Abstract
Presynaptic neurexins (NRXs) bind to postsynaptic neuroligins (NLs) to form Ca2+-dependent protein complexes that bridge neural synapses. NRXs bind NLs through their membrane-proximal LNS domain, which contains a single site of alternative splicing (splice site 4) that gives rise to two isoforms: +4, which includes a 30-residue splice insertion, and Δ which lacks this insertion. Here we present crystal structures of the neuroligin-binding Δ isoforms of the LNS domains from β-NRX1 and β-NRX2, crystallized in the presence of Ca2+ ions. The Ca2+-binding site in the 3.0Å β-NRX2 structure is disordered. In contrast, the 1.7Å β-NRX1 structure reveals a single Ca2+ ion, with one of its coordinating ligands donated by a glutamic acid from an adjacent β-NRX1 molecule, ∼12Å from the splice insertion site. NMR studies of β-NRX1 Δ and β-NRX1+4 show that the splice-inserted sequence forms an unstructured loop in isolation, and remains at least partially disordered in a β-NRX1+4 / NL complex. These results suggest a likely mode of Ca2+-dependent interaction with NLs, and raise the possibility that β-NRX insertion sequence 4 may function in roles independent of neuroligin binding.
Introduction
Neurexins and neuroligins are implicated as central organizing molecules for excitatory glutamatergic and inhibitory GABAergic synapses in the vertebrate nervous system (Brose, 1999; Chih et al., 2005; Dean and Dresbach, 2006; Graf et al., 2004). They function as cell adhesion molecules, with postsynaptic neuroligins (NL) binding to presynaptic neurexins (NRX) to form transsynaptic complexes (Missler et al., 1998). Members of each protein family can trigger formation of a hemisynapse: neuroligins trigger presynaptic differentiation (Scheiffele et al., 2000) and neurexins trigger postsynaptic differentiation (Graf et al., 2004). Interference with the neurexin/neuroligin system leads to defects in synaptic transmission (Chih et al., 2005; Missler et al., 2003; Varoqueaux et al., 2006), and human genetic studies have identified neuroligin genes as strong candidates for a causative role in autism spectrum disorders (Garber, 2007; Jamain et al., 2003; Tabuchi et al., 2007).
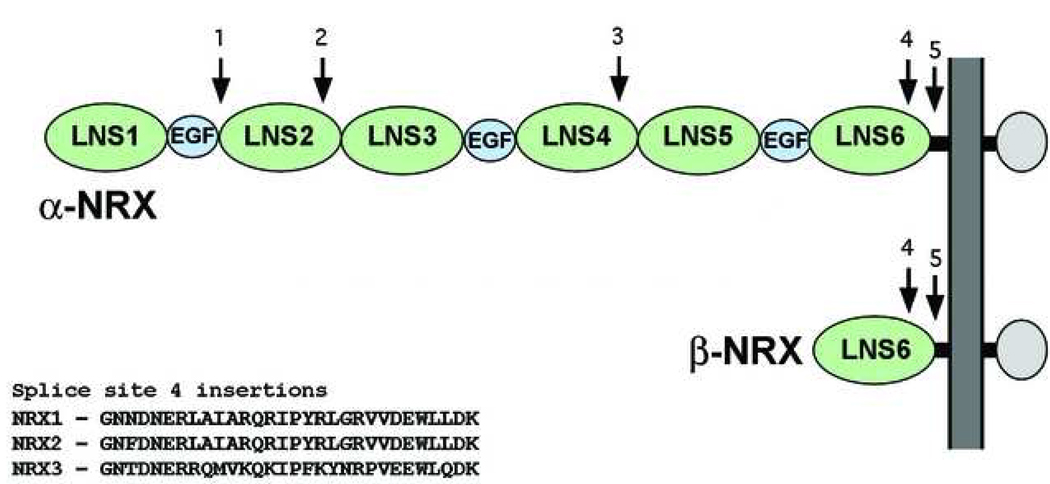
Neurexins and neuroligins are diversified by alternative mRNA splicing such that up to four variant mature proteins are produced for each neuroligin gene, and up to thousands are produced for each neurexin gene. The neurexin family includes three members in mammals, NRX1–3 (Ichtchenko et al., 1996). Each family member is encoded by a gene that includes two independent promoters, which are either used to express a long α-NRX, or a truncated β-NRX with a unique N-terminus (Fig. 1) (Ushkaryov and Sudhof, 1993). Both forms undergo alternative splicing at five different sites (termed sites 1–5) in α-NRX and two sites (sites 4 and 5) in β-NRX (Ichtchenko et al., 1996; Tabuchi and Sudhof, 2002; Ushkaryov and Sudhof, 1993).
