Abstract
The goal of this study was to develop a kinetic analysis that could predict the behavior of embryonic stem cell-derived neural progenitor cells (ESNPCs) in response to treatment with neurotrophin-3 (NT-3). Previous studies have shown that NT-3 activates the mitogen-activated protein (MAP) kinase cascade in embryonic stem cells and promotes differentiation of ESNPCs into neurons. MAP kinase activation after NT-3 stimulation was confirmed experimentally, and a kinetic analysis was developed using rate constants obtained from the literature. Concentrations of select signaling components were estimated for ESNPCs using real-time reverse transcription polymerase chain reaction by comparing mRNA levels to those of cell types with known protein concentrations. This assumption was validated using Western blots, and incorporated into the analysis. This analysis was used to predict the minimum NT-3 concentration necessary to promote neuronal differentiation of ESNPCs based on the activation of MAP kinase. These predictions were then tested experimentally to confirm the validity of the analysis. Finally, expression of the transcription factor mammalian achate schute homolog 1 and β-tubulin III (an early neuronal marker) was examined in response to the different NT-3 doses to confirm the link between MAP kinase activation and neuronal differentiation. Overall, this study provides insight into the kinetics of the intracellular processes that promote ESNPC differentiation to neurons.
Introduction
Embryonic stem (ES) cells have generated much interest due to their pluripotent nature that allows them to generate replacement cells for damaged or diseased tissue.1,2 Generating different types of kinetic analysis can give insight into and predict ES cell behavior.3 Such analysis can offer the benefit of reducing the number of experiments that must be performed while giving insight into the intracellular processes that govern cell behavior. Many different attempts have been made to produce such models with varying results. Lauffenburger and colleagues have developed simple, three parameter models to characterize the rates of self-renewal versus differentiation for murine stem cells seeded on different extracellular matrices while being exposed to two different growth factors.4 This work was further extended to study intracellular signaling events and identify molecules that were influencing these processes.5 However, they did not use this information to generate a kinetic analysis. They later used Bayesian analysis to generate the signaling networks, giving insight into the intracellular processes and signaling cascades that determine ES cell differentiation. This analysis then was used to generate probability plots to predict ES cell behavior.6 Zandstra et al. have developed ligand–receptor signaling threshold (LIST) models that predicted ES cell differentiation based on the number of ligand–receptor complexes found on the cell surface.7,8 All of these aforementioned models focused on the effects of leukemia inhibitory factor (LIF), hyperinterleukin-6, and fibroblast growth factor, which help maintain ES cells in their undifferentiated state.
To predict ES cell behavior in response to a growth factor that promotes differentiation, Lai et al. developed a kinetic-based analysis of sonic hedgehog (Shh) signaling.9 This model accurately replicated the effects of Shh treatment on ES cells by demonstrating switch-like behavior. This analysis was further extended into a tissue-patterning model that predicted development based on the distribution of Shh signaling in the developing chick spinal cord.10 Such models provide valuable insight into the differentiation process.
Neurotrophins, a family of growth factors that regulates development and survival of the nervous system, have been shown to play important roles in ES cell differentiation and survival.11–14 Of particular interest is neurotrophin-3 (NT-3) due to its ability to promote the differentiation of ES cells into neurons through activation of the mitogen-activated protein (MAP) kinase cascade.15,16 NT-3 also induces production of basic helix–loop–helix proteins, such as the transcription factor mammalian achate schute homolog 1 (Mash1).17 Upon translocation to the nucleus, Mash1 then activates genes that promote neuronal differentiation as well as upregulation of Mash1 transcription.18 This study hypothesized that the activation of the MAP kinase cascade would promote translocation of Mash1 to the nucleus and result in neuronal differentiation of the ES cells. It has been previously shown that ES cells transfected to express Mash1 differentiated into motoneurons and helped hemiplegic mice regain function after injury.19 Additionally, these cells promoted functional recovery as indicated by Basso, Beattie, and Bresnahan scores after complete spinal cord transection in mice when compared to the injury-only controls.20 Thus, developing a kinetic analysis of the effect of NT-3 signaling on the differentiation of ES cells into neurons could help generate tissues for treatment of central nervous system defects, such as spinal cord injury (SCI).
NT-3 activates the MAP kinase cascade by binding to its high-affinity receptor, tyrosine kinase receptor C (TrkC).21 The reactions that make up this cascade are shown in Figure 1. The receptor dimerizes and autophosphorylates, allowing the adaptor protein Src homolog and collagen domain protein (Shc) to bind to the receptor and become phosphorylated. The growth factor receptor bound protein (Grb) can then bind to activated Shc. Grb normally binds the son of sevenless homolog protein (SOS) to form a Grb–SOS complex, even in the absence of cascade activation. As a result, binding usually occurs between Grb–SOS complexes and activated Shc to form Grb–SOS–Shc complexes. These complexes, in turn, catalyze the conversion of the inactive form of Ras (Ras-GDP) to its active form (Ras-GTP). Ras-GTP activates Raf, a serine/threonine kinase, that phosphorylates Map/Erk kinase (Mek). Mek can be phosphorylated in two different sites, resulting in Mek-P and Mek-PP. Activated Mek, in turn, phosphorylates extracellular signal regulated kinase (Erk), which can also be phosphorylated on one or both residues. The activated Erk can bind to transcription factors, such as Mash1, that regulate cellular processes. The amount of activated Mek and Erk can be additionally regulated by protein phosphatase 2 that removes the added phosphate groups from the enzymes, resulting in loss of activation. The signaling cascade is additionally regulated by G-protein activating protein (GAP), which deactivates Ras-GTP. These steps help ensure that the activation of the MAP kinase cascade is reversible.
FIG. 1.
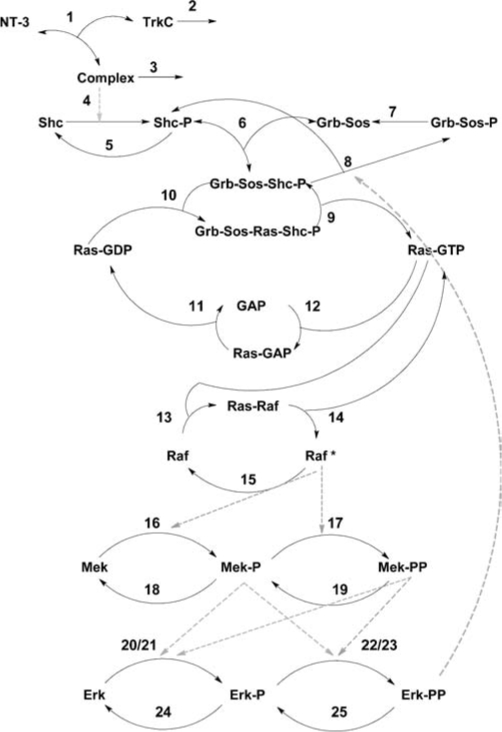
Schematic showing the individual components and reactions of the MAP kinase cascade. Each reaction is numbered, and the rate constants corresponding to each constant are given in Table 1.
Previous groups have thoroughly characterized the MAP kinase signaling cascade and its role in promoting the differentiation of rat phenochromocytoma (PC12) cells. These cells initially start as round, undifferentiated cells. When nerve growth factor (NGF) is applied, these cells undergo a differentiation process and extend neurites.22,23 In order for these cells to differentiate, MAP kinase cascade activation must be sustained. Many different kinetic models have been developed to characterize the behavior of PC12 cells in response to neurotrophin stimulation,24–27 and these models were used as a starting point for this study. Additionally, the concentrations of the different proteins that make up the MAP kinase signaling cascade are known for the PC12 cell line, and this information could be used to estimate the concentrations of the signaling cascade components in stem cells or embryonic stem cell–derived neural progenitor cells (ESNPCs).
