Abstract
The applications of block copolymers are myriad, ranging from electronics to functionalized resins to therapeutics. The ring-opening metathesis polymerization (ROMP) is an especially valuable reaction for block copolymer assembly because each block can be generated with length control. We sought to use this polymerization to expand the repertoire of block copolymers by implementing a strategy that involves post-polymerization modification of a backbone bearing selectively reactive groups. To this end, we demonstrate that ROMP can be used to synthesize a block copolymer scaffold that possesses three types of functional groups – a succinimidyl ester, an α-chloroacetamide group, and a ketone – each of which can be modified independently. Thus, a single scaffold can be elaborated to afford a wide range of block copolymers. Exploiting this synthetic approach and the length control offered by ROMP, we assemble block copolymers capable of traversing the membrane and entering mammalian cells.
Introduction
Polymers composed of two or more blocks derived from chemically different monomer units are termed block copolymers. The unique properties of block copolymers have led to their use in many applications, including the development of thermoplastic elastomers, nanofabrication, imaging agents, and drug delivery.1–3 The majority of block copolymer applications depend on imbuing each block with distinct properties (e.g. hydrophobicity or hydrophilicity), such that the resulting polymer can form a macromolecular assembly.4 This blueprint has resulted in preparation of many self-assembling block copolymers. We envisioned soluble, monomeric block copolymers also could be valuable. For example, biologically active block copolymers could be designed in which each domain serves a different biological function. To advance both traditional and non-traditional applications of block copolymers, we sought to develop a divergent synthetic method that could be used to rapidly vary block copolymer composition.
The preparation of well-defined block copolymers requires a living polymerization, that is, a reaction in which chain growth proceeds without competition from chain transfer or termination. Two approaches are typically employed to generate block copolymers: sequential addition of monomers, or coupling of two end-functionalized polymer chains. The first strategy is versatile and widely utilized; it has been applied to synthesize block copolymers using numerous types of polymerization reactions, including anionic, cationic, free-radical, and metal-catalyzed.2,4 Sequential living polymerization reactions can be used to provide a variety of architectures such as linear di- and tri-blocks, star block, and cyclic block copolymers.5–8
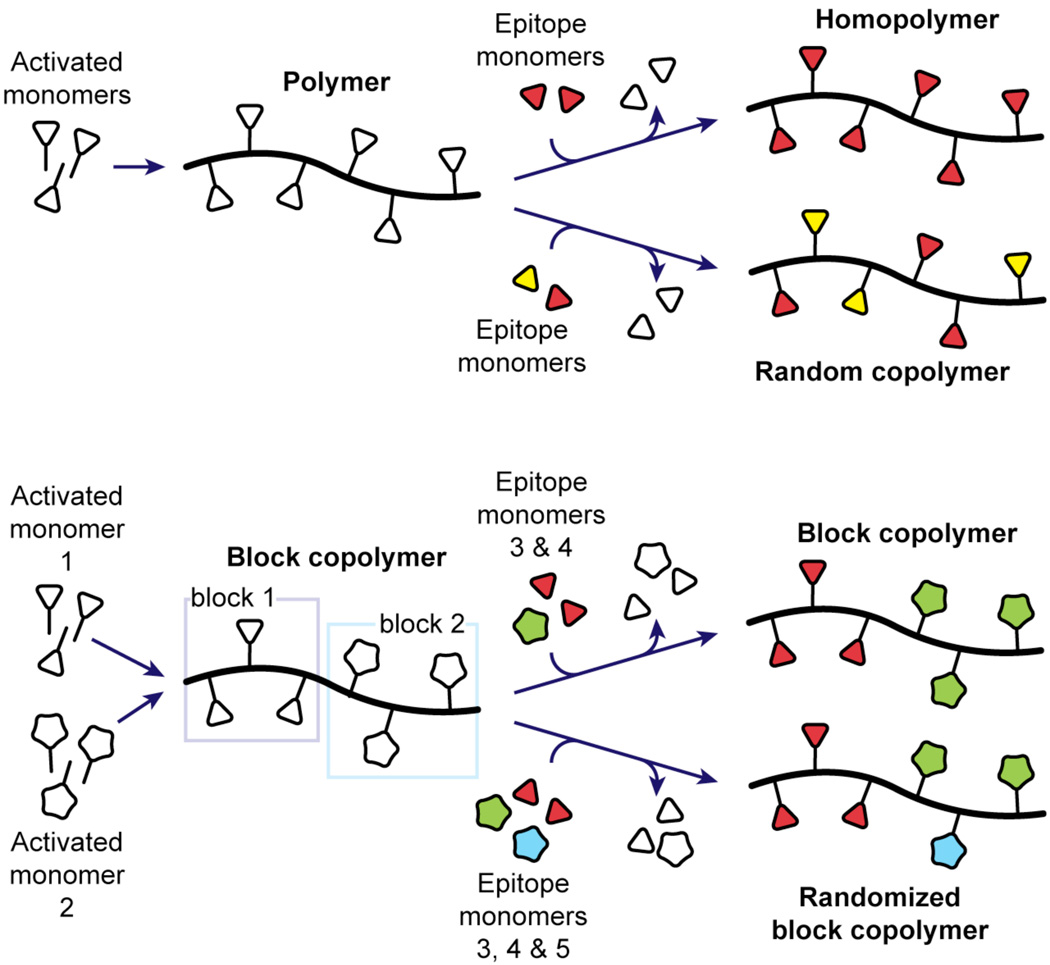
Another powerful approach for polymer preparation is to assemble a general polymer template that can be modified (Figure 1). This approach has been applied to synthesize homopolymers and random copolymers with biological activities.9–15 We envisioned that access to block copolymers that can be modified using nucleophiles present in biological molecules would facilitate the divergent synthesis of new types of bioactive substances, including those that function in protein recruitment and oligomerization, as targeted therapeutics, as probes to study cell–cell recognition or signal transduction, and as intracellular multivalent ligands.
Figure 1.
Examples of linear polymer architectures, including two derived from a general homopolymer scaffold (top) and two derived from a general block copolymer scaffold (bottom).
The ring-opening metathesis polymerization (ROMP) has excellent attributes for block copolymer synthesis.16,17 Because ROMP can be living, and initiation rates can exceed propagation rates, defined polymers of low polydispersity can be generated.18,19 With ruthenium carbene initiators, polymer length can be regulated by varying the monomer to initiator (M:I) ratio.20 Similarly, defined block copolymers can be synthesized by conducting living ROMP, such that when one monomer is consumed, a second is added.21 Finally, ROMP also provides the means to cap polymer products with unique end groups.22–36
We reasoned that block copolymers with diverse properties could be generated from a single scaffold composed of blocks that can be selectively and non-reversibly engineered to introduce unique groups. To this end, we synthesized two monomers that can undergo a living ring-opening metathesis polymerization and bear groups that can be subsequently transformed chemoselectively with mild conditions that are biologically compatible. Moreover, termination of the polymerization reaction installs a ketone, which serves as a third site for selective modification. With a route to this general scaffold, we identified conditions for the introduction of new functional groups onto each reactive segment. The utility of this approach was demonstrated by the selective conversion of our general copolymer scaffold into a block copolymer that is internalized by mammalian cells.