Figure 1.
The α- and β-neurexins are transcribed from alternative promoters of the same genes. α-neurexins contain an N-terminal signal peptide, which is removed from the mature protein, followed by neurexin repeats, consisting of LNS (green) and EGF-like domains (blue). Both share a common transmembrane segment (TM), and a short cytoplasmic region. The β-neurexins contain a different signal peptide and a short β-neurexin specific sequence and share membrane proximal regions, including the final LNS domain, LNS6, with their α counterparts. Positions of the five alternative splice sites of α-neurexins are numbered and indicated by arrows.
Neuroligins, the post-synaptic binding partners of neurexins, are also diversified by alternative splicing. Five members of the NL family are known in humans (Bolliger et al., 2001; Ichtchenko et al., 1995; Ichtchenko et al., 1996; Jamain et al., 2003) and at least four genes have been discovered in rodents to date, Nlgns 1–3 (Varoqueaux et al., 2006), and a more distant member Nlgn6 (Genbank accession number EF692521). Two potential sites of alternative splicing, A and B, are found in the Nlgn genes. Nlgn3 has two alternative exons that can be inserted at site A; the B splice insertion site is only used in Nlgn1. This gives rise to a total of 10 different isoforms for Nlgn1–3 (four for Nlgn1, two for Nlgn2, and four for Nlgn3). NL subtypes are differentially localized in vivo: NL1 is found predominantly at excitatory synapses, NL2 at inhibitory synapses and NL3 at both. (Budreck and Scheiffele, 2007; Song et al., 1999; Varoqueaux et al., 2004). Recent protein interaction and cell culture studies indicate that alternative splicing also confers functional properties for both NRXs and NLs: For example, an insert at splice site 4 of β-NRXs selectively promotes GABAergic postsynaptic assembly (Chih et al., 2006; Graf et al., 2006), whereas an insert at site B of NL1 promotes selective function at glutamatergic synapses (Chih et al., 2006). These findings and the considerable molecular diversity of NLs and NRXs and the differential effects of some isoforms has led to the hypothesis that alternative splicing in NRXs and NLs might underlie a selective adhesive code for CNS synapses.
Initial insights into the structural basis of β-NRX/NL interaction have been acquired through solution small angle X-ray scattering (SAXS) experiments (Comoletti et al., 2007). This study yields insight into the overall shape of the complex, and shows that neuroligins likely dimerize via a four-helix bundle motif characteristic of the related cholinesterases. One β-NRX molecule binds to each NL protomer to form a 2:2 complex, with the two β-NRX molecules separated by ∼107Å. Nevertheless, atomic-level mechanisms of β-NRX/NL interactions remain unclear.
The neuroligin-binding region of β-neurexin ectodomains is composed of a single Laminin-Neurexin-Sex hormone (LNS) domain. Two LNS domain crystal structures from neurexins have been previously determined. The first of these (Rudenko et al., 1999), corresponding to the neuroligin-binding β–NRX1 LNS domain from mouse, revealed a structure similar to that of agrin, another LNS domain-containing protein. Agrin is also alternatively spliced, and the positions of alternative splice sites are conserved between agrin and β-NRX1 (Rudenko et al., 1999). Alternative splicing of agrin is thought to play a regulatory role in organization of neuromuscular junctions (Ferns et al., 1993). A recent functional mutagenesis study has confirmed that binding to neuroligins and synaptogenic activity is mediated by the same face of the β-NRX1 LNS domain that contains the alternative splice sites (Graf et al., 2006).