Previous work has demonstrated the potential of using ESNPCs as a treatment for SCI.28 These neural progenitors are formed from undifferentiated mouse ES cells using a 4−/4+ retinoic acid treatment protocol.29 Additional work has shown that NT-3 promotes differentiation of these cells into neurons.30 It was hypothesized that stimulating ESNPCs with NT-3 would result in activation of the MAP kinase cascade, leading to an increase in the fraction of cells differentiating into neurons. The current study confirms the activation of the MAP kinase cascade by NT-3 in ESNPCs experimentally, and kinetic parameters from the literature were used to quantitatively model the reactions occurring inside of the cell after NT-3 stimulation. Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) was used to estimate the concentrations of selected signaling proteins for ESNPCs by comparing their mRNA levels to those found in PC12 cells, and these values were incorporated into the kinetic analysis. The analysis was then used to predict the minimum concentration of NT-3 needed to activate the MAP kinase cascade and promote neuronal differentiation. These predictions were tested experimentally, and further work investigated the effect of different NT-3 doses on the regulation of the transcription factor Mash1 and the resulting ESNPC differentiation into neurons. This study has produced a kinetic analysis that gives insight into the intracellular processes that promote ESNPC differentiation into neurons, which could be important when producing engineered tissues from such cells to serve as treatment for SCI. Specifically, this analysis gives insight into the minimum NT-3 concentrations needed to promote neuronal differentiation, which is an important consideration when designing engineered tissues for SCI repair.
Materials and Methods
All reagents and media for cell culture were purchased from Invitrogen (Grand Island, NY) unless otherwise indicated. All reagents, primers, and kits used for quantitative real-time RT-PCR were purchased from Qiagen (Valencia, CA). The quantitative phosphoErk1/2 enzyme-linked immunosorption assay (ELISA) was purchased from Active Motif (Carlsbad, CA).
Cell culture
Rat PC12 cells
PC12 cells were cultured in suspension inside of T75 flasks (Fischer Scientific, Pittsburgh, PA) containing F-12K media supplemented with 15% horse serum (HS), 2.5% fetal bovine serum (FBS), and 1% antibiotic/antimycotic, and passaged weekly. To collect RNA for estimating protein concentrations, individual flasks of cells were centrifuged at 1015 g to pellet cells, and the cells were rinsed with phosphate-buffered saline (PBS; pH 7.4). The resulting cells were lysed with Buffer RLT from the RNeasy kit.
Mouse ES cells
RW4 mouse ES cells obtained from D. Gottlieb were cultured on gelatinized T25 flasks (Fisher Scientific) in complete media (Dulbecco's modified Eagle's medium containing 10% FBS, 10% newborn calf serum, and 0.3 M nucleoside mix) containing 1000 U/mL LIF and 10−4M β-mecaptoethanol (BME) as previously described.30,31 To induce ESNPC formation, the cells were plated on agar-coated Petri dishes (MidSci, St. Louis, MO) and induced to form embryoid bodies (EBs) using the 4−/4+ retinoic acid differentiation protocol.29 In this process, the cells were cultured for 4 days in complete media without LIF and BME followed by additional 4 days of culture in complete media with 100 nM retinoic acid. During this process, the media was changed every other day.
The resulting cell aggregates are referred to as 4−/4+ EBs and contain ESNPCs. Previous work from our lab has characterized the phenotypes of these cells.30 Approximately, 70% of these cells express the nestin marker, indicating that the majority of these cells are ESNPCs. Approximately, 60% of the cells stain positive for SSEA-1, which is expressed by undifferentiated mouse ES cells. These results suggest that there is coexpression of these two markers in some cells. None of the cells expressed the following three mature cell phenotype markers, including β-tubulin III (neurons), O4 (oligodendrocytes), and glial fibrillary acidic protein (GFAP, astrocytes), suggesting that these progenitor cells have not undergone terminal differentiation.
To collect RNA for estimating protein concentrations and Western blotting, individual flasks of unstimulated PC12 cells and plates containing 4−/4+ EBs were added to 15 mL centrifuge tubes and allowed to settle. The cells were rinsed with PBS. For the RT-PCR experiments, the resulting cells were lysed with Buffer RLT from the RNeasy kit. For Western blotting, the resulting cells were lysed for 15 min using RIPA buffer (25 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS, all from Sigma, St. Louis, MO) containing protease inhibitor cocktail (Pierce, Rockford, IL). The resulting solution was then centrifuged at 14,000 g to pellet the cell debris before performing protein quantification. For cell seeding experiments, the EBs were allowed to settle and media aspirated. The resulting cells were resuspended in 5 mL of 0.25% trypsin-EDTA and incubated at 37°C for 20 min. The reaction was quenched with 5 mL of complete media. The cells were counted and seeded at a density of 20,000 cells per well of a 96-well plate and 100,000 cells per well for a 24-well plate.
Quantitative analysis of Erk1/2 activation
To determine the time course of phosphoErk1/2 activation, dissociated cells obtained from 4−/4+ EBs were seeded onto gelatin-coated 96-well plates and serum starved overnight. To confirm MAP kinase activation after NT-3 stimulation, ESNPCs were treated with 25 ng/mL of NT-3 (Peprotech, Rocky Hill, NJ). For the experiments to confirm the kinetic analysis, the cells were stimulated by adding 200 μL of complete media containing 2 and 10 ng/mL of NT-3 to each individual well. Cells were fixed using 100 μL of 8% formaldehyde at different time points after stimulation. The relative amounts of phosphoErk1/2 and Erk1/2 were then determined quantitatively using an Erk1/2 ELISA following the manufacturer's instructions, including an overnight incubation with primary antibodies. The plates were developed and read using a multiwell plate reader (MultiSkan RC Labsystems, Helsinki, Finland). The levels of phosphoErk were normalized to the amount of phosphoErk present when no NT-3 is present. Thus, all experimental data are reported in terms of relative levels of phosphoErk expression and represent Erk active due to stimulation by NT-3.
Quantitative real-time RT-PCR for estimating protein concentrations
To obtain RNA for real-time RT-PCR analysis, cell lysates obtained from unstimulated PC12 cells and 4−/4+ EBs were homogenized using a Qiashredder kit, followed by RNA isolation using an RNEasy kit. From the isolated RNA, cDNA was synthesized using the Quantitech Reverse Transcription kit according to the manufacturer's instructions. Using the Quantitech SYBR Green PCR master mix combined with gene-specific QuantiTect primer assays, quantitative realtime PCR was performed using an Applied Biosystems' 7000 Real-Time PCR System using the following PCR cycle: 50°C (2 min), 95°C (10 min), followed by 40 cycles of 95°C (15 s), 55°C (30 s), and 72°C (30 s) with fluorescent signal detected at 72°C. Target genes were normalized to β-actin expression to account for variations in cDNA concentration between samples. The QuantiTect primer assays are validated to have a PCR efficiency of 100%.
To estimate the protein concentrations, the differences in gene expression levels between the two cell lines were calculated using the comparative delta crossover threshold (Ct) method, which allows for determining relative expression of RNA between two samples. In this method, the Ct refers to the cycle number when the fluorescence released by the PCR reaction begins to increase exponentially. Lower Ct values indicate higher concentrations of RNA present. The difference between relative Ct values for each sample is used to estimate the fold difference between samples. For this analysis, it was assumed that the levels of mRNA expression in each cell line would provide an accurate correlation with protein expression levels for that cell line. These results were confirmed qualitatively using Western blots (Fig. 2). For determining relative levels of Mash1 expression, 4−/4+ EBs were dissociated, and the resulting cells were seeded onto gelatinized plates and then stimulated with the different NT-3 doses. Cell lysates were harvested 2 h after stimulation, and the RNA was extracted as previously described. The differences in gene expression levels were compared to untreated cells using the delta Ct method.
