Abstract
Time-resolved Stokes-shift experiments measure the dynamics of biomolecules and of the perturbed solvent near them on subnanosecond time scales, but molecular dynamics simulations are needed to provide a clear interpretation of the results. Here we show that simulations using standard methods quantitatively reproduce the main features of TRSS experiments in DNA and provide a molecular assignment for the dynamics. The simulations reproduce the magnitude and unusual power-law dynamics of the Stokes shift seen in recent experiments [D. Andreatta, et al., J. Am. Chem. Soc. 127, 7270 (2005)]. A polarization model is introduced to eliminate cross correlations between the different components contributing to the signal. Using this model, well-defined contributions of the DNA, water and counterion to the experimental signal are extracted. Water is found to have the largest contribution and to be responsible for the power-law dynamics. The counterions have a smaller, but non-negligible contribution with a time constant of 220 ps. The contribution to the signal of the DNA itself is minor and fits a 30 ps stretched exponential. Both time-averaged and dynamic distributions are calculated. They show a small subset of ions with a different coupling, but no other evidence of substates or dynamic heterogeneity.
I. INTRODUCTION
The fact that water is essential to biology is almost axiomatic. However, studying the water near biomolecules is more difficult than studying the biomolecules themselves.1 Time-resolved Stokes shift (TRSS) and related experiments are one of the few methods available with the potential to measure the dynamics of water near proteins or DNA on subnanosecond timescales. Current TRSS results appear to indicate strongly perturbed water dynamics in these systems.2–4 However, TRSS actually measures the dynamics of the electric field acting on a chromophore embedded in the biomolecule. Building a molecular picture from electric-field dynamics alone is difficult, and uncertainty remains as to whether the unusual TRSS dynamics should be attribute to perturbed water, to motion of the biomolecule itself, to counterion movement or to some inextricably coupled combination of these motions.
Computer simulations appear to provide a clear route to building a molecular picture of dynamics in biomolecules. However, although biomolecular simulation methods have been extensively tested and refined by comparison to structural data, rigorous comparisons to experimental data on dynamics are uncommon. Questions remain about the ability of current simulation methods to quantitatively describe dynamics, especially at long times. In addition, the molecular interpretation of the simulation data is complicated by strong coupling between the components of the systems and the resulting strong cross-correlations in their dynamics.5–8
This paper tackles these interrelated problems through a detailed comparison and analysis of TRSS data and simulation in oligonucleotides. In particular, we attempt to understand the unexpected TRSS result reported by some of us in DNA.9,10 In experiments covering six decades in time, from 40 fs to 40 ns, we not only saw the subpicosecond dynamics expected from “normal” water, but we also saw relaxation extending smoothly out to the end of the experimental time range. This relaxation did not show distinctive time scales that could be easily attributed to multiple, independent processes. Rather, it could be fit over the entire time range by a single power law. In this paper, we will first show that this behavior is accurately replicated in simulations and will then use the simulation data to disentangle the contributions from water, counterions and DNA.
The TRRS experiment relies on a chromophore in which electronic excitation creates an instantaneous change in the charge distribution. In favorable chromophores, the change in charge distribution can be modeled as the creation of a dipole at a point within the chromophore. The electric field generated by this new dipole extends into the environment surrounding the chromophore and exerts a force on every nearby charged site. The environment relaxes in response to these forces in a manner determined by the intrinsic dynamics of the material. As the charges in the environment reorganize, they create a reaction electric field at the position of the original dipole. This field stabilizes the excited state of the chromophore relative to the ground state, and as a result, the chromophore’s fluorescence is shifted to lower energy. The TRSS experiment literally consists of measuring the mean fluorescence frequency as a function of time after excitation, but the results can be interpreted as the electric-field dynamics of the material surrounding the probe. In many ways, the TRSS experiment can be thought of as a localized version of a dielectric-relaxation experiment. We have recently reviewed experimental TRSS results in DNA.4 That review cited some of the conclusions from this paper.
Much of our current knowledge of the interaction between DNA and water comes from simulation. The time-averaged distribution of water and counterions around DNA has been extensively investigated.11,12 The dynamics of water near DNA have also been characterized in terms of theoretical quantities such as diffusion coefficients, orientational correlation functions and hydrogen-bond lifetimes.11,13 However, to the best of our knowledge, the only direct simulations of TRSS experiments in DNA so far have been those of Pal et al.7 and of Furse and Corcelli.8
Pal et al. found multiple time scales in general agreement with experiment, but they did not have simulations long enough to compare to the longest times seen in experiment. In assigning the response to different components of the system, they pointed out the existence of strong cross correlations that complicate the interpretation. Nevertheless, they made qualitative arguments that water was the dominant contributor, even for the long time response, with a significant additional contribution from counterions. The DNA contribution was found to be minor.
Furse and Corcelli simulated a probe bound in the groove of DNA, rather than one replacing a base.8 They dealt with cross correlations with a “linear-response” approach that was originally proposed by Nilsson and Halle in the context of TRSS experiments in proteins.5. They came to the opposite conclusion of Pal et al.: The water plays a negligible role in the slow response and the DNA response is most important at long times. The conflicting interpretations of Pal et al. and Furse and Corcelli will be discussed in more depth later, after our own analysis has been presented.
Although proteins and DNA are different in many ways, the issues involved in interpreting both experiments and simulations are similar. Slow TRSS responses have been repeatedly reported in proteins.14–25 Simulations have alternatively assigned the slow dynamics to water,6,26 to the protein,5,6 or to coupled water-protein motions.27 Much of this disagreement arises from differences in interpretation, specifically, how to divide the total TRSS dynamics among the components of the system. This question will be addressed here in the context of DNA, but the results may have implications for interpreting TRSS results in proteins as well.
After briefly describing the methodology in Sec. II, the paper starts with a detailed comparison of TRSS experiments and simulation in Sec. III. Rather than comparing fitting parameters, we make a direct and quantitative comparison of the raw results. Many common assumptions are either avoided or made more explicit. TRSS experiments in DNA are complicated by the fact that the decay functions are strongly nonexponential and by the fact that either simulation or experiment may not be long enough to capture all the relaxation. Special attention is paid to the effect of different assumptions about the long-time relaxation. Quantitative agreement is found between experiment and simulation (with some degradation at the longest times), including the existence of broadly distributed, power-law-like relaxation extending out to the nanosecond time scale.
Section IV looks at the effective distance away from the probe that is sampled in the TRSS experiment. These results show that the simulation box is sufficiently large. Electric-field fluctuations from more distant material are small, and the method used to treat them is not important.
Section V tackles the issue of how to assign the TRSS response to water, counterions or DNA and in particular how to deal with the strong cross correlations between these components. We propose a “polarization model.” This method is motivated by the idea that the coupling between components is due to a linear polarization of one component by another, similar to the coupling between charges and dielectrics in macroscopic electrostatics. A simple linear transformation of coordinates will then cause the cross-correlations to go to zero at all times. This method works extremely well. Using this method, water is found to be the dominant contributor to the TRSS at all times. Its relaxation spans the entire measured time range and is responsible for the overall power-law behavior seen in experiment. Counterions have a secondary, but non-negligible contribution with a well-defined relaxation time of about 200 ps. The DNA relaxation time is near 30 ps, but the amplitude of its contribution is very small.
Whenever dynamics are non-exponential, it is always important to ask if the total sample contains multiple long-lived configurations with different relaxation times. The existence of such configurations has been invoked to explain charge-transfer dynamics in DNA.28–32 Section VI looks for evidence of such DNA substates, but finds none.
The reader will soon discover that this paper does not attempt to make a meticulous duplication of the experimental system in the simulation. In fact, the simulation is one that was produced independently from the experiments and that has been extensively characterized in previous publications.33–35 The premise is that the important features of the TRSS results are broadly characteristic of DNA and not highly dependent on system-specific details. Specifically, the simulation and experiment are on oligomers of different length and different sequence; the simulation is on native DNA, whereas the experimental DNA is modified by the incorporation of a probe; the probe electronic structure is not modeled in detail, but is represented by a simple point dipole. Fortunately, the agreement between simulation and experiment supports the assumption that these differences are not important. The fact that these issues play a minor role is also a significant conclusion of the paper.
II. METHODS
TRSS Experiment
The TRSS data set examined here is the most extensive one available on DNA, covering the time range from 40 fs to 40 ns.9,10 The TRSS experiments were performed on a 17 base-pair oligonucleotide (5′-GCATGCGC(cou)CGCGTACG-3′) and its complement. The central base-pair is replaced by a coumarin-102 group (cou)36 on one strand and an abasic site on the opposite strand. Results have been combined from three different techniques: time-correlated single-photon counting, fluorescence up-conversion, and transient absorption. Details of the data collection and analysis are described in previous publications.9,10 The reproducibility and error limits of the TRSS method are discussed in Ref. 37. Further details on the TRSS experiment can be found in the literature.38–41
Molecular Dynamics Simulation
The simulation trajectory analyzed here is one that has been examined previously33–35 and that has recently been extended to 60 ns.42 The simulation methods are standard and are described in detail in the previous papers. For the current analysis, the first 14 ns are regarded as an equilibration time, and only the last 46 ns are used. The simulation configuration was saved every picosecond, which sets the limit on the fastest process observable in the simulation. The simulation is on the Dickerson dodecamer 5′-CGCGAATTCGCG-3′.
