Abstract
Histone acetyltransferases (HATs) and histone deacetylases (HDACs) function antagonistically to control histone acetylation states that are crucial to many cellular processes. We describe here genome-wide mapping experiments that reveal that both HATs (CBP, p300, PCAF, Tip60, MOF) and HDACs (HDAC1, HDAC2, HDAC3, HDAC6) on chromatin are positively correlated with gene expression and histone acetylation. We provide evidence that Tip60 and HDAC6 are targeted to transcribed regions of active genes by phosphorylated RNA Pol II. Our results indicate that MLL-mediated H3K4 methylation primes chromatin to facilitate histone acetylation. Our data suggest that the majority of HDACs in the human genome function to reset chromatin by removing acetylation in active genes; the dynamic cycle of acetylation and deacetylation by transient HAT/HDAC binding prevents Pol II from binding to the genes primed by H3K4 methylation and poises them for future activation.
Introduction
Histone acetylation plays key roles in modulating chromatin structure and function (Shahbazian and Grunstein, 2007). The acetylation state of a given chromatin locus is controlled by two classes of antagonizing histone modifying enzymes, HATs and HDACs, which add or remove acetyl groups to/from target histones, respectively. Acetylaton is generally associated with transcriptional activation and several HATs have been identified as transcription co-activators including GCN5, PCAF, CBP, p300, Tip60, and MOF, reviewed in (Roth et al., 2001; Yang, 2004a). In contrast, histone deacetylation is generally associated with transcriptional repression and HDACs have been identified as transcriptional co-repressors (Kadosh and Struhl, 1997; Rundlett et al., 1998; Taunton et al., 1996). These enzymes are highly conserved from yeast to human. Based on their homology with yeast orthologs and other phylogenic analyses, the 18 HDACs in humans can be grouped into four classes: Class I (HDAC1, 2, 3, and −8 with a homology to Rpd3), Class II (HDAC4, 5, 6, 7, 9, and 10 with a homology to Hda1), Class III (Sirt1, 2, 3, 4, 5, 6, and 7 with a homology to Sir2), and Class IV (HDAC11) (de Ruijter et al., 2003; Yang and Gregoire, 2005; Yang and Seto, 2008). Similar to HATs, HDACs have critical functions in many cellular pathways and their misregulation has been linked to multiple cancers. Chemicals that inhibit HDAC activity are currently among the most promising drugs in anti-cancer therapies.
Co-repressor HDACs are traditionally considered to repress/inhibit transcription by associating with gene promoters and are replaced by stimulating co-activator HATs for subsequent activation upon signal transduction (Berger, 2007; Xu et al., 1999). Histone acetylation has been suggested to play roles in both transcriptional initiation and elongation. Acetylation of nucleosomes surrounding transcription start sites (TSSs) may stabilize the binding of other chromatin remodeling factors at promoter regions (Hassan et al., 2001) and/or destabilize nucleosome structure (Boeger et al., 2003; Reinke and Horz, 2003), which may lead to decreased nucleosome occupancy immediately upstream of TSSs and facilitate RNA Pol II binding (Schones et al., 2008). Nucleosomes also present formidable barriers to the passage of Pol II during transcriptional elongation (Orphanides et al., 1998). Global acetylation in transcribed regions is required for increased levels of basal transcription in yeast (Govind et al., 2007; Vogelauer et al., 2000). Consistent with these genetic and biochemical results, genome-wide location analysis in yeast found that HAT binding is correlated with transcriptional activation, reviewed by (Shahbazian and Grunstein, 2007). However, the mode of association of the transcriptional co-repressors, HDACs, with gene expression in yeast has been a topic of debate. Some studies suggested they are associated with gene repression (Kadosh and Struhl, 1997; Robert et al., 2004; Xie et al., 1999), while others found they are elevated in active genes (Kurdistani et al., 2002; Wang et al., 2002). Currently, little is known about the genome-wide profiles of HDACs in higher eukaryotic organisms. While genome-wide profiling of HATs in mammalian systems has been performed, the data is limited. Genome-wide analysis of TAF1 binding confirmed its promoter localization (Kim et al., 2005); another HAT, p300, may be associated with both promoters and enhancers (Heintzman et al., 2007)(Visel et al., 2009).
Our previous genome-wide mapping has revealed numerous combinatorial patterns of histone modifications in human CD4+ T cells (Barski et al., 2007; Wang et al., 2008) and suggested that modification patterns can indicate differentiation potential of cells (Cui et al., 2009; Wei et al., 2009), reviewed in (Wang et al., 2009). To understand the mechanisms of pattern establishment and the specificity of various HATs and HDACs, we decided to systematically determine the genomic locations of these enzymes in human CD4+ T cells. Surprisingly, we found that the binding for all HATs and HDACs analyzed is positively correlated with gene expression, Pol II binding and acetylation levels. The p300 and CBP HATs are associated with enhancers and promoters, whereas MOF, PCAF and Tip60 are elevated in transcribed regions in addition to promoters of active genes. Our data suggest that HDAC6, previously believed to function mainly in the cytoplasm, is targeted to chromatin of active genes. Interestingly, HDAC6 and Tip60 may be recruited to active genes possibly through direct interaction with phosphorylated RNA Pol II. Inhibition of HDAC activities revealed two major roles of HDACs: (1) removal of acetyl groups at active genes added by HATs during transcriptional initiation and elongation to maintain an adequate level of acetylation; and (2) removal of acetyl groups added by transient binding of HATs at inactive gene promoters to maintain a reduced level of acetylation and to prevent Pol II from binding. Knockdown of WDR5, an essential subunit of the MLL complexes, indicated that H3K4 methylation primes the chromatin of a subset of silent genes and facilitates histone acetylation, thus providing large-scale support for the crosstalk between histone modifications. Our results suggest that the dynamic cycle of acetylation and deacetylation by the transient binding of HATs and HDACs, together with prior H3K4 methylation, may poise the primed genes for future activation.
