exposure to C. albicans, P. aeruginosa or both. In vitro data presented are the means + SEM of cytokine production by CD4 T cells isolated from a single mouse with a total of four individual mice per group. SEM represents the variation in cytokine production for the four individual mice. Statistical significance determined by the students t-test *, p<0.0001, ***, p<0.01 compared with values from C. albicans or P. aeruginosa exposed mice. (D) Production from splenic CD4 T cells restimulated in vitro with antigens from C. albicans, P. aeruginosa or both after in vivo exposure to C. albicans or P. aeruginosa or both. In vitro data presented are the means + SEM of cytokine production by CD4 T cells isolated from a single mouse with a total of four individual mice per group. SEM represents the variation in cytokine production for the four individual mice. Statistical significance determined by the students t-test; **, p<0.05 compared with values in C. albicans exposed mice. For all panels in this figure, results are representative of two individual experiments.
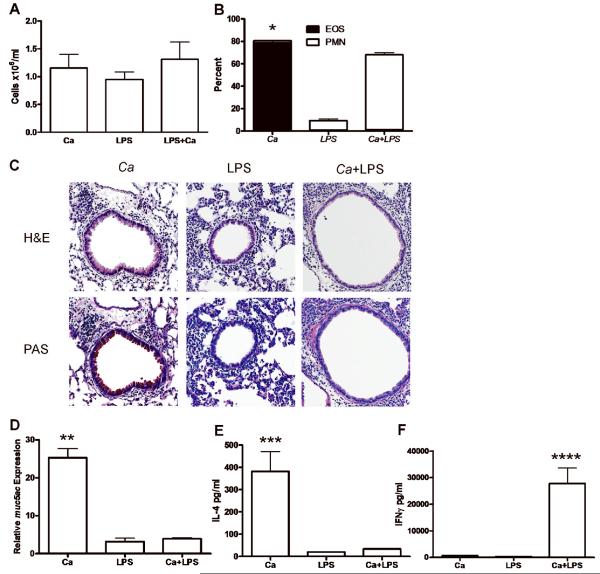
The adaptive immune response to C. albicans is Th1 and not Th2 in the presence of LPS
As in other Gram negative bacteria, the P. aeruginosa outer membrane contains abundant lipopolysaccaride (LPS) [30–32]. LPS has been shown to modulate CD4 Th1/Th2 responses in a dose dependent manner [33]. To determine if Th1-type responses elicited by exposure to P. aeruginosa lysates were similar to those elicited by purified LPS, we examined the immune response against airway exposure to C. albicans antigens in combination with bacterial LPS. We selected our dose of LPS to be similar to the dose present in the cell lysates of P. aeruginosa and similar to levels used in other models of Gram negative infection [34–36]. As expected, C. albicans, LPS and coexposure to LPS and C. albicans together all induced airway inflammation (Figure 5A). Analysis of differential cell counts showed that exposure to LPS alone caused lung inflammation characterized largely by the presence of macrophages at the time of lung harvest with few neutrophils and no eosinophils (Figure 5B). However, while C. albicans stimulated an eosinophilic inflammatory cell infiltration, the addition of LPS to C. albicans resulted in an immune response in which neutrophilic inflammation was predominent (Figure 5B). Peribronchial lung tissue inflammation was observed on H&E staining after C. albicans, LPS and coexposure to LPS and C. albicans; however, epithelial mucus staining was only observed after C. albicans alone, was not present after LPS exposure and was abolished by the addition of LPS to C. albicans (Figure 5C). Mucus production was further quantified by measuring muc5ac gene expression, which demonstrated high levels of expression selectively in the C. albicans exposed mice, but not in LPS-exposed and LPS and C. albicans exposed mice, confirming the lung histology findings (Figure 5D). These results suggest that C. albicans induces a Th2 immune response in vivo with eosinophilic inflammation and mucus production while bacterial LPS is sufficient to alter this immune response to Th1 as characterized by neutrophil accumulation and a lack of mucus production. To confirm this, CD4 T cells were restimulated ex vivo after in vivo exposure to C. albicans, LPS or LPS and C. albicans coexposure. As expected, CD4 T cells from the C. albicans exposed mice produced high levels of IL-4 and low levels of IFNγ indicating a Th2 bias (Figure 5E). Conversely, CD4 T cells from mice exposed to both LPS and C. albicans produced low levels of IL-4 and high levels of IFNγ supporting the generation of a Th1 phenotype (Figure 5F). LPS alone did not result in significant IL-4 or IFNγ production, indicating that the production of these cytokines is antigen-specific (Figure 5E, 5F). We conclude that the absence of LPS in the fungi favors a Th2-biased immune response.