FIG. 2.
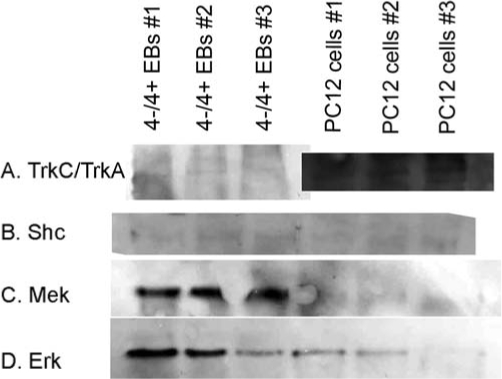
Western blot analysis of receptor, Shc, Mek, and Erk expression in PC12 cells and 4−/4+ embryoid bodies. (A) TrkA/TrkC receptor (molecular weight: 145 and 140 kDa, respectively)—both Western blots were developed for the same amount of time. (B) Shc (molecular weight: 46 kDa). (C) Mek (molecular weight: 45 kDa). (D) Erk (molecular weight: 42 kDa).
Confirmation of RT-PCR results by Western blotting
Cell lysates were obtained as described above, and the protein concentration was quantified using a bicinchoninic acid (BCA) assay (Pierce) and a set of known protein concentration standards. Based on the results of the BCA, 500 ng of protein lysate were loaded into each well of a 10% Tris-HCl ReadyGel (BioRad, Hercules, CA), and the proteins were separated using gel electrophoresis. The proteins were then transferred to nitrocellulose membranes (BioRad). The membranes were blocked in Tris-buffered saline (TBS; pH 7.4) containing 5% bovine serum albumin and 0.1% Tween-20 (both from Sigma) for 2 h followed by an overnight incubation with primary antibody. All primary antibodies, including TrkA, Shc, Mek, and Erk (Cell Signaling Technology, Boston, MA), were used at a 1:1000 dilution except for TrkC (Santa Cruz Biotechnology, Santa Cruz, CA), which was used at 1:200 dilution. The next day, the membranes were washed three times with TBS containing 1% Tween-20. The membranes were then incubated with secondary antibody (rabbit IgG depending on primary, 1:1000 dilution; Cell Signaling Technology) for 1 h followed by an additional three washes with TBS containing 1% Tween-20. The blots were then developed using LumiGlo reagent according to the manufacturer's instructions (Cell Signaling Technology), and the resulting signal was detected on Classic Blue BX film (MidSci) using a developer (Fisher Industries, Geneva, IL). The blots for TrkC and TrkA receptor were developed for the same amount of time to allow for a comparison between the two samples. Molecular weight was determined by comparison to Precision Plus Protein Standards (BioRad).
Generating a kinetic analysis of MAP kinase signaling
Kinetic analysis development
The reactions that make up the MAP kinase signaling cascade are shown in Figure 1; the reactions are labeled. Table 1 lists the constants associated with each reaction. Forward and reverse rate constants are listed when applicable. For enzymatic reactions, Michaelis–Menten kinetics were assumed with Km and the Vmax or Kcat given. Based on these reactions, differential equations modeling the overall reaction rates for each of the components of the MAP kinase signaling cascade were developed and coded into Matlab (Mathworks, Natick, MA). This set of equations was integrated numerically using the ode45 solver with the initial concentrations of each protein listed in Table 2. NT-3 concentration was assumed to be constant based on the time course analyzed (∼3h) to allow for ease of computation, and receptor recycling was not considered due to the short time course.
Table 1.
Rate Constants Used in Kinetic Analysis
| Reaction number | Reaction constant | Value | Units | Reference |
|---|---|---|---|---|
| 1 | Kf | 2.78E8 | M−1 min−1 | 21 |
| Kr | 0.73 | min−1 | 21 | |
| 2 | Kdeg | 0.0069 | min−1 | 25 |
| 3 | Kdeg | 0.012 | min−1 | 25 |
| 4 | Kcat | 12 | min−1 | 38 |
| Km | 5E5 | molecule cell−1 | ||
| 5 | Vmax | 6.12E4 | molecule cell−1 min−1 | 39 |
| Km | 2.04E5 | molecule cell−1 | ||
| 6 | Kf | 0.002 | molecule min−1 | 26 |
| Kr | 3.81 | min−1 | ||
| 7 | Vmax | 75 | molecule cell−1 min−1 | 26 |
| Km | 2E4 | molecule cell−1 | ||
| 8 | Kcat | 1.6 | min−1 | 26 |
| Km | 6E5 | molecule cell−1 | ||
| 9 | K | 15 | min−1 | 26 |
| 10 | Kf | 0.0163 | molecule min−1 | 26 |
| Kr | 10 | min−1 | ||
| 11 | K | 720 | min−1 | 26 |
| 12 | Kf | 0.005 | molecule min−1 | 26 |
| Kr | 60 | min−1 | ||
| 13 | Kf | 0.0012 | molecule min−1 | 26 |
| Kr | 3 | min−1 | 26 | |
| 14 | K | 27 | min−1 | 26 |
| 15 | Vmax | 9.7E4 | molecule cell−1 min−1 | 26 |
| Km | 6000 | molecule cell−1 | ||
| 16 | Kcat | 50 | min−1 | 26 |
| Km | 9000 | molecule cell−1 | ||
| 17 | Kcat | 9000 | min−1 | 38 |
| Km | 2.74E4 | molecule cell−1 | ||
| 18 | Vmax | 9.2E5 | molecule cell−1 min−1 | 38 |
| Km | 6E5 | molecule cell−1 | ||
| 19 | Vmax | 2.7E4 | molecule cell−1 min−1 | 26 |
| Km | 9E3 | molecule cell−1 | ||
| 20/22 | Kcat | 8.3 | min−1 | 26 |
| Km | 9E4 | molecule cell−1 | 26 | |
| 21/23 | Kcat | 9 | min−1 | 38 |
| Km | 2.78E4 | molecule cell−1 | 38 | |
| 24 | Vmax | 1.8E4 | molecule cell−1 min−1 | 38 |
| Km | 9000 | molecule cell−1 | 38 | |
| 25 | Vmax | 4E5 | molecule cell−1 min−1 | 26 |
| Km | 6E5 | molecule cell−1 | 26 |
Table 2.
Estimating Protein Concentrations of Signaling Components (Number Per Cell)
| Protein | HeLa cells | PC12 cells | Fold differencea | ESNPCsb |
|---|---|---|---|---|
| TrkA/TrkC | 50,000 (32) | 130,000 (24) | 2.40 ± 0.4 | 54,200 |
| Shc | 101,000 (32) | 30,000 (38) | 0.70 ± 0.3 | 42,860 |
| Grb-Sos | 51,000 (32) | 20,000 (26) | — | Used PC12 data |
| Ras | 35,000 (32) | 20,000 (26) | — | Used PC12 data |
| GAP | 12,000 (32) | 15,000 (26) | — | Used PC12 data |
| Raf | 40,000 (32) | 10,000 (26) | — | Used PC12 data |
| Mek | 22,000,000 (32) | 100,800 (38) | 0.22 ± 0.4 | 458,200 |
| Erk | 21,000,000 (32) | 750,000 (26) | 0.78 ± 0.3 | 961,500 |
Comparing expression levels in PC12 cells to those in ESNPCs. Standard deviation is also given.