The other LNS-domain crystal structure known for neurexins is that of the second LNS domain of α-neurexin 1, which is not known to interact with neuroligins. This structure revealed a Ca2+ ion bound near the positions of the alternative splice sites (Sheckler et al., 2006). Bound Ca2+ was not found in the original structure of β-NRX1; however the crystallization conditions were devoid of calcium. Sequence analysis reveals conservation of the Ca2+ ligating residues, suggesting that β-neurexins may also bind calcium at this position. Mutation of two of these putative calcium binding residues was found to ablate neuroligin interaction (Graf et al., 2006). Neurexins have three known extracellular binding partners in the mammalian brain: neuroligins, dystroglycan and neurexophilins (Ichtchenko et al., 1995; Petrenko et al., 1996; Sugita et al., 2001). Neuroligins exhibit Ca2+-dependent binding to α- and β-neurexins, dystroglycan shows Ca2+-dependent binding preferentially to α-neurexins, and neurexophilin binding is Ca2+-independent and specific to α-neurexin.
Here we report crystal structures for the Δ splice forms of the membrane-proximal neuroligin-binding LNS domains from β-NRX1 and β-NRX2, determined in the presence of Ca2+ ions. The 1.7Å resolution structure from β-NRX1 reveals a bound Ca2+ ion, ligated by side chains from residues D137 and N208, and carbonyl oxygen atoms from V154 and I206, corresponding to a similar Ca2+ binding site identified in structures of another LNS domain. The structure presented shows that the Ca2+ ion is incompletely coordinated by the β-NRX1 molecule, and coordination is completed by a glutamic acid side chain from another molecule. This is similar to findings for the ligand-binding A domains from integrins, which contain a metal ion-dependent adhesion site (MIDAS) organized around an incompletely coordinated Mg2+ ion (Lee et al., 1995). The Ca2+ binding site of the β-neurexin ectodomain involves elements of the β9-β10 loop that also contains splice insertion 4. This region of the molecule is disordered in the 3.0Å β-NRX2 structure, indicating substantial molecular mobility in the neuroligin binding region. NMR chemical shifts and nuclear Overhauser effect measurements for the Δ and +4 splice forms of β-NRX1 suggest that the peptide segment inserted at splice insertion 4 is disordered both in the absence of neurexin-binding partners and in the presence of at least one binding partner, neuroligin. The apparent lack of participitation of the splice insertion sequence in binding to NL suggests that β-NRX insertion sequences might play roles independent of neuroligin binding.
Results
Production of recombinant neurexins and crystallization
We focused on the single LNS domain of the β-neurexins ectodomain segment, which contains the neuroligin-binding site of β-neurexins, as well as a single site of alternative splicing, splice insertion site 4. We thus produced each of the neurexins in two splice forms: one that included splice insertion sequence 4 (referred to as +4), and another lacking this insertion (referred to as Δ). Extracellular domains of β-NRX were expressed as GST-fusion proteins in E.coli. We used a Mosquito crystallization robot to screen through commercially prepared sets of crystallization reagents. We found crystallization “hits” for each of the neurexins lacking splice insertion sequences: β-NRX1Δ, β-NRX2Δ, and β-NRX3Δ. Crystals of β-NRX1Δ and β-NRX2Δ were optimized, and we present structures here at 1.7Å and 3.0Å resolution for β-NRX1Δ and β-NRX2Δ, respectively. Despite extensive screening (at least 1000 conditions for each protein at each 4°C and 20°C) no crystals could be obtained for any of the neurexins that included a splice insertion sequence.