Assumptions are needed on how to place the test dipole within the simulated system. We placed r0 at the center-of-mass of one of the two central adenines. The axis system for measuring the electric-field direction was linked to the adenine and followed its motion: the x-axis is along the long axis of the base, the y-axis runs along the short axis and the z-axis is perpendicular to the plane of the base (Figure 1, see Supporting Information). The details of these assumptions prove to be unimportant for the final results. We emphasize that this dipole is a hypothetical object for measuring the electric field and was not present during the generation of the trajectory.
Figure 1.
Left: Adenine used in the simulation showing the axis system with the origin at the position of the test point r0. Right: Coumarin used in the experiment showing the position and direction of its change in dipole between the ground and excited states δμ.
The electric field was calculated by summing over the contributions from all the atoms within the primary simulation cell using the charges from the simulation force field. The boundaries of the cell were drawn so as not to split any water molecules. (Splitting the molecules at the boundary creates free charges and extraneous fields. In principle these fields average to zero, but in practice they add a great deal of noise to the results.) The atomic charges on the adenine containing r0 were set to zero to avoid measuring the intramolecular vibrations of the adenine itself. This approach is consistent with the experiments, in which the vibronic structure of the probe is not included in the Stokes shift. Results from each of the two central adenines were compared and are identical to within the noise level (see Supporting Information). Averages of the two are shown here.
Because the results span many decades in time, they are presented on a logarithmic time scale. The correlation functions have been averaged over bins that are uniform on this scale. A bin extends from t to 1.01t.
Linking Experiment to Simulation
The TRSS experiment yields the fluorescence frequency ω(t) as a function of time after excitation. This frequency is equivalent to the difference between ground- and excited-state energies. We model it by ω(t) = ω0 − E(r0, t)·δμ/η, where δμ is the change in dipole moment between the ground and excited states and E(r0, t) is the electric field at r0.
Other methods have been used to model TRSS experiment in biomolecules. Nilsson and Halle,5 Golosov and Karplus6 and Furse and Corcelli8 used a sum of changes in atomic charges times the electric potential at each atom of the probe chromophore. In principle, this procedure captures quadrupolar and higher-order components of the change-in-charge distribution and incorporates the gradient and higher spatial derivatives of the electric field. However, the change-in-charge distribution for coumarin is known to be close to dipolar.43 Our simpler description of the TRSS purely in terms of the electric-field dynamics at a single point in space is further justified by the success of the simulation.
In their simulations of TRSS in DNA, Pal et al. substituted the total ground-state interaction energy of the probe for the transition energy.13 These quantities are similar, but also have significant differences. The ground-state energy includes short-ranged Lennard-Jones interactions that contribute little to the transition energy. In coumarin, the ground-state charge distribution is significantly different from the ground-excited state charge difference.43 In particular, the ground-state charge distribution is not purely dipolar, so the dynamics of electric field gradients are incorporated into their simulations. Although the qualitative results will be similar, these differences should be kept in mind when making detailed comparisons between our results and theirs.
The TRSS experiment is a non-equilibrium measurement. The system is driven out of equilibrium by an extrinsic perturbation—the coumarin dipole moment—and the return to equilibrium is observed. The results can be characterized by two quantities: a normalized response function
| (1) |
and a magnitude
| (2) |
Both components are expressed in terms of the component of the electric field along δμ, Eδμ = E·δμ.
In contrast, the simulation is an equilibrium measurement. The system stays near equilibrium, and the dynamics of the thermal fluctuations are observed. These fluctuations are characterized by the normalized electric-field correlation function
| (3) |
and the magnitude of the electric-field fluctuations
| (4) |
Both quantities are defined in terms of the deviation of the electric field from its mean
| (5) |
The average has been labeled to emphasize that it must be taken over an infinitely long trajectory.
The comparison of the TRSS experiments to the simulations is based on linear-response theory.44 So long as the perturbation to the system is small, the experimental non-equilibrium response function is equal to the simulated equilibrium correlation-function, R(t) = C(t). In addition, the Stokes-shift magnitude is related to the mean-squared magnitude of the electric-field fluctuations
| (6) |
III. COMPARISON OF SIMULATION TO EXPERIMENT
Comparison of Magnitudes and the Lack of Asymmetry
An important comparison is between the simulated and experimental values for the total Stokes shift ΔS. This calculation requires knowledge of the change in dipole moment between the ground and excited states δμ. Unfortunately, values for this quantity from different experimental methods and from different calculations vary by more than a factor of two.45,46 We use the value | δμ | = 3.8 D from microwave absorption of coumarin 102 in dioxane.47 This value is very similar to the one from solvatochromism of coumarin 10248 and lies in the middle of the range of other reported values.46,49–54 The direction of the dipole-moment change is close to the long axis of the coumarin ring system (Figure 1).46,55
In principle, the DNA helix is a very anisotropic environment. As might be expected, Table 1 shows that there is a static electric field at the probe point, which is oriented midway between the y- and z-axes. However, the root-mean-squared (rms) fluctuations in this field are nearly as large as its average magnitude. The size of the fluctuation is only slightly anisotropic; it varies only ±10% along the different coordinate axes. The dynamics of these fluctuations are even less anisotropic. Correlation functions for the field along different axes are nearly indistinguishable (see Supporting Information). Thus, in terms of the factors that affect the TRSS experiment, the interior of DNA is nearly isotropic. As a result, selecting the correct orientation of the probe dipole in the simulation is not terribly important. The rest of the paper will focus on the x-component of the electric field, unless otherwise specified.
Table 1.
Averages and root-mean-squared fluctuations in the components of the electric field at the center of mass of the central adenine from simulation and the Stokes shift of coumarin 102 calculated from these values.
The simulated value of ΔS is calculated from the root-mean-squared (rms) magnitude of the fluctuations in the field using eqs 5 and is compared to the experimental value in Table 1. The Stokes shift calculated from the x-component of the electric field is within 25% of the experimental value. This agreement is quite good considering the problems involved: the uncertainty in the magnitude of δμ discussed above, the uncertainties in extrapolating to infinite time in both experiment and simulation and the different treatment of vibronic effects, as discussed below. Simulation and experiment are in agreement on the magnitude of the Stokes shifts to the extent that these uncertainties allow.
Comparison of Dynamics
There are three factors that complicate the comparison of the experimental time-resolved Stokes shifts to the simulated electric-field correlation function. The first factor is the value of the equilibrium Stokes shift S(∞). In many systems, relaxation is complete within the fluorescence lifetime, and S(∞) is equal to the steady-state Stokes shift. Alternatively, when the Stokes shift reaches a constant value within the measurement time window, the value of S(∞) can be fit to the long time portion of the TRSS data. Unfortunately, neither of these conditions holds in DNA, as shown in Figure 2. The TRSS is still increasing at the longest measurable time.
Figure 2.
Stokes shifts S(t) of coumarin in DNA from simulation (red line) and from experiment (black symbols). Circles: time-correlated single photon counting; squares: fluorescence up-conversion; triangles: transient absorption. Parameters used to interpret the simulation (δS0 = −875cm−1, b = 2600 cm−1) have been adjusted to optimize the fit before 5 ns. The dashed, horizontal lines show S(∞) corresponding to the correlation functions shown in Figure 3: highest/red, S(∞) = 2045 cm−1; middle/blue, S(∞) = 1888 cm−1; lowest/green, S(∞) = 1725 cm−1.
The second complicating factor is the value of the initial Stokes shift S(0). In principle, the Stokes shift could be defined as the shift of the emission from the excitation wavelength. However with this definition, the Stokes shift would capture a substantial amount of vibrational dynamics of the probe and its local environment. Here, the TRSS results are reported as Stokes shifts relative to the steady-state fluorescence frequency of the same material in a frozen glass, S(t) = ωglass − ω(t), where ω(t) and ωglass are the mean fluorescence frequencies in the liquid and glass respectively. Both intramolecular and intermolecular vibrational dynamic still occur rapidly in the glass, so their effect is removed from the Stokes shift. Diffusive motions, which are relatively slow in the liquid (slower than ~40 fs), freeze out in the glass. Only their effects remain in the experimental Stokes shifts. Thus the experimental results, which are taken after 40 fs, should extrapolate to S(0) ≈ 0.
Although it is possible to remove vibrational and inertial motions from the experimental measurements, it is not as easy to do the same in simulation. In the simulation, vibrations of the covalent bonds to hydrogen are fixed, but other vibrational motions of the probe and DNA are not. In addition, fast, librational motions of the water are included in the simulation, but are largely eliminated from the experiments. Thus, the apparent value of S(0) is not the same in the simulation and the experiment, because of their differing treatments of vibrational motions. The difference in these two values δS0 must be fit when comparing the simulations and experiments.
The third problem arises from the need for a good value of the mean electric field in order to calculate the electric-field correlation function from the simulation (eq. 3–eq. 5). The standard approach is to measure the average over the simulation period T and assume that value is the same as an average over infinite time, <E>T = <E>∞. This assumption is perfectly valid if the simulation period is much longer than the longest relaxation time. However, if there is a possibility of relaxations as long as or longer than T, we must consider the possibility that <E>T ≠ <E>∞. If we do not consider the possibility that there is an error in measuring the mean, the correlation function is artificially forced to zero at the end of the simulation.
A convenient way to parameterize the error in the measurement of the mean field is through its ratio to the mean size of the fluctuations in the field
| (7) |
The true correlation function C∞ is then relate to the correlation function calculated with the mean over a finite time CT by
| (8) |
Combining these three unknown parameters: the difference in vibrational contributions δS0, the equilibrium Stokes shift S(∞) and the error in measuring the mean r; the experimental Stokes shift is related to the finite-time correlation function by
| (9) |
| (10) |
Only two parameters need to be adjusted: the effects of changing S(∞) or r are interchangeable.