Results
Mapping the genome-wide distribution of HATs and HDACs
We have performed ChIP-Seq experiments for five HATs (p300, CBP, MOF, PCAF and Tip60) and four HDACs (HDAC1, HDAC2, HDAC3 and HDAC6) using chromatin prepared from human primary resting CD4+ T cells as described previously (Barski et al., 2007). These HATs and HDACs were chosen because they are expressed in human T cells and specific antibodies are available for them. To stabilize HATs/HDACs on chromatin, we first treated cells with bifunctional membrane-permeating cross-linker, disuccinimidyl glutarate (DSG), to preserve the protein-protein association (Tian et al., 2005) before formaldehyde crosslinking. All the antibodies were validated using immunoblotting assays (Figure S1 and Table S1) and the ChIP DNA samples resulting from these antibodies were confirmed using qPCR assays. To gain further confidence in our ChIP-Seq data for these modifying enzymes, we compared our data with published binding sites for p300 in the literature (Table S2) and confirmed selected sites using qPCR assays (Figures S2 and S3).
HATs are enriched in active genes and correlated with gene expression
CBP and p300 are structurally highly homologous and functionally redundant (Roth et al., 2001). Both enzymes are tumor suppressors and function as integrators for many signal transduction pathways through interaction with numerous critical transcription factors and other functional proteins. Our data indicate that p300 is highly enriched in promoters (Figures 1A) and potential enhancers identified by intergenic DNase I hypersensitive (HS) sites (Figure 1B). The latter is consistent with recent reports that acetylation islands (Roh et al., 2007) and p300 binding sites (Visel et al., 2009) can be used to predict functional enhancers. As expected, CBP demonstrated a global distribution pattern very similar to p300 in promoters and enhancers (Figures 1A & 1B). These data are consistent with a role for these enzymes in transcriptional initiation (Cho et al., 1998). We found that 8707 promoters are associated with one or both of these enzymes. Although p300 and CBP are highly homologous, 222 promoters were associated with only p300 and 2747 were associated with only CBP, whereas 5738 promoters associated with both (Figure S4A). The differential target genes of p300 and CBP may explain their functional differences.
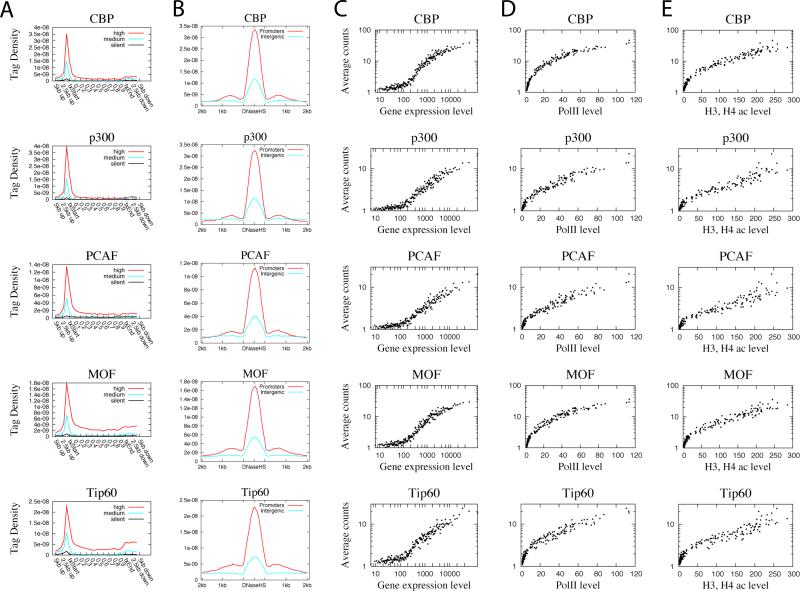
Figure 1. All HATs are correlated with gene expression, Pol II and acetylation levels.
A. Profiles of HATs binding across 5’ gene ends, 3’ gene ends and gene body regions of the 1000 most active, intermediately active and least active genes were examined using ChIP-Seq. txStart: transcription start site. txEnd: transcription end site.
B. Profiles of HATs binding across intergenic (5kb away from any gene) or promoter (defined as +/− 1kb surrounding TSS) DNase HS sites. DNase HS sites were obtained from (Boyle et al., 2008).
C. Correlation between HAT binding and gene expression levels. Genes were grouped to 100 gene (one dot in the figure) sets according to expression level. The HAT binding level in promoter region was calculated for the same 100 gene sets. The y-axis indicates the HAT binding level and the x-axis indicates the expression level.
D. Correlation between HAT binding and RNA Pol II binding levels among the 100 gene sets grouped according to expression levels as defined in panel C. The y-axis indicates the HAT binding level and the x-axis indicates the Pol II level.
E. Correlation between HAT binding and histone acetylation levels among the 100 gene sets grouped according to expression levels as defined in panel C. The acetylation level was calculated by pooling all reads for 18 histone acetylations mapped previously (Wang et al., 2008). The y-axis indicates the HAT binding level and the x-axis indicates the acetylation level.
PCAF (p300/CBP associated factor) and GCN5 are another pair of highly homologous HATs in mammals, reviewed by (Yang, 2004b). In addition to histone acetylation activity, the bromodomains of GCN5 and PCAF can bind acetylated N-terminal tails of histone H3 and H4 (Dhalluin et al., 1999). Thus, they act as both acetylation “producers” and “effectors”. Similar to p300 and CBP, PCAF was highly enriched in promoters of active genes (Figure 1A). PCAF appears to be relatively more enriched in gene body regions as compared to CBP and p300 (Figure 1A), which is consistent with the proposed function of PCAF in transcription elongation (Cho et al., 1998). The binding of PCAF was also elevated in intergenic DNase HS sites (Figure 1B), suggesting that it may also be involved in enhancer activities. Interestingly, 76.4% and 92.9% of genes associated with PCAF in promoter regions were also bound by p300 and CBP, respectively, in the promoter regions (Figures S4B and S4C).