Biophysical analysis of neurexins and complexes with neuroligins
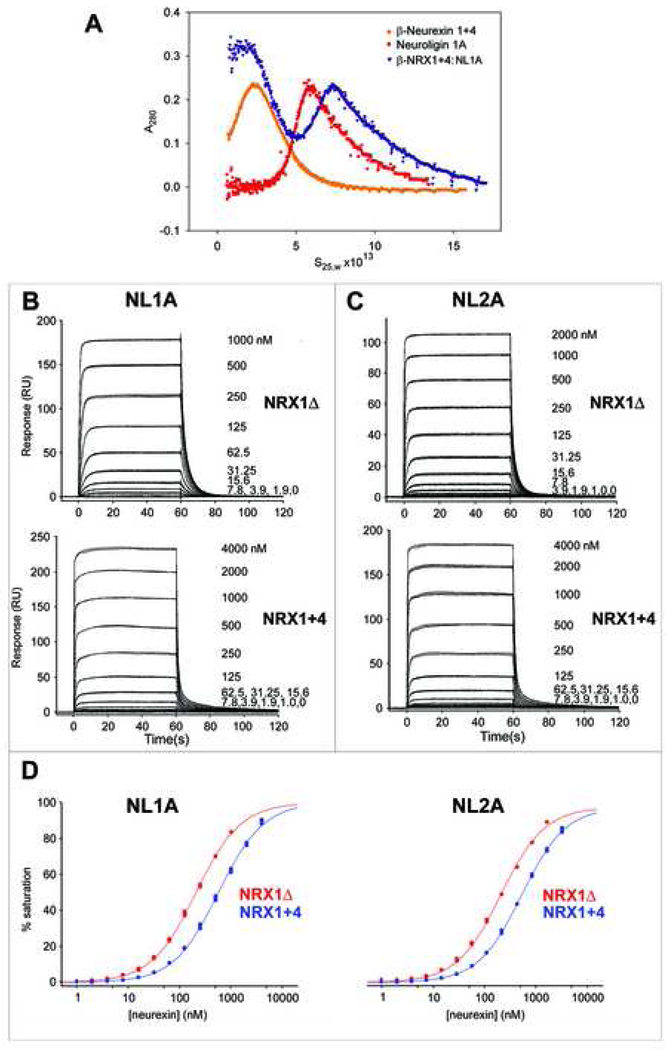
In order to determine the oligomeric states of the neurexins and neuroligins used in our studies, and to characterize their association as complexes, we used multiple methods of biophysical analysis. First, equilibrium analytical ultracentrifugation (AUC) was used to characterize the association states of the LNS regions from β-NRX1Δ, β-NRX1+4, and β-NRX2Δ. The solubility of the β-NRX2+4 protein was too low for accurate AUC determinations, yielding precipitate in later stages of the run. The results of these experiments (data not shown) show that the LNS domains from β-NRX1Δ, β-NRX1+4, and β-NRX2Δ, all behave as monomers, and reveal no evidence of higher order associations. These results agree with earlier AUC experiments on neurexins (Comoletti et al., 2006). Second, we produced β-NRX1:NL1A ectodomain complexes by incubating a slight molar excess of either β-NRX1Δ or β-NRX1+4 ectodomains with the soluble ectodomain from NL1A, and separated complexes from unbound β-NRX1 by gel filtration chromatography. In these experiments, using a calibrated Superdex S200 column, β-NRX:NL complexes eluted at a volume corresponding to molecular mass ∼190kDa, suggesting a 2:2 complex. Sedimentation velocity experiments on isolated β-NRX1+4:NL1A complex (Fig. 2A) yielded a sedimentation coefficient of ∼7.3s, consistent with a 2:2 β-NRX:NL complex as previously reported (Comoletti et al., 2006).
Figure 2.
SPR binding analysis of β-NRX1Δ and 1+4 with NL1A and NL2A. Sensograms of the interactions of β-NRX1Δ and β-NRX1+4 with immobilized NL1A (A) and NL2A (B). Binding was tested at the concentration range indicated in each panel. (C) Binding isotherms for β-NRX1Δ (shown in red) and 1+4 (shown in blue) interacting NL1A and 2A. The responses vs. concentration plots were fit using a 1:1 interaction model. The KD for each interaction is listed in table 1. The two plots are drawn to scale.