Estimated from the experimental data and concentrations for PC12 cells.
Sensitivity analysis of the estimated protein concentrations
As mentioned in the Introduction, many groups have implemented kinetic analysis of the MAP kinase signaling pathway and performed extensive sensitivity analysis on the effects of changing the various rate constants.24,25,32 Thus, for this study, it was chosen to focus on the effect of estimating the protein concentrations on the outcomes of the kinetic analysis. To determine the sensitivity of the model to each of the estimated protein concentrations, the standard deviation for each experiment was used to calculate new estimated values to be used in the kinetic analysis by adding it to or subtracting it from the fold difference. Thus, two new estimated values were generated for each protein in addition to the original estimate. Simulations were then run for both the 2 and 10 ng/mL NT-3 being present, using all three estimated protein concentrations (estimated value using the fold difference plus the standard deviation, the actual estimated value, and the estimated value using the fold difference minus the standard deviation) and compared to determine the effect of changing each protein concentration on the kinetic analysis. This analysis was important to confirm that the error intrinsic to the experimental data did not have an effect on the predicted values chosen for further study.
Quantifying neuronal differentiation of ESNPCs after NT-3 stimulation
Fluorescence-activated cell sorting (FACS) was used to quantify the fraction of cells differentiating into neurons after stimulation with different NT-3 doses. ESNPCs were seeded into two-dimensional culture in 24-well plates as described above and stimulated with 0, 2, 10, and 25 ng/mL of NT-3. After 3 days of culture, the media was changed to neural basal media containing B27 supplement diluted 1:50 and the same dose of NT-3. The media was not changed for the rest of the study. After 14 days of culture, the cells were isolated from the plate by adding 250 μL of trypsin-EDTA (0.25%) to each well and incubating for 20 min at 37°C. The reaction was quenched by adding 250 μL of complete media. The cells were then centrifuged at 1015 g, and the media was aspirated.
Staining for β-tubulin III, which is expressed by early neurons, was performed as previously described.33 All steps were performed at 4°C, and all solutions were made using PBS. The cells were fixed for 20 min in 1% formaldehyde followed by permeabilization using a 0.5% saponin solution for 20 min. The cells were then blocked in solution of 5% goat serum (Sigma) and 0.1% saponin. The cells were then incubated with the β-tubulin III antibody (Covance, Berkeley, CA; 1:1000) for 30 min followed by incubation with an AlexaFluor 488 goat anti-mouse IgG (Invitrogen; 1:200) secondary antibody for an additional 30 min. The cells were then washed twice with PBS containing 2% calf serum and 0.1% sodium azide (Sigma) and sorted on a FACS Calibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) as previously described. As a control, cells stained with only the same secondary antibody were used to eliminate nonspecific background staining. All analysis was performed using CellQuest software (Becton Dickinson).
Statistical analysis
For the quantitative ELISAs, 3 wells (n = 3) of a 96-well plate were evaluated at each time point and dose of NT-3 to determine the levels of phosphoErk and total Erk present. For determining expression of Mash1 and β-tubulin III, three sets of 6-well replicates (n = 3) seeded in 24-well plates were collected and analyzed as described above. ANOVA with Scheffe post hoc tests were used to compare groups. Significance was considered to be p < 0.05.
Results
Generation and validation of a phosphoERK kinetic analysis in ESNPCs
Before generating a kinetic analysis of the signaling cascade, it was important to characterize MAP kinase activation in ESNPCs after NT-3 stimulation. Accordingly, a quantitative phosphoErk ELISA was performed to confirm that Erk activation does occur in ESNPCs stimulated with 25 ng/mL of NT-3. This dose was chosen based on a previous work because it promoted increased differentiation of ESNPCs seeded into fibrin scaffolds into neurons compared to cultures without growth factors present in the media.33 As seen in Figure 3A, the levels of phosphoErk present increase after NT-3 stimulation until it reaches a peak at around 40 min after stimulation when ∼40% of the Erk is activated. After this point, the levels of activated ERK decrease to ∼20% at 90 min. Similar amounts of phosphoErk were observed at the later time points, suggesting that this is the steady state level of activation. Based on these experimental results, a kinetic analysis of the activation of Erk by the MAP kinase signaling cascade in ESNPCs was developed. The different reactions (Fig. 1) that compose the cascade were taken from the literature, and rate constants for each step were obtained from the literature as seen in Table 1. The concentration of each species in the cascade was then calculated at the various time points using ordinary differential equations based on the rates of the different reactions of the signaling cascade.
FIG. 3.
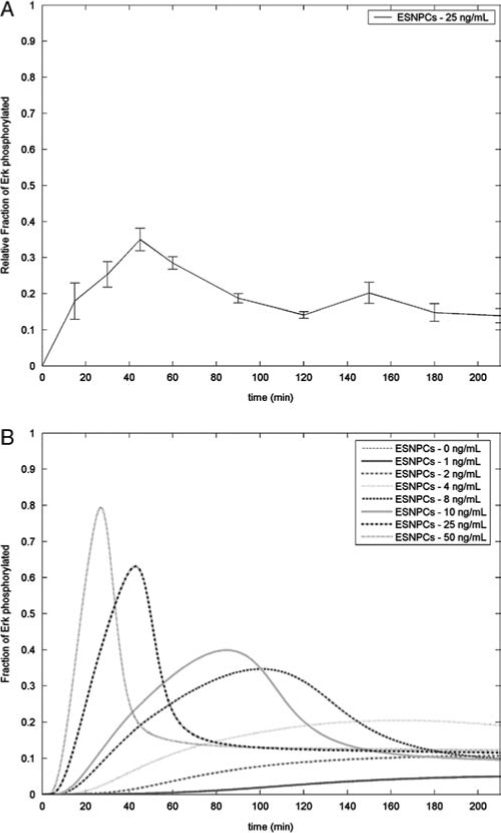
Kinetic analysis of MAP kinase cascade both experimentally and computationally. (A) Experimental characterization of the Erk activation in ESNPCs following stimulation with 25 ng/mL of NT-3. Error bars represent standard deviation. (B) Patterns of Erk activation following stimulation by different NT-3 doses as predicted by the kinetic analysis of the MAP kinase signaling cascade.
Table 2 contains the initial concentrations of the different components of the MAP kinase signaling cascade for HeLa and PC12 cells obtained from the literature. The amount of each protein present was compared between these two cell lines to determine which proteins exhibited large variations in concentration between these two cell lines. Based on these data, it was determined that the concentrations of receptor, Shc, Erk, and Mek vary the most between cell lines and thus will have a greater impact on the mathematical model. Based on the differences observed in expression level, the concentrations of these proteins (receptor, Shc, Erk, and Mek) in ESNPCs were estimated using a combination of quantitative real-time RT-PCR analysis and the PC12 data from the literature. The fold difference was calculated based on the results of the real-time RT-PCR and then used to estimate the concentration of selected proteins in the 4−/4+ EBs. Thus, it was assumed that the mRNA level for each gene expressed was directly proportional to the amount of protein being expressed by the cell line. Western blotting was used to qualitatively confirm this assumption and the results of the RT-PCR experiments. The blots for TrkA/TrkC receptor, Shc, Mek, and Erk expression are shown in Figure 2. The real-time RT-PCR results indicated that the 4−/4+ EBs expressed the TrkC receptor at much lower levels than the PC12 cells expressed the TrkA receptor. The RT-PCR results indicated that the levels of Shc and Erk were slightly higher in the ESNPCs when compared to the PC12 cells. Mek was expressed at much higher levels in the ESNPCs compared to the PC12 cells.