MOF and Tip60 belong to the MYST family of HATs that are less characterized and more diverse as compared to the CBP/p300 and GCN5/PCAF HAT families (Yang, 2004b). Tip60 is the catalytic subunit of an evolutionary conserved complex, which contains at least 12 subunits including TRRAP (Doyon et al., 2004). It has been implicated in multiple roles in transcription, DNA damage repair, and cell cycle regulation as well as being linked to many diseases. MOF is the catalytic subunit of an evolutionary conserved MSL complex that is involved in dosage compensation and gene expression in Drosophila, though the functional role of the human MSL complex remains to be determined (Rea et al., 2007). Tip60 can acetylate H2AK5, H3K14, and H4K5, K8, K12, K16 (Ikura et al., 2000), whereas both Drosophila and human MOF are specific for H4K16 acetylation (Dou et al., 2005). We found that MOF was detected at high levels at the promoter region and was also relatively more enriched in gene body region as compared to CBP and p300 (Figure 1A), which is different from the Drosophila MOF that shows a bimodal binding at promoters and 3’ ends of genes (Kind et al., 2008). Similarly, Tip60 exhibits strong binding at both the promoter and within the gene body of active genes (Figure 1A). These data are consistent with the elevated H4K16 acetylation along gene bodies of active genes (Wang et al., 2008) and with a role for Tip60 in transcriptional elongation. MOF and Tip60 were detected at 8025 and 6797 genes, respectively, with an overlap of 5462 genes between them (Figure S4D). MOF and Tip60 also showed similar binding patterns at both the promoter and intergenic DNase HS sites (Figure 1B).
The higher levels of HATs in active genes suggest that they may be positively correlated with gene activity. This was indeed confirmed by the scatter plot analysis of average gene expression and average HAT ChIP-Seq read counts at promoters (Figure 1C). Interestingly, our data indicate that all the HATs are positively correlated with RNA Pol II binding (Figure 1D). As expected, we also observed positive correlation between HAT binding and histone acetylation levels (Figure 1E).
In summary, our data indicate that p300 and CBP are mainly targeted to promoters and enhancers, whereas PCAF, MOF and Tip60 are also associated with transcribed regions of active genes in addition to promoters. These data are consistent with the notion that p300 and CBP are involved in transcriptional initiation, while PCAF, MOF and Tip60 may also be involved in transcriptional elongation. We also find that even though homologous HATs can bind to distinct sets of genes, many genes are associated with multiple HATs (for example in the NFκB and CD69 genes shown in Figures S2 and S3), suggesting that they may function together either collaboratively, sequentially or independently at the same target genes.
HDACs are associated with active genes and positively correlated with transcription
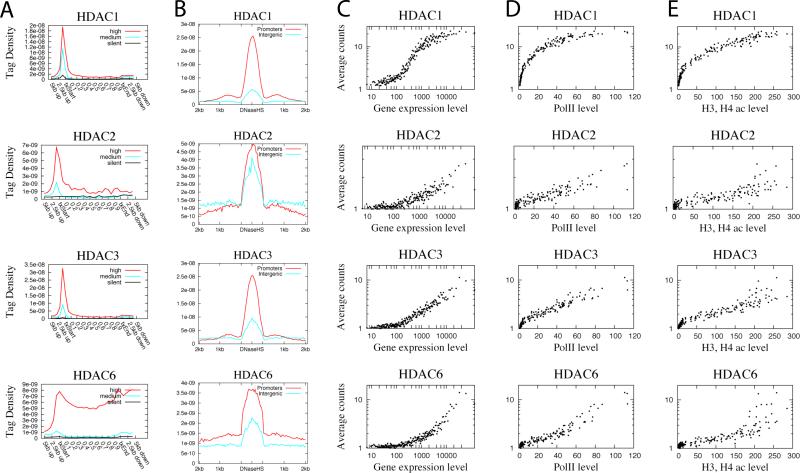
HDACs are generally believed to act as transcriptional co-repressors (Berger, 2007; Li et al., 2007). It has been traditionally believed that HDACs are bound to repressed genes and are replaced by HATs upon gene activation (Xu et al., 1999). However, little is known about their target gene specificity in the human genome. Our data revealed that all the HDACs analyzed, including Class I HDACs, HDAC1, HDAC2 and HDAC3 and Class II HDAC, HDAC6, were enriched in active genes but not silent genes (Figure 2A). HDAC1 and HDAC3 were mainly detected in promoter regions, whereas HDAC2 and HDAC6 were elevated in both the promoter and gene body regions of active genes. We also examined their presence at potential enhancers and found that HDAC1, 3 and 6 were mainly enriched in promoter DNase HS sites, whereas HDAC2 was highly elevated in both the intergenic and promoter DNase HS sites (Figure 2B).
Figure 2. HDACs are positively correlated with gene expression, Pol II binding and acetylation levels.
A. Profiles of HDAC binding across 5’ gene ends, 3’ gene ends and gene body regions of 1000 most active, intermediately active and silent genes.
B. Profiles of HDAC binding across intergenic or promoter DNase HS sites.
C. Correlation between HAT binding and gene expression levels. See Figure 1C for details.
D. Correlation between HAT binding and RNA Pol II binding levels. See Figure 1D for details.
E. Correlation between HAT binding and histone acetylation levels. See Figure 1E for details.
Our observation of the association of HDAC6 with chromatin was unexpected because it is believed to be predominantly cytoplasmic, reviewed in (Boyault et al., 2007). The in vivo histone deacetylase activity of HDAC6 has not been demonstrated though it has strong activity in vitro. Our data that HDAC6 was detected in nuclear extracts (Figure S1) and highly enriched in chromatin of 4706 active genes (Figure 2) strongly suggest that it functions to deacetylate histones in vivo.
The enrichment of these HDACs in active genes suggests that they may be positively associated with gene expression. Indeed, we found that all HDACs tested were positively correlated with mRNA expression levels (Figure 2C) and Pol II levels (Figure 2D). Interestingly, HDAC levels also appeared to positively correlate with histone acetylation levels (Figure 2E).
In summary, our data confirmed the expectation that HATs, as transcription co-activators, are highly enriched in active genes (Figure 1 and Figure S5). Surprisingly, we found that HDACs are also highly enriched in active genes (Figure 2). The majority of HDACs in the human genome are associated with active genes and only a minor fraction are detected in silent genes (Figure S5). This observation suggests that one major function of HDACs is to remove the acetyl group added by HATs in active genes and to reset chromatin modification following gene activation, as suggested previously for Hos2 (Wang et al., 2002).
