Abstract
After DNA damage, caspases cleave and activate proteins involved in cell death by apoptosis but also cleave and inactivate proteins implicated in DNA repair. Here we report a rapid onset of Rad51 cleavage by caspase 3 in BRCA2-defective mouse and human cells. This rapid cleavage was reduced markedly by transfer of full-length human BRCA2 into BRCA2-defective mouse or human cells, which also blocked the association of caspase 3 and Rad51 proteins. Overall caspase 3 activity was increased in BRCA2-defective cells, but the time course was much slower than that for Rad51 cleavage. We further showed that caspase 3 cleavage of Rad51 resulted in a functional decrease in Rad51 strand exchange activity and that inhibition of caspase 3 activity increased Rad51 protein levels and Rad51 foci. These findings indicate that BRCA2 inhibits Rad51 cleavage and subsequent apoptosis.
INTRODUCTION
DNA damage in cells results in either DNA repair or cell death by apoptosis. Regulation of the cell’s decision between repair and apoptosis involves cell cycle checkpoints and DNA repair mechanisms and is essential for the maintenance of genomic integrity (1). Ionizing radiation, which produces double-strand DNA breaks, activates caspases, a family of aspartic acid-specific cysteine proteases (2–4). One member, caspase 3, cleaves the DNA repair proteins poly(ADP-ribose) polymerase (PARP) (5), DNA-PK (6), ATM (7) and RAD51 (8, 9). It specifically cleaves RAD51 at the DVLD-187 site (aspartic acid-valine-leucine-aspartic acid) (9). Thus caspase 3 both activates cell death and inhibits DNA repair.
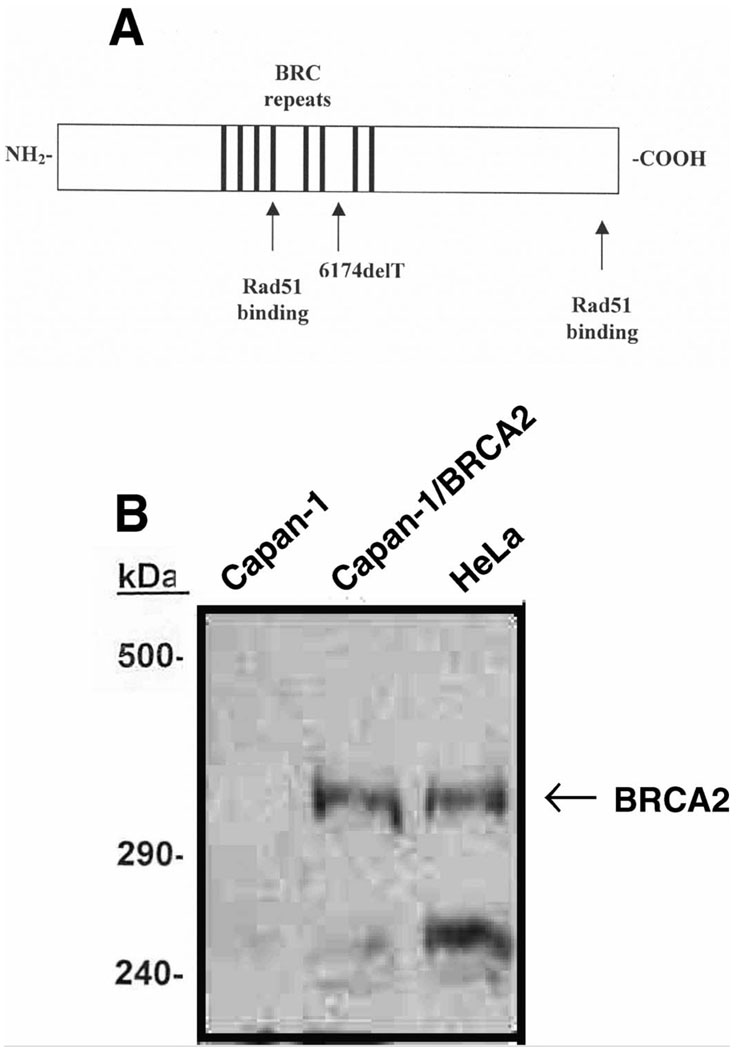
Two protein domains of BRCA2 bind Rad51: (1) a central domain containing BRC repeats (10–12) and (2) a C-terminal domain regulated by CDK-dependent phosphorylation (13). BRC peptides bind Rad51 monomers and block formation of nucleoprotein filaments required for recombinase activity (10–12). BRCA2 and Rad51 co-localize to nuclear foci where DNA repair likely occurs (14–16), but localization of Rad51 to foci is reduced in cell lines lacking BRCA2 (17–19). These findings support a model in which Rad51 and BRCA2 interact with each other and with damaged DNA (the structure of the BRCA2 gene is shown in Fig. 1A).