Splice insertion sequence 4 weakens β-NRX1 interactions with NL1A and NL2A
Surface plasmon resonance (SPR) binding assays were used to determine the effect of the insert at splice site 4 of β-NRX1 in β-NRX1/NL interactions (Fig. 2 and Table 1). Purified neuroligins were tethered to the chip surface. This experimental configuration avoids the avidity effects that would result from using the dimeric neuroligins as the mobile analyte. Avidity enhancement by analyte multimerism can contribute to substantial artificial enhancement of measured KDs (Myszka, 1999). Preliminary binding tests showed that β-NRX1Δ4 and especially β-NRX1+4 injected at 2 and 4 µM respectively bound nonspecifically to the dextran layer of the sensor chip. Addition of BSA in the running buffer was sufficient to suppress such nonspecific interactions.
Table 1.
KDs for the interaction of β-NRX1Δ and 1+4 with NL1A and 2A
| KD(nM) | KD ratio relative to Δ isoform | ||
|---|---|---|---|
| NL | β-NRX | ||
| 1A | 1Δ | 206.9(8)1 | 1 |
| 1+4 | 571(2) | 2.8 | |
| 2A | 1Δ | 238.3(7) | 1 |
| 1+4 | 621.2(8) | 2.6 |
The number in brackets represents the error of the fit in the last significant digit.
Fig. 2B shows high-quality sensograms for the interaction of β-NRX1Δ4 and β-NRX1+4 with NL1A. The triplicate injections for each concentration were highly reproducible indicating that β-NRX1Δ4 and β-NRX1+4 binding to NL1A was specific. The binding reactions reached equilibrium at all concentrations tested. The responses at equilibrium were plotted against the concentration of neurexin to calculate the KD for each interaction using a 1:1 binding model (Fig.2D). The β-NRX1Δ4:NL1A interaction had a KD of 206.9(8) nM compared to the β-NRX1+4:NL1A interaction, which had a KD of 571(2) nM. (The number in parentheses for all KDs represents the error of the fit in the last significant digit.) Thus, the presence of the insert at splice site 4 of β-NRX1 weakens binding almost three-fold.
To investigate whether the change in affinity in the presence of the insert at splice site 4 of β-NRX1 is isoform specific, the interactions of β-NRX1Δ4 and +4 were also examined with NL2A (Fig. 2C). The sensograms exhibit the same general behavior as with NL1A, with highly reproducible responses that reach equilibrium at all concentrations tested. From the equilibrium-binding analysis in Fig.2D, the KD for the β-NRX1Δ4/NL2A interaction was 238.3(7) nM, while the KD for the NRX1+4/NL2A was 621.2(8) nM. Just as we found for NL1A, the presence of the insert at splice site 4 of β-NRX1 weakened binding approximately 2.6-fold, suggesting that the presence of splice insertion 4 causes a moderate decrease in NL binding affinity. A previous study reported that the presence of splice insert 4 reduces the relative affinity of β-NRX1 for NL1A by 2.1-fold, but enhanced the relative affinity of β-NRX1 for NL2A (Comoletti et al., 2006). This study was also based on SPR binding assays, however the limited quality of the SPR data prevented the authors from calculating the absolute affinities for these interactions; estimation of relative KDs from such “comparative” SPR analyses is known to be unreliable (van Holde et al., 2005).
NMR experiments show that splice insertions at β-neurexin splice site 4 are disordered, even in complex with neuroligin
Our inability to obtain crystals of neurexins that include splice insertion sequences suggested that the insertions might be structurally disordered, and thereby hamper crystallization. We therefore undertook solution NMR experiments using 15N-labeled neurexins 1Δ and 1+4, identical but for the presence of the splice insertion sequence.
First, we recorded Fast Heteronuclear Single Quantum Coherence (FHSQC) 1H-15N spectra to assess the foldedness and uniqueness of the conformations of β-NRX1Δ and β-NRX1+4. Second, we measured the steady-state {1H}-15N nuclear Overhauser effect (NOE) to establish the degree of conformational restriction of the polypeptide backbone. Because the amide N-H bond has an essentially fixed length, the steady-state NOE depends only on the motion of each peptide moiety and typically is >0.5 for well-ordered sites in globular proteins.