Figure 2 shows a comparison between the experimental TRSS data and the simulation results after optimizing these parameters. The agreement is extremely good, especially before 5 ns. TRSS data are often compared on a more qualitative basis, for example, by comparing the number or size of time constants from fits to each data set. We believe that the direct comparison of Stokes shifts is the most rigorous method of comparing experiment and simulation. The good agreement see in Figure 2 comes despite the numerous detailed differences between the real and model systems. We conclude that the important features of the TRSS experiment are determined by general features of the DNA structure. We also note here that the agreement after 5 ns is not as good as before 5 ns, but we postpone discussion of this feature until later in the paper.
Comparison of Correlation Functions and the Existence of Very Slow Relaxations
Although the Stokes shifts are a rigorous basis for comparing simulation and experiment, the infinite-time correlation function is more useful in interpreting the results. To convert the Stokes shifts to C∞ requires the specification of one additional parameter: r and S(∞) can no longer be combined into a single constant.
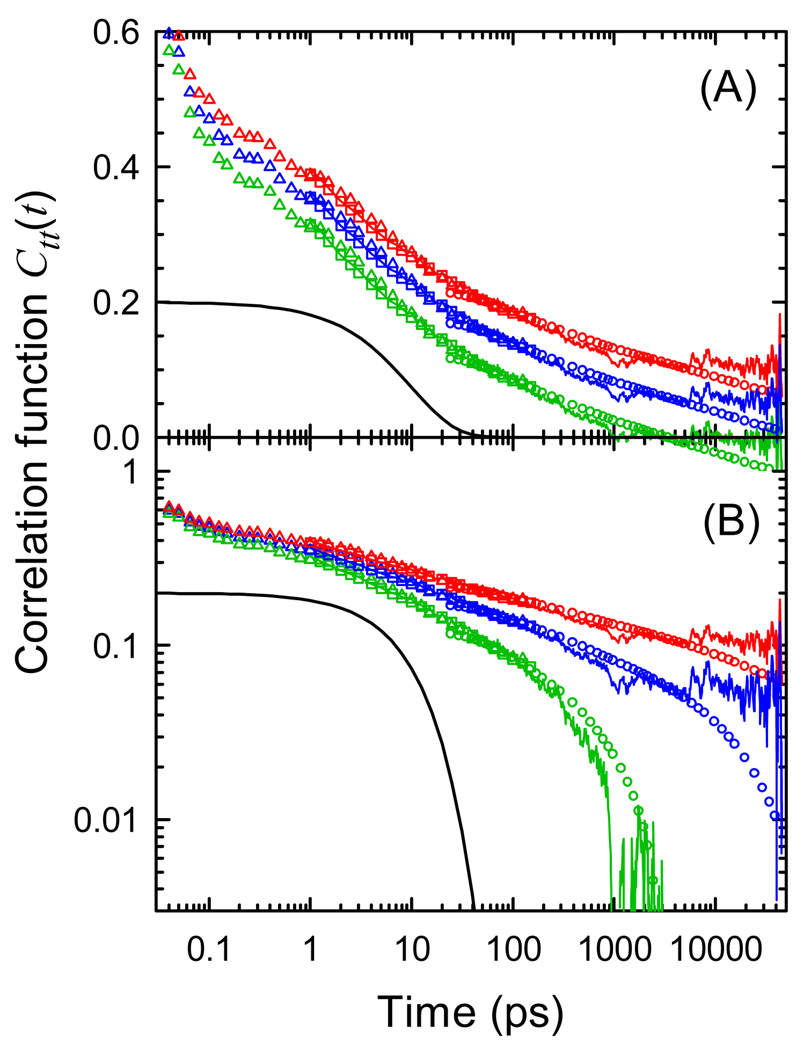
Figure 3 shows the correlation functions corresponding to the Stokes shifts in Figure 2 on both semi-log and log-log plots. Three values of r are shown. The values of S(∞) implied by these values of r are shown as dashed lines in Figure 2. In all cases, the relaxation is strongly nonexponential. Whereas most of an exponential relaxation occurs over one decade in time, these correlation functions extend over at least five decades and possibly more. The early portion of the relaxation is well modeled as a power law, that is, as a straight line in Figure 3B, in all cases shown.
Figure 3.
Examples of correlation functions consistent with the Stokes shifts in Figure 2 on (A) a semi-log plot and (B) a log-log plot. Symbols: Experimental data (see Figure 2). Noisy lines: simulation. Highest/red: r = 0.35, S(∞) = 2045 cm−1. Middle/blue: r = 0.12, S(∞) = 1760 cm−1, Lowest/green: r = 0, S(∞) = 1725 cm−1. The smooth black curve is a single exponential decay shown for contrast with the broadly distributed correlation decays.
In the first case (lowest, green), it is assumed that there is no long time relaxation and that the mean field obtained from the simulation has no error (r = 0). This choice forces the simulated correlation function to go to zero at the end of the measurement window. However, the resulting value of S(∞) is lower than the long-time experimental points (see Figure 2). As a result, the experimental correlation function dips to negative values at long times (Figure 3A).
The second case (highest, red) is motivated by the desire to maintain power-law behavior at long times, as well as at short times. We must assume a significant error in the mean field measured over the simulation time (r = 0.35), but this assumption is consistent with the fact that the correlation function extends well past the measurement time window.
The third case (middle, blue) is intermediate between the first two. It places S(∞) near the largest measured experimental value. This value is suggested by the correlation functions assigned to water later in the paper.
The choice between these alternative interpretations is subjective. We could argue that the simplest result is not to infer a change in the time dependence if the data do not demand it. In that case, the power-law is best. Alternatively, we could argue that the simplest result is not to infer the existence of long relaxations of unknown origin. In that case, the result that converges to zero most quickly is best. Lastly, we could insist that the simplest result is the one that does not infer anything beyond what is experimentally measured. In that case, the result that puts S(∞) closest to the largest measured Stokes shift is best.
Ultimately, none of these arguments is definitive. Until more information is available, all these possibilities should be recognized as plausible interpretations. If the results are reduced with a fitting function: multiexponential, stretched exponential, power-law, etc.; one of these interpretations will be chosen arbitrarily and the others discarded.
Determining the longest relaxation time in DNA is complicated by these issues as well as by the increasing difference between experiment and simulation at long times. The experimental results favor an increasing Stokes shift at long times; the simulation results favor a leveling of the Stokes shift at long times. The random error in the experimental measurements has been estimated from replicated measurements to be less than ±30 cm−1,37 and the random error in the simulation can be judged by the noise in the results. By these criteria, the differences between the simulation and experiment are real. On the other hand, the differences in Stokes shift in experiment and simulation are quite small in absolute size. The difference could easily arise from small systematic error in either measurement.
It should be noted that even under the most conservative interpretation, relaxation persists out to at least 5 ns. For comparison, many TRSS experiments in proteins report that relaxation is complete within a few hundred picoseconds,17–19,23,25 although there are also reports of relaxation extending beyond 1 ns.15,16,20–22,24
IV. EFFECTIVE RANGE OF INTERACTION
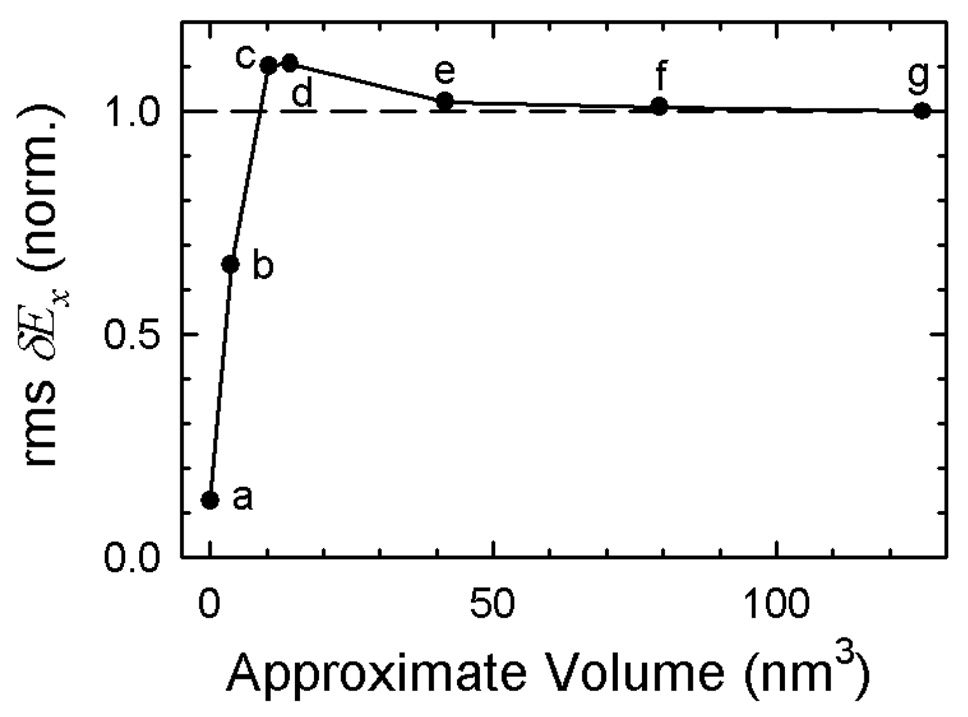
To explore the effective range of the TRSS experiment in DNA, the size of the electric-field fluctuation was calculated as a function of the amount of material around the probe point included in the electric-field calculation. The results are shown in Figure 4 organized by the approximate volume of the material included. The exact description of the material included at each point is given in the Supporting Information.