FIG. 1.
Structure and expression of BRCA2. Panel A: Human BRCA2 contains 3418 amino acids. The central region contains a series of eight conserved BRC repeats that bind RAD51. The 6174delT mutation produces a frameshift and premature termination that disrupts the last two BRC repeats and introduces a premature stop codon. Panel B: Western blot of 35S-labeled cells with anti-human C-terminal BRCA2 antibody. Lane 1: untransfected Capan-1 cells; lane 2: Capan-1 cells transfected with human BRCA2 cDNA; lane 3: HeLa cells.
We investigated the effects of BRCA2 expression on the stability of Rad51. We have hypothesized that BRCA2 binds Rad51 and enhances the stability of Rad51 protein by protecting it from proteolytic cleavage. Here we report a rapid onset of Rad51 cleavage by caspase 3 in BRCA2-defective mouse and human cells. This rapid cleavage is markedly reduced by transfer of full-length human BRCA2 into BRCA2-defective mouse or human cells. Overall caspase 3 activity is increased in BRCA2-defective cells, but the time course is much slower than that observed for Rad51 cleavage. We further show that caspase 3 cleavage of Rad51 results in a functional decrease in Rad51 strand exchange activity. BRCA2 appears to inhibit the usual association between caspase 3 and Rad51, and inhibition of caspase 3 activity increases Rad51 protein levels and increases Rad51 foci. These findings indicate that BRCA2 inhibits Rad51 cleavage and subsequent apoptosis.
MATERIALS AND METHODS
Cell Culture
Capan-l, BRCA2lexl/lex2 [Brca2− mouse embryonic fibroblast (MEF) cells, obtained from Dr. Paul Hasty, Lexicon Genetics] and MCF-7 cells were maintained at 37°C in 95% air/5% CO2. Capan-l cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 20% Fetal Clone (Hyclone), 1% antibiotic/antimycotic solution, and 1% glutamine. BRCA2lex1/lexl (Brca2−) cells were grown in DMEM supplemented with 10% Fetal Clone, 1% antibiotic/antimycotic solution, and 1% glutamine. MCF-7 cells were grown in DMEM supplemented with 10% certified fetal bovine serum (Gibco-BRL), 1% antibiotic/antimycotic, 1% glutamine, 1% insulin transferrin selenium A (ITSA), and 1% nonessential amino acids.
Transfections and Generation of Stable Cell Lines
Full-length human BRCA2 cDNA (confirmed by DNA sequence) was cloned into pREP4 to produce an RSV-regulated expression vector. Transient and stable transfections were performed in Capan-l cells by lipofection using Targefect F-l (Targeting Systems) and in Brca2− cells by a calcium phosphate-mediated protocol. Stable transfectants were pooled (at least 100 clones for each transfectant pool). BRCA2 expression was confirmed by Western blot analysis as below using an antibody raised against the C-terminus of BRCA2 (BRCA2, amino acids 3245–3418, Pharmingen). Wild-type and non-cleavable GFP-tagged RAD51 constructs were a generous gift from Yinyin Huang (Dana-Farber Cancer Institute, Harvard Medical Institute).
Western Blotting Analyses
Cell lysates were subjected to polyacrylamide gel electrophoresis (12% gel) and transferred to PVDF filters (Millipore, Immobilon-P) overnight at 30 V. The membrane was stained with Ponceau S prior to blocking to assess loading and transfer efficiency. The membrane was blocked using 5% nonfat dry milk, 0.2% Tween 20 in Tris-buffered saline for 1 h at room temperature. After blocking and subsequent washing, the blot was exposed to the primary antibody. We used affinity-purified rabbit anti-human RAD51 (Ab-1, Oncogene Research Products) at a dilution of 1:2500, affinity-purified rabbit anti-human BRCA2 (anti-BRCA2, amino acids 3245–3418, Pharmingen) at a dilution of 1:1000, and affinity-purified rabbit anti-human caspase 3 antibody (Pharmingen) at a dilution of 1:2000 as primary antibodies. The blot was washed with 0.2% Tween in Tris-buffered saline for 1 h at room temperature before being exposed to a horseradish peroxidase-conjugated secondary antibody (Jackson Immuno Research) at a dilution of 1:7500 at room temperature for 1 h. The blot was again washed extensively. Bands were visualized using chemiluminescence detection employing Luminol (3-aminophthalydrazide) and p-coumaric acid. Immunoprecipitation-Western blot analyses were performed by first performing the immunoprecipitation in 1.5 ml of RIPA buffer, using Protein A sepharose for antibody capture, and then performing Western blotting as described above for each antibody.