Results and Discussion
Monomer Design and Synthesis
Our polymer synthesis plan required us to identify monomers with two key attributes. First, they must bear groups that are stable under the polymerization conditions yet capable of participation in subsequent modification reactions. Second, each precursor monomer must possess a group that allows for selective derivatization of each block. Substituted norbornene derivatives are excellent monomers for ROMP, and select derivatives can be used to assemble polymeric scaffolds that can be elaborated.10–12,15,36–40 We and others had established the utility of ROMP-derived polymers bearing the succinimidyl ester group.10–12,41–43 Though other electrophile-containing scaffolds have been employed,11,15,44–47 their modification reactions are reversible or utilize transition metals; we wanted a process that would form a stable covalent linkage and use mild reaction conditions. Consequently, we focused on the design of a monomer unit with reactivity orthogonal to the succinimidyl ester.
Succinimidyl esters are especially reactive toward amines, a nucleophile present in many biologically active epitopes. Thiolates, which are also employed frequently in biological conjugation processes, are even more nucleophilic than amines. We postulated that this difference could serve our purpose. Specifically, we sought to identify an electrophile with tempered reactivity that could withstand exposure to an amine yet react with a thiolate. An α-chloroacetamide appeared to meet this criteria, and we reasoned that a block displaying this functionality would be stable to reaction of an amine with a succinimidyl ester-bearing block.43,48–50 Subsequent treatment with a thiolate would result in the displacement of the chloro groups. In this way, epitopes or functional groups of interest could be appended selectively to each block.
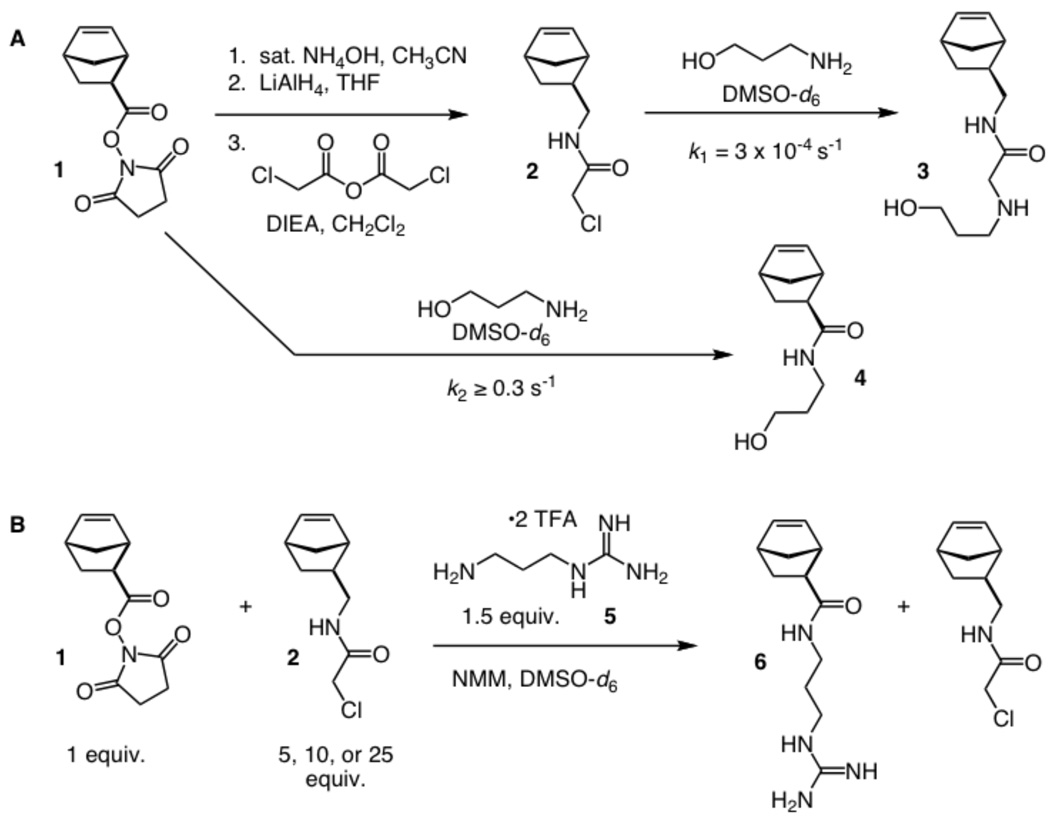
To assess the feasibility of the synthetic strategy, we generated two norbornene derivatives. The amine-reactive succinimidyl ester-substituted norbornene 1 is readily accessible.10 The α-chloroacetamide-substituted monomer 2 was assembled in three steps from norbornene derivative 1 (Scheme 1). With these monomers in hand, we determined their relative rates of reaction using 1H NMR spectroscopy. Both monomers (10 mM in deuterated DMSO) were combined with 3-amino-1-propanol (10 equiv) in an NMR tube (Scheme 1A). Pseudo first-order rate constants were determined at 305 K (32 °C) by integrating diagnostic 1H NMR signals and plotting the concentration of starting material over reaction time. The resulting values were found to be ≥ 0.3 s−1 for the succinimidyl ester-substituted norbornene 1 and 3 × 10−4 s−1 for the α-chloroacetamide-substituted norbornene 2 (Figure 2). The rate of reaction for the amine and succinimidyl ester group is a lower limit, because the process was complete by the time of the first scan. Accordingly, the minimum pseudo first order rate constant was estimated based on 99.9% reaction at 20 s, a time point that allows for the scan delay and sample mixing time. This analysis indicates that the succinimidyl ester is at least 1000-fold more reactive toward amines than is the α-chloroacetamide group.
Scheme 1.
Production of the α-chloroacetamide-containing monomer (A). Pseudo first-order rate constants for the reactions of the monomers with 3-amino-1-propanol are given. Due to the 1000-fold rate difference, amines react selectively with the succinimidyl ester-containing monomer (B).