Figure 4.
The root-mean-squared fluctuation of the x-component of the electric field inside DNA as a function of the volume around the probe point that is included in the calculation of the field. Descriptions of each volume are in the text and in the Supporting Information.
Point (g) is the full calculation including all the material within the rectangular primary simulation cell; point (f) is the central cubic region of the simulation cell; point (e) is a sphere just large enough to contain the entire DNA chain. The size of the fluctuations does not change as the outer regions of the simulation cell are excluded. These results show that the simulation box is large enough for these calculations and justifies the neglect of the reaction field from material outside the primary simulation cell for calculating the electric-field fluctuations at r0.
This result does not contradict the fact that long range electrostatic interactions are important for the mean dynamics of the system. In Figure 4, the same underlying trajectory is always used, and that trajectory is calculated with Ewald sums to account for long range electrostatics. The difference is only in the final calculation of the electric-field fluctuations. Fluctuations are reduced rapidly as more material is included, and as a result, their range is expected to be shorter than the range of the mean electrostatic interaction.
Point (a) measures only the fluctuations generated by the thymine paired to the probe adenine. The contribution of the opposing base to the electric-field fluctuations is quite small.
Point (d) represents a good estimate of the effective range for electric-field fluctuations in DNA. It includes the fluctuations from the material within a 15 Å sphere centered on the probe point. This sphere accommodates the nearest-neighbor base-pairs flanking the central AT pair as well as the linking sugars and phosphates and nearby water and counterions. The fluctuations from this material are slightly larger than those from the full cell. Presumably the material beyond this sphere is polarized by the field from the material within the sphere. This polarization creates a reaction field that opposes and damps the fluctuations to a small extent.
Point (c) is the same as (d), except that the atoms of the DNA itself have been excluded, leaving only the water and counterions. Point (b) is the inverse; the water and counterions of (d) are excluded, leaving only a portion of the DNA proper. The sum of (b) and (c) is larger than (d), indicating that there are strong anti-correlations between the field from the water and the field from the DNA. This result is an example of the problem of assigning the fluctuations to individual components of the system. This problem is discussed in more detail in the next section.
V. DECOMPOSITION OF THE TOTAL ELECTRIC-FIELD INTO COMPONENTS
Bare Components Show Strong Cross-Correlations
It is natural to ask which molecular species within the entire DNA system are primarily responsible for the TRSS response. We divide the entire system into three components: the DNA itself (d), the surrounding water (w) and the sodium counterions (i). The total (t) electric field is the sum of the electric field from each of these components
| (11) |
Autocorrelation functions for these components are defined as
| (12) |
and cross correlations are defined as
| (13) |
With this normalization, the total autocorrelation is always one at time zero, Ctt(0) = 1. A value less than one at 1 ps indicates the loss of correlation due to subpicosecond processes. The component correlation functions are normalized so that they sum to the total:
| (14) |
For the moment, we will not consider corrections to the mean of the field (i.e., r = 0).
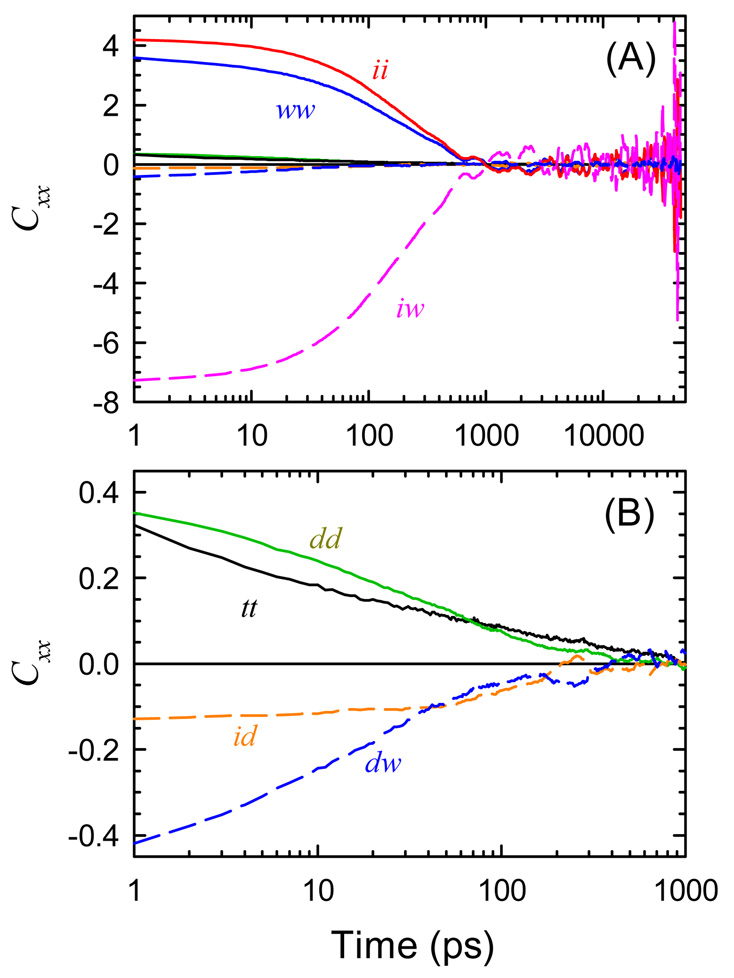
All six component correlation functions and the total correlation function are shown in Figure 5. The cross correlations between the three components are negative and large. The largest cross correlation is 20 times larger than the total correlation function. Pal, et al. reported qualitatively similar results in their analysis of TRSS in DNA.7 In the presence of such strong cross correlations, it is meaningless to discuss the dynamics of one component separately from those of another.
Figure 5.
Auto- (solid curves) and cross-correlations (dashed curves) of the uncorrected components of the electric field: DNA (d), water (w) and ions (i). (B) An expansion in both time and amplitude of the data in (A). The cross correlations are large compared to the autocorrelations.
Polarization Model Eliminates Cross-Correlations
A possible origin of the strong cross-correlations in this system is suggested by classical electrostatics. The field from one component polarizes the other components. As examples, the ions can polarize the water molecules by aligning them, or the ions can polarize the DNA by forcing it to bend or twist. The bare electric field created by any component is partially cancelled by the reaction field from the material that it polarizes, leading to strong, negative cross-correlations.
Motivated by this idea, a simple model is proposed. The total field from each component (Ew, Ed, Ei) is assumed to consist of an intrinsic contribution (EW, ED, EI) and additional terms due to that component’s polarization by the other components. These latter contributions are represented by an effective linear susceptibility χnm representing the portion of the field En induced by field Em. Thus,
| (15) |
Solving for the intrinsic fields in terms of the total fields yields
| (16) |
Equation 16 defines a simple linear transformation of the coordinates used to describe the system. With the normalization of the diagonal elements given, the total field is still the sum of these new components
| (17) |
and the total correlation function is still the sum of the component auto- and cross-correlations
| (18) |
By adjusting the values of the three susceptibilities, it is always possible to force the three cross correlations to zero at a single time. Figure 6 shows the correlation functions where this choice has been made at 5 ps. The significant result is that the cross correlations are zero not only at this time, but are nearly zero across the entire measurement window. Despite its simplicity, the polarization model accurately describes the total electric field in terms of three nearly independent components,
| (19) |
In the absence of cross-correlations, it becomes meaningful to discuss the water, ion, and DNA dynamics as distinct and independent contributions to the total TRSS response.
Figure 6.
Autocorrelations (A) and cross-correlations (B) of the intrinsic components of the electric field derived from the polarization model: total (t), DNA (D), water (W) and ions (I). The vertical scale in (B) is expanded 10 times relative to (A). Using coordinates from the polarization model, the cross-correlations become negligible. There is no adjustment of the mean (r = 0) for any component.
The polarization model in eq 15 is not the most general linear transformation possible: the polarization of the ion cloud by the water (χiw) or DNA (χid) and the polarization of the DNA by the water field (χdw) are not included. However, giving these terms nonzero values causes the model to fail. Thus, the absence of these effects is a result drawn from the simulation.
The reason that only three polarization terms are needed can be rationalized on the basis of the time scales of the responses. The fast coordinates follow the slow coordinates closely, but the slow coordinates do not respond effectively to fluctuations of the fast coordinates. Thus, the slow ions polarize the fast water, but the ions do not respond to the rapidly changing field from the water. The time scale separation between the components is not perfect, but it is quite good. The tail of CWW extends past the end of CII, but this tail is only a small fraction of the total water response. In principle, there may be some response of the ions to the slowest fluctuations of the water, but this effect is too small to resolve.
Manning has recently predicted that fluctuations in the counterion distribution drive bending fluctuations in DNA.56 This prediction is consistent with our finding of an ion-DNA coupling χdi.
Modeling the Component Relaxations
Having decomposed the simulation results into three components, we return to the issue of a possible error in the mean electric field. However, now we can consider a correction to the mean of each component separately, instead a correction to the total field.