Tip60 and HDAC6 may be targeted to active genes through interaction with phosphorylated CTD of RNA Polymerase II
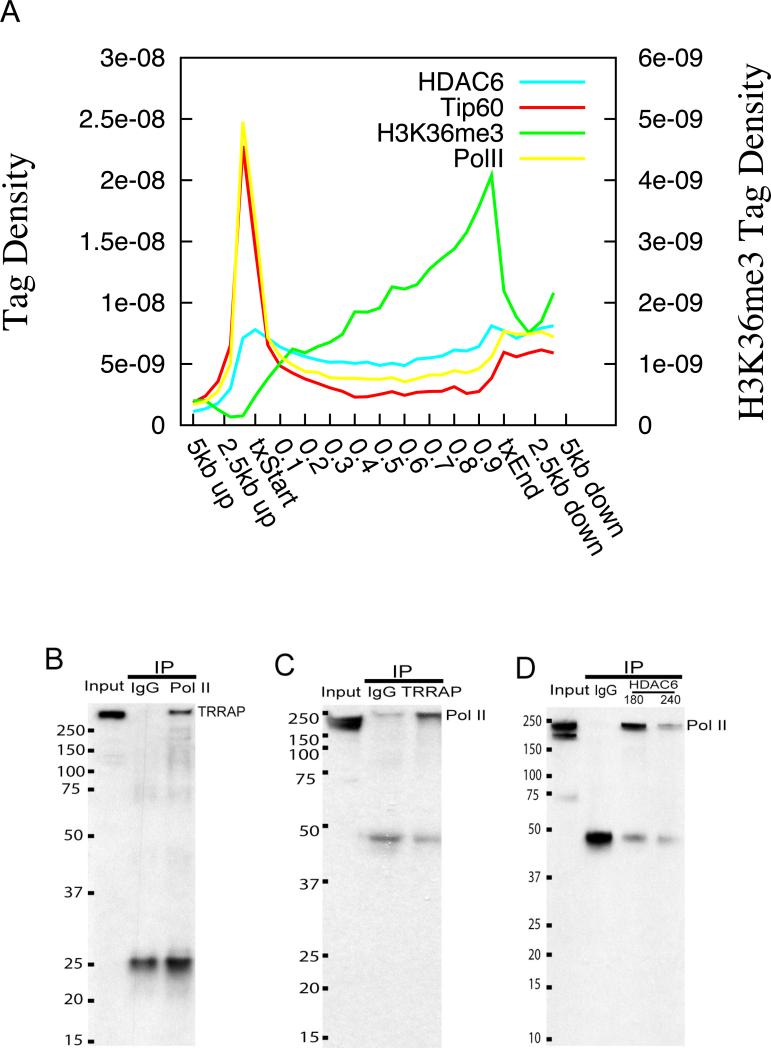
We noticed that both Tip60 and HDAC6 are highly elevated in the transcribed regions of active genes. The targeting of the yeast Rpd3 deacetylase complexes to the transcribed gene body regions has been suggested to be mediated by H3K36 methylation through the chromodomain of the Eaf3 subunit (Carrozza et al., 2005; Joshi and Struhl, 2005). However, one human homolog of Eaf3, the MORF4 protein, lacks the chromodomain (Carrozza et al., 2005), suggesting that the mammalian HDAC complexes may use different targeting mechanisms. Indeed, our data indicate that the distribution profiles of both Tip60 and HDAC6 in the gene body region appeared to parallel the signals of Pol II more than the signals of H3K36me3 (Figure 3A). Therefore, we hypothesized that Tip60 and HDAC6 may physically interact with Pol II, thus being recruited to transcribed regions in the human genome. To test this hypothesis, we examined whether they can be co-immunoprecipitated with Pol II. However, the antibodies against Tip60 failed to pull down Pol II (data not shown). We reasoned that the antibody binding might inhibit the interaction between Tip60 and Pol II. Therefore, we examined the reciprocal interaction of Pol II with TRRAP, which is a core subunit of the Tip60 complex (Ikura et al., 2000). As shown in Figure 3B, TRRAP was co-immunoprecipitated with Pol II. Interestingly, we found that only the phosphorylated Pol II was co-immunoprecipitated with TRRAP (Figure 3C). Similarly, we only detected the phosphorylated form but not the unphosphorylated form of Pol II from the immunoprecipitates using anti-HDAC6 antibodies (Figure 3D), though we failed to detect HDAC6 from Pol II immunoprecipitates (data not shown). We believe this interaction is specific otherwise we expect to also detect the non-phosphorylated form of Pol II (lower band 220KDa). Therefore, we conclude that Tip60 and HDAC6 may be recruited to the transcribed regions of active genes through a different mechanism from yeast by directly or indirectly interacting with elongating Pol II.
Figure 3. Tip60 and HDAC6 are recruited to active genes through interaction with phosphorylated RNA Polymerase II.
A. Distribution profiles of HDAC6, Tip60, Pol II and H3K36me3 across the active genes were plotted. The left y-axis indicates tag densities for HDAC6, Tip60 and Pol II. The right axis indicates tag densities for H3K36me3.
B. Co-immunoprecipitation of TRRAP with RNA Pol II. Nuclear extracts (Input) and immunoprecipitates using control IgG or Pol II antibodies were resolved by SDS-PAGE and blotted with antibodies against TRRAP. The size markers are indicated on the left of the panel. The band corresponding to TRRAP is indicated on the right.
C. Co-immunoprecipitation of phosphorylated RNA Pol II with TRRAP. The band corresponding to phosphorylated Pol II is indicated on the right.
D. Co-immunoprecipitation of phosphorylated RNA Pol II with HDAC6. The band corresponding to phosphorylated Pol II is indicated on the right.
HATs and HDACs are targeted to activated genes upon TCR signaling
If Tip60 and HDAC6 are recruited by elongating Pol II, the change in Pol II binding should lead to a corresponding change in Tip60 and HDAC6 binding. To test this hypothesis, we activated resting T cells using anti-CD3 and anti-CD28 antibodies and analyzed the changes in gene expression, Pol II, Tip60 and HDAC6 binding. As shown in Figure 4A, the fold changes in read counts of all these enzymes demonstrated a positive correlation with that of gene expression. Interestingly, both the changes in Tip60 and HDAC6 binding also exhibited a positive correlation with that of Pol II upon TCR signaling (Figure 4B). These data are consistent with the observation that Tip60 and HDAC6 physically interact with Pol II, further supporting the notion that Tip60 and HDAC6 are recruited to transcribed region of active genes by interacting with elongating Pol II.
Figure 4. Recruitment of Tip60 and HDAC6 by TCR signaling is correlated with the recruitment of RNA Pol II.