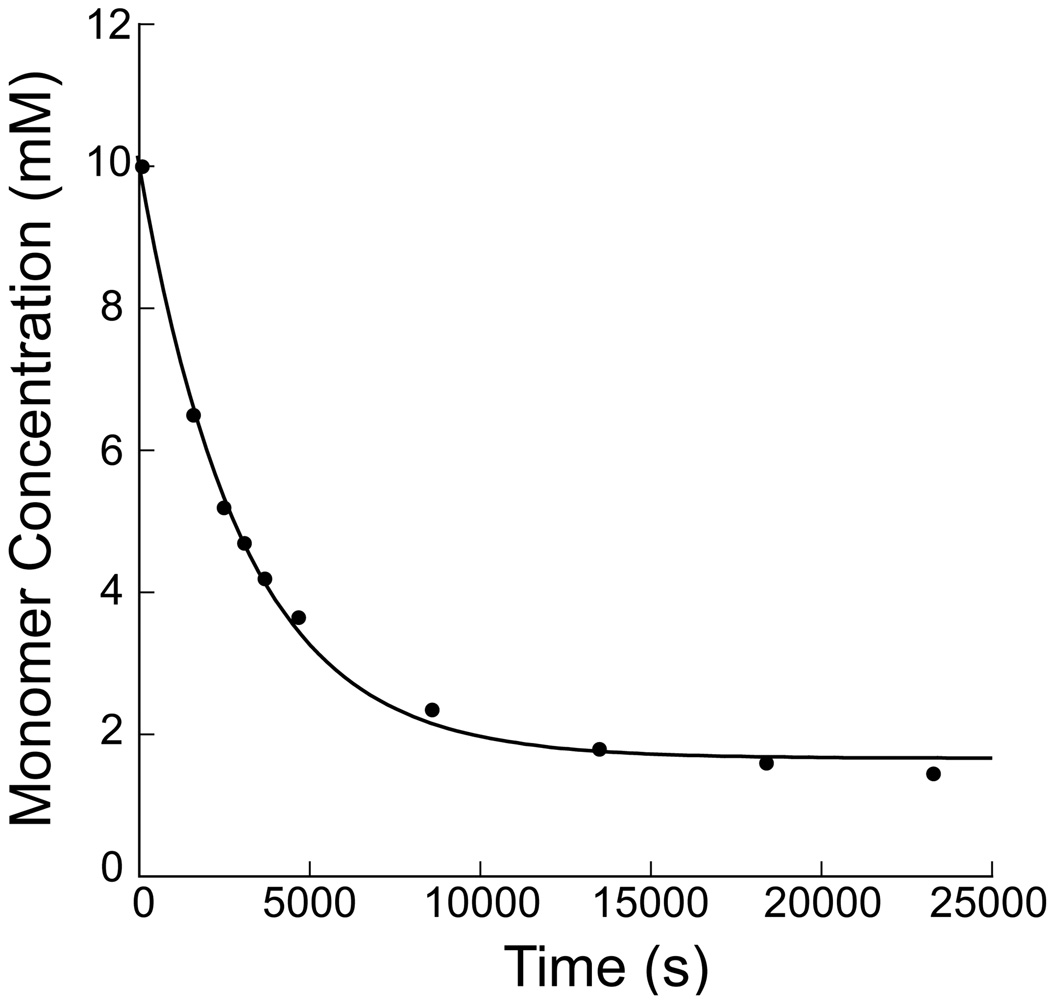
Figure 2.
The pseudo first order rate constant, kobs = 3 × 10−4 s−1 , for the reaction between monomer 2 (10 mM) and 3-amino-2-propanol (100 mM) was determined by monitoring the decrease in α-chloroacetamide methylene group resonance over time by NMR spectroscopy (see supporting information). The monomer concentration was calculated and plotted versus time (above). The kobs was determined using a first order decrease fit [y = A∞−((A∞−A0)*exp(−kobs*t)), where y = monomer concentration, t = time, A0 = initial concentration of 2, and A∞ = concentration of 2 at t∞).
If the monomers have the requisite selectivity, a competition reaction with an amine should transform the succinimidyl ester into an amide, yet leave the α-chloroacetamide monomer untouched. We tested for this anticipated chemoselectivity by treating the monomer with a model amine, N-(3-aminopropyl)-guanidine 5 (Scheme 1B).51 Specifically, the amine (1.5 equiv) was added to a solution of the succinimidyl ester-substituted norbornene (1 equiv), the α-chloroacetamide-substituted norbornene (5, 10, or 25 equiv), and N-methylmorpholine in deuterated DMSO. The ratios and conditions were chosen to simulate those employed to modify block copolymers of different lengths. After 8 h, 1H NMR spectroscopy revealed that all conditions tested resulted in complete conversion of the succinimidyl ester-substituted norbornene to the expected amide, while no reaction of the α-chloroacetamide-substituted norbornene was observed. These experiments support the feasibility of our block copolymer modification strategy.