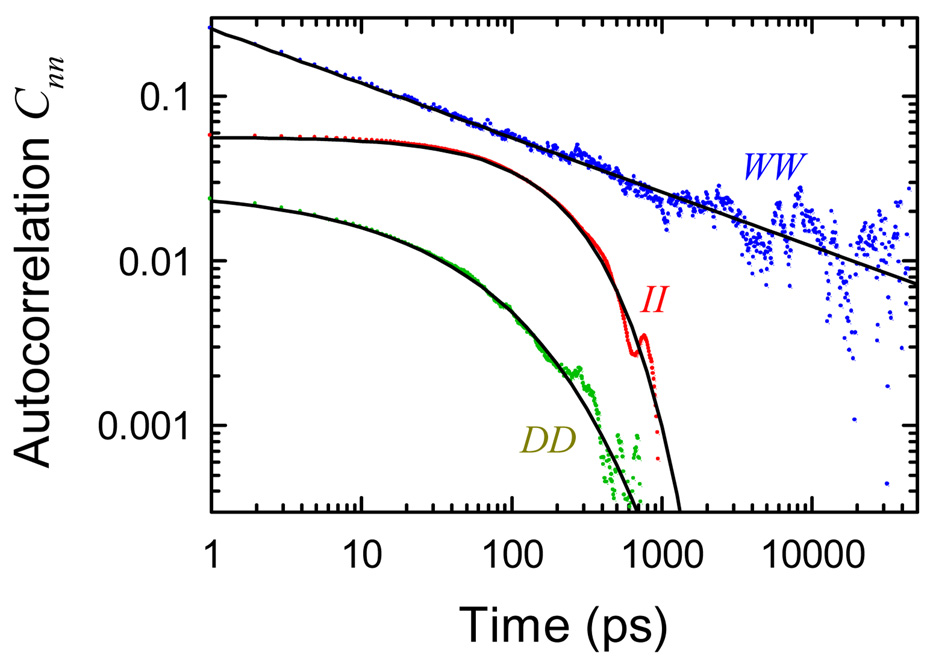
For the ion and DNA components, this issue is easily resolved. Both CII and CDD decay to zero well before the end of the measurement window. Moreover, they do so rapidly (Figure 7). Shifting the value of the mean only adds a flat pedestal to the autocorrelation at long times. There is no reason to add a correction to these components.
Figure 7.
Autocorrelations of the intrinsic components of the electric field (dots): DNA (D), water (W) and ions (I); and fits (solid lines). See eq 20–eq 22. The mean in CWW has been adjusted (r = 0.12) to give the best power law for the water.
In contrast, CWW decays slowly throughout the early part of the measurement and nearly fits a power-law, except at long times. Adding a small correction to the mean (r = 0.12) makes CWW a power-law through out the measured time range (Figure 7).
After this correction, all three autocorrelation function can be fit with simple forms:
| (20) |
| (21) |
| (22) |
These fits are shown in Figure 7. The DNA autocorrelation function is a strongly stretched exponential. The ion autocorrelation is almost exponential, but including a slight stretching definitely improves the fit. Both these fits over times greater than 1 ps extrapolate to values very close to the correct ones at zero time, which are not visible in the figure: CII(0) = 0.058 and CDD(0) = 0.043. Thus, very little of the subpicosecond dynamics are due to either the ions or the DNA. All of the subpicosecond dynamics can be attributed to the water. This reasoning augments the conclusion that the water is primarily responsible for the highly dispersed nature of the dynamics.
VI. DISTRIBUTIONS AND HETEROGENEITY
Static Distributions and the Possibility of Substates
An important issue is whether the system can be divided into substates, i.e., two or more distinct and relatively long lived configurations.57 Examples of possible substates include B vs B′ forms of the DNA58,59 or helices with and without a groove-bound ion.35 Zewail and Barton have seen two different electron-transfer lifetimes in DNA, which they attribute to distinct, long-lived base-stacking conformations.28–30 Several theoretical models of charge-transfer in DNA invoke “gating,” which invokes a special conformation to promote charge transfer.31,32 Because TRSS and charge transfer are both strongly dependent on electric-field dynamics, it might be hoped that these simulations would show clearer evidence for these substates. Evidence of substates with a 500 ps lifetime has been found in this sequence using rms deviations of the DNA itself from its average structure.34 However, it is not clear if these structural conformations couple to charge transfer or to TRSS experiments, or if other substates could be defined by looking at the ion or water structure.
A place to begin looking for evidence of such substates is to examine the static probability distributions for the electric field
| (23) |
If the electric field is due to the sum of a large number of independent contributions, this distribution will be a Gaussian; if it is dominated by a small number of substates, a non-Gaussian distribution will result.
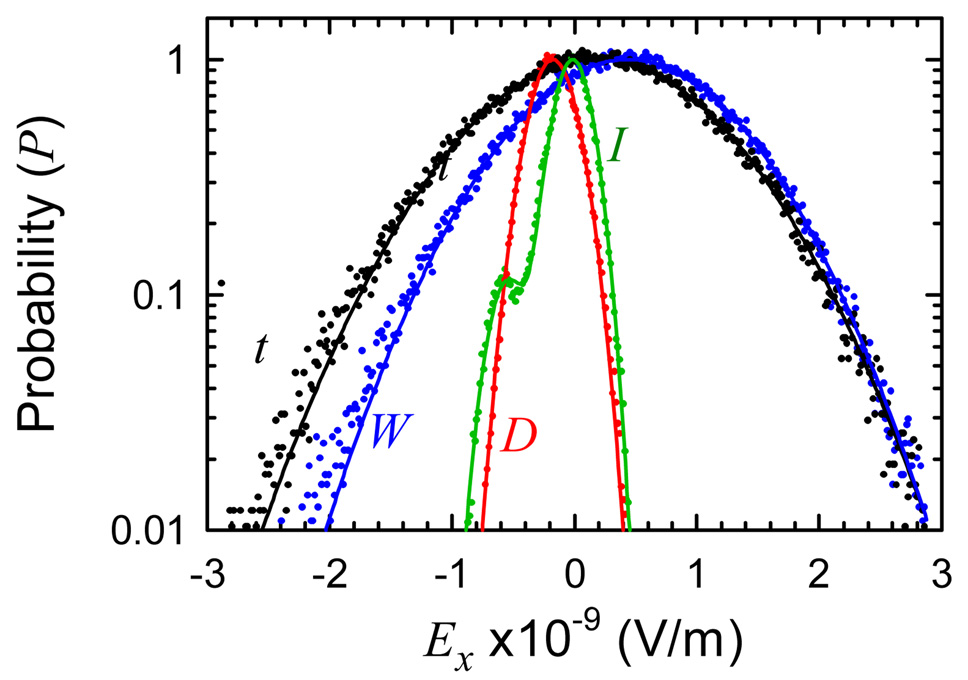
Figure 8 shows the distributions for the total electric field and for the components derived from the polarization model. The width of the water field distribution accounts for most of the width of the total-field distribution, whereas the DNA- and ion-field distributions are much narrower. This result is expected based on the relative size of the correlation functions at zero time. The total, water and DNA fields have very nearly Gaussian distributions, as shown by the fits in Figure 8. In particular, the DNA does not show two components that could be attributed to distinct substates.
Figure 8.
Semi-log plot of the time-averaged distribution of electric-field values (dots) and fits (curves). Total (t, black) and components from the polarization model: water (W, blue), DNA (D, red) and ions (I, green). Curves are single Gaussians, except for the ion distribution, which is the sum of two Gaussians (see Supporting Information).
However, the ion-field distribution does show a distinct side peak. This distribution has been fit by the sum of two Gaussians. Two ion subpopulations can be discerned, with the smaller containing about 10% of the total population. The ion contribution to the total electric field is too small for these subpopulations to be resolved in the total-field distribution.
Dynamic Distributions Show No Dynamic Heterogeneity
The distribution defined in eq 23 can detect static heterogeneity, that is, subpopulations that have different average values of the electric field. It is also possible to have rate heterogeneity—different subpopulations that relax at different speeds, even though their time-averaged distributions may be the same.
For example, consider the DNA correlation function, which is distinctly nonexponential. It is reasonable to ask if there are fast and slow substates, each with an exponential relaxation. The nonexponential decay is then an artifact of averaging together the signals from multiple subpopulations. These subpopulations need to be stable during the electric-field relaxation, but could exchange at longer times. If such dynamic heterogeneity exists, it might be more realistic to fit the correlation function with a multiexponential, rather than a stretched exponential or power-law.
Dynamic heterogeneity can be detected by looking at the dynamic distribution
| (24) |
This function looks at the distribution of changes in the electric field that occur during a time t. It is similar to the van Hove correlation function. At time zero, G(δE, 0) is a delta function. At infinite time, it becomes the autocorrelation of the static distribution P(δE = E − <E>). If this final distribution is Gaussian and there is no dynamic heterogeneity, the distribution will also be Gaussian at intermediate times. On the other hand, if one subpopulation moves toward the equilibrium distribution more rapidly than the others, the intermediate distributions will be non-Gaussian.