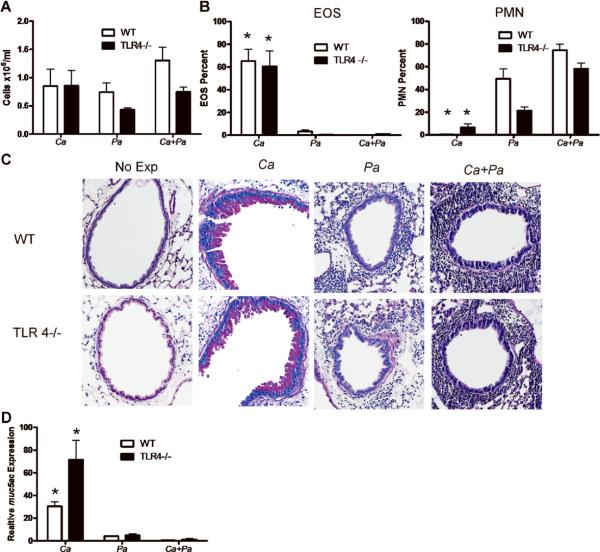
P. aeruginosa alters the adaptive immune response to C. albicans independent of TLR4 but dependent on MyD88
To determine if the Th1-biasing effect of P. aeruginosa on C. albicans immune response was simply due to the presence of LPS we used TLR4 deficient mice. TLR4 is a receptor for pathogen-associated molecular patterns (PAMPs) expressed by cells of the innate immune system that initiates intracellular signaling in response to LPS [37]. Since fungi do not contain LPS, C. albicans exposure in TLR4 deficient mice resulted in a Th2 biased inflammatory response similar to that seen in WT mice (Figure 6A, 6B). TLR4 deficiency did not affect airway inflammation induced by C. albicans as determined by total cell number and percent of eosinophils in BALF, indicating that the immune response to C. albicans is TLR4-independent. However, as expected, the total cell number and percent neutrophils in BALF of TLR4 deficient mice exposed to P. aeruginosa were reduced compared with the numbers in WT mice (Figure 7A, 7B), indicating that the type of immune response generated to P. aeruginosa is partially due to the presence of LPS. Interestingly, the absence of TLR4 did not prevent P. aeruginosa exposure from switching the immune response to C. albicans from eosinophilia to neutrophilia (Figure 7A, 7B). Furthermore, TLR4 deficiency did not prevent P. aeruginosa from inhibiting mucus production triggered by C. albicans (Figure 7C, 7D). These results indicate that the effect that P. aeruginosa has on C. albicans immune response is not due exclusively to the presence of LPS.
FIG. 6.
The adaptive immune response to C. albicans (Ca) is Th1 and not Th2 in the presence of LPS. (A) BALF cell numbers after exposure to either C. albicans, LPS (5 μg) or both following the protocol as described in Figure 1. There is no significant difference between groups. (B) Percentage cell type in BALF by cell type as determined by manual differential cell count of neutrophils (PMN) and eosinophils (EOS). Values represent the mean ± SEM of 4 mice per group and are representative of experiments performed twice. *, p<0.0001 EOS in C. albicans compared with EOS in LPS or C. albicans and LPS exposed mice. (C) Lungs were inflation fixed, sectioned and stained with H&E and PAS. Representative images are 100x magnification. (D) Real time RT-PCR using total lung tissue to detect muc5ac after oropharyngeal exposure to either C. albicans, LPS or both. Expression of muc5ac is presented relative to expression in non-exposed C57BL/6J mice. **, p<0.001 muc5ac in C. albicans compared with LPS or Ca and LPS exposed mice. (E) Splenic CD4 T cell production of IL-4 by ex vivo restimulated CD4 T cells after in vivo exposure to C. albicans, LPS or both, as determined by ELISA. In vitro data presented are the means ± SEM of one representative experiment of two with four mice per group. ***, p<0.05 IL-4 in C. albicans compared with values in LPS or C. albicans and LPS exposed mice. (F) Splenic CD4 T cell production of IFNγ by ex vivo restimulated CD4 T cells after in vivo exposure to C. albicans, LPS or both, as determined by ELISA. In vitro data presented are the means ± SEM of one representative experiment of two presenting combined data from four mice per group. ****, p<0.05 IFNγ in C. albicans compared with values in C. albicans or LPS exposed mice.
FIG. 7.
P. aeruginosa (Pa) alters the adaptive immune response to C. albicans (Ca) independently of TLR4. (A) BALF cell numbers after exposure of C57BL/J6 (WT) or TLR4−/− mice to either C. albicans or P. aeruginosa or both using the protocol described in Figure 1. (B) BALF cell percentage by cell type as determined by manual differential cell count of neutrophils (PMN) and eosinophils (EOS). Values represent the mean ± SEM of 4 mice per group and are representative of experiments performed twice. Statistical significance was determined by two-way ANOVA; *, p<0.0001 compared with values in P. aeruginosa or C. albicans and P. aeruginosa exposed WT or TLR4−/− mice. (C) Lungs were inflation fixed, sectioned and stained with PAS. Representative images are 100x magnification. (D) Real time RT-PCR using total lung tissue to detect muc5ac after oropharyngeal exposure to either C. albicans, P. aeruginosa or both. Expression of muc5ac is presented relative to expression in non-exposed C57BL/6J mice. Statistical significance was determined by two-way ANOVA to be p<0.0001. *, p<0.0001 compared with corresponding groups in P. aeruginosa or C. albicans and P. aeruginosa exposed WT or TLR4−/− mice.
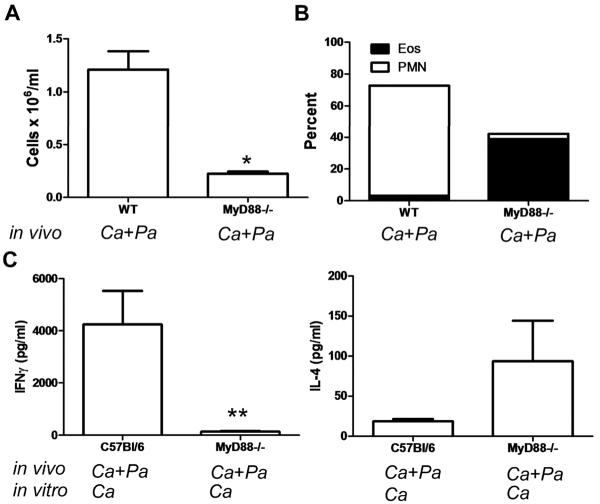
To determine if other members of the TLR family were important in immunodeviating the adaptive immune response to C. albicans by P. aeruginosa, we exposed wild type (WT) and MyD88 deficient (MyD88−/−) mice to a combination of C. albicans and P. aeruginosa lysates. As expected, airway inflammation was severely compromised in MyD88−/− mice compared with WT mice (Figure 8A). Interestingly however, while WT mice have a predominance of airway neutrophils, MyD88 −/− mice have an eosinophilic predominant airway inflammatory response suggesting that the immunodeviation induced by P. aeruginosa is dependent on MyD88 signals (Figure 8B). Additionally, CD4 T cells from MyD88−/− mice restimulated in vitro with C. albicans antigens appear to produce more IL-4 and less IFNγ compared to CD4 T cells from WT mice (Figure 8C). Taken together, these results suggest that the immunodeviation of the adaptive response to C. albicans from Th2 to Th1 by P. aeruginosa is independent of LPS and TLR 4 but requires other MyD88-dependent signals.