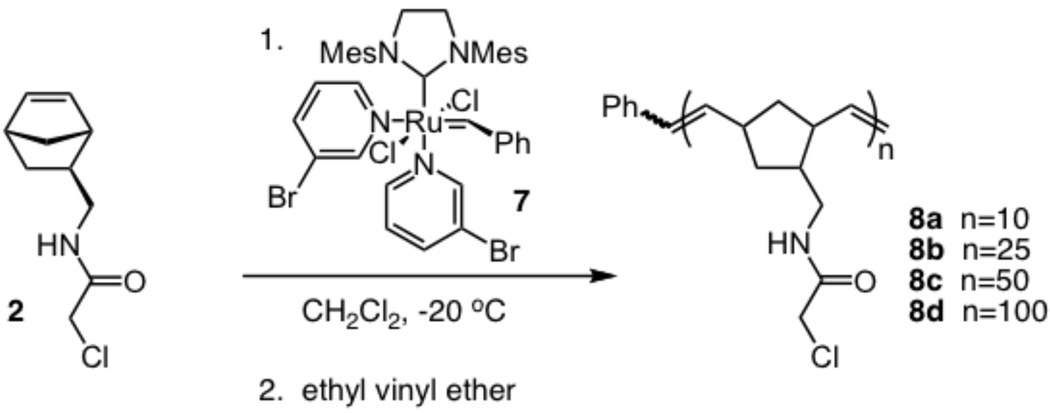
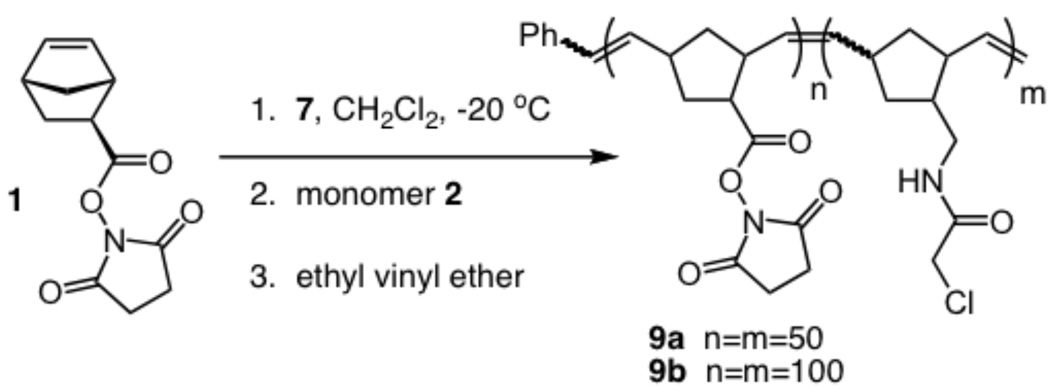
Given that the α-chloroacetamide group has the necessary stability for block copolymer synthesis, we tested whether a monomer bearing this group would give rise to a living polymerization.52 A distinguishing characteristic of a living polymerization is the linear dependence of the number average molecular weight (Mn) on the monomer to initiator ratio (M:I). We therefore assessed the products resulting from exposure of ruthenium complex 753,54 to increasing M:I ratios (10:1, 25:1, 50:1, and 100:1) (Scheme 2, n = M:I). Complete consumption of the substituted norbornene 2 was observed within 20 min at −20 °C. The reactions were terminated with ethyl vinyl ether20 to yield the homopolymers 8a-d. The resulting polymers were characterized by 1H NMR and gel permeation chromatography (GPC) afforded the Mn and polydispersity index (PDI), respectively. A linear relationship between the Mn values and the M:I ratios (see supporting information) was observed. These results indicate the polymerization is living, and they suggest that compound 2 can be used for block copolymer generation. Accordingly, norbornene 1 was exposed to ruthenium initiator 7 (Scheme 3, n = M:I). After thinlayer chromatography indicated that the monomer was consumed, the α-chloroacetamide-containing monomer (2) was added. The polymerization was allowed to proceed, and after the second monomer was consumed, the reaction was terminated with an enol ether to form polymers 9a-b. Characterization of these materials by GPC indicated that they have narrow PDI values of 1.2 and 1.3, respectively and the expected mass of the polymer. These observations indicate that the α-chloroacetamide-substituted norbornene can undergo living polymerization and can be utilized in the generation of defined block copolymers.
Scheme 2.
Synthesis of α-chloroacetamide-substituted homopolymers.
Scheme 3.
Synthesis of general block copolymers
Our next objective was to determine whether each block could be modified independently. To this end, succinimidyl ester-substituted norbornene monomer 1 was exposed to initiator 7 at a M:I ratio of 10:1 and the α-chloroacetamide-substituted monomer 2 was subsequently introduced using a M:I of 50:1. The polymerization reaction was terminated with 10-methoxydec-9-ene-2-one11 to afford copolymer 10, which bears a ketone end-cap. Integration of the alkene, α-chloroacetamide methylene, and phenyl proton signals indicates that the degree of polymerization (DP) of the succinimidyl ester block is 8 and that of the α-chloroacetamide block is 45. Moreover, the block copolymer product retains a narrow PDI of 1.12, as determined by GPC analysis.
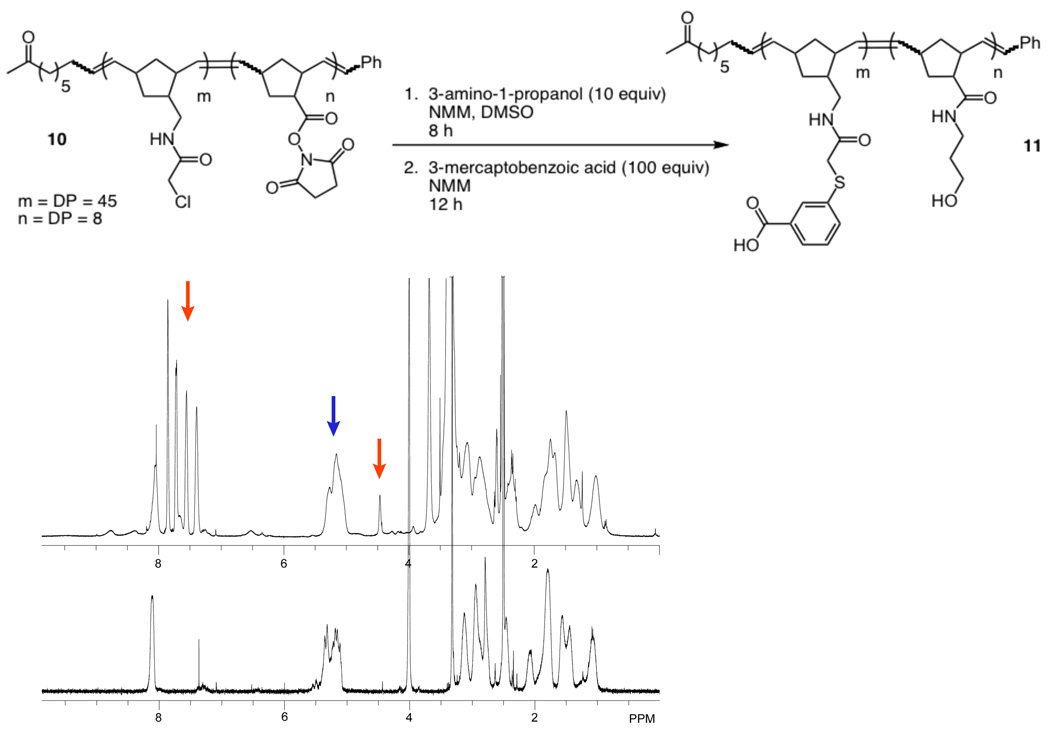
To test whether the block copolymer possesses the desired reactivity, we employed two different prototype nucleophiles: the amine 3-amino-1-propanol and the thiol 3-mercaptobenzoic acid. These compounds have 1H NMR signals that are distinct from each other and the polymer backbone. The functionalized block copolymer was generated by first exposing compound 10 to 3-amino-1-propanol (10 equiv), such that the succinimidyl ester monomer units (DP = 8) undergo reaction (Figure 3). Subsequent treatment with 3-mercaptobenzoic acid (100 equiv) resulted in the alkylation of the α-chloroacetamide monomer units (DP = 45) to afford polymer 11. Integration of the 1H NMR spectrum confirmed the presence of an average of eight conjugated 3-amino-1-propanol moieties and forty-five 3-mercaptobenzoic acid groups. These data indicate that compound 10 has the desired selective reactivity; consequently, it can be used as a scaffold to expedite the synthesis of diverse block copolymers.