These results were consistent with the protein levels observed using Western blots (Fig. 2). Higher levels of TrkA receptor expression were observed in PC12 cells compared to TrkC receptor expression in the ESNPCs. Both blots were exposed for the same amount of time to allow for visualization of the TrkC receptor, which resulted in more background being observed for the TrkA portion of the blot. The level of Mek expression was also much higher in the ESNPCs when compared to the PC12 cells. Western blotting also indicated that the ESNPCs expressed slightly more Shc and Erk compared to the PC12 cells, supporting the RT-PCR results.
These concentrations were then used as initial values for running the kinetic analysis. The kinetic analysis was performed on concentrations ranging from 0 to 50 ng/mL NT-3 (Fig. 3B). When no NT-3 is present, the model predicted that no Erk will be activated. At the lower doses (1, 2, and 4 ng/mL), the levels of activated Erk increased slowly over time, while only a small percentage (∼5–20%) of the total Erk is activated. At higher doses (8–50 ng/mL), the time course of Erk activation follows a similar pattern to the experimental data shown in Figure 3A, where a large peak is observed followed by a decrease to the steady state levels. Thus, the model mimics the activation of Erk when the ESNPCs are stimulated with 25 ng/mL of NT-3.
Sensitivity analysis was performed to determine the effect of changing the different protein concentrations based on the fold difference estimated using real-time RT-PCR. The results are shown in Figure 4 for each of the four protein concentrations estimated (TrkC receptor, Shc, Mek, and Erk). Changing the concentrations of the downstream components (Mek and Erk) had little effect on the pattern of Erk activation. Changing the concentration of proteins at the beginning of the cascade, such as TrkC receptor and Shc, did have an effect, and when the concentration of TrkC or Shc was increased, activation occurred more rapidly. However, the sensitivity analysis demonstrated that changing these concentrations based on the error as quantified by the standard deviation for these experiments did not result in the appearance of an activation peak when only 2 ng/mL of NT-3 was present or the disappearance of the activation peak when 10 ng/mL of NT-3 was present, suggesting that these two concentrations will evoke different responses from the ESNPCs.
FIG. 4.
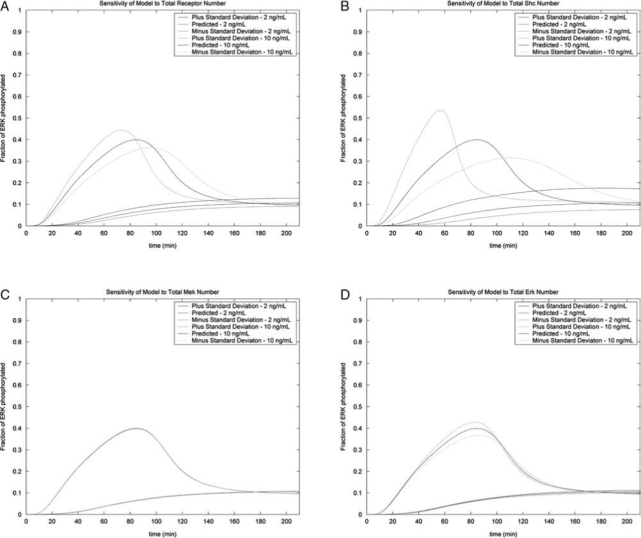
Testing the sensitivity of the analysis in response to changing the different protein concentrations estimated from experimental data. Concentrations were calculated using the fold difference plus or minus the standard deviation of the experiment, and these values were used in the analysis. Responses to 2 and 10 ng/mL are shown in the plots. (A) Sensitivity of the analysis in response to changing receptor concentration. (B) Sensitivity of the analysis in response to changing Shc concentration. (C) Sensitivity of the analysis in response to changing Mek concentration. (D) Sensitivity of the analysis in response to changing Erk concentration.
Prediction of the minimum NT-3 concentration necessary for activation of Erk
Based on the kinetic analysis, two different NT-3 concentrations were selected for further investigation to determine the minimum concentration needed to generate an Erk activation peak. It was hypothesized that stimulation of ESNPCs with 2 ng/mL of NT-3 would not result in a large activation peak, while treatment with 10 ng/mL of NT-3 would produce such a peak. To test this hypothesis, quantitative phosphoErk ELISAs were used to determine the time course of Erk activation for both sets of conditions. The results are shown in Figure 5 along with the activation curves predicted by the kinetic analysis. When the ESNPCs were stimulated with 2 ng/mL of NT-3, only a small fraction of the total Erk was activated (∼10%), and this result was consistent with the activation predicted by the kinetic analysis. When the ESNPCs were stimulated with 10 ng/mL of NT-3, a large peak with ∼50% of the total Erk being activated was observed, with peak activation occurring around 90 min after stimulation. Significantly, more Erk was activated from 45 min until 180 min when the ESNPCs were stimulated with 10 ng/mL NT-3 compared to the amount activated by 2 ng/mL of NT-3 over the same time points. This peak began to decrease after 180 min as predicted by the analysis. The experimental data showed a slower decrease in amount of phosphoErk compared to curve predicted by the model. The activation of Erk occurred more slowly when the ESNPCs were stimulated with 10 ng/mL of NT-3 compared to 25 ng/mL of NT-3, which was also predicted by the kinetic analysis. These results confirmed that the kinetic analysis replicated the trend of intracellular signaling events occurring in the ESNPCs after stimulation with different doses of NT-3.
FIG. 5.
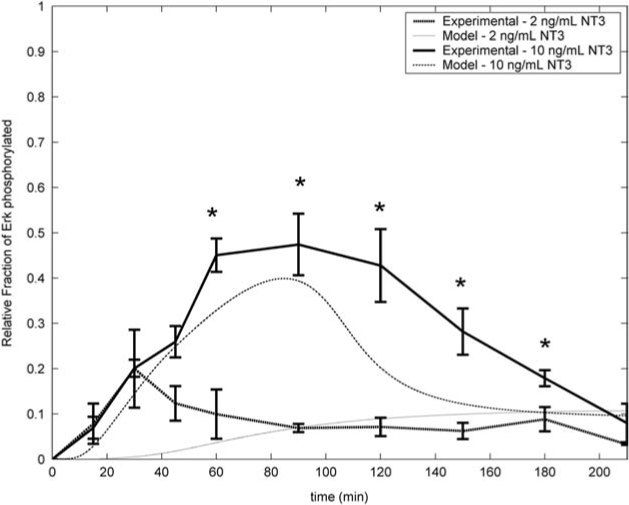
Comparison of the predicted and experimental patterns of Erk activation in ESNPCs following stimulation with 2 and 10 ng/mL of NT-3. *p < 0.05 compared to activation induced by 2 ng/mL NT-3 at the same time point. Error bars indicate standard deviation.
Regulation of the transcription factor Mash1 in response to NT-3 stimulation
To further examine the link between the activation of Erk and the observed neuronal differentiation of the ESNPCs, quantitative real-time RT-PCR was used to look at the expression of the transcription factor Mash1 in response to different doses of NT-3 relative to the basal levels of expression, as previous studies have shown that Mash1 plays an important role in promoting the differentiation of stem cells into neurons. It was assumed that the amount of mRNA present in the ESNPCs correlated to the levels of Mash1 protein expression. The relative amounts of Mash1 mRNA present after NT-3 stimulation are shown in Figure 6A. When the ESNPCs were stimulated with 2 ng/mL of NT-3, the levels of Mash1 expression remained similar to the basal levels (p > 0.05). When the ESNPCs were stimulated with 10 or 25 ng/mL of NT-3, the levels of Mash1 tripled, which is consistent with the activation of Erk observed previously, supporting our hypothesis. The amount of Mash1 observed after stimulation with these two doses was increased compared to ESNPCs treated with 2 ng/mL of NT-3. These results were promising; however, the half-life of mRNA can vary substantially depending on the specific protein, and mRNA concentrations may not reflect the amount of protein produced. Thus, the next step was to confirm that these increased levels of Mash1 expression correlated with neuronal differentiation as indicated by the expression of the β-tubulin III protein.