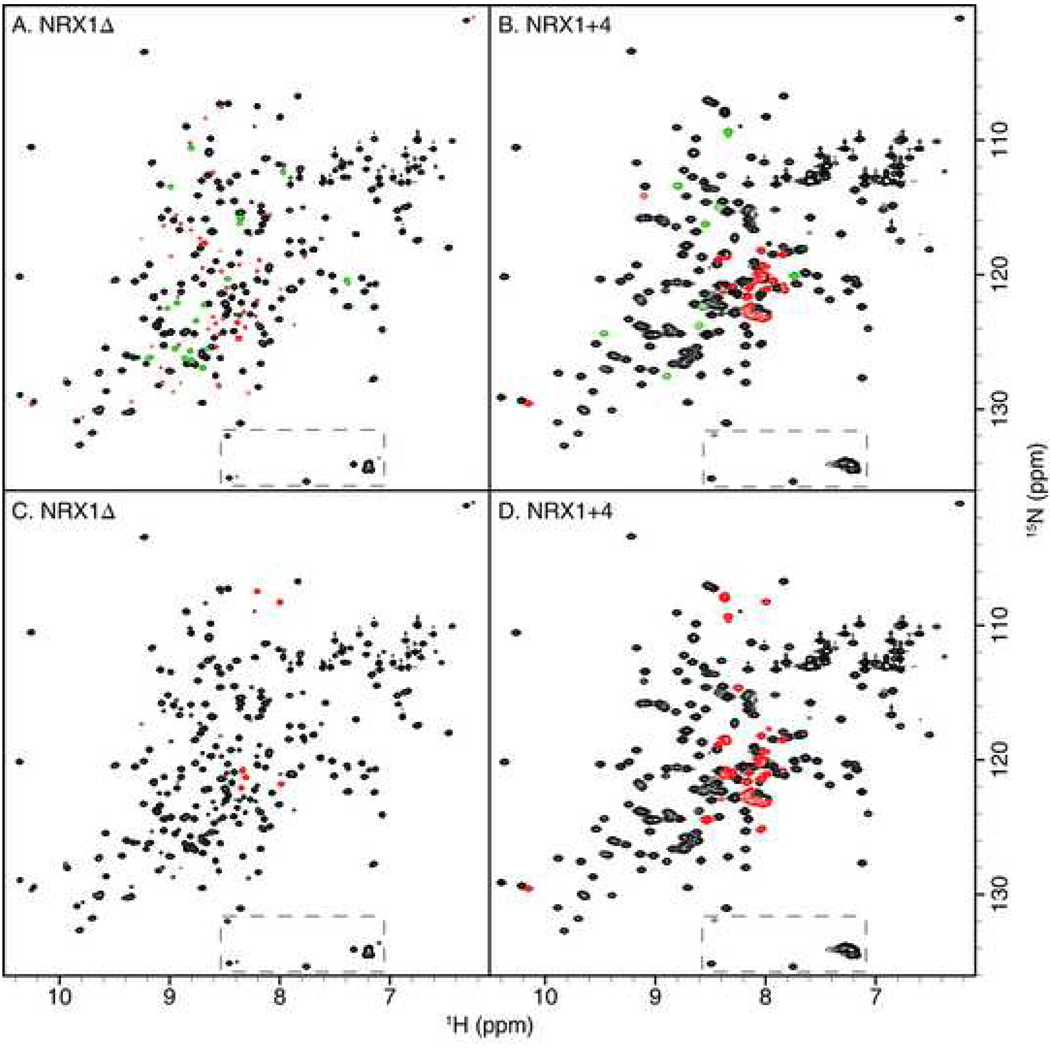
FHSQC spectra for β-NRX1Δ and β-NRX1+4 are shown in Fig. 3. In Figure 3A and 3B, the crosspeaks have been color-coded to indicate whether the peaks are common to both spectra, unique to β-NRX1Δ, or unique to β-NRX1+4. In Figure 3C and 3D, the crosspeaks have been color-coded to indicate whether the {1H}-15N NOE is less than