FIG 8.
P. aeruginosa (Pa)-induced immunodeviation of the adaptive immune response to C. albicans (Ca) is MyD88 dependent. (A) (A) BALF cell number after exposure of C57BL/J6 (WT) or MyD88−/− mice to C. albicans and P. aeruginosa using the protocol described in Figure 1. Statistical significance determined by the students t-test; *, p<0.001. (B) BALF cell percentage by cell type as determined by manual differential cell count of neutrophils (PMN) and eosinophils (EOS). Values represent the mean ± SEM of 4 mice per group. Statistical significance was determined by one-way ANOVA to be p<0.0001. (C) IL-4 and IFNγ production as determined by ELISA using splenic CD4 T cells from C56Bl/6 or MyD88−/− mice restimulated in vitro with C. albicans antigens after in vivo exposure to both C. albicans and P. aeruginosa. In vitro data presented are the means + SEM of cytokine production by CD4 T cells isolated from a single mouse from a total of five individual mice per group. SEM represents the variation in cytokine production for the five individual mice. Statistical significance determined by the students t-test; **, p<0.01 compared with values in C57Bl/6 mice exposed to C. albicans and P. aeruginosa.
DISCUSSION
COPD, bronchiectasis and CF are often complicated by chronic airway infections [15, 26, 38–42]. P. aeruginosa is a commonly encountered bacteria and is typically treated aggressively with antibiotics, particularly at time of acute exacerbation [15–18]. Frequently, P. aeruginosa colonization of the airway is accompanied by colonization with different species of fungi such as C. albicans and A. fumigatus [24, 25, 27, 39, 43–46]. In vitro work has demonstrated that P. aeruginosa has a number of interactions with C. albicans [47, 48]. Additional studies have suggested that the presence of C. albicans within the airway is a risk factor for contracting P. aeruginosa lung infection. Unlike bacterial infection, little effort is made clinically to treat fungal airway colonization and the fungi are often considered innocuous bystanders [16, 17, 26, 27]. However, the presence of multiple microbes could affect the adaptive immune response to specific microbes. Little is known about the in vivo interaction between bacterial and fungal products and how it may impact on the immune response to these products. We have previously shown that direct airway exposure to A. fumigatus is sufficient to generate a Th2-biased immune response characterized by the accumulation of eosinophils and mucus production [5]. In this study, we show a similar type of response after exposure to antigens from C. albicans. Thus, direct exposure of lung to fungal antigens is capable of inducing an antigen-specific Th2 immune response. In contrast, we also show here that direct exposure of lung to bacterial antigens induces a Th1 type of immune response characterized by airway neutrophilia without mucus secretion.
Th2 immune responses are associated with infections by extracellular pathogens, while Th1 responses are associated primarily with intracellular pathogens [8]. However, in our studies we use killed pathogen lysates instead of infectious organisms, indicating that the type of immune response in the lung is not determined only by the specific cellular route of infection. Instead, the results shown here suggest that the presence of components specifically expressed in fungi, but not in bacteria are likely to be the cause of the Th2-biased response to fungal antigens. Alternatively, the absence in fungi of a specific component that promotes a Th1 response could result in a default Th2 immune response. Since the administration of P. aeruginosa together with C. albicans induces a Th1 response, the later mechanism is more likely to be the case.
LPS is a component of P. aeruginosa and other Gram-negative bacteria that rapidly triggers the innate immune response, but can also indirectly modulate Th1/Th2 bias probably indirectly through innate cytokine production [33]. Fungi lack LPS and our data show that the absence of TLR4 has no effect on the immune response induced by C. albicans airway exposure, further demonstrating that no contaminant LPS contributes to the Th2 immune response to fungal antigen. Thus, the lack of LPS in fungi could determine the lack of Th1 and default to Th2 response. Although somewhat controversial, LPS has been shown to affect infection with live C. albicans by increasing fungal-related mortality [49–51]. We show here that through direct exposure to the lung, LPS can also modify the immune response to C. albicans lysates by deviating the immune response from Th2 to Th1. However, the Th1 bias observed when both P. aeruginosa and C. albicans antigens were administered together was not impaired in TLR4 deficient mice, indicating that components of P. aeruginosa other than LPS promote the Th1 bias when fungi are present. Our results in MyD88−/− indicate that TLRs other than TLR4 are involved. The airway epithelium expresses a variety of TLRs and may itself be an important component in the innate immune response to pathogens [52–54]. In addition to TLR4 the TLRs, TLR2, TLR5 and TLR9 play a central role in the stimulation of proinflammatory cytokine production from airway epithelium in response to P. aeruginosa [55–57]. Thus, signals from TLR2, TLR5 and TLR9 may be responsible for the Th1 bias seen after exposure to these products. P. aeruginosa also possesses a variety of virulence factors important in organism pathogenesis, including proteases, lipases and type III secretion factors that exploit the host environment and allow the organism to escape from tissues [58–61]. P. aeruginosa also has virulence factors that help it evade the immune response, notably polysaccarides such as alginate [62, 63]. These virulence factors are most relevant in live infections with P. aeruginosa and are unlikely to play a significant role in our studies as the P. aeruginosa lysates were heat-killed there by inactivating the enzymatic activity required to impart virulence. Our results demonstrate that the Th1 response to P. aeruginosa was less robust in TLR4 deficient mice while the response to P. aeruginosa and C. albicans in these mice was not affected. This result suggests that the adaptive immune response to the combined microbial products is directed against C. albicans antigens rather than P. aeruginosa antigens, further demonstrating the ability of P. aeruginosa to immunodeviate the response to C. albicans independent of LPS and TLR4.