Measurement of Caspase 3 Activity
Aliquots of cells frozen at the indicated times after irradiation were processed for enzymatic assay as described elsewhere (22). Cell lysates were centrifuged at 900g for 10 min at 4°C. Protease assays included 178 µl of reaction buffer (100 mM Hepes, pH7.5, 20% v/v glycerol, 5 mM dithiothreitol, and 0.5 mM EDTA), 2 µl of 10 mM acetyl-Asp-Glu-Val-Asp-p-nitroanilide (Ac-DEVD-pNA) in Me2SO (100 µM final concentration), and 20 µl of cell lysate. Caspase 3 activity was determined by enzymatic conversion of DEVD-p-nitroaniline to p-nitroaniline measured at 405 nm.
Protein Purification and DNA Strand Exchange Studies
Vectors were constructed that expressed human RAD5l and caspase 3 in E. coli using the pGex5 GST vector that allows processing of the GST after purification with factor Xa. GST-RAD51 was purified from over expressing bacteria by spermidine precipitation and dialysis (23) and purification on a glutathione sepharose column followed by Xa treatment and elution with buffer. GST-caspase 3 was purified on glutathione sepharose followed by Xa cleavage and elution with buffer (100 mM NaCl and 2 mM DTT) and was allowed to cool slowly to room temperature. Strand exchange assays were performed as described elsewhere (24–26), with the following modifications: 30 µM single-stranded phiX174 DNA was incubated at 37°C for 3 min before Rad51 or Rad51 plus caspase 3 were added. In strand exchange assays with RAD51, 10 µM protein was used. For assays with both RAD51 and caspase 3, equimolar amounts of both proteins were preincubated together in reaction buffer for 30 min at 37°C prior to addition to the single-stranded DNA. The mixture of proteins with single-stranded DNA was incubated for 5 min before 30 µM linear duplex RF1 ΦX174 DNA (New England Biolabs) was added with a final reaction buffer of 50 mM triethanolamine-HCl (pH 7.5), 80 mM Koac, 0.5 mM Mg (OAc)2, 2 mM ATP, 1 mM DTT, and 100 µg/ml bovine serum albumin. Then 100 mM ammonium sulfate was added after equilibration of the prior reagents. These were then incubated a further 2 h at 37°C. ATP was omitted from the control as a no ATP control, and 20 µM DEVD was added to the protein mixture in the DEVD control. Reactions were terminated by addition of 0.2 volume stop buffer (100 mM Tris-HCl, pH 7.5, 100 mM MgCl2, 25 mg/ml SDS, 10 mg/ml proteinase K, 2.5 µg/ml ethidium bromide) and analyzed on 1% agarose gels. To stabilize joint molecules, ethidium bromide (1 µg/ml) was added to the gel and gel running buffer, photographed and quantified by digital photography.
Immunofluorescence Microscopy
The cells were seeded and allowed to grow on cover slips in six-well tissue culture plates. After treatment, the cells were fixed in 2% paraformaldehyde, neutralized with 1 M glycine-Tris, pH 7.4, and washed in PBS for 5 min. Then the cells were probed with an affinity-purified rabbit anti-human RAD51 antibody (Ab-l, Oncogene Research Products) diluted 1:5000 in dilution buffer (5% normal donkey serum, 1% BSA-Fraction V and 0.1% Triton X-l00 in PBS), washed three times for 5 min in PBS, probed with Cy3-conjugated anti-rabbit secondary antibody (Jackson Laboratory) diluted 1:8000 in dilution buffer, and washed three times for 5 min in PBS. The fixed cells were analyzed with a Zeiss Axiophot fluorescence microscope.
RESULTS
Ionizing Radiation Induces a Rapid Caspase 3 Cleavage of Rad51 in BRCA2-Defective Cells
Huang (9) reported previously that caspase 3 produced a slow cleavage of RAD51 in irradiated U-937 cells. Given the interaction between RAD51 and BRCA2, we hypothesized that BRCA2 might affect caspase 3 cleavage of Rad51. For these studies we used cells of two BRCA2-defective cell lines: (1) Capan-l cells, a human pancreatic carcinoma cell line lacking the C-terminus of BRCA2, and (2) BRCA2lexl/lex2 mouse embryo fibroblast cells, also lacking the Brca2 C-terminus (Brca2−) (20). Capan-l cells also contain defective p53, but the Brca2− cells are primary cells and contain intact p53. The sequences of human and mouse RAD51 are almost identical, differing by only 4 amino acids (21); therefore, interactions between mouse RAD51 and human BRCA2 would still be likely to occur.