Figure 3.
Top: Synthesis of the substituted block copolymer. Left: 1H NMR spectra of 10 (bottom) and 11 (top). Percentile conversion (99%) was determined by integration of aromatic and hydroxyl signals (red arrows) of the conjugated functional groups of 11 against the alkene signal (blue arrow) of the polymer backbone.
Copolymers possessing an intracellular delivery block
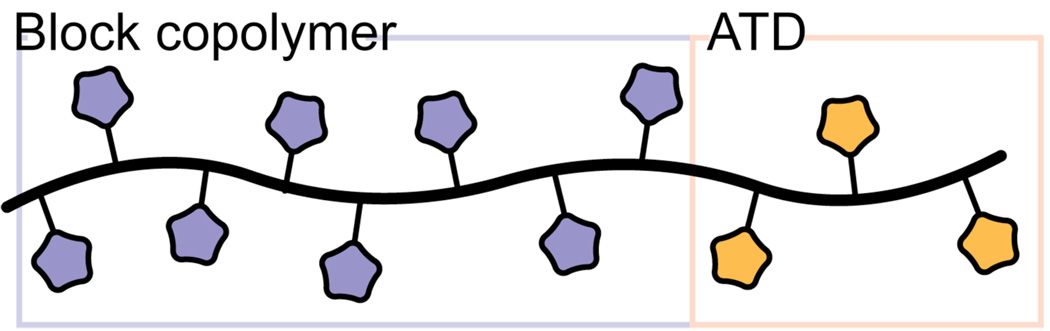
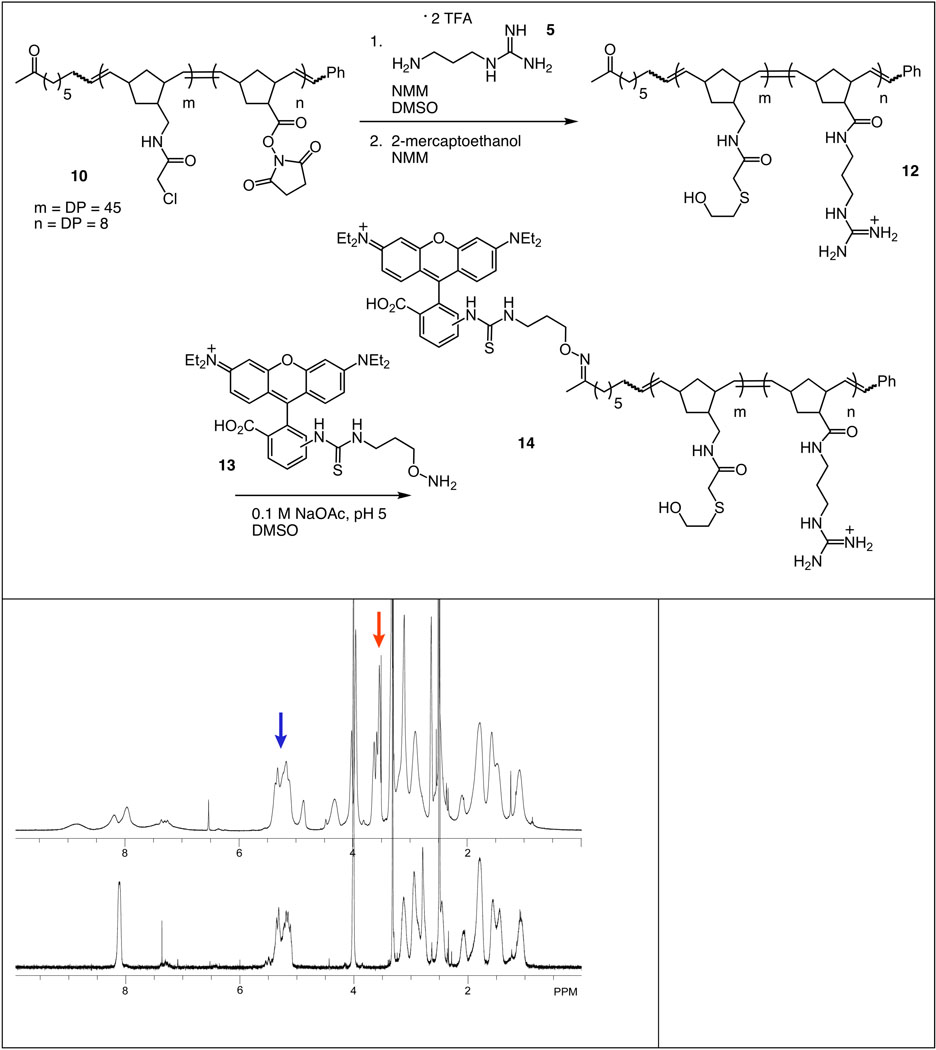
To illustrate the utility of the synthetic route, we used it to synthesize materials capable of crossing the cell membrane. Polymeric ligands are valuable probes of cell surface receptor–ligand interactions,55–63 but most polymers are not cell permeable. Bioactive polymers that can function within the cell could serve as powerful tools to investigate and control new types of biological processes. We reported previously that ROMP can be used to synthesize a polymeric artificial translocation domain (ATD),51 and we postulated that merging the ATD strategy with the synthetic method described herein would yield cell-permeable block copolymers (Figure 4). To test this idea, we used the general scaffold 10 to form a copolymer in which one block was designed to function as an ATD. The ATD was generated by exposure of the polymer to N-(3-aminopropyl)guanidine 551 to convert the succinimidyl esters to amides, thereby leaving the α-chloroacetamide-substituted block available for subsequent functionalization (Figure 5). We incorporated mercaptoethanol into the second block, because its distinct 1H NMR signals facilitated the characterization of the polymer product 12. The data indicate that a conversion of 99% was attained. For subsequent visualization using fluorescence microscopy, a fluorophore was appended to the polymer via selective reaction of the terminal ketone moiety with rhodamine B derivative 1351 to form an oxime (82% conversion). Thus, block copolymer 14 is the product of three different chemoselective reactions, providing an unprecedented level of chemical control over polymer modification and diversification.
Figure 4.
The design for cell-permeable multivalent arrays involves a block copolymer containing an artifical translocation domain (ATD) (yellow) and a block (blue) substituted with groups that serve a separate function.
Figure 5.
Top: Synthesis of an end-labeled block copolymer. Left: 1H NMR spectra of 10 (bottom) and 12 (top). Percentile conversion (98%) was determined by integration of the mercaptoethanol methylene signal (red arrow) 12 against the alkene signal (blue arrow) of the polymer backbone. The guanidinium signals are not distinguishable from those of the norbornene signals of the polymer backbone.