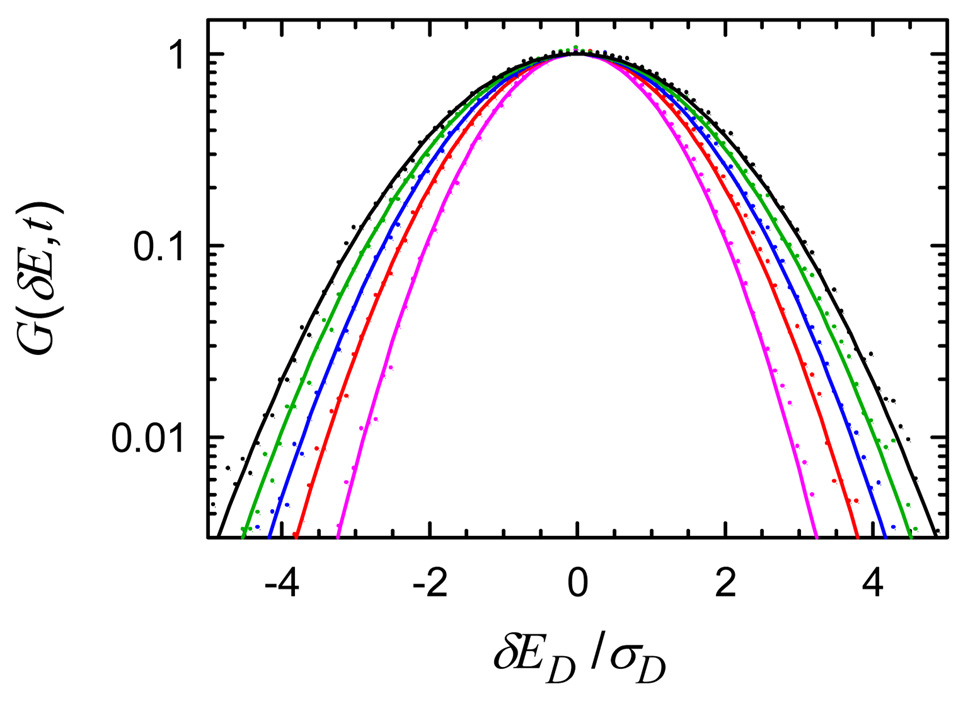
The dynamic distribution of the DNA field is shown in Figure 9. It remains Gaussian at all measured times, and thus, there is no dynamic heterogeneity in this coordinate. The water and total dynamic distribution functions also are Gaussian at all times after 1 ps (Supporting Information).
Figure 9.
Dynamic distribution G(δE, t) for the DNA component of the electric field (dots) with Gaussian fits (curves). The widths increase with time (1 ps/violet, 10 ps/red, 30 ps/blue, 100 ps/green, 1 ns/black). The electric field has been scaled by σD, the standard deviation of the static distribution P(ED).
The fact that G(δE, t) is a Gaussian at all times shows the electric-field dynamics conform to the Gaussian-dynamics approximation.60,61 This approximation neglects all correlation functions beyond the second cumulant. We conclude that the second-order correlation functions shown in Figure 7 contain all the information available on the electric-field dynamics.
In the Gaussian-dynamics approximation, the TRSS experiment also gives a fluorescence line shape that is unchanging with time. TRSS measurements in DNA have found that the line shape is very constant,38 even though this is not a general result in TRSS measurements. Once again, experiment and simulation are in agreement.
The ion dynamic distribution is more complex (Figure 10). It has been modeled by
| (25) |
At long times, this function properly represents the autocorrelation of the two Gaussians seen in the static distribution (Figure 8). Broadening of the central Gaussian is due to motion within each of the subpopulations. The side peaks increase in amplitude due to exchange between the subpopulations. Equation 25 links the widths of the central and side peaks, so that at long times they reach the ratio expected from the static distribution.
Figure 10.
Dynamic distribution G(t, δE) of the ion component of the electric field (dots) with multi-Gaussian fits (curves). The widths increase with time (1 ps/violet, 10 ps/red, 30 ps/blue, 100 ps/green, 300 ps/dashed violet, 1 ns/black). The electric field has been scaled by σI, the standard deviation of the static distribution P(EI).
Only two time-dependent quantities appear in this fit, a(t) and σ(t). Correlations functions can be formed to represent the dynamics within populations
| (26) |
and the exchange between populations
| (27) |
These correlation functions are compared to the standard electric-field correlation function in Figure 11. All three correlation functions are essentially identical. Thus, there is no dynamic heterogeneity in the ion-component of the electric field. Apparently there are two types of ion configuration that each couple differently to the TRSS probe, but the dynamics for these two configurations are the same.
Figure 11.
Ion correlation functions calculated from the electric field CII(t) (curve), from the width of the dynamics distribution Cσ(t) (crosses) and from the amplitude of the side peak in the dynamic distribution Ca(t) (circles). Values at 10 ns are used for the infinite time values in forming Ca and Cσ
VII. DISCUSSION
Water Dominates the TRSS Response in DNA
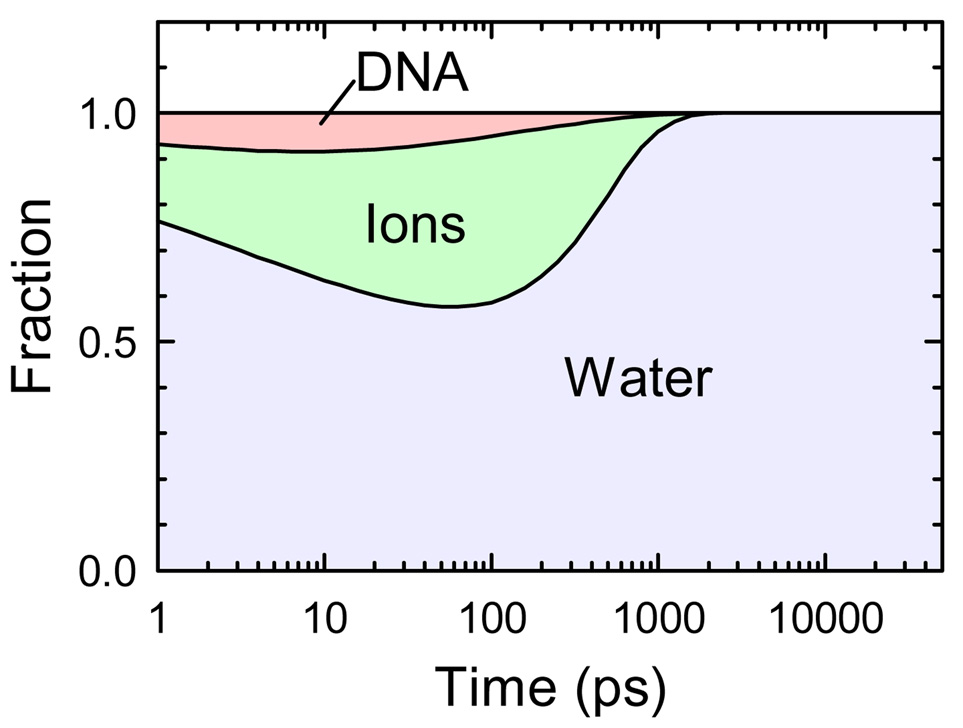
A central issue in interpreting TRSS experiments in a complex system like DNA is which portion of the system is being measured. The simulations presented here show that TRSS is predominantly measuring the dynamics of water. In terms of the contributions to the total rms fluctuations, water is responsible for 90%, the ions for 6% and the DNA for 4%. Two-thirds of the electric-field correlation is lost before 1 ps, and this loss is almost entirely due to water dynamics. Pure water relaxation is subpicosecond,62 so this two-thirds can be considered “normal”. The remaining third represents anomalously slow dynamics.
At times longer than 1 ps, the fraction of the remaining correlation due to each component is illustrated in Figure 12. At all times, water accounts for a majority of the correlation and is primarily responsible for the slow dynamics. At times near 100 ps, the ion dynamics contribute significantly, but at all times, the contribution of the DNA itself is negligible.
Figure 12.
Fractional contributions of each component to the total electric-field correlation remaining at each time. The curves are constructed from the fits in Figure 7.
Polarization-Model versus Linear-Response Decomposition
Although it is straightforward to decompose the total electric field into several components (eq 17), the way to decompose the total correlation function into corresponding components is a more subtle problem. With three field components, the total correlation function decomposes into six correlation functions: three autocorrelations and three cross-correlations. The presence of cross correlations prevents the interpretation of any of these functions as the independent dynamics of one component of the system.
The polarization model introduced here is one solution to this problem. The coordinates are transformed to make the cross-correlations zero. The total correlation function becomes the sum of three autocorrelations (eq 19)). Each autocorrelation function defines the independent dynamics of one component of the system.
The new coordinates are not strictly defined by the chemical species involved. Thus, EI represents the stochastic fluctuations of the electric-field from the ions plus the mean polarization field from the DNA and water that develops in response to the ion field. The polarization response is negative, so the ion field is heavily shielded; EI is much weaker than the total ion field Ei. EW represents the total field of the water minus the mean polarization of the water by the ions and DNA. This leaves just the fluctuations of the water field that are stochastically independent of the other components. Again, the corrected field EW is much weaker than total field Ew.