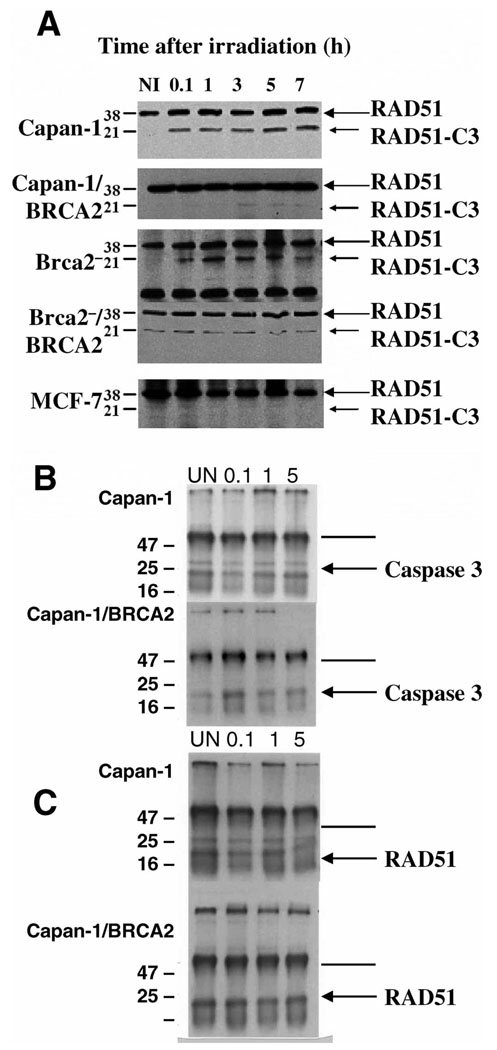
Pooled stable transfectants expressing BRCA2 (protein expression analyzed in Fig. 1B) were isolated and Rad51 cleavage was analyzed in cells with or without wild-type BRCA2. The level of expression in the BRCA2 transfectants was comparable to that seen in epithelial cells. Irradiation of both Capan-l and Brca2− cells induced a rapid cleavage of RAD5l within 6 min (Fig. 2A, panels 1 and 3). However, transfection of either mouse or human BRCA2-defective cells with wild-type human BRCA2 resulted in decreased cleavage of RAD51, with BRCA2-tranasfected Brca2− cells showing a consistently low basal activity (Fig. 2A, panels 2 and 4). Control MCF-7 cells, which lack caspase 3 activity, exhibited no detectable cleavage of RAD51 (Fig. 2A, panel 5). These results indicate that full-length, wild-type BRCA2 is necessary for RAD51 to avoid significant caspase 3 cleavage and remain intact.
FIG. 2.
Panel A: Caspase 3 cleavage of RAD51 after irradiation. Western blot showing caspase 3-mediated cleavage of RAD51 (large arrow, 38 kDa) to RAD51-C3 (small arrow, 21 kDa) after exposure to 10 Gy of ionizing radiation (time in hours). NI is the nonirradiated control; 0.1 represents an aliquot processed 6 min after irradiation (i.e., as rapidly as possible). Polyclonal RAD51 antibody detects the 38-kDa RAD51 and 21-kDa cleavage product. Panel 1, Capan-1 cells; panel 2, Capan-1 cells transfected with a full-length human BRCA2 cDNA; panel 3, Brca2− cells; panel 4, Brca2− cells transfected with BRCA2 cDNA; panel 5, MCF-7 cells, which reportedly lack caspase 3 activity (9). Samples were standardized by loading equal concentrations of protein. Panel B: Caspase 3 immunoprecipitation (IP)-Western blot showing association of Rad51 with caspase 3 before and after irradiation (time in hours). Immunoprecipitation with Rad51 antibody, immunoprecipitates fractionated by SDS-PAGE and blotted with a 1:2000 dilution of caspase 3 antibody. Upper panel, Capan-1 cells; lower panel, BRCA2-transfected Capan-1 cells, standardized by loading equal protein. Arrow indicates 32-kDa human caspase 3 protein. Panel C: Rad51 IP-Western blot. Immunoprecipitation with caspase 3 antibody, then blotting with Rad51 antibody (1:2500). Arrow indicates the 36-kDa human Rad51 protein. In panels B and C, the darker line denotes heavy-chain and the lighter line light-chain immunoglobulin bands.