A-C: The changes in Pol II, Tip60 and HDAC6 binding after TCR signaling determined using ChIP-Seq (y-axis) are plotted against the changes in expression levels (x-axis). Genes were grouped to 100 gene set as one dot in the figure.
D, E: The changes in Tip60 and HDAC6 binding levels after TCR signaling are plotted against the changes in Pol II binding level.
HDACs function to control acetylation levels in active genes
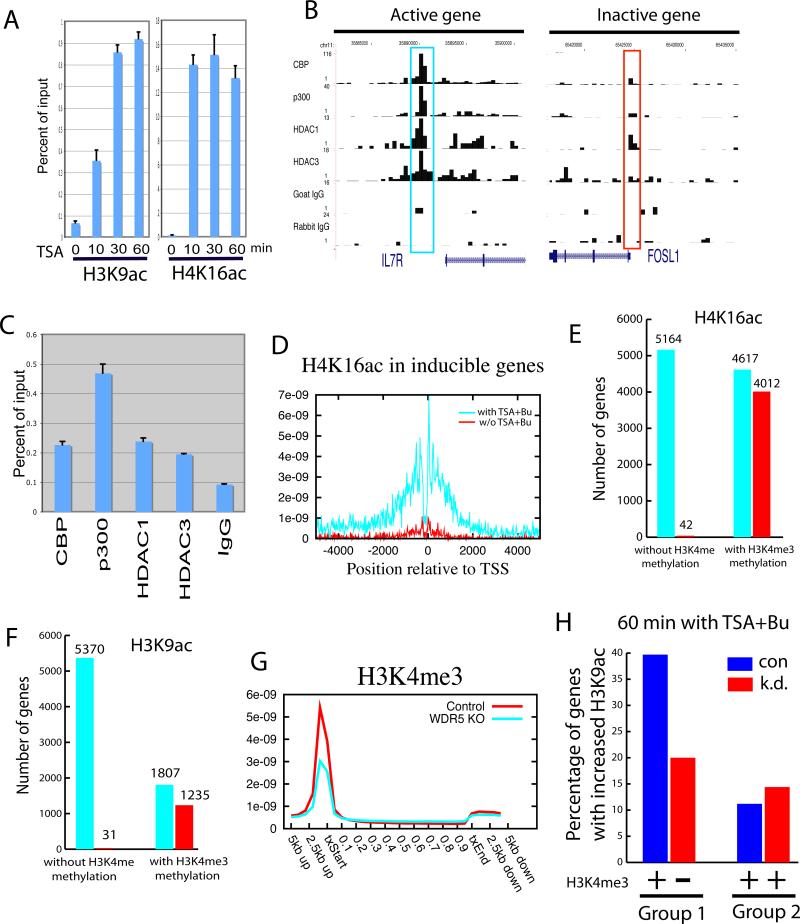
The HDACs recruited to active genes by Pol II may function to remove the acetylation added during gene activation by HATs. To test this hypothesis, we inhibited HDAC activities using the HDAC inhibitors trichostatin A (TSA) and Butyrate and examined the genome-wide changes in H3K9ac and H4K16ac using ChIP-Seq. Indeed, we observed remarkable increases in both H3K9 and H4K16 acetylation in active genes when resting T cells were treated with TSA + Butyrate, as exemplified for the CD4 locus (Figure 5A). Because both Pol II and HDACs are enriched much more in active genes than inactive genes, particularly in the promoter and downstream of TSS regions (Figures 1, 2 and 5B, 5C), inhibition of HDAC activities should result in increased acetylation more in active genes than in inactive genes. Indeed, our data revealed that the highest increase in both H3K9ac and H4K16ac was detected in active genes, especially in regions surrounding the TSS (Figures 5D and 5E). Therefore, we conclude that HDACs recruited to active genes are functional and act to control the acetylation level together with the HATs.
Figure 5. Inhibition of HDAC activities leads to further increases of acetylation levels in active genes.
A. HDAC inhibitor treatment causes remarkable increases in histone acetylation in active genes. Resting T cells were treated with TSA and Butyrate and the distribution of H3K9ac and H4K16ac was analyzed using ChIP-Seq. The acetylation pattern of the CD4 locus is shown as custom tracks on the UCSC genome browser. TSA+Bu: the cells were treated with TSA and Butyrate.
B. Pol II is highly enriched in the promoter and downstream of TSSs of active genes. The profiles of Pol II binding surrounding TSSs are shown for active, intermediately active and silent genes.
C. HDAC6 II is highly enriched in the promoter and downstream of TSSs of active genes. The profiles of Pol II binding surrounding TSSs are shown for active, intermediately active and silent genes.
D. The most increase in H3K9 acetylation is detected in the promoter and downstream of TSSs of active genes. Act:0h, Act:2h, Sil:0h, and Sil:2h: the normalized acetylation levels of the active genes and silent genes with TSA treatment (2 hrs) or no treatment (0 hr).
E. The most increase in H4K16 acetylation is detected in the promoter and downstream of TSSs of active genes.
HDAC inhibitor treatment reveals poised genes
HDACs have been suggested to function as transcriptional co-repressors and associate with repressed genes (Berger, 2007; Li et al., 2007). However, our ChIP-Seq data did not reveal high levels of binding of HDACs to silent genes (Figure 2), suggesting that they may not stably bind to their target genes of repression. To understand the functional roles of HDACs in gene silencing, we examined the histone acetylation patterns of silent genes with TSA + Butyrate treatment. Interestingly, we found that a subset of silent genes exhibited a remarkable increase in acetylation, as demonstrated for the FOSL1 gene (Figure 6A). However, examination of HAT and HDAC binding using the ChIP-Seq data revealed no high level binding of HDACs in inactive genes (Figure 2A and Figure 6B, the region highlighted in the red box). Therefore, the remarkable increase in acetylation in the presence of HDAC inhibitors suggests that these genes are subject to constant acetylation and deacetylation cycles, likely through transient and unstable binding of HATs and HDACs. To test this hypothesis, we examined the binding of HATs and HDACs to the FOSL1 promoter using qPCR analysis of ChIP DNA. We indeed detected low but reproducible binding of both HATs and HDACs to this region (Figure 6C). The rapid increase in acetylation in as short as 10 minutes in the presence of HDAC inhibitors (Figure 6A) suggests that HATs frequently but transiently visit the promoters to add acetyl groups to histones and HDACs keep removing these acetylation signals to maintain the repression of these genes. This dynamic cycle of acetylation and deacetylation by transient binding of HATs and HDACs may poise the genes for future activation. To test whether this is the case, we identified 167 genes, which were induced by TCR signaling in CD4+ T cells, and examined their histone acetylation profiles with the HDAC inhibitor treatment. We indeed found a remarkable increase in both H4K16ac (Figure 6D) and H3K9ac (Figure S6) in the presence of HDAC inhibitors. In summary, our data indicate that HATs can transiently and frequently bind to the non-expressed genes to acetylate the histones; HDACs also bind transiently and frequently to remove the acetyl group and keep the genes inactive. The concerted action of HATs and HDACs poises these genes for future activation.