To date, the components of fungi that can promote the Th2 immune response remains unclear. Some studies have proposed that the presence of fungal-associated allergenic proteinase is responsible for the Th2 type of immune response to these microorganisms [64, 65]. A. fumigatus antigens devoid of proteases are not sufficient to cause a Th2 immune response, but rather only with the addition of proteases does direct airway inoculation result in allergic disease [64]. Similarly, non-allergenic proteins such as ovalbumin can be made allergenic by the addition of fungal protease as an adjuvant [64]. However, our studies were performed with fixed or heat-killed extracts without active proteases, suggesting that enzymatic activity was not required to generate an immune response. Thus, although we cannot discard the involvement of proteases during fungal infection, they are not absolutely required for the induction of a Th2 immune response. The C-type lectin receptor (CLR) class of pattern recognition receptors (PRR), including dectin 1, are also important in regulating mammalian immune responses to a variety of fungi, including C. albicans [8]. Dectin 1 recognizes β-glucan, the complex carbohydrate common in fungal cell walls [66, 67] and plays in important role in host defense against germinating fungal yeast [8, 68, 69]. Its role in the generation of an in vivo Th2 immune response to fungal lung exposure remains uncertain.
Additional factors that qualitatively influence the type of immune response are the route of fungal exposure, live versus killed organism and the phase of fungal growth [9, 10, 69, 70]. In vivo fungal administration of conidia via the gut, via the lung or systemically has been shown to induce a Th1 immune response, which is necessary in the clearance of live organism [8]. If the conidia are killed, then the adaptive immune response is characterized by IL-4-producing CD4 T cell clones [70] suggesting that the host can distinguish between conidia that have the potential to grow and cause further damage and those that cannot [70]. Mannoproteins on the fungal cell wall have been shown to stimulate robust Th1 responses and thereby prevent disseminated infection [8, 71]. Our work and that of others have clearly documented a Th2 immune response with fungal administration directly to lung and may be a consequence of the hyphal components of our fungal lysates [64, 65].
Polymicrobial infections are common in chronic lung disease and a better understanding of the host's immune response to pathogens is essential in developing new therapies and guiding current treatment. Here we show the novel finding that in the lung P. aeruginosa is able to immunomodulate the host response to C. albicans from Th2 to Th1 in an LPS/TLR4 independent but MyD88 dependent mechanism.
MATERIAL AND METHODS
Reagents
A. fumigatus lysates, generously provided by Dr. Kieren Marr, Department of Medicine, Portland, Oregon, were generated by culture of Af293 isolate (5 days, 37°C) in RPMI 1640 (+10% FCS). The hyphal mat was harvested, sequentially washed with PBS, and disrupted by vortexing with glass beads. The slurry was subjected to paraformaldehyde (1%) for microbial inactivation. Protein concentration was measured using the Bradford assay (BioRad Laboratories, Hercules, CA). The product was concentrated using an endotoxin-free dialysis membrane with 10 kDa pore size (Pierce Biotechnology, Rockford, IL) to achieve a final protein concentration of 800–1000 μg/mL. P. aeruginosa strain PA14 [72] and C. albicans strain SC5314 [73] were grown in 100 ml of DMEM medium (#10-13-CV, Mediatech, Manassas, VA) in 250 ml flasks at 37°C for 16 h at 200 rpm. Flasks were inoculated from 5 ml overnight cultures. Cells were harvested by centrifugation, washed one time, then resuspended in 100 mM phosphate buffer (pH 7). Bacterial cells were lysed by ballistic disintegration via 6 1-min pulses with 0.2 mm glass beads using a Beadbeater (Biospec Products, Bartlesville, OK) with 1 min incubations on ice in between each pulse. C. albicans cells were similarly lysed except that 0.5 mm beads were used. Both bacterial and fungal lysates were heated at 65°C for 10 minutes to kill any remaining cells. The absence of any viable cells was confirmed by plating 100 μl of the lysate onto solid medium and inoculation of 500 μl of the lysate into liquid medium followed by incubation at 37°C for 24 and 48h. Viability experiments were performed using Luria broth medium for P. aeruginosa, and Yeast Extract/Peptone/Dextrose for C. albicans. The protein concentration of lysates was determined by the Bradford Assay (BioRad, Hercules, CA) and the lysate was diluted in phosphate buffer to a final concentration of 1 mg/ml. Lysates were stored frozen at −80°C prior to use.
Mice
6–8-week-old male C57BL/6J mice were purchased from The Jackson Laboratories (Bar Harbor, ME). 6–8-week-old male TLR4–/–and MyD88 −/− mice both on the C57BL/6J background were bred at the University of Vermont. All mice were housed in the Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility at the University of Vermont. The Institutional Animal Care and Use Committee granted approval for all studies. Mice were anesthetized with i.p. pentobarbital sodium (70–90 mg/kg) prior to nonsurvival surgery performed on day 19 after initial microbial product exposure.
Microbial product airway exposure protocol
Mice were briefly anesthetized (isoflurane by inhalation) and oropharyngeally exposed under direct vocal cord visualization to 5 μg of protein of either A. fumigatus, C. albicans, P. aeruginosa or C. albicans and P. aeruginosa cellular lysates or P. aeruginosa-derived LPS (Sigma, St. Louis, MO) or PBS in a volume of 40 μl on days 0, 7 and 14. This is a model of acute exposure to microbial products. Although a three administration protocol was used for the studies in this manuscript, a similar type of immune response of lesser magnitude was obtained after a single exposure to microbial products.
BALF
Tracheas were cannulated with 2 cm of 22 gauge polyethelyene tubing attached to a 23 gauge needle, 1 ml of cold PBS was instilled into the lungs and the BALF fluid was collected. Cells were pelleted and counted using an Advia cell counter (Bayer, Terrytown, NY) and supernatant was stored at −80 C for cytokine analysis. Cells (50 × 103) were cytospun onto glass slides and stained with Hema-3 (Biochemical Sciences, Swedesboro, NJ). Two hundred cells per slide were counted and scored as macrophages, eosinophils, neutrophils or lymphocytes based on characteristic morphology and staining.