We next asked whether ionizing radiation induced a rapid caspase 3/Rad51 association and subsequent cleavage in BRCA2-defective cell lines or whether radiation induced caspase 3 cleavage of prebound Rad51. Our results showed that caspase 3 is associated with Rad51 in human BRCA2-defective Capan-1 cells prior to irradiation (Fig. 2B and C, upper panel) but is not detected in BRCA2-transfected Capan- 1 cells (Fig. 2B and C, lower panel). This association was seen both by immunoprecipitation with Rad51 antibody followed by immunoblotting with caspase 3 antibody (Fig. 2B) and by immunoprecipitation with caspase 3 antibody followed by immunoblotting with Rad51 (Fig. 2C). These association assays could be performed in human Capan-1 cells but not in mouse Brca2− (null) cells, because no mouse caspase 3 antibody is available.
BRCA2 Inhibits Generalized Caspase 3 Activity
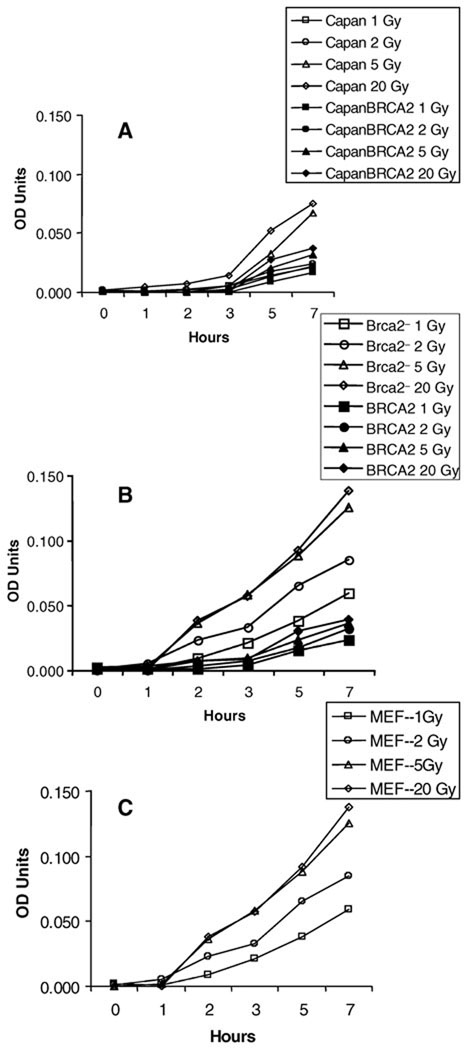
Several potential models could explain how loss of BRCA2 might increase caspase 3 cleavage of Rad51: (1) a non-specific rapid increase in caspase 3 activity after exposure to ionizing radiation; (2) a specific increase only in cleavage of Rad51 or a few yet unknown substrates; (3) an initial rapid increase in Rad51 cleavage followed by a more generalized increase in caspase 3 activity and/or apoptosis. Thus we wanted to compare the time course of caspase 3 activity after irradiation of BRCA2-defective cells with that of cells complemented with BRCA2. Capan-1 cancer cells (Fig. 3A) and mouse Brca2− fibroblasts (Fig. 3B) were analyzed using a chromogenic substrate; the results showed a slow, dose-dependent increase in overall caspase 3 activity after irradiation. Because there was a clear difference in caspase 3 activity in BRCA2-transfected and untransfected cells, we also measured caspase 3 activity in irradiated mouse embryo fibroblasts without a defect in Brca2 (Fig. 3C). The results show that Brca2− cells do not have grossly different caspase 3 responses to radiation when compared to control mouse embryo fibroblasts but that overexpression of BRCA2 appears to decrease radiation-induced caspase 3 activity in a dose-dependent manner.
FIG. 3.
Caspase 3 activity in irradiated BRCA2-defective and transfected cells. panel A: Caspase 3 activity in irradiated Capan-1 cells by identical assay. Panel B: Caspase 3 activity in irradiated Brca2− cells. Panel C: Caspase 3 activity in irradiated mouse embryo fibroblasts. OD on the Y axis is caspase 3 activity as measured by enzymatic conversion of DEVD-p-nitroaniline to p-nitroaniline measured at 405 nm. Points are triplicate determinations.