Figure 6. HATS and HDACs function at silent genes primed by H3K4 methylation.
A. Rapid increase of histone acetylation in silent genes caused by HDAC inhibitor treatment. ChIP assays were performed using H3K9ac and H4K16ac antibodies with chromatin from cells treated in the absence or presence of HDAC inhibitors. The ChIP DNA was analyzed using qPCR.
B. The ChIP-Seq signals for the DIPA gene (active) and FOSL1 gene (silent) were displayed. The signals highlighted in blue are not statistically significant as determined using peak-finding programs.
C. qPCR assays revealed low but reproducible binding of HATs and HDACs in the silent FOSL1 promoter (see Panel B).
D. H4K16ac was strongly elevated in the TCR-inducible genes by TSA + Butyrate treatment. H4K16 acetylation was profiled using ChIP-Seq in resting T cells in the presence or absence of TSA + Butyrate treatment. The H4K16ac profiles were plotted for the 167 silent genes that are induced by TCR signaling.
E. The presence of H3K4 methylation indicates the potential of histone H4K16 acetylation. All genes (9,781) not associated with H4K16ac were separated into the genes with H3K4 methylation (5,164) and those without (4,617). The number of genes that became acetylated with the HDAC inhibitor treatment was examined for each group (shown by the red bar).
F. The presence of H3K4 methylation indicates the potential of histone H3K9 acetylation. All genes (7,177) not associated with H3K9ac were grouped and analyzed similarly as in Panel E. The number of genes that became acetylated are shown by the red bar.
G. Knock-down of WDR5 decreased H3K4me3 signals in the promoter regions. H3K4me3 distribution in wild type and Knock-down cells were determined using ChIP-Seq and the profiles were analyzed as in Figure 1.
H. Inhibition of H3K4me3 modification decreased H3K9ac with the HDAC inhibitor Treatment. H3K4me3 and H3K9ac profiles were analyzed in the control (con) and WDR5 knockdown (k.d.) cells. Group 1 genes lost H3K4me3 signals in the knockdown cells; Group 2 genes did not show significant decrease in H3K4me3 signals in the knockdown cells. y-axis indicates the percentage of genes that showed an increase of at least 2-fold in H3K9ac with the HDAC inhibitor treatment in both of these two gene groups.
H3K4 methylation facilitates histone acetylation
To attempt to understand the differences between the silent genes that became acetylated and those that did not in the presence of the HDAC inhibitors, we examined the methylation patterns and found that the former was associated with H3K4 methylation and H2A.Z, whereas the latter was not (Figure S7). To confirm this on a genome-wide scale, we identified a total of 9,781 promoters that were not associated with significant H4K16ac and examined their acetylation after HDAC inhibitor treatment. Interestingly, 87% of the 4,617 promoters associated with H3K4 methylation acquired significant levels of H4K16ac, whereas only 0.8% of the 5,164 promoters not associated with H3K4 methylation became acetylated at H4K16 (Figure 6E). Similarly, among the 7,177 promoters that did not exhibit H3K9ac before the TSA treatment, 68% of the 1,807 promoters with H3K4 methylation became acetylated, whereas only 0.6% of the 5,370 promoters without H3K4 methylation became acetylated at H3K9 after the TSA treatment (Figure 6F). These results strongly suggest that H3K4 methylation facilitates the histone acetylation events.
To test this hypothesis directly, we knocked down WDR5, which is an essential subunit of the MLL complexes that are responsible for H3K4me2 and H3K4me3 modifications (Wysocka et al., 2005). The H3K4me3 level was significantly decreased in HeLa cells transfected with a vector expressing an shRNA targeting WDR5 as compared to cells transfected with a control vector (Figures S8A and S8B). ChIP-Seq analysis revealed that the overall H3K4me3 binding decreased about 2-fold in the knockdown cells (Figure 6G). To determine whether inhibition of H3K4 methylation leads to changes in histone acetylation, the HeLa cells transfected with the siRNA or control vector were treated with TSA + Butyrate for 60 min, and the genome-wide distribution of H3K9ac and H4K16ac was analyzed using ChIP-Seq. First, we identified 325 genes (Group 1) that were associated with H3K4me3 in wild type HeLa cells but lost the modification in the knockdown cells. As a control, we identified 409 genes (Group 2) that showed less than 10% change in the H3K4me3 level in the knockdown cells. Among the Group 1 genes, 40% exhibited an increase of at least 2-fold in H3K9ac in the control cells with the HDAC inhibitor treatment, whereas only around 19% these genes demonstrated a similar increase in H3K9ac in the WDR5 knockdown cells, which is a 50% decrease in gene number (Figure 6H). In contrast, about 11% of the Group 2 genes demonstrated an increase in H3K9ac with the HDAC inhibitor treatment in the control and 14% of the Group 2 genes showed an increase in H3K9ac in the WDR5 knockdown cells (Figure 6H). A decrease in H4K16ac was also observed in Group 1 but not Group 2 genes with the HDAC inhibitor treatment (Figure S8C). These data indicate that prior H3K4 methylation primes a subset of non-expressed genes for histone acetylation in vivo. The fact that only 50% of the genes demonstrated decreases in H3K9ac could be related with the possibility that H3K4me1 or H3K4me2 instead of H3K4me3 are present in some of the Group 1 genes.