Quantitative PCR analysis of gene expression
The right lung from each experimental mouse was removed and snap frozen in liquid nitrogen. It was then ground to a fine powder using a chilled motar and pestle and total RNA was extracted using trizol and further purified using an RNeasy kit (Qiagen, Valencia, CA). First-strand cDNA was synthesized from 1 μg RNA using random primers and Superscript II reverse transcriptase (Invitrogen, San Diego, CA). Quantitative PCR was performed on cDNA with Assays on Demand predesigned muc5ac and hypoxanthine-guanine phosphoribosyltransferase (HPRT) primer/probe sets (Applied Biosystems, Foster, CA), using TaqMan Universal PCR Master Mix and the Applied Biosystems PRISM 7700 Sequence Detection System. The level of muc5ac expression was normalized to HPRT housekeeping gene levels and relative muc5ac mRNA levels were determined according to the comparative cycle threshold method (Applied Biosystems, ABI Prism 7700 Sequence Detection System, User Bulletin #2). Briefly, the threshold cycle (CT) was determined for muc5ac and HPRT in each sample. The ΔCT was calculated for each sample by subtracting the CT of HPRT from the CT of muc5ac. The baseline ΔCT was assigned to non-AHA exposed WT mice. The ΔΔCT values were calculated by subtracting the ΔCT of the baseline from the ΔCT of the experimental samples. The ΔΔCT values were transformed into relative values using the equation, expression= 2−ΔΔCT
Lung histology
Lungs were processed for histological analysis as described previously [74] and stained with H&E and PAS according to routine procedures.
Cell preparation and activation
Single-cell suspensions were generated from spleen by passing the disassociated tissue through a 70-μm mesh, and lymphocytes were enriched by separation with Lymphocyte Separation Medium (MP Biomedicals, Solon, OH). Single cell suspensions were generated from lung by infusion of Type 1 collagenase (1 ml, 1 mg/ml, Gibco, Carlsbad, CA) into the cannulated trachea followed by mincing of lung tissue and incubation in 5 ml of additional type 1 collagenase for 15 minutes at 37 C. Tissue was further dissociated by repeated aspiration with a 14 gauge hollow needle and 60 ml syringe followed by filtering with a 70-μm mesh. Cells were then spun at 600 rotations per minute for 6 minutes, washed with Hank's balanced salt solution (HBSS, Mediatech Inc., Manassas, VA) and resuspended in 5 ml of HBSS. CD4 T cells were isolated from the cellular suspensions by positive selection using CD4 magnetic microbeads (Miltenyi Biotec, Auburn, CA), according to the manufacturer's protocol and purity was confirmed (typically >93%) by surface CD4 and T-cell receptor staining and FACS analysis. Isolated CD4 T cells (2 × 106cells/ml) were incubated in the presence of syngeneic antigen presenting cells (APC, 4 × 106cells/ml) and activated either by A. fumigatus cellular lysates (5 μg/ml), C. albicans cellular lysates (5 μg/ml), or P. aeruginosa cellular lysates (5 μg/ml) for 72 hours.
APCs were obtained from untreated mice by splenic T cell depletion with negative selection using Abs to CD4 (GK1.5) [75], CD8 and Thy-1 [76], and treatment with rabbit complement. The remaining cells were then treated with mitomycin C, as previously described [77], prior to incubation with the CD4 T cells.
Determination of cytokine production in BALF and by CD4+ T cells
ELISAs were performed on the BALF and the cell culture supernatant for IL-4, IL-5, IL-13, IL-17 and IFNγ using Duosets (R&D Systems, Minneapolis, MN), according to manufacturer's recommendations.
Determination of Pulmonary function
Mice were anesthetized, tracheotomized and mechanically ventilated for the measurement of respiratory impedence (Zrs) using the forced oscillation technique as previously described [78]. Mice were ventilated at a rate of 2.5 Hz, with a tidal volume of 0.2 ml and 3 cmH2O positive end-expiratory pressure (flexiVent; SCIREQ, Montreal, PQ, Canada). Impedence data were collected during regular ventilation to establish a baseline for each mouse, and inhaled methacholine (Sigma-Aldrich) in saline was then delivered via aerosol in successively increasing doses (0, 3.125, 12.5, and 50 mg/ml). Multiple linear regression was used to fit Zrs spectra derived from measured pressure and cylinder displacement to the constant phase model of the lung [79].
where Z is input impedence of the respiratory system, Rn is Newtonian resistance composed primarily of flow resistance of conducting pulmonary airways, I is the inertence of the gas in central airways, G represents dissipation of viscous energy in respiratory tissues, Hti represents elastic energy storage in the tissues, f is frequency, , ωis 2πf, and αcouples G and Hti. Using this model, the dose response of Rn to methacholine was determined as an index of AHR.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 4.0 (GraphPad Software, San Diego, CA). Statistical significance was determined by the Student's t test unless otherwise indicated. For all analyses p ≤ 0.05 was considered statistically significant.
ACKNOWLEDGEMENTS
This work was supported by the Center for Biomedical Research Excellence Program of the National Center for Research Resources Grant P20RR15557, the Cystic Fibrosis Foundation and BRIN award from the NCRR.