Caspase 3 Cleavage of RAD51 Reduces Strand Exchange Activity
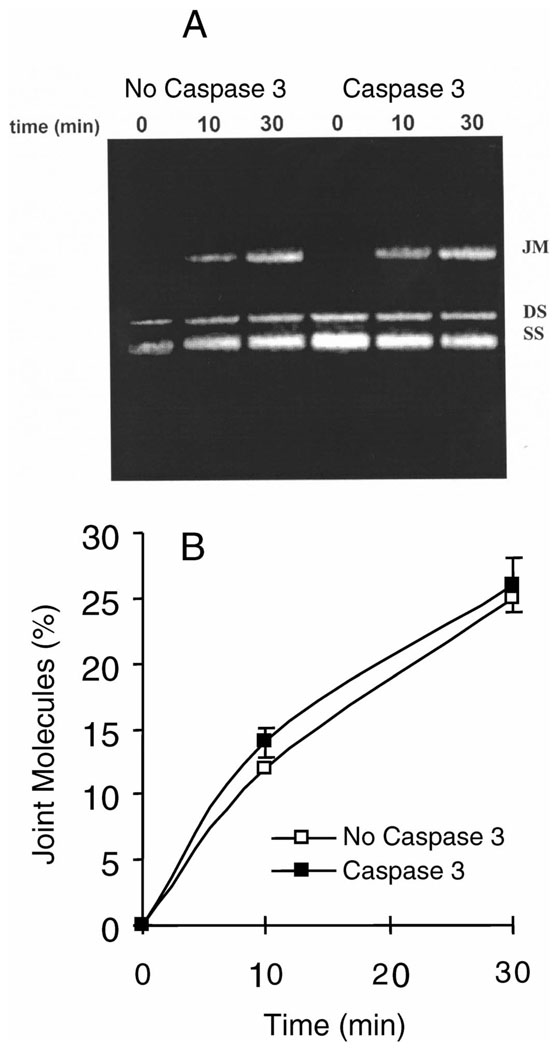
After we observed the immediate cleavage of RAD51 by caspase 3 after exposure to ionizing radiation when wild-type BRCA2 was not present, we determined the effects of caspase 3 cleavage of RAD51 on the efficiency of DNA repair. For these experiments, strand exchange assays were performed; these in vitro assays model the strand invasion step of homologous recombination that is catalyzed by RAD51. Purified RAD51, Rad51 D-A mutant, and caspase 3 proteins were prepared. The efficiency of RAD51 in mediating strand exchange in the absence and presence of caspase 3 was assessed by determining the extent of homologous DNA pairing in single-stranded and linear double-stranded ΦX174 molecules. Figure 4A and B shows that Rad51 protein-mediated production of joint molecules is inhibited by caspase 3 protein, but this inhibition is reversed by adding the caspase inhibitor DEVD. After we assessed the decrease in strand exchange activity resulting from RAD51 being cleaved by caspase 3, we wanted to know how the efficiency of strand exchange would be affected if RAD5l was not vulnerable to caspase 3. Strand exchange assays were performed (Fig. 5A, B) with a recombinant, non-cleavable form of RAD51 (denoted as D-A RAD51) in which the DVLD-187 sequence was mutated to AVLA, thereby making the site unrecognizable by caspase 3. The assays were performed in the same manner as those with wild-type RAD51. The results indicated that Rad51 D-A protein-mediated joint molecule production is not inhibited by caspase 3 protein.
FIG. 4.
Homologous DNA pairing and strand exchange by Rad51 and caspase 3. ΦX174 viral single-stranded DNA (SS) is paired with the linear homologous duplex ΦX174 DNA (DS) to form joint molecules (JM). Panel A: Ethidium bromide-stained agarose gel showing bands after different times of incubation of Rad51 protein with single-strand and double-strand DNA in the presence and absence of caspase 3, or when caspase 3 and its inhibitor DEVD were both present in the reactions. −ATP indicates the control reaction in the absence of added ATP. Panel B: Quantification of percentage conversion of duplex DNA to joint molecules by Rad51. Experiments were repeated three times with equivalent results. Means and SE are shown.
FIG. 5.
Homologous DNA pairing and strand exchange shown graphically. Panel A: Ethidium bromide-stained agarose gel showing bands after incubation of caspase-resistant mutant Rad51 D-A protein with single-strand and double-strand DNA in the presence and absence of caspase 3. Panel B: Quantification of percentage conversion of duplex DNA to joint molecules by caspase 3-resistant Rad51 D-A. Experiments were repeated three times with equivalent results. Means and SE are shown.