FIG. 6.
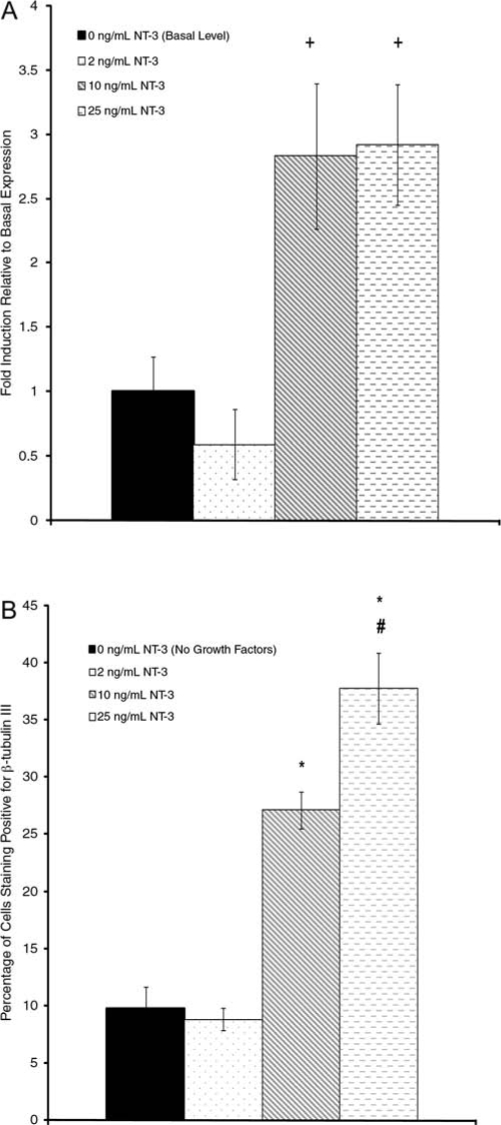
Examining the effects of different NT-3 doses of expression of Mash1 and neuronal differentiation. (A) Relative levels of Mash1 expression after stimulation with different NT-3 doses compared to the basal expression levels. (B) Fraction of ESNPCs differentiating into neurons after stimulation with different NT-3 doses. +p < 0.05 compared to ESNPCs treated with 2 ng/mL of NT-3. *p < 0.05 compared to untreated ESNPCs and ESNPCs treated with 2 ng/mL of NT-3. #p < 0.05 compared to ESNPCs treated with 10 ng/mL of NT-3. Error bars indicate standard deviation.
Quantification of neuronal differentiation of ESNPCs after NT-3 stimulation
To demonstrate the effect of NT-3 on the differentiation state of ESNPCs, FACS was used to quantify the fraction of cells that differentiated in neurons as indicated by β-tubulin III staining following treatment with the different NT-3 doses (Fig. 6B). When no NT-3 was applied to the cells, about 10% of the cells differentiated into neurons. When 2 ng/mL of NT-3 was applied the ESNPCs, similar levels of neuronal differentiation were obtained compared to the untreated ESNPCs (p = 0.96). However, when higher doses of NT-3 (10 and 25 ng/mL) were applied, ∼ 30–40% of the ESNPCs differentiated into neurons, which was significantly more than the untreated ESNPCs and ESNPCs treated with 2 ng/mL of NT-3. Additionally, the fraction of neurons generated by the 25 ng/mL NT-3 (∼38%) dose was significantly greater than the fraction produced by the 10 ng/mL NT-3 dose (∼28%). These data are consistent with both the activation of the MAP kinase cascade and the relative expression levels of Mash1 in this study.
Discussion
The goal of this work was to produce a kinetic analysis of NT-3 signaling in ESNPCs that provides insight into the intracellular processes that promote neuronal differentiation. Many different growth factors and their respective signaling pathways have been implicated in neuronal differentiation, such as the Wnt, bone morphogenic protein, and Shh.34–36 However, this study chose to focus on NT-3 because of its ability to promote neuronal differentiation of stem cells seeded inside of 3D scaffolds for tissue engineering applications and to take advantage of previous work characterizing the MAP kinase cascade.14,24–27,30,32 To achieve this goal of producing a kinetic analysis, it was hypothesized that the mechanism of NT-3–induced differentiation in ESNPCs was due to activation of the MAP kinase cascade, which would promote the production of the transcription factor Mash1. Upregulation of Mash1 has been shown to promote differentiation of mouse ES cells into neurons.19,20 The experimental data in Figure 3A support this hypothesis, showing the activation of MAP kinase cascade in ESNPCs in response to NT-3 stimulation as indicated by levels of phosphoErk present. Based on this experiment, a kinetic analysis of MAP kinase signaling for ESNPCs was implemented using rate constants taken from the literature and select protein concentrations estimated experimentally from mRNA levels. To determine which protein concentrations should be estimated, the differences in concentrations for the signaling pathway components were compared between two cell lines that had already been characterized: HeLa and PC12 cells. If the concentrations were within the same order of magnitude, then the values for the PC12 cells were used in the analysis. Otherwise, real-time RT-PCR was used to estimate the concentration of protein present in the ESNPCs based on mRNA levels.
To estimate the protein concentrations, real-time RT-PCR was used to determine the relative expression of Trk receptor, Shc, Mek, and Erk in PC12 cells and 4−/4+ EBs containing ESNPCs, and theses data were combined with previous data from the literature to approximate the protein concentrations in the ESNPCs. These results were confirmed qualitatively using Western blots. The expression of the TrkA receptor in PC12 was approximately 2.4 times greater than the expression of the TrkC receptor in the ESNPCs. This result is expected because PC12 cells express high levels of the TrkA receptor, which allows them to be responsive to NGF. Conversely, the ESNPCs expressed higher levels of mRNA encoding Shc, Mek, and Erk compared to the PC12 cells. This result followed the same trend like the HeLa cells, which also express more Mek and Erk compared to the PC12 cells. Sensitivity analysis revealed that the concentrations of the upstream components (TrkC receptor and Shc) had a greater effect on the outcome of the kinetic analysis compared to the downstream components (Mek and Erk).
The results of the kinetic analysis can be seen in Figure 3B. The system appears to demonstrate a switch-like behavior where at low concentrations, only small amounts of Erk become activated, and at higher concentrations, a large peak of activated Erk is observed. This shift in activation occurs in between the 4 and 8 ng/mL doses of NT-3. To confirm this kinetic analysis experimentally, two values were chosen on each side of this switch point to test this prediction. A dose of 2 ng/mL of NT-3 was predicted to not activate large amount of Erk, while 10 ng/mL of NT-3 was predicted to produce a peak of Erk activation after stimulation. Sensitivity analysis suggested that this behavior would be observed even if the estimated protein concentrations for the ESNPCs were perturbed as shown in Figure 4. These results were confirmed experimentally by comparing them to the predicted values from the analysis (Fig. 5).