This strategy is advantageous, as the synthesis of polymer 12 illustrates. Specifically, the presence of the guanidinium groups would require that a monomer bearing guanidinium groups undergo controlled polymerization. Monomers bearing guanidinium groups are not soluble in organic solutions, yet the ruthenium catalyst that we employed is not water-soluble. Additionally, even if conditions could be found to carry out the polymerization (i.e., mixed solvent systems) control over polymer polydispersity would be compromised. Indeed, all of the previous routes to guanidinium-substituted polymers have employed post-polymerization modification.42,62,64,65 Our general strategy also could be used to make block polymers in which intact proteins are attached to the polymer via reaction of a cysteine side chain,66 and these are materials that could not be accessed by monomer polymerization.
Block Copolymer Internalization
To determine whether block copolymer 14 can be internalized by cells, we used confocal microscopy. HeLa cells were incubated at 37 °C in a humidified, 5% CO2 atmosphere for 1 h with the fluorophore-labeled block copolymer 14 (2 µM) (Figure 6). Fluorescence microscopy revealed that the block copolymer is internalized, as indicated by its localization in vesicles and the cytoplasm. Compared with the ATD alone,51 the block copolymer is taken up similarly. In contrast, a mercaptoethanol-substituted homopolymer lacking the ATD failed to undergo internalization (see supporting information). These data demonstrate that the ATD block can promote uptake of macromolecules, including block copolymers. We showed previously that a guanidinium-substituted homopolymeric ATD is taken up by cells via endocytosis, a process that occurs at physiological temperature but not at low temperature.51 We correspondingly assessed whether the block copolymer uptake is temperature-dependent. When cells were incubated for 1 h at 4 °C with copolymer 14 (5 µM), no internalization was observed (data not shown). Thus, like 1, the guanidinium-substituted homopolymer and the block copolymer gain entry via an endocytic pathway of entry. These results highlight the utility of our synthetic route for generating copolymers with new functions.
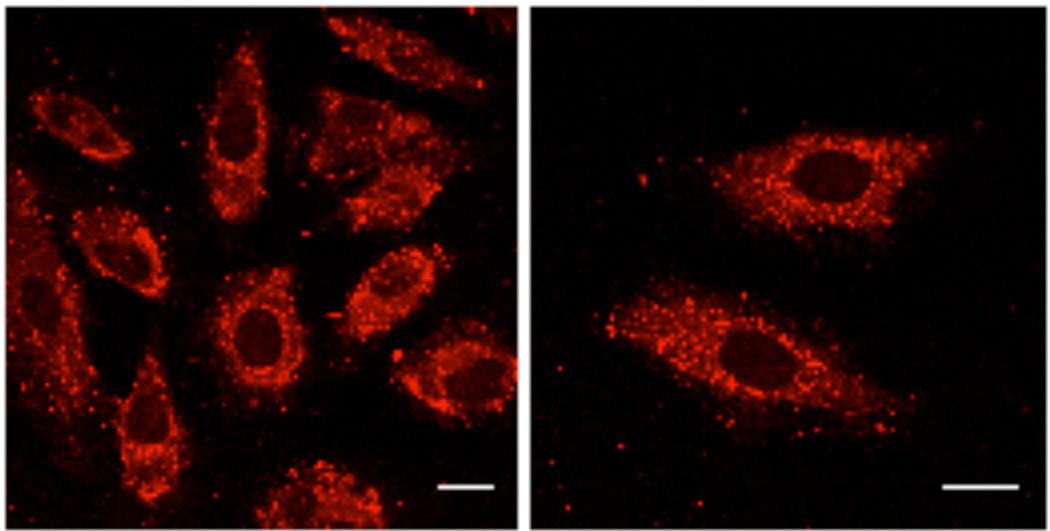
Figure 6.
Confocal microscopy images of live HeLa cells incubated with the rhodamine-labeled, mercaptoethanol-substituted block copolymer (14, 2 mM) for 1 h at 37 °C. The block copolymer is internalized by cells and localized in both endocytic vesicles (punctate fluorescence) as well as in the cytoplasm (diffuse fluorescence). Bar = 25 mm.
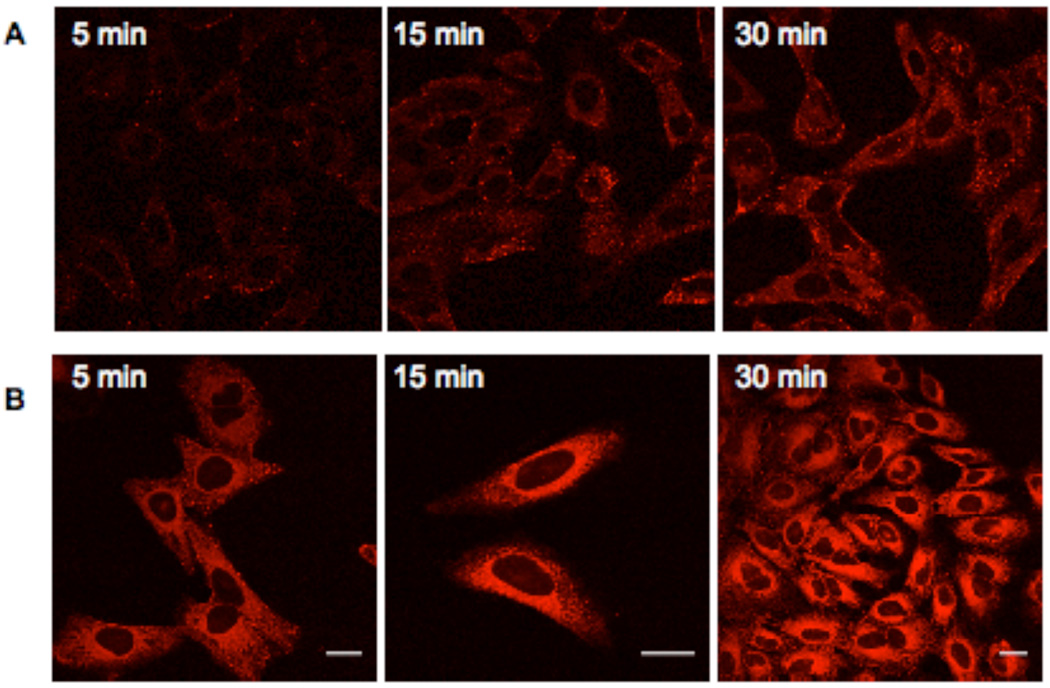
To ascertain the time scale of internalization and escape from the endosomes, cells were treated with the rhodamine-labeled copolymer 14 (2 or 10 µM) for varying amounts of time (5 min to 1 h) (Figure 7). The copolymer (2 µM) could be detected not only in vesicles near the membrane but also in the cytoplasm after only 5 min (Figure 7A). These data indicate that uptake is rapid. Additionally, the mercaptoethanol-substituted block copolymer was localized predominantly in the cytoplasm, which reveals that it is not restricted to endosomal compartments (Figure 7B). Accordingly, not only can the guanidinium ATD can serve as a delivery agent for macromolecules, and the block copolymer backbone reported here can be used to create cell-permeable multivalent ligands.