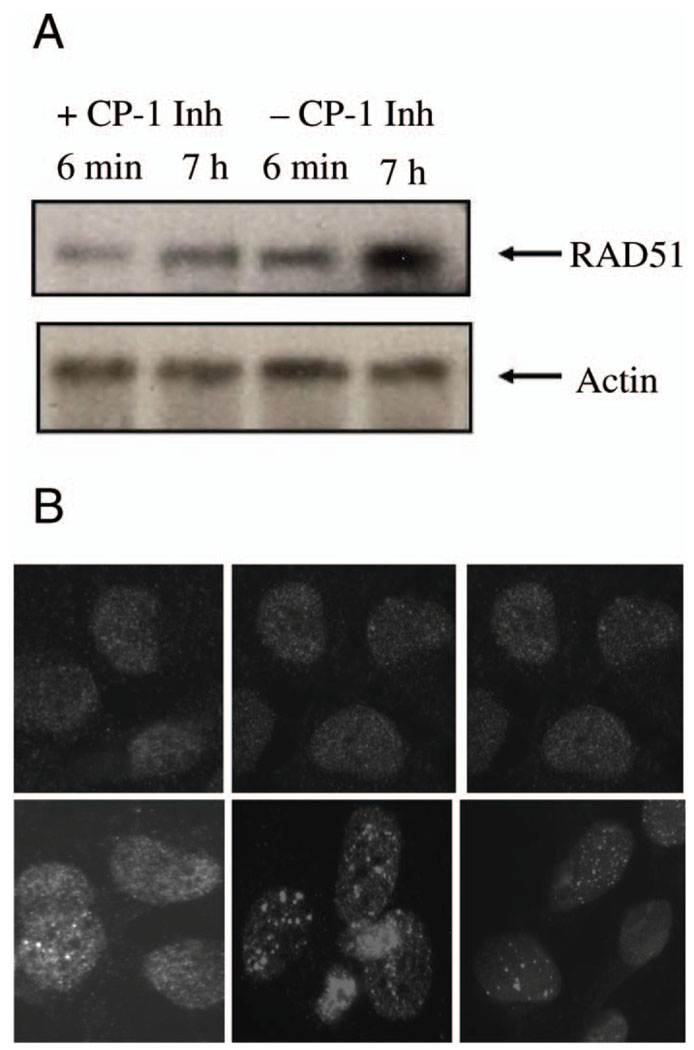
Inhibition of Caspase 3 Activity In Vivo Increases Rad51 Protein Levels and Rad51 Foci
To determine whether caspase 3 cleavage of Rad51 plays a role in BRCA2-defective cells in vivo, we analyzed Rad51 activity and function in Capan-1 cells treated with caspase inhibitors. The results in Fig. 6A show that the levels of Rad51 protein in irradiated cells increase to a much greater extent in Capan-1 cells treated with the caspase 3 inhibitor DEVD than in cells treated with a caspase 1 inhibitor. This indicates that caspase 3 activity functionally decreases RAD51 protein levels in these cells. We then determined the extent of Rad51 focus formation in irradiated Capan-1 cells in the presence of caspase inhibitors and observed more numerous and larger foci in cells treated with a caspase 3 inhibitor (Fig. 6B, lower middle panel) than in caspase 1 inhibitor-treated cells (Fig. 6B, lower left panel) or in control cells (Fig. 6B, lower right panel).
FIG. 6.
Effect of caspase 3 inhibition on Rad51 levels and function in vivo. Panel A: Western blots showing Rad51 (upper panels) and control actin (lower panels) protein levels 6 min and 7 h after irradiation with 10 Gy. Results in the presence of caspase 1 inhibitor (+CP-1 Inh) are compared with those observed with the caspase 3 inhibitor DEVD (+CP-3 Inh). Panel B: Rad51 immunofluorescence experiment showing the presence of Rad51 protein and postirradiation focus formation. Upper panels are results without radiation and lower panels are results 4 h after irradiation with 10 Gy. The left panels are untreated cells, center panels are cells treated with caspase 3 inhibitor, and right panels are cells treated with caspase 1 inhibitor. Upper panels were photographed for a longer period to show Rad51 protein.