Abbreviations
- AHR
Airway hyperresponsiveness
- Rn
Airway resistance
- Zrs
Respiratory impedence
REFERENCES
- 1.Cieslewicz G, Tomkinson A, Adler A, Duez C, Schwarze J, Takeda K, Larson KA, et al. The late, but not early, asthmatic response is dependent on IL-5 and correlates with eosinophil infiltration. J Clin Invest. 1999;104:301–308. doi: 10.1172/JCI7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochkur SI, Jacobsen EA, Protheroe CA, Biechele TL, Pero RS, McGarry MP, Wang H, et al. Coexpression of IL-5 and eotaxin-2 in mice creates an eosinophil-dependent model of respiratory inflammation with characteristics of severe asthma. J Immunol. 2007;178:7879–7889. doi: 10.4049/jimmunol.178.12.7879. [DOI] [PubMed] [Google Scholar]
- 3.Shen HH, Ochkur SI, McGarry MP, Crosby JR, Hines EM, Borchers MT, Wang H, et al. A causative relationship exists between eosinophils and the development of allergic pulmonary pathologies in the mouse. J Immunol. 2003;170:3296–3305. doi: 10.4049/jimmunol.170.6.3296. [DOI] [PubMed] [Google Scholar]
- 4.Chaplin DD. 1. Overview of the human immune response. J Allergy Clin Immunol. 2006;117:S430–435. doi: 10.1016/j.jaci.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Allard JB, Poynter ME, Marr KA, Cohn L, Rincon M, Whittaker LA. Aspergillus fumigatus generates an enhanced Th2-biased immune response in mice with defective cystic fibrosis transmembrane conductance regulator. J Immunol. 2006;177:5186–5194. doi: 10.4049/jimmunol.177.8.5186. [DOI] [PubMed] [Google Scholar]
- 6.Portnoy JM, Barnes CS, Kennedy K. Importance of mold allergy in asthma. Curr Allergy Asthma Rep. 2008;8:71–78. doi: 10.1007/s11882-008-0013-y. [DOI] [PubMed] [Google Scholar]
- 7.Zeldin Y, Kidon MI, Magen E, Bibi H, Cohen A, Waisel Y, Kivity S. Impact of specific allergen sensitization on the prevalence of asthma in patients with allergic rhinitis from adjacent distinct geographic areas. Ann Allergy Asthma Immunol. 2008;101:30–34. doi: 10.1016/S1081-1206(10)60831-9. [DOI] [PubMed] [Google Scholar]
- 8.Pamer EG. Immune responses to commensal and environmental microbes. Nat Immunol. 2007;8:1173–1178. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- 9.Rivera A, Ro G, Van Epps HL, Simpson T, Leiner I, Sant'Angelo DB, Pamer EG. Innate immune activation and CD4+ T cell priming during respiratory fungal infection. Immunity. 2006;25:665–675. doi: 10.1016/j.immuni.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Hohl TM, Rivera A, Pamer EG. Immunity to fungi. Curr Opin Immunol. 2006;18:465–472. doi: 10.1016/j.coi.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Vandenbulcke L, Bachert C, Van Cauwenberge P, Claeys S. The innate immune system and its role in allergic disorders. Int Arch Allergy Immunol. 2006;139:159–165. doi: 10.1159/000090393. [DOI] [PubMed] [Google Scholar]
- 12.Bauer S, Hangel D, Yu P. Immunobiology of toll-like receptors in allergic disease. Immunobiology. 2007;212:521–533. doi: 10.1016/j.imbio.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Renz H, Blumer N, Virna S, Sel S, Garn H. The immunological basis of the hygiene hypothesis. Chem Immunol Allergy. 2006;91:30–48. doi: 10.1159/000090228. [DOI] [PubMed] [Google Scholar]
- 14.Moss RB. Pathophysiology and immunology of allergic bronchopulmonary aspergillosis. Med Mycol. 2005;43(Suppl 1):S203–206. doi: 10.1080/13693780500052255. [DOI] [PubMed] [Google Scholar]
- 15.Papi A, Bellettato CM, Braccioni F, Romagnoli M, Casolari P, Caramori G, Fabbri LM, et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173:1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- 16.Latzin P, Fehling M, Bauernfeind A, Reinhardt D, Kappler M, Griese M. Efficacy and safety of intravenous meropenem and tobramycin versus ceftazidime and tobramycin in cystic fibrosis. J Cyst Fibros. 2007 doi: 10.1016/j.jcf.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Blumer JL, Saiman L, Konstan MW, Melnick D. The efficacy and safety of meropenem and tobramycin vs ceftazidime and tobramycin in the treatment of acute pulmonary exacerbations in patients with cystic fibrosis. Chest. 2005;128:2336–2346. doi: 10.1378/chest.128.4.2336. [DOI] [PubMed] [Google Scholar]
- 18.Anzueto A, Sethi S, Martinez FJ. Exacerbations of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2007;4:554–564. doi: 10.1513/pats.200701-003FM. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Kosorok MR, Farrell PM, Laxova A, West SE, Green CG, Collins J, et al. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. Jama. 2005;293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- 20.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:991–998. doi: 10.1164/rccm.200509-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tumkaya M, Atis S, Ozge C, Delialioglu N, Polat G, Kanik A. Relationship between airway colonization, inflammation and exacerbation frequency in COPD. Respir Med. 2007;101:729–737. doi: 10.1016/j.rmed.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Happel KI, Dubin PJ, Zheng M, Ghilardi N, Lockhart C, Quinton LJ, Odden AR, et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J Exp Med. 2005;202:761–769. doi: 10.1084/jem.20050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro J, Rainisio M, Harms HK, Hodson ME, Koch C, Mastella G, Strandvik B, et al. Factors associated with poor pulmonary function: cross-sectional analysis of data from the ERCF. European Epidemiologic Registry of Cystic Fibrosis. Eur Respir J. 2001;18:298–305. doi: 10.1183/09031936.01.00068901. [DOI] [PubMed] [Google Scholar]