HDAC inhibitor treatment leads to increased Pol II binding at the primed genes
The transcription of the silent genes, which are modified (primed) by H3K4 methylation but not histone acetylation, may be repressed by HDACs. Histone deacetylation may inhibit transcription at the initiation and/or elongation step. If these genes are bound by Pol II at promoters and the inhibition is at the elongation step, HDAC inhibitor treatment, which induces a significant increase in acetylation, should increase their mRNA levels. Thus, we profiled the mRNAs in cells in the absence and presence of the HDAC inhibitors. Our data indicate that the HDAC inhibitor treatment induced only 2 genes out of the 3927 “true” silent genes that were associated with neither histone acetylation nor H3K4 methylation. Interestingly, only 15 among the 1382 genes primed by H3K4 methylation were induced by the treatment, although most of them became highly acetylated in the presence of HDAC inhibitors. These results suggest that the repression of transcription by HDACs may occur at an earlier step. To test this hypothesis, we analyzed Pol II binding in the absence and presence of the HDAC inhibitors using ChIP-Seq. Remarkably, we found that the HDAC inhibitor treatment resulted in a significant increase in Pol II binding in the gene promoters primed by H3K4 methylation but not in the silent genes (Figures 7A and 7B). Among the 1358 inactive promoters that were associated with H3K4 methylation but not Pol II, 420 (31%) and 814 (60%) became bound by Pol II after treatment with the HDAC inhibitor for 2 and 12 hrs, respectively. In contrast, only 0.4% and 0.7% of the silent promoters not associated with prior H3K4 methylation became bound by Pol II under these conditions. Even though the primed promoters became bound by Pol II following HDAC inhibitor treatment, they were not transcribed, suggesting that transcriptional activation requires further signals following Pol II binding in the promoter region. Nonetheless, these results indicate that preventing Pol II binding is an important step in the transcriptional repression by HDACs.
Figure 7. HDACs inhibit Pol II binding to the promoters primed by H3K4 methylation.
A. Promoters primed by H3K4 methylation become bound by Pol II following HDAC inhibitor treatment. Pol II binding levels were calculated by normalizing the Pol II ChIP-Seq sequence reads in the promoter regions with the number of promoters.
B. HDAC inhibitor treatment induces Pol II binding to the LMO4 promoter that was primed by prior H3K4 methylation.
C. High levels of both HATs and HDACs are associated with active genes. HDACs function to reset chromatin by removing acetyl groups added by HATs recruited by elongating Pol II.
D. Low levels of both HATs and HDACs are associated with inactive genes primed by H3K4 methylation. HDACs function to repress transcription by preventing Pol II binding by removing acetyl groups added by transient binding of HATs.
E. No detectible levels of either HATs or HDACs are associated with silent genes devoid of any significant H3K4 methylation.
Discussion
Drugs with the ability to inhibit HDAC activity are currently among the most promising drugs in anti-cancer and other diseases. However, the drug development community faces significant challenges in selection and design of drugs for clinical candidates, partially due to the lack of known targets of HATs and HDACs (Kazantsev and Thompson, 2008). Our genome-wide distributions of HATs and HDACs provide new clues for understanding the mechanisms for how HDAC inhibitors work. For example, the efforts in understanding the functional mechanisms of the drugs that inhibit HDAC6 have focused on cytoplasmic proteins including heat shock proteins or α-tubulin (Kazantsev and Thompson, 2008). Hence, the novel nuclear roles of HDAC6 associated with both primed and active genes need to be considered for any further drug design and development.
Distinct functions of HATs and HDACs in active and inactive genes
Based on the binding patterns of HATs, HDACs, histone acetylation and methylation as well as the results from HDAC inhibitor treatment, we propose that there are three major modes of association of HATs and HDACs in the genome:
(1) Active genes (Figure 7C)
These genes are expressed and associated with histone methylation including H3K4me1, 2, 3 as well as H2A.Z in promoters and H2BK5me1, H3K9me1, H3K27me1, H3K36me3, H3K79me1, 2, 3 and H4K20me1 in gene body regions (Barski et al., 2007; Wang et al., 2008). We find that the highest level of both HATs and HDACs are detected in these genes and their binding is positively correlated with expression level and Pol II level. What is the function of HDACs in these active genes? One possibility is that chromatin modifications such as acetylation need to be reset following transcription, as proposed previously (Wang et al., 2002). HDACs are recruited to these genes to reset the chromatin modification states and maintain an adequate level of histone acetylation, following the activities of Pol II and HATs. Indeed, our data indicated that inhibition of HDAC activity through TSA + Butyrate treatment caused increases in acetylation in the active genes with the level of increase being correlated with the HDAC binding, suggesting that HDAC recruitments to active genes function to remove acetyl groups in active genes. Excessive histone acetylation in transcribed regions can destabilize chromatin and increase cryptic initiation of transcription. It has been shown that the Rpd3-containing complexes recruited by the H3K36 methylation signals following the RNA Pol II passage can deacetylate histones and prevent cryptic transcriptional initiation in yeast (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). Our data showing that HDAC6 physically interacts with the elongating form of RNA Pol II and its distribution parallels the Pol II but not the H3K36me3 signal suggest that the deacetylase is directly recruited to actively transcribed regions by RNA Pol II and may bypass H3K36me3. Further experiments will be needed to clarify the potentially different mechanisms employed by different organisms.
(2) Primed genes (Figure 7D)
Although these genes do not associate with significant histone acetylation and are not expressed, they are associated with H3K4 methylation and H2A.Z. Even though our ChIP-Seq assays did not reveal significant binding of either HATs or HDACs at these genes, the more sensitive qPCR assays indicated binding of these enzymes, albeit at relatively low levels. These data suggest that HATs may bind transiently to these promoters and add acetyl groups, which are then removed by the transient binding of HDACs. Indeed, inhibition of HDAC activities resulted in remarkable increases in acetylation in these promoters, suggesting that the chromatin in these promoters is subject to dynamic regulation by HATs and HDACs. The rapid removal of the acetyl group by HDACs is consistent with a role in transcriptional repression. Therefore, the cycle of transient acetylation and deacetylation may keep these genes unexpressed but at the same time maintain the promoters in a potentiated state for future activation upon receiving external signals. There are at least two potential mechanisms by which HDACs repress transcription from these promoters: inhibition of Pol II binding to the promoters and inhibition of Pol II elongation in the transcribed regions. Our data revealed that HDAC inhibitor treatment did not lead to either significantly increased Pol II levels in transcribed regions nor mRNA abundance; instead, it resulted in a remarkable increase in Pol II levels in the promoter regions, suggesting that a major step of transcriptional repression of these genes by HDACs is the inhibition of Pol II binding to their promoters.