Figure 7.
HeLa cells were treated with the rhodamine-labeled, mercaptoethanol-substituted block copolymer (14) at 2 µM (A) or 10 µM (B) concentrations for varying amounts of time at 37 °C. The block copolymer (2 µM, A) is present in both vesicles and the cytoplasm after incubation as brief as 5 min; fluorescence increases in both locations over time. The cytoplasmic localization is more pronounced when the cells are treated with a higher concentration of block copolymer (10 µM, B). Bar = 25 µm.
Conclusions
We have generated a block copolymer scaffold via ROMP that is amenable to selective functionalization with nucleophilic groups. This single reactive scaffold can be employed to create a large number of unique polymers and copolymers with a wide variety of properties. To illustrate the utility of the synthetic strategy, we converted the scaffold to cell-permeable block copolymers. Indeed, the general block copolymer synthetic strategy described herein could be used to generate agents for many applications. For example, we envision this approach can yield block copolymers that can serve as delivery vehicles for targeted therapeutics. In addition, it can yield block copolymers that promote the formation of intracellular protein assemblies that elicit desirable biological responses.
Supplementary Material
Acknowledgement
This research was supported by NIGMS of the NIH (GM049975). We thank K.A. Dickson, T.-Y. Chao, and R. T. Raines for helpful discussions. Confocal microscopy was performed at the W.M. Keck Laboratory for Biological Imaging at the University of Wisconsin–Madison. EMK acknowledges the NIH Chemistry–Biology Interface Training Program for a predoctoral fellowship (GM008505). The NMR facility is supported in part by the NSF (CHE-0342998 and CHE-9629688) and the NIH (RR13866).
Footnotes
Supporting Information Available. Experimental preparations and characterization of compounds. This information is available free of charge via the Internet at http://pubs.acs.org/.
Reference Section
- 1.Duncan R. Nat. Rev. Drug Discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 2.Lodge TP. Macromol. Chem. Phys. 2003;204:265–273. [Google Scholar]
- 3.Shenhar R, Norsten TB, Rotello VM. Adv. Mater. 2005;17:657–669. [Google Scholar]
- 4.Hadjichristidis N, Pitsikalis M, Iatrou H. Block Copolymers I: Synthesis of block copolymers. Adv. Polym. Sci. 2005;189:1–124. [Google Scholar]
- 5.Szwarc M, Levy M, Milkovich R. J. Am. Chem. Soc. 1956;78:2656–2657. [Google Scholar]
- 6.Iatrou H, Hadjichristidis N. Macromolecules. 1992;25:4649–4651. [Google Scholar]
- 7.Fujimoto T, Zhang HM, Kazama T, Isono Y, Hasegawa H, Hashimoto T. Polymer. 1992;33:2208–2213. [Google Scholar]
- 8.Yin R, Amis EJ, Hogenesch TE. Macromol. Symp. 1994;85:217–238. [Google Scholar]
- 9.Choi SK, Mammen M, Whitesides GM. J. Am. Chem. Soc. 1997;119:4103–4111. [Google Scholar]
- 10.Strong LE, Kiessling LL. J. Am. Chem. Soc. 1999;121:6193–6196. [Google Scholar]
- 11.Pontrello JK, Allen MJ, Underbakke ES, Kiessling LL. J. Am. Chem. Soc. 2005;127:14536–14537. doi: 10.1021/ja053931p. [DOI] [PubMed] [Google Scholar]
- 12.Carrillo A, Gujraty KV, Rai PR, Kane RS. Nanotechnology. 2005;16:S416–S421. doi: 10.1088/0957-4484/16/7/016. [DOI] [PubMed] [Google Scholar]
- 13.Gujraty KV, Joshi A, Saraph A, Poon V, Mogridge J, Kane RS. Biomacromolecules. 2006;7:2082–2085. doi: 10.1021/bm060210p. [DOI] [PubMed] [Google Scholar]
- 14.Ladmiral V, Mantovani G, Clarkson GJ, Cauet S, Irwin JL, Haddleton DM. J. Am. Chem. Soc. 2006;128:4823–4830. doi: 10.1021/ja058364k. [DOI] [PubMed] [Google Scholar]
- 15.Yang SK, Weck M. Macromolecules. 2008;41:346–351. [Google Scholar]
- 16.Bielawski CW, Grubbs RH. Prog. Polym. Sci. 2007;32:1–29. [Google Scholar]
- 17.Singh R, Czekelius C, Schrock RR. Macromolecules. 2006;39:1316–1317. [Google Scholar]
- 18.Trnka TM, Grubbs RH. Acc. Chem. Res. 2001;34:18–29. doi: 10.1021/ar000114f. [DOI] [PubMed] [Google Scholar]
- 19.Schrock RR, Hoveyda AH. Angew. Chem. Int. Ed. 2003;42:4592–4633. doi: 10.1002/anie.200300576. [DOI] [PubMed] [Google Scholar]
- 20.Schwab P, France MB, Ziller JW, Grubbs RH. Angew. Chem. Int. Ed. 1995;34:2039–2041. [Google Scholar]
- 21.Cannizzo LF, Grubbs RH. Macromolecules. 1988;21:1961–1967. [Google Scholar]
- 22.Mitchell JP, Gibson VC, Schrock RR. Macromolecules. 1991;24:1220–1221. [Google Scholar]
- 23.Albagli D, Bazan GC, Schrock RR, Wrighton MS. J. Am. Chem. Soc. 1993;115:7328–7334. [Google Scholar]
- 24.Schrock RR. Tetrahedron. 1999;55:8141–8153. [Google Scholar]
- 25.Gibson VC, Okada T. Macromolecules. 2000;33:655–656. [Google Scholar]
- 26.Gordon EJ, Gestwicki JE, Strong LE, Kiessling LL. Chem. Biol. 2000;7:9–16. doi: 10.1016/s1074-5521(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 27.Owen RM, Gestwicki JE, Young T, Kiessling LL. Org. Lett. 2002;4:2293–2296. doi: 10.1021/ol0259239. [DOI] [PubMed] [Google Scholar]