The kinetic analysis accurately predicted what doses of NT-3 would generate an activation peak, and it also predicted that 25 ng/mL NT-3 would activate the MAP kinase cascade more rapidly than 10 ng/mL of NT-3. The analysis predicted that 25 ng/mL of NT-3 would produce a sharp peak in phosphoErk around 45 min after stimulation with ∼60% of the total Erk being phosphorylated. The experimental data showed that this peak did occur at the 45 min time point when ∼40% of the total Erk was phosphorylated. Similarly, for the 10 ng/mL of NT-3 dose, the analysis predicted that activation would occur more slowly around 90 min after NT-3 stimulation with ∼40% of the total Erk activated. The experimental data showed peak activation occurring at 90 min with ∼50% of the total Erk activated. This peak was flatter compared to the sharp peak that was observed for the 25 ng/mL of NT-3. The predicted peak for the 10 ng/mL of NT-3 dose decreased more rapidly than the curve obtained from the experimental data. This result could be due to the error present during quantification of the activated phosphoErk or from the experimental error during the RT-PCR experiments used to estimate the protein concentrations. For the 2 ng/mL dose, the prediction was that the level of Erk activation would rise slowly to ∼10%, while the experimental data showed Erk activation ranging between 10% and 20%. These experimental values showed slightly faster activation initially than the predicted values, but were consistent with the predicted values at later time points.
Previous work has demonstrated that NT-3–mediated activation of the MAP kinase cascade and Mash1 transcription factor resulted in neuronal differentiation of neural stem cells derived from mouse embryos.16,17 The results showed that when a large Erk activation peak was observed, an increase in Mash1 production as indicated by mRNA levels was also observed, suggesting a correlation. These data are consistent with experimental results observed by other groups.17 One possible mechanism to explain this observation is that the phosphoErk can activate Mash1, allowing it to translocate to the nucleus where it promotes further Mash1 transcription, along with transcription of genes that promote neuronal differentiation. Additionally, quantitative analysis using FACS confirmed the influence of the different NT-3 doses on the differentiation of ESNPCs into neurons, further supporting this hypothesis. When higher levels of Mash1 mRNA were observed, the fraction of cells that differentiated into neurons also increased with 25 ng/mL of NT-3 being the most effective dose for generating neurons from the ESNPCs.
It is important to note that our characterization of these cells using FACS is limited to determining which cells express a certain phenotypic marker. Additional characterization of the differentiation state of these cells could be performed by evaluating cell morphology or through the use of functional assays for neuronal activity. Additionally, our hypothesis could be further confirmed by using a TrkC inhibitor to abolish the effect of NT-3–induced activation of the MAP kinase cascade. If such an inhibitor was used, it would be expected that no MAP kinase activation would be observed and that the differentiation of the ESNPCs into neurons would be prevented, confirming our hypothesis. The use of such inhibitors provides another potential method of validating our kinetic analysis. These limitations are important to consider when designing future studies involving kinetic analysis for predicting stem cell differentiation.
This work demonstrates how a kinetic analysis can be developed using experimental data and rate constants obtained from the literature. Such an analysis was implemented and used to predict the minimum concentration of NT-3 needed to induce neuronal differentiation from ESNPCs. Such information is useful for designing engineered tissues. For example, the minimum NT-3 concentration necessary provides an initial starting point when designing scaffold materials containing NT-3 for promoting neuronal differentiation of stem cells. Such analysis can reduce the number of experiments necessary for optimizing such scaffolds and reduce the use of excessive amounts of growth factor.
While this present study was limited to looking at the effects of a single growth factor, this kinetic analysis could be expanded for a variety of applications. The present analysis showed how changing the initial protein concentrations present in a cell line can alter the kinetics of a signaling cascade. Thus, to expand this analysis to other cell lines, such as neural stem cells or mesenchymal stem cells, would require obtaining such experimental data. To generate such an analysis for a different growth factor and signaling cascade would require more effort and would require obtaining receptor/ligand kinetics and rate constants for the individual steps of the cascade. This specific kinetic analysis could be extended to include the presence of additional growth factors, such as platelet-derived growth factor (PDGF). PDGF has been shown to have synergistic effects when used in combination with NT-3 treatment for promoting differentiation of ESNPCs into both neurons and oligodendrocytes.30 More recently, PDGF has also been shown to activate the Erk signaling pathway, promoting oligodendrocyte differentiation.37 Thus, the current kinetic analysis could be extended to include the presence of PDGF and its receptor along with the transcription factors associated with the specific lineages, such as Mash1 and Olig2, for predicting the fraction of cells differentiating into neurons and oligodendrocytes, respectively. One of the limitations of the current analysis is that it does not take into account the presence of other growth factors and signaling cascades that could influence ESNPC differentiation.
In conclusion, this study has developed a kinetic analysis that replicates the activation of the MAP kinase cascade in ESNPCs derived from ES cells. The analysis was successfully used to predict the concentration of NT-3 that would be required to promote neuronal differentiation of these cells as confirmed by both expression of the transcription factor Mash1 and expression of a neuronal marker (β-tubulin III). Such models help give understanding into how intracellular signaling can be used to predict stem cell behavior. This specific analysis can be applied to promoting ESNPC differentiation for tissue engineering applications for the treatment of SCI. Additionally, this approach provides one way of looking at the effects of specific growth factors on stem cell differentiation.
Acknowledgments
This work was supported by NIH R01 NS051454. The authors thank Shengzhou Wu and the Hope Center for Neurological Disorders, which is supported by NIH Neuroscience Blueprint Interdisciplinary Center Core grant (P30 NS057105), for use of the real-time PCR machine, and Allison Rader and Jacob Riehm for technical assistance.
References
- 1.Elisseeff J.H. Embryonic stem cells: potential for more impact. Trends Biotechnol. 2004;22:155. doi: 10.1016/j.tibtech.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Polak J.M. Bishop A.E. Stem cells and tissue engineering: past, present, and future. Ann NY Acad Sci. 2006;1068:352. doi: 10.1196/annals.1346.001. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill A. Schaffer D.V. The biology and engineering of stem-cell control. Biotechnol Appl Biochem. 2004;40:5. doi: 10.1042/BA20030195. [DOI] [PubMed] [Google Scholar]
- 4.Prudhomme W.A. Duggar K.H. Lauffenburger D.A. Cell population dynamics model for deconvolution of murine embryonic stem cell self-renewal and differentiation responses to cytokines and extracellular matrix. Biotechnol Bioeng. 2004;88:264. doi: 10.1002/bit.20244. [DOI] [PubMed] [Google Scholar]