- 25.Doern GV, Brogden-Torres B. Optimum use of selective plated media in primary processing of respiratory tract specimens from patients with cystic fibrosis. J Clin Microbiol. 1992;30:2740–2742. doi: 10.1128/jcm.30.10.2740-2742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parameswaran GI, Murphy TF. Infections in chronic lung diseases. Infect Dis Clin North Am. 2007;21:673–695. viii. doi: 10.1016/j.idc.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulpa P, Dive A, Sibille Y. Invasive pulmonary aspergillosis in patients with chronic obstructive pulmonary disease. Eur Respir J. 2007;30:782–800. doi: 10.1183/09031936.00062206. [DOI] [PubMed] [Google Scholar]
- 28.Yasuo M, Fujimoto K, Tanabe T, Yaegashi H, Tsushima K, Takasuna K, Koike T, et al. Relationship between calcium-activated chloride channel 1 and MUC5AC in goblet cell hyperplasia induced by interleukin-13 in human bronchial epithelial cells. Respiration. 2006;73:347–359. doi: 10.1159/000091391. [DOI] [PubMed] [Google Scholar]
- 29.Kim YK, Oh SY, Jeon SG, Park HW, Lee SY, Chun EY, Bang B, et al. Airway exposure levels of lipopolysaccharide determine type 1 versus type 2 experimental asthma. J Immunol. 2007;178:5375–5382. doi: 10.4049/jimmunol.178.8.5375. [DOI] [PubMed] [Google Scholar]
- 30.Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger M. Inflammatory mediators in cystic fibrosis lung disease. Allergy Asthma Proc. 2002;23:19–25. [PubMed] [Google Scholar]
- 32.Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 33.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. 2002;196:1645–1651. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altemeier WA, Matute-Bello G, Gharib SA, Glenny RW, Martin TR, Liles WC. Modulation of lipopolysaccharide-induced gene transcription and promotion of lung injury by mechanical ventilation. J Immunol. 2005;175:3369–3376. doi: 10.4049/jimmunol.175.5.3369. [DOI] [PubMed] [Google Scholar]
- 35.Daubeuf B, Mathison J, Spiller S, Hugues S, Herren S, Ferlin W, Kosco-Vilbois M, et al. TLR4/MD-2 monoclonal antibody therapy affords protection in experimental models of septic shock. J Immunol. 2007;179:6107–6114. doi: 10.4049/jimmunol.179.9.6107. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez D, Keller AC, Faquim-Mauro EL, de Macedo MS, Cunha FQ, Lefort J, Vargaftig BB, et al. Bacterial lipopolysaccharide signaling through Toll-like receptor 4 suppresses asthma-like responses via nitric oxide synthase 2 activity. J Immunol. 2003;171:1001–1008. doi: 10.4049/jimmunol.171.2.1001. [DOI] [PubMed] [Google Scholar]
- 37.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 38.Aaron SD, Angel JB, Lunau M, Wright K, Fex C, Le Saux N, Dales RE. Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163:349–355. doi: 10.1164/ajrccm.163.2.2003122. [DOI] [PubMed] [Google Scholar]
- 39.Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology. 2007;153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 40.Hurst JR, Wedzicha JA. The biology of a chronic obstructive pulmonary disease exacerbation. Clin Chest Med. 2007;28:525–536. v. doi: 10.1016/j.ccm.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Mayer-Hamblett N, Aitken ML, Accurso FJ, Kronmal RA, Konstan MW, Burns JL, Sagel SD, et al. Association between pulmonary function and sputum biomarkers in cystic fibrosis. Am J Respir Crit Care Med. 2007;175:822–828. doi: 10.1164/rccm.200609-1354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagel SD, Sontag MK, Wagener JS, Kapsner RK, Osberg I, Accurso FJ. Induced sputum inflammatory measures correlate with lung function in children with cystic fibrosis. J Pediatr. 2002;141:811–817. doi: 10.1067/mpd.2002.129847. [DOI] [PubMed] [Google Scholar]
- 43.Afessa B, Morales IJ, Scanlon PD, Peters SG. Prognostic factors, clinical course, and hospital outcome of patients with chronic obstructive pulmonary disease admitted to an intensive care unit for acute respiratory failure. Crit Care Med. 2002;30:1610–1615. doi: 10.1097/00003246-200207000-00035. [DOI] [PubMed] [Google Scholar]
- 44.Bulpa PA, Dive AM, Garrino MG, Delos MA, Gonzalez MR, Evrard PA, Glupczynski Y, et al. Chronic obstructive pulmonary disease patients with invasive pulmonary aspergillosis: benefits of intensive care? Intensive Care Med. 2001;27:59–67. doi: 10.1007/s001340000768. [DOI] [PubMed] [Google Scholar]
- 45.Murali PS, Pathial K, Saff RH, Splaingard ML, Atluru D, Kurup VP, Fink JN. Immune responses to Aspergillus fumigatus and Pseudomonas aeruginosa antigens in cystic fibrosis and allergic bronchopulmonary aspergillosis. Chest. 1994;106:513–519. doi: 10.1378/chest.106.2.513. [DOI] [PubMed] [Google Scholar]
- 46.Ritz N, Ammann RA, Casaulta Aebischer C, Schoeni-Affolter F, Schoeni MH. Risk factors for allergic bronchopulmonary aspergillosis and sensitisation to Aspergillus fumigatus in patients with cystic fibrosis. Eur J Pediatr. 2005;164:577–582. doi: 10.1007/s00431-005-1701-4. [DOI] [PubMed] [Google Scholar]
- 47.Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 48.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 49.Akagawa G, Abe S, Yamaguchi H. Mortality of Candida albicans-infected mice is facilitated by superinfection of Escherichia coli or administration of its lipopolysaccharide. J Infect Dis. 1995;171:1539–1544. doi: 10.1093/infdis/171.6.1539. [DOI] [PubMed] [Google Scholar]
- 50.Burd RS, Raymond CS, Dunn DL. Endotoxin promotes synergistic lethality during concurrent Escherichia coli and Candida albicans infection. J Surg Res. 1992;52:537–542. doi: 10.1016/0022-4804(92)90125-j. [DOI] [PubMed] [Google Scholar]