- 28.Grubbs RH. Tetrahedron. 2004;60:7117–7140. [Google Scholar]
- 29.Riegler S, Slugovc C, Trimmel G, Stelzer F. Macromol. Symp. 2004;217:231–246. [Google Scholar]
- 30.Slugovc C. Macromol. Rapid Commun. 2004;25:1283–1297. [Google Scholar]
- 31.Hilf S, Berger-Nicoletti E, Grubbs RH, Kilbinger AFM. Angew. Chem. Int. Ed. 2006;45:8045–8048. doi: 10.1002/anie.200602323. [DOI] [PubMed] [Google Scholar]
- 32.Hilf S, Kilbinger AFM. Macromol. Rapid Commun. 2007;28:1225–1230. [Google Scholar]
- 33.Hilf S, Grubbs RH, Kilbinger AFM. J. Am. Chem. Soc. 2008;130:11040–11048. doi: 10.1021/ja8022863. [DOI] [PubMed] [Google Scholar]
- 34.Matson JB, Grubbs RH. Macromolecules. 2008;41:5626–5631. [Google Scholar]
- 35.Hilf S, Hanik N, Kilbinger AFM. J. Polym. Sci., Part A: Polym. Chem. 2008;46:2913–2921. [Google Scholar]
- 36.Miki K, Kuramochi Y, Oride K, Inoue S, Harada H, Hiraoka M, Ohe K. Bioconjugate Chem. 2009;20:511–517. doi: 10.1021/bc800449s. [DOI] [PubMed] [Google Scholar]
- 37.Roberts KS, Sampson NS. Org. Lett. 2004;6:3253–3255. doi: 10.1021/ol048935y. [DOI] [PubMed] [Google Scholar]
- 38.Bertin PA, Gibbs JM, Shen CKF, Thaxton CS, Russin WA, Mirkin CA, Nguyen ST. J. Am. Chem. Soc. 2006;128:4168–4169. doi: 10.1021/ja056378k. [DOI] [PubMed] [Google Scholar]
- 39.Binder WH, Kluger C. Macromolecules. 2004;37:9321–9330. [Google Scholar]
- 40.Kluger C, Binder WH. J. Polym. Sci., Part A: Polym. Chem. 2007;45:485–499. [Google Scholar]
- 41.Barrett AGM, Cramp SM, Roberts RS, Zecri F. J. Org. Lett. 2000;2:261–264. doi: 10.1021/ol991208w. [DOI] [PubMed] [Google Scholar]
- 42.Allen MJ, Raines RT, Kiessling LL. J. Am. Chem. Soc. 2006;128:6534–6535. doi: 10.1021/ja061383p. [DOI] [PubMed] [Google Scholar]
- 43.Carrillo A, Yanjarappa MJ, Gujraty KV, Kane RS. J. Poly. Sci. A: Polymer Chemistry. 2006;44:928–939. [Google Scholar]
- 44.Barrett AGM, Hopkins BT, Kobberling J. Chem. Rev. 2002;102:3301–3323. doi: 10.1021/cr0103423. [DOI] [PubMed] [Google Scholar]
- 45.Xiao SJ, Brunner S, Wieland M. J. Phys. Chem. B. 2004;108:16508–16517. [Google Scholar]
- 46.Harned AM, Zhang MJ, Vedantham P, Mukherjee S, Herpel RH, Flynn DL, Hanson PR. Aldrichim. Acta. 2005;38:3–16. [Google Scholar]
- 47.Liaw DJ, Huang CC, Hong SM, Chen WH, Lee KR, Lai JY. Polymer. 2006;47:4613–4621. [Google Scholar]
- 48.Lindley H. Biochem. J. 1960;74:577–584. doi: 10.1042/bj0740577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barglow KT, Cravatt BF. Chem. Biol. 2004;11:1523–1531. doi: 10.1016/j.chembiol.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 50.Rajakumar P, Rasheed AMA, Rabia AI, Chamundeeswari D. Bioorg. Med. Chem. Lett. 2006;16:6019–6023. doi: 10.1016/j.bmcl.2006.08.124. [DOI] [PubMed] [Google Scholar]
- 51.Kolonko EM, Kiessling LL. J. Am. Chem. Soc. 2008;130:5626–5627. doi: 10.1021/ja8001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grubbs RH. Angew. Chem. Int. Ed. 2006;45:3760–3765. doi: 10.1002/anie.200600680. [DOI] [PubMed] [Google Scholar]
- 53.Love JA, Morgan JP, Trnka TM, Grubbs RH. Angew. Chem. Int. Ed. 2002;41:4035–4037. doi: 10.1002/1521-3773(20021104)41:21<4035::AID-ANIE4035>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 54.Choi TL, Grubbs RH. Angew. Chem. Int. Ed. 2003;42:1743–1746. doi: 10.1002/anie.200250632. [DOI] [PubMed] [Google Scholar]
- 55.Mortell KH, Gingras M, Kiessling LL. J. Am. Chem. Soc. 1994;116:12053–12054. [Google Scholar]
- 56.Maynard HD, Okada SY, Grubbs RH. J. Am. Chem. Soc. 2001;123:1275–1279. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]
- 57.Kiessling LL, Gestwicki JE, Strong LE. Angew. Chem. Int. Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y, Sampson NS. Curr. Opin. Struct. Biol. 2006;16:544–550. doi: 10.1016/j.sbi.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 59.Smith D, Pentzer EB, Nguyen ST. Polym. Rev. 2007;47:419–459. [Google Scholar]
- 60.Puffer EB, Pontrello JK, Hollenbeck JJ, Kink JA, Kiessling LL. ACS Chem. Biol. 2007;2:252–262. doi: 10.1021/cb600489g. [DOI] [PubMed] [Google Scholar]
- 61.Binder JB, Raines RT. Curr. Opin. Chem. Biol. 2008;12:767–773. doi: 10.1016/j.cbpa.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangold SL, Carpenter RT, Kiessling LL. Org. Lett. 2008;10:2997–3000. doi: 10.1021/ol800932w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Courtney AH, Puffer EB, Pontrello JK, Yang ZQ, Kiessling LL. Proc. Natl. Acad. Sci. USA. 2009;106:2500–2505. doi: 10.1073/pnas.0807207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolonko EM, Kiessling LL. J. Am. Chem. Soc. 2008;130 doi: 10.1021/ja8001716. 5626-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alfred SF, Lienkamp K, Madkour AE, Tew GN. J. Polym. Sci., Part A: Polym. Chem. 2008;46:6672–6676. [Google Scholar]
- 66.Bontempo D, Heredia KL, Fish BA, Maynard HD. J. Am. Chem. Soc. 2004;126:15372–15373. doi: 10.1021/ja045063m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.