Nilsson and Halle5 followed by Golosov and Karplus6 and Furse and Corcelli8 used a different method to decompose the total electric-field correlation function in their analyses of TRSS experiments on proteins and DNA. We will call this method the Linear-Response Decomposition (LRD). They split the total autocorrelation into cross-correlations between each component field and the total field. In the current notation, the LRD divides the total correlation function into three pieces
| (28) |
The method is equivalent to adding one-half of each cross-correlation to each of the corresponding autocorrelations,
| (29) |
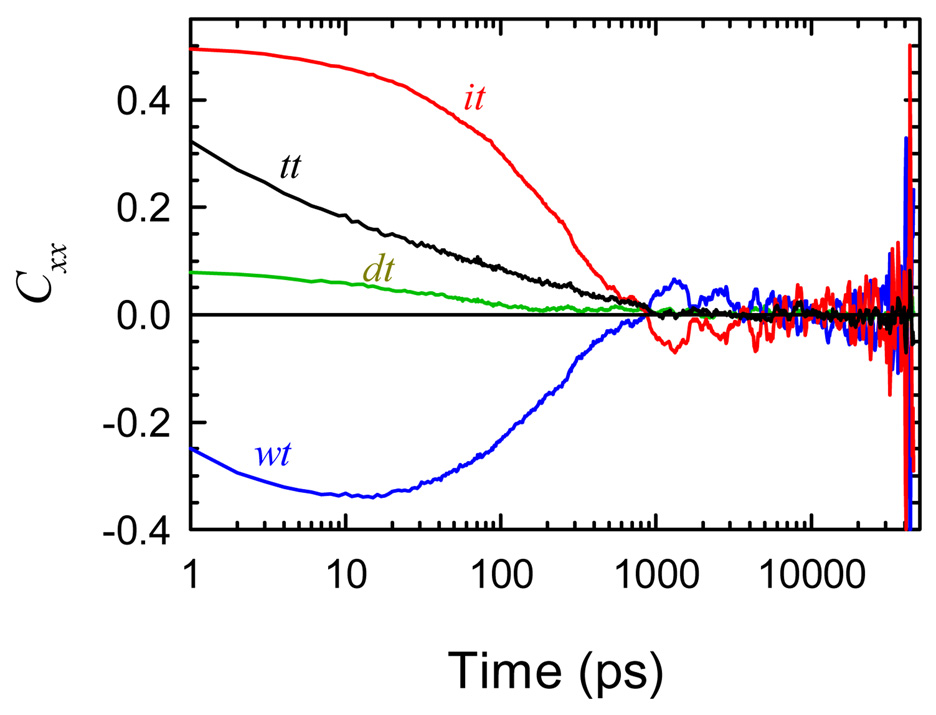
The decomposition of our results by the LRD method is shown in Figure 13. The results are radically different from those of the polarization model (Figure 6 and Figure 7). The time dependence and relative magnitudes of the component correlation functions are completely different. The water component Cwt is even strongly negative. Thus choosing between these methods is critical to making the correct interpretation of TRSS experiments.
Figure 13.
The total electric field correlation (tt) and its decomposition into components by the linear-response method. The results are dramatically different from the decomposition by the polarization model (Figure 6A).
Both the polarization-model and linear-response decompositions are mathematically correct means of reducing six correlation functions to three correlation functions. The difference is in the physical interpretation that can be given to the resulting functions. The key feature in decomposing a system into components is that the components should behave independently, at least to a sufficient level of approximation. The polarization model is based on this criterion.
The justification for the LRD is based on linear-response theory.63 Just as the total correlation function can be associated with the nonequilibrium TRSS experiment, the LRD correlation functions can be associated with hypothetical nonequilibrium experiments. For example Cwt could be measured by creating the probe dipole at t = 0 and perturbing the entire system as in the TRSS experiment, but in the probing step, only measuring the contribution to the signal due to the electric field from the water. Unfortunately, it is not practical to perform such an experiment, so the linear-response arguments do not yield a practical advantage. Rather, the LRD functions will contain interactions between components. For example, Cwt contains contributions in which the ions directly respond to the initial perturbation, the water polarization is changed by the movement of the ions, and the resulting water field is detected. It is not clear that this process should be described simply as “the water response.”
We note that the ability to perform the LRD decomposition is mathematically guaranteed, and so it is universally applicable. The applicability of the polarization model is contingent on the linearity of the couplings within the system. Its applicability must be tested on each system. Conversely, the success of the polarization model in itself makes an important statement about the physics of the system. We have spoken of the linear couplings as coming from electrostatic polarization effects. This is a plausible interpretation, but not a rigorous result of the model.
Comparison to other TRSS simulations in DNA
Pal, et al. also concluded that the DNA itself had little contribution to the TRSS response and that the slow response was due to a combination of the water and counterions.13 Our decomposition goes further and shows that water is much more important than the counterions. They describe a strong role for ions based on the large size of their bare electric field, but note that the water-ion cross correlations strongly counter this field. Our analysis confirms this idea and provides a quantitative extraction of the independent water and ion contributions.
Furse and Corcelli have analyzed simulations of the TRSS response of a probe bound in the minor groove of DNA.8 This system was measured experimentally by Pal, Zhao and Zewail.64 Contrary to our conclusion (and to Pal et al.’s conclusion), they find that the slow TRSS response is dominated by DNA motion and that the water contribution is negligible. Their conclusion and ours are based on different methods: we focus on the polarization-model decomposition, whereas they rely on the LRD. However, the LRD of our simulation (Figure 13) yields results that are dramatically different from Furse and Corcelli’s. This disparity suggests that there are important physical differences between our systems.
One potentially important difference is that our probe is in the base stack of DNA, whereas in Furse and Corcelli’s system, the probe is in the minor groove. Our results in Sec. IV show that the range sampled in a TRSS experiments is relatively short. It is possible that we see strong effects from highly perturbed water in the grooves of DNA. In Furse and Corcelli’s system, the probe molecule displaces the minor-groove water and is further away from the major-groove water. As a result, their system may only sense the relatively unperturbed water further away from the DNA.
VIII. SUMMARY AND CONCLUSIONS
This paper has made a detailed and quantitative comparison of TRSS experiments and computer simulation in DNA. We have taken special care to avoid bias due to the limited simulation time. These biases are particularly difficult to avoid when the dynamics are broadly distributed in time, as they are in DNA. We also avoided the use of fitting functions and made direct comparisons between the raw data from simulation and experiment.
The resulting agreement is excellent. The time-dependence of the Stokes shift, the magnitude of the Stokes shift and the time-independence in the fluorescence line shape are all accurately reproduced by the simulations. The simulations confirm the unusual features seen in DNA experiments—dynamics spread over many decades in time with no clear separation into discrete timescales. The simulations confirm the existence of relaxation in DNA extending out to 5 ns and possibly longer.
The same behavior is seen in the simulations and experiment despite many differences in the details of the systems. We can conclude that the essential phenomena are not very sensitive to these details, including oligomer length, sequence, perturbations due to introducing the probe, deviations from linear response theory, long range interaction with the solvent and higher multipole moments of the probe.
The major contribution of simulations is that they can give a detailed molecular interpretation to the dynamics observed in the TRSS experiment. However, the simple assignment of dynamic to specific components of the system – water, ions, DNA – can be defeated by cross correlations. In agreement with Pal, et al,13 we find that these cross correlations are extremely strong in DNA. We developed a polarization model based on the idea that these cross correlations are primarily due to linear dielectric-polarization effects. This model defines new coordinates that consist of the intrinsic electric-field fluctuations of a component shielded by the polarization that it creates in the other components. When viewed in terms of these new coordinates, cross correlations are nearly eliminated, and it is reasonable to discuss three independent contributions to the TRSS signal. We argued that this model gives more interpretable results than the linear-response method used in several recent simulations of TRSS in proteins and DNA.5,6,8
Although many questions remain, a basic picture does begin to emerge. The TRSS signal in DNA is dominated by the fluctuations inherent to the water. In particular, the anomalous dynamics seen in DNA are due to the water. The water contribution is most simply described by a fractional power law. The important water is within 15 Å of the probe. It is a small step to infer that the anomalous dynamics are due to water that is perturbed by confinement in the grooves and/or electrostatic perturbation by the phosphates.
This conclusion seems at variance with a substantial amount of evidence that the Dickerson dodecamer used in the simulation contains a “spine of hydration,” in which the water in the minor groove is more structured than in other sequences.65–67 The effect on dynamics is not as well documented, but presumably the increased structure is associated with slower dynamics. A resolution of this apparent contradiction awaits a more detailed understanding of the water dynamics sensed in the TRSS experiment.
In contrast to the water, the DNA and counterion contributions have well-defined relaxation timescales of 30 ps and 200 ps respectively. The counterions make a secondary, but non-negligible contribution to the TRSS response near its relaxation time. This result is consistent with the experimental observation of a changing TRSS response upon changing the counterion.37 However, the DNA contribution is quite small and unlikely to be experimentally observable. DNA substates have been seen in structural measurements applied to simulation34,58,59 or have been hypothesized to explain charge-transfer in DNA.28–32 We find no evidence for substates in our results. Apparently, substates definable on a structural basis differ little in their electric-field dynamics.
The behavior of water in biochemical systems, and particularly its dynamics, are topics of growing importance. The current study shows that water near DNA is strongly perturbed and is quite unlike bulk water. Many questions remains about this perturbed water, but the combination of TRSS experiments and computer simulations can be an effective tool in answering these questions.
Supplementary Material
Acknowledgements
We thank Profs. R. J. Cave and E. W. Castner for providing unpublished details from their calculations on coumarins and Prof. S. A. Corcelli and K. E. Furse for providing us with a preprint of their work. This work was supported by the National Institutes of Health (GM-61292) with additional support from the National Science Foundation (CHE-0809306).