Our data indicating that silent promoters with prior H3K4 methylation became substantially acetylated after the treatment with HDAC inhibitors suggest that H3K4 methylation may prime the chromatin structure and facilitate the acetylation process. Indeed, knocking down WDR5, which down-regulated H3K4 methylation, significantly decreased histone acetylation upon HDAC inhibitor treatment. How does H3K4 methylation facilitate histone acetylation? Previous in vitro studies have suggested that H3K4 methylation of nucleosome core particles can increase the efficiency of acetylation by p300 (Wang et al., 2001). There are several potential mechanisms by which H3K4 methylation can promote histone acetylation, reviewed by (Berger, 2007). First, it recruits ATP-dependent chromatin remodeling complexes to Chd1 (Pray-Grant et al., 2005) and NURF (Wysocka et al., 2006), which opens chromatin to allow other histone modification enzymes to bind. Second, it is recognized directly by histone acetylation complexes (Martin et al., 2006). In addition, the MOF acetylase is recruited as a subunit of the MLL1 complex that recognizes H3K4me2 and converts it to H3K4me3 (Dou et al., 2005; Wysocka et al., 2005). Our data provide large-scale experimental support that H3K4 methylation facilitates histone acetylation and different histone modifications can act sequentially or cooperatively.
(3) Silent genes (Figure 7E)
These genes can be separated into two groups: one is associated with wide-spread H3K27me3 signals, which are added by Polycomb group proteins, and the other does not associate with any of the 40 modifications examined. Nonetheless, neither of them shows detectible acetylation/deacetylation activities as revealed by inhibition of HDAC activity and genome-wide mapping of acetylation patterns. Therefore, these genes are the least metabolically active in histone acetylation/deacetylation cycles.
In summary, our results reveal that there are two major roles for HDACs. One is their function in active genes, where high levels of HDACs act to remove the acetyl group added by high levels of HATs during the process of transcriptional initiation and elongation and reset the chromatin structure required for the next round of transcription. The other is their function is at primed genes, where transient binding of HDACs removes the acetyl group resulted from transient binding of HATs, maintains a low level of acetylation and prevents Pol II binding, thereby maintaining promoters in an inactive state. In addition, we have demonstrated that H3K4 methylation by the MLL complexes primes a subset of non-expressed genes for histone acetylation, providing experimental support that these genes may be poised by chromatin modification for future expression. Our data define the division of labor for different HATs and HDACs and provide a rich resource for further understanding of their function in transcriptional regulation and genome function.
EXPERIMENTAL PROCEDURES
T Cell Purification, Chromatin Preparation, ChIP, and ChIP-Seq
Human CD4+ T cells were purified from blood as described before (Barski et al., 2007; Wang et al., 2008). Resting T cells or cells activated using CD3 and CD28 antibody beads (Schones et al., 2008) were treated with 2 mM disuccinimidyl glutarate (Sigma) for 45 minute at room temperature to preserve the protein-protein interactions, prior to the cross-linking with 1% formaldehyde at room temperature for 10 minute. Chromatin preparation, ChIP and Solexa sequencing were performed as described (Barski et al., 2007; Wang et al., 2008). Chromatin templates from different donors were pooled and stored at −80 °C. Six to eight μg of antibodies were used for each ChIP experiment.
Co-Immunoprecipitation (Co-IP), Immnoblotting, and Antibody Specificity
Jurkat cell nuclear extracts were prepared as described (Wang et al., 2008). Co-IP assays were performed as described (Cho et al., 2007). The specificity of each antibody was tested by Western blotting with Jurkat nuclear extracts. The antibodies for Co-IP and ChIP-seq were listed in Table S1.
HDAC Inhibitor Assay in resting CD4+ T cells and HeLa cells
CD4+ T cells were resuspended in complete RPMI1640 medium and incubated in the presence of 100 ng/ml Trichostatin A (TSA, Sigma) and 2 mM Sodium Butyrate for 0 h, 10 min, 30min, 1 hr, 2 hrs, 4 hrs, and 8 hrs. HeLa cell were transfected with a vector expressing an shRNA targeting WDR5 for 3 days before the HDAC inhibitor treatment. Native chromatin from these cells was prepared by Microccocal nuclease digestion as described (Barski et al., 2007; Wang et al., 2008). ChIP assays were performed using chromatin from these cells with anti-H3K9ac and -H4K16ac antibodies for ChIP-Seq and qPCR analysis.
Data Analysis
Sequence reads of mostly 25 bp were obtained using the Solexa Analysis Pipeline. All reads were mapped to the human genome (hg18) and only uniquely matching reads were retained. The output of the Solexa Analysis Pipeline was converted to browser extensible data (BED) files detailing the genomic coordinates of each read. Please see Supplemental information for identification of ChIP-enriched islands using SICER (Zang et al., Bioinformatics, in press), HAT/HDAC profiles across gene body and DNase HS regions, and correlation analysis between HAT/HDAC binding and gene expression.
Data deposition: we have deposited the short reads, summary BED files, identified peaks, and expression data sets in the GEO database (GSE15735).
Supplementary Material
Acknowledgements
The mRNA profiles were analyzed on Affymetrix gene chips by the NHLBI Genomics Core Facility. D.E.S. was supported by the Lenfant Biomedical Fellowship of NHLBI. C. Z. and W. P. was partially supported by the University Facilitating Fund of the George Washington University. The research was supported by the Division of Intramural Research Program of the National Institute of Heart, Lung and Blood Institute, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26:5468–5476. doi: 10.1038/sj.onc.1210614. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol Cell Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rea S, Xouri G, Akhtar A. Males absent on the first (MOF): from flies to humans. Oncogene. 2007;26:5385–5394. doi: 10.1038/sj.onc.1210607. [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of direct genomic targets downstream of the nuclear factor-kappaB transcription factor mediating tumor necrosis factor signaling. J Biol Chem. 2005;280:17435–17448. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr Opin Genet Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. Embo J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004a;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004b;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.