- 5.Prudhomme W. Daley G.Q. Zandstra P. Lauffenburger D.A. Multivariate proteomic analysis of murine embryonic stem cell self-renewal versus differentiation signaling. Proc Natl Acad Sci USA. 2004;101:2900. doi: 10.1073/pnas.0308768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolf P.J. Prudhomme W. Daheron L. Daley G.Q. Lauffenburger D.A. Bayesian analysis of signaling networks governing embryonic stem cell fate decisions. Bioinformatics. 2005;21:741. doi: 10.1093/bioinformatics/bti056. [DOI] [PubMed] [Google Scholar]
- 7.Zandstra P.W. Lauffenburger D.A. Eaves C.J. A ligand-receptor signaling threshold model of stem cell differentiation control: a biologically conserved mechanism applicable to hematopoiesis. Blood. 2000;96:1215. [PubMed] [Google Scholar]
- 8.Viswanathan S. Benatar T. Rose-John S. Lauffenburger D.A. Zandstra P.W. Ligand/receptor signaling threshold (LIST) model accounts for gp130-mediated embryonic stem cell self-renewal responses to LIF and HIL-6. Stem Cells. 2002;20:119. doi: 10.1634/stemcells.20-2-119. [DOI] [PubMed] [Google Scholar]
- 9.Lai K. Robertson M.J. Schaffer D.V. The sonic hedgehog signaling system as a bistable genetic switch. Biophys J. 2004;86:2748. doi: 10.1016/S0006-3495(04)74328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha K. Schaffer D.V. Signal dynamics in sonic hedgehog tissue patterning. Development. 2006;133:889. doi: 10.1242/dev.02254. [DOI] [PubMed] [Google Scholar]
- 11.Lachyankar M.B. Condon P.J. Quesenberry P.J. Litofsky N.S. Recht L.D. Ross A.H. Embryonic precursor cells that express Trk receptors: induction of different cell fates by NGF, BDNF, NT-3, and CNTF. Exp Neurol. 1997;144:350. doi: 10.1006/exnr.1997.6434. [DOI] [PubMed] [Google Scholar]
- 12.Averbuch-Heller L. Pruginin M. Kahane N. Tsoulfas P. Parada L. Rosenthal A. Kalcheim C. Neurotrophin 3 stimulates the differentiation of motoneurons from avian neural tube progenitor cells. Proc Natl Acad Sci USA. 1994;91:3247. doi: 10.1073/pnas.91.8.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C.S. Tee L.Y. Dusenbery S. Takata T. Golden J.P. Pierchala B.A. Gottlieb D.I. Johnson E.M., Jr. Choi D.W. Snider B.J. Neurotrophin and GDNF family ligands promote survival and alter excitotoxic vulnerability of neurons derived from murine embryonic stem cells. Exp Neurol. 2005;191:65. doi: 10.1016/j.expneurol.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Levenberg S. Burdick J.A. Kraehenbuehl T. Langer R. Neurotrophin-induced differentiation of human embryonic stem cells on three-dimensional polymeric scaffolds. Tissue Eng. 2005;11:506. doi: 10.1089/ten.2005.11.506. [DOI] [PubMed] [Google Scholar]
- 15.Barnabe-Heider F. Miller F.D. Endogenously produced neurotrophins regulate survival and differentiation of cortical progenitors via distinct signaling pathways. J Neurosci. 2003;23:5149. doi: 10.1523/JNEUROSCI.23-12-05149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim M.S. Nam S.H. Kim S.J. Kang S.Y. Lee Y.S. Kang K.S. Signaling pathways of the early differentiation of neural stem cells by neurotrophin-3. Biochem Biophys Res Commun. 2007;357:903. doi: 10.1016/j.bbrc.2007.04.045. [DOI] [PubMed] [Google Scholar]
- 17.Ito H. Nakajima A. Nomoto H. Furukawa S. Neurotrophins facilitate neuronal differentiation of cultured neural stem cells via induction of mRNA expression of basic helix-loop-helix transcription factors Mash1 and Math1. J Neurosci Res. 2003;71:648. doi: 10.1002/jnr.10532. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama R. Ohtsuka T. Hatakeyama J. Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda R. Kurokawa M.S. Chiba S. Yoshikawa H. Hashimoto T. Tadokoro M. Suzuki N. Transplantation of motoneurons derived from MASH1-transfected mouse ES cells reconstitutes neural networks and improves motor function in hemiplegic mice. Exp Neurol. 2004;189:280. doi: 10.1016/j.expneurol.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 20.Hamada M. Yoshikawa H. Ueda Y. Kurokawa M.S. Watanabe K. Sakakibara M. Tadokoro M. Akashi K. Aoki H. Suzuki N. Introduction of the MASH1 gene into mouse embryonic stem cells leads to differentiation of motoneuron precursors lacking Nogo receptor expression that can be applicable for transplantation to spinal cord injury. Neurobiol Dis. 2006;22:509. doi: 10.1016/j.nbd.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Tebar A. Dechant G. Gotz R. Barde Y.A. Binding of neurotrophin-3 to its neuronal receptors and interactions with nerve growth factor and brain-derived neurotrophic factor. EMBO J. 1992;11:917. doi: 10.1002/j.1460-2075.1992.tb05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tischler A.S. Greene L.A. Nerve growth factor-induced process formation by cultured rat pheochromocytoma cells. Nature. 1975;258:341. doi: 10.1038/258341a0. [DOI] [PubMed] [Google Scholar]
- 23.Greene L.A. Tischler A.S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aksan I. Kurnaz M.L. A computer-based model for the regulation of mitogen activated protein kinase (MAPK) activation. J Recept Signal Transduct Res. 2003;23:197. doi: 10.1081/rrs-120025203. [DOI] [PubMed] [Google Scholar]
- 25.Qiu D. Mao L. Kikuchi S. Tomita M. Sustained MAPK activation is dependent on continual NGF receptor regeneration. Dev Growth Differ. 2004;46:393. doi: 10.1111/j.1440-169x.2004.00756.x. [DOI] [PubMed] [Google Scholar]
- 26.Brightman F.A. Fell D.A. Differential feedback regulation of the MAPK cascade underlies the quantitative differences in EGF and NGF signalling in PC12 cells. FEBS Lett. 2000;482:169. doi: 10.1016/s0014-5793(00)02037-8. [DOI] [PubMed] [Google Scholar]
- 27.Yamada S. Taketomi T. Yoshimura A. Model analysis of difference between EGF pathway and FGF pathway. Biochem Biophys Res Commun. 2004;314:1113. doi: 10.1016/j.bbrc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 28.McDonald J.W. Liu X.Z. Qu Y. Liu S. Mickey S.K. Turetsky D. Gottlieb D.I. Choi D.W. Transplanted embryonic stem cells survive, differentiate and promote recovery in injured rat spinal cord. Nat Med. 1999;5:1410. doi: 10.1038/70986. [DOI] [PubMed] [Google Scholar]
- 29.Bain G. Kitchens D. Yao M. Huettner J.E. Gottlieb D.I. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- 30.Willerth S.M. Faxel T.E. Gottlieb D.I. Sakiyama-Elbert S.E. The effects of soluble growth factors on embryonic stem cell differentiation inside of fibrin scaffolds. Stem Cells. 2007;25:2235. doi: 10.1634/stemcells.2007-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willerth S.M. Arendas K.J. Gottlieb D.I. Sakiyama-Elbert S.E. Optimization of fibrin scaffolds for differentiation of murine embryonic stem cells into neural lineage cells. Biomaterials. 2006;27:5990. doi: 10.1016/j.biomaterials.2006.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoeberl B. Eichler-Jonsson C. Gilles E.D. Muller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20:370. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 33.Willerth S.M. Faxel T.E. Gottlieb D.I. Sakiyama-Elbert S.E. The effects of soluble growth factors on embryonic stem cell differentiation inside of fibrin scaffolds. Stem Cells. 2007;25:2235. doi: 10.1634/stemcells.2007-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frebel K. Wiese S. Signalling molecules essential for neuronal survival and differentiation. Biochem Soc Trans. 2006;34:1287. doi: 10.1042/BST0341287. [DOI] [PubMed] [Google Scholar]
- 35.Pozniak C.D. Pleasure S.J. A tale of two signals: Wnt and Hedgehog in dentate neurogenesis. Sci STKE 2006. 2006:pe5. doi: 10.1126/stke.3192006pe5. [DOI] [PubMed] [Google Scholar]
- 36.Salie R. Niederkofler V. Arber S. Patterning molecules; multitasking in the nervous system. Neuron. 2005;45:189. doi: 10.1016/j.neuron.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Hu J.G. Fu S.L. Wang Y.X. Li Y. Jiang X.Y. Wang X.F. Qiu M.S. Lu P.H. Xu X.M. Platelet-derived growth factor-AA mediates oligodendrocyte lineage differentiation through activation of extracellular signal-regulated kinase signaling pathway. Neuroscience. 2008;151:138. doi: 10.1016/j.neuroscience.2007.10.050. [DOI] [PubMed] [Google Scholar]
- 38.Bhalla U.S. Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 39.Kholodenko B.N. Demin O.V. Moehren G. Hoek J.B. Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem. 1999;274:30169. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]