Footnotes
Supporting Information Available: Complete description of the coordinate system, results for individual bases and for other dipole directions, complete specification of regions used in Figure 4 and the dynamic distribution function of the total electric field. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ball P. Chem. Rev. 2008;108:74. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 2.Pal SK, Zewail AH. Chem. Rev. 2004;104:2099. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 3.Bagchi B. Chem. Rev. 2005;105:3197. doi: 10.1021/cr020661+. [DOI] [PubMed] [Google Scholar]
- 4.Berg MA, Coleman RS, Murphy CJ. Phys. Chem. Chem. Phys. 2008;10:1229. doi: 10.1039/b715272h. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson L, Halle B. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13867. doi: 10.1073/pnas.0504181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golosov AA, Karplus M. J. Phys. Chem. B. 2007;111:1482. doi: 10.1021/jp065493u. [DOI] [PubMed] [Google Scholar]
- 7.Pal S, Maiti PK, Bagchi B, Hynes JT. J. Phys. Chem. B. 2006;110:26396. doi: 10.1021/jp065690t. [DOI] [PubMed] [Google Scholar]
- 8.Furse KE, Corcelli SA. J. Am. Chem. Soc. 2008;130:13103. doi: 10.1021/ja803728g. [DOI] [PubMed] [Google Scholar]
- 9.Andreatta D, Sen S, Pérez Lustres JL, Kovalenko SA, Ernsting NP, Murphy CJ, Coleman RS, Berg MA. J. Am. Chem. Soc. 2006;128:6885. doi: 10.1021/ja0582105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreatta D, Pérez Lustres JL, Kovalenko SA, Ernsting NP, Murphy CJ, Coleman RS, Berg MA. J. Am. Chem. Soc. 2005;127:7270. doi: 10.1021/ja044177v. [DOI] [PubMed] [Google Scholar]
- 11.Makarov V, Pettitt BM, Feig M. Acc. Chem. Res. 2002;35:376. doi: 10.1021/ar0100273. [DOI] [PubMed] [Google Scholar]
- 12.Auffinger P, Hshem Y. Curr. Opin. Struct. Biol. 2007;17:325. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Pal S, Maiti PK, Bagchi B. J. Chem. Phys. 2006;125:234903. doi: 10.1063/1.2403872. [DOI] [PubMed] [Google Scholar]
- 14.Changenet-Barret P, Choma CT, Gooding EF, DeGrado WF, Hochstrasser RM. J. Phys. Chem. B. 2000;104:9322. [Google Scholar]
- 15.Toptygin D, Savtchenko RS, Meadow ND, Brand L. J. Phys. Chem. B. 2001;105:2043. [Google Scholar]
- 16.Pal SK, Mandal D, Sukul D, Sen S, Bhattacharyya K. J. Phys. Chem. B. 2001;105:1438. [Google Scholar]
- 17.Cohen BE, McAnaney TB, Park ES, Jan YN, Boxer SG, Jan LY. Science. 2002;296:1700. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 18.Mandal D, Sen S, Sukul D, Bhattacharrya K, Mandal AK, Banerjee R, Roy S. J. Phys. Chem. B. 2002;106:10741. [Google Scholar]
- 19.Petushkov VN, van Stokkum IHM, Gobets B, van Mourik F, Lee J, van Grondelle R, Visser A. J. Phys. Chem. B. 2003;107:10934. [Google Scholar]
- 20.Lampa-Pastirk S, Beck WF. J. Phys. Chem. B. 2004;108:16288. doi: 10.1021/jp0654359. [DOI] [PubMed] [Google Scholar]
- 21.Guha S, Sahu K, Roy D, Mondal SK, Roy S, Bhattacharyya K. Biochemistry. 2005;44:8940. doi: 10.1021/bi0473915. [DOI] [PubMed] [Google Scholar]
- 22.Xu J. J. Am. Chem. Soc. 2006;128:1214. doi: 10.1021/ja055746h. [DOI] [PubMed] [Google Scholar]
- 23.Qiu WH, Kao YT, Zhang LY, Yang Y, Wang LJ, Stites WE, Zhong DP, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13979. doi: 10.1073/pnas.0606235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toptygin D, Gronenborn AM, Brand L. J. Phys. Chem. B. 2006;110:26292. doi: 10.1021/jp064528n. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Wang L, Kao Y-T, Qiu W, Yang Y, Okobiah O, Zhong D. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18461. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandyopadhyay S, Chakraborty S, Balasubramanian S, Bagchi B. J. Am. Chem. Soc. 2005;127:4071. doi: 10.1021/ja042847r. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Hassanali AA, Kao YT, Zhong D, Singer SJ. J. Am. Chem. Soc. 2007;129:3376. doi: 10.1021/ja0685957. [DOI] [PubMed] [Google Scholar]
- 28.Wan CZ, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6014. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill MA, Becker H-C, Wan C, Barton JK, Zewail AH. Angew. Chem. Int. Ed. Engl. 2003;42:5896. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill MA, Barton JK. J. Am. Chem. Soc. 2004;126:11471. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 31.Berlin YA, Buring AL, Siebbles LDA, Ratner MA. J. Phys. Chem. A. 2001;105:5666. [Google Scholar]
- 32.Barnett RN, Cleveland CL, Joy A, Landman U, Schuster GB. Science. 2001;294:567. doi: 10.1126/science.1062864. [DOI] [PubMed] [Google Scholar]
- 33.Young MA, Jayaram B, Beveridge DL. J. Phys. Chem. 1998;102:7666. [Google Scholar]
- 34.Young MA, Ravishanker G, Beveridge DL. Biophys. J. 1997;73:2313. doi: 10.1016/S0006-3495(97)78263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young MA, Jayaram B, Beveridge DL. J. Am. Chem. Soc. 1997;119:59. [Google Scholar]
- 36.Coleman RS, Madaras ML. J. Org. Chem. 1998;63:5700. [Google Scholar]
- 37.Sen S, Gearheart L, Rivers E, Lui H, Coleman RS, Murphy CJ, Berg MA. J. Phys. Chem. B. 2006;110:13248. doi: 10.1021/jp056327+. [DOI] [PubMed] [Google Scholar]
- 38.Sen S, Paraggio NA, Gearheart LA, Connor EE, Issa A, Coleman RS, Wilson DM, III, Wyatt MD, Berg MA. Biophys. J. 2005;89:4129. doi: 10.1529/biophysj.105.062695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somoza MI, Andreatta D, Coleman RS, Murphy CJ, Berg MA. Nucleic Acids Res. 2004;32:2495. doi: 10.1093/nar/gkh577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brauns EB, Madaras ML, Coleman RS, Murphy CJ, Berg MA. Phys. Rev. Lett. 2002;88:158101. doi: 10.1103/PhysRevLett.88.158101. [DOI] [PubMed] [Google Scholar]
- 41.Brauns EB, Madaras ML, Coleman RS, Murphy CJ, Berg MA. J. Am. Chem. Soc. 1999;121:11644. [Google Scholar]
- 42.Ponomarev SY, Thayer KM, Beveridge DL. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14771. doi: 10.1073/pnas.0406435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds L, Gardecki JA, Frankland SJV, Horng ML, Maroncelli M. J. Phys. Chem. 1996;100:10337. [Google Scholar]
- 44.Carter EA, Hynes JT. J. Chem. Phys. 1991;94:5961. [Google Scholar]
- 45.Chowdhury A, Locknar SA, Premvardhan LL, Peteanu LA. J. Phys. Chem. A. 1999;103:9614. [Google Scholar]
- 46.Cave RJ, Castner EW. J. Phys. Chem. A. 2002;106:12117. [Google Scholar]
- 47.Samanta A, Fessenden RW. J. Phys. Chem. 2000;104:8577. [Google Scholar]
- 48.Moog RS, Davis WW, Ostrowski SG, Wilson GL. Chem. Phys. Lett. 1999;299:265. [Google Scholar]
- 49.Moylan CR. J. Phys. Chem. 1994;98:13513. [Google Scholar]
- 50.Parkanyi C, Antonious MS, Aaron J-J, Buna M, Tine A, Cisse L. Spectrosc. Lett. 1994;27:439. [Google Scholar]
- 51.Ravi M, Soujanya T, Samanta A, Radhakrichnan TP. J. Chem. Soc., Faraday Trans. 1995;91:2739. [Google Scholar]
- 52.Memkovich NA, Reis H, Baumann W. J. Luminesc. 1997;71:255. [Google Scholar]
- 53.McCarthy PK, Blanchard GJ. J. Phys. Chem. 1993;97:12205. [Google Scholar]
- 54.Garzillo C, Improta R, Peluso A. J. Mol. Struct. 1998;426:145. [Google Scholar]
- 55.Cave RJ, Castner EW. private communication. [Google Scholar]
- 56.Manning GS. Biophys. J. 2006;90:3208. doi: 10.1529/biophysj.105.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frauenfelder H, Parak F, Young RD. Annu. Rev. Biophys. Biophys. Chem. 1988;17:451. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- 58.Trieb M, Rauch C, Wellenzohn B, Wibowo F, Loerting T, Liedl KR. J. Phys. Chem. B. 2004;108:2470. [Google Scholar]
- 59.Beveridge DL, Dixit SB, Barreiro G, Thayer KM. Biopolymers (Nucleic Acid Sciences) 2004;73:380. doi: 10.1002/bip.20019. [DOI] [PubMed] [Google Scholar]
- 60.Loring RF, Yan YJ, Mukamel S. J. Chem. Phys. 1987;87:5840. [Google Scholar]
- 61.Loring RF, Yan YJ, Mukamel S. J. Phys. Chem. 1987;91:1302. [Google Scholar]
- 62.Jimenez R, Fleming GR, Kumar PV, Maroncelli M. Nature. 1994;369:471. [Google Scholar]
- 63.Bernhard W, Callen HB. Rev. Mod. Phys. 1959;31:1017. [Google Scholar]
- 64.Pal SK, Zhao LA, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8113. doi: 10.1073/pnas.1433066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopka ML, Fratini AV, Drew HR, Dickerson RE. J. Mol. Biol. 1983;163:129. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- 66.Feig M, Pettitt BM. Biopolymers. 1998;48:199. doi: 10.1002/(SICI)1097-0282(1998)48:4<199::AID-BIP2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 67.Halle B, Denisov VP. Biopolymers. 2000;48:210. doi: 10.1002/(sici)1097-0282(1998)48:4<210::aid-bip3>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.