DISCUSSION
Previous studies have suggested that maintenance of intact RAD51 contributes to abrogation of apoptosis (9). This suggests that RAD5l must avoid caspase 3 cleavage for the cell to survive an apoptosis-initiating event. Our results lead to the following conclusions: (1) Ionizing radiation produces a rapid cleavage of Rad51 by caspase 3 that is inhibited by BRCA2 expression; (2) BRCA2 appears to block the association of Rad51 and caspase 3, which is unaffected by ionizing radiation; (3) caspase 3 cleavage of Rad51 reduces its strand exchange activity in vitro; (4) inhibition of caspase 3 in vivo results in increased Rad51 protein and Rad51 focus formation after irradiation. Our finding that ionizing radiation induced rapid Rad51 cleavage but slowed generalized caspase 3 activation in BRCA2-defective cells is consistent with a model in which BRCA2 protects Rad51 from caspase 3 cleavage and thus inhibits overall caspase 3 activation and apoptosis. Specific homologous recombination assays and MMC sensitivity survival assays will be needed in isogenic cells to show that these findings are important for homologous recombination in vivo. Because prior work clearly shows that BRCA2 has a direct role in DNA repair in concert with Rad51, our studies indicate that BRCA2 may have multiple pleiomorphic functions including inhibition of caspase 3 cleavage of Rad51 and inhibition of apoptosis.
ACKNOWLEDGMENTS
Funding for this research was provided by two grants from the NIH: NCI CA85269 (JTH), and CA9694402 (ETB).
REFERENCES
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396 doi: 10.1038/25292. 643–639. [DOI] [PubMed] [Google Scholar]
- 2.Wolf BB, Green DR. Suicidal tendencies: apoptotic cell death by caspase family proteinases. J. Biol. Chem. 1999;274:20049–20052. doi: 10.1074/jbc.274.29.20049. [DOI] [PubMed] [Google Scholar]
- 3.Villa PO, Kaufmann SH, Earnshaw WC. Caspases and caspase inhibitors. Trends Biochem. Sci. 1997;22:388–393. doi: 10.1016/s0968-0004(97)01107-9. [DOI] [PubMed] [Google Scholar]
- 4.Widmann C, Gibson S, Johnson GL. Caspase-dependent cleavage of signaling proteins during apoptosis. A turn-off mechanism for antiapoptotic signals. J. Biol. Chem. 1998;273:7141–7147. doi: 10.1074/jbc.273.12.7141. [DOI] [PubMed] [Google Scholar]
- 5.Casciola-Rosen L, Nicholson DW, Chong T, Rowan KR, Thornberry NA, Miller DK, Rosen A. Apopain/CPP32 cleaves proteins that are essential for cellular repair: a fundamental principle of apoptotic death. J. Exp. Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nueda A, Hudson F, Mivechi NF, Dynan WS. DNA-dependent protein kinase protects against heat-induced apoptosis. J. Biol. Chem. 1991;274:14988–14996. doi: 10.1074/jbc.274.21.14988. [DOI] [PubMed] [Google Scholar]
- 7.Smith GC, DiFagagna F, Lakin ND, Jackson SP. Cleavage and inactivation of ATM during apoptosis. Mol. Cell. Biol. 1999;19:6076–6084. doi: 10.1128/mcb.19.9.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flygare J, Armstrong RC, Wennborg A, Orsan S, Hellgren D. Proteolytic cleavage of HsRad51 during apoptosis. FEBS Lett. 1998;427:247–251. doi: 10.1016/s0014-5793(98)00433-5. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Nakada S, Ishiko T, Utsugisawa T, Datta R, Kharbanda S, Yoshida K, Talanian RV, Weichselbaum R, Yuan ZM. Role for caspase-mediated cleavage of Rad51 in induction of apoptosis by DNA damage. Mol. Cell. Biol. 1999;19:2986–2997. doi: 10.1128/mcb.19.4.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem. Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 14.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. USA. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 16.Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos. Trans. R. Soc. Lond. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 18.Godthelp BC, Artwert F, Joenje H, Zdzienicka MZ. Impaired DNA damage-induced nuclear RAD51 foci formation uniquely characterizes Fanconi anemia group D1. Oncogene. 2002;21:5002–5005. doi: 10.1038/sj.onc.1205656. [DOI] [PubMed] [Google Scholar]
- 19.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 20.Morimatsu M, Donoho G, Hasty P. Cells deleted for Brca2 COOH terminus exhibit hypersensitivity to gamma-radiation and premature senescence. Cancer Res. 1998;58:3441–3447. [PubMed] [Google Scholar]
- 21.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and RecA. Nat. Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 22.Kojima H, Endo K, Moriyama H, Tanaka Y, Alnemri ES, Slapak CA, Teicher B, Kufe D, Datta R. Abrogation of mitochondrial cytochrome c release and caspase-3 activation in acquired multidrug resistance. J. Biol. Chem. 1998;273:16647–16650. doi: 10.1074/jbc.273.27.16647. [DOI] [PubMed] [Google Scholar]
- 23.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–403. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 24.Baumann P, West SC. Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A, and hRad52. J. Mol. Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 25.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 26.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]