- 51.Henry-Stanley MJ, Hess DJ, Erickson EA, Garni RM, Wells CL. Effect of lipopolysaccharide on virulence of intestinal candida albicans. J Surg Res. 2003;113:42–49. doi: 10.1016/s0022-4804(03)00156-2. [DOI] [PubMed] [Google Scholar]
- 52.Chaudhuri N, Whyte MK, Sabroe I. Reducing the toll of inflammatory lung disease. Chest. 2007;131:1550–1556. doi: 10.1378/chest.06-2869. [DOI] [PubMed] [Google Scholar]
- 53.Greene CM, McElvaney NG. Toll-like receptor expression and function in airway epithelial cells. Arch Immunol Ther Exp (Warsz) 2005;53:418–427. [PubMed] [Google Scholar]
- 54.Raymond T, Schaller M, Hogaboam CM, Lukacs NW, Rochford R, Kunkel SL. Toll-like receptors, Notch ligands, and cytokines drive the chronicity of lung inflammation. Proc Am Thorac Soc. 2007;4:635–641. doi: 10.1513/pats.200706-067TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez MI, Prince A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr Opin Pharmacol. 2007;7:244–251. doi: 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Adamo R, Sokol S, Soong G, Gomez MI, Prince A. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialoGM1 and toll-like receptor 2 as well as toll-like receptor 5. Am J Respir Cell Mol Biol. 2004;30:627–634. doi: 10.1165/rcmb.2003-0260OC. [DOI] [PubMed] [Google Scholar]
- 57.Soong G, Reddy B, Sokol S, Adamo R, Prince A. TLR2 is mobilized into an apical lipid raft receptor complex to signal infection in airway epithelial cells. J Clin Invest. 2004;113:1482–1489. doi: 10.1172/JCI20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doring G, Obernesser HJ, Botzenhart K, Flehmig B, Hoiby N, Hofmann A. Proteases of Pseudomonas aeruginosa in patients with cystic fibrosis. J Infect Dis. 1983;147:744–750. doi: 10.1093/infdis/147.4.744. [DOI] [PubMed] [Google Scholar]
- 59.Kharazmi A. Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol Lett. 1991;30:201–205. doi: 10.1016/0165-2478(91)90026-7. [DOI] [PubMed] [Google Scholar]
- 60.Overhage J, Bains M, Brazas MD, Hancock RE. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol. 2008 doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stepinska M, Trafny EA. Diverse type III secretion phenotypes among Pseudomonas aeruginosa strains upon infection of murine macrophage-like and endothelial cell lines. Microb Pathog. 2007 doi: 10.1016/j.micpath.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 62.Gacesa P. Bacterial alginate biosynthesis--recent progress and future prospects. Microbiology. 1998;144(Pt 5):1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 63.Morici LA, Carterson AJ, Wagner VE, Frisk A, Schurr JR, Honer zu Bentrup K, Hassett DJ, et al. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J Bacteriol. 2007;189:7752–7764. doi: 10.1128/JB.01797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kheradmand F, Kiss A, Xu J, Lee SH, Kolattukudy PE, Corry DB. A protease-activated pathway underlying Th cell type 2 activation and allergic lung disease. J Immunol. 2002;169:5904–5911. doi: 10.4049/jimmunol.169.10.5904. [DOI] [PubMed] [Google Scholar]
- 65.Kiss A, Montes M, Susarla S, Jaensson EA, Drouin SM, Wetsel RA, Yao Z, et al. A new mechanism regulating the initiation of allergic airway inflammation. J Allergy Clin Immunol. 2007;120:334–342. doi: 10.1016/j.jaci.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 66.Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, Sudo K, et al. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007;8:39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- 67.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gersuk GM, Underhill DM, Zhu L, Marr KA. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J Immunol. 2006;176:3717–3724. doi: 10.4049/jimmunol.176.6.3717. [DOI] [PubMed] [Google Scholar]
- 69.Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, Pamer EG. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 2005;1:e30. doi: 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rivera A, Van Epps HL, Hohl TM, Rizzuto G, Pamer EG. Distinct CD4+-T-cell responses to live and heat-inactivated Aspergillus fumigatus conidia. Infect Immun. 2005;73:7170–7179. doi: 10.1128/IAI.73.11.7170-7179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 72.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 73.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poynter ME, Irvin CG, Janssen-Heininger YM. A prominent role for airway epithelial NF-kappa B activation in lipopolysaccharide-induced airway inflammation. J Immunol. 2003;170:6257–6265. doi: 10.4049/jimmunol.170.12.6257. [DOI] [PubMed] [Google Scholar]
- 75.Dialynas DP, Quan ZS, Wall KA, Pierres A, Quintans J, Loken MR, Pierres M, et al. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5: similarity of L3T4 to the human Leu-3/T4 molecule. J Immunol. 1983;131:2445–2451. [PubMed] [Google Scholar]
- 76.Jones B. Evidence that the Thy-1 molecule is the target for T cell mitogenic antibody against brain-associated antigens. Eur J Immunol. 1983;13:678–684. doi: 10.1002/eji.1830130813. [DOI] [PubMed] [Google Scholar]
- 77.Cohn L, Homer RJ, MacLeod H, Mohrs M, Brombacher F, Bottomly K. Th2-induced airway mucus production is dependent on IL-4Ralpha, but not on eosinophils. J Immunol. 1999;162:6178–6183. [PubMed] [Google Scholar]
- 78.Tomioka S, Bates JH, Irvin CG. Airway and tissue mechanics in a murine model of asthma: alveolar capsule vs. forced oscillations. J Appl Physiol. 2002;93:263–270. doi: 10.1152/japplphysiol.01129.2001. [DOI] [PubMed] [Google Scholar]
- 79.Hantos Z, Daroczy B, Suki B, Nagy S, Fredberg JJ. Input impedance and peripheral inhomogeneity of dog lungs. J Appl Physiol. 1992;72:168–178. doi: 10.1152/jappl.1992.72.1.168. [DOI] [PubMed] [Google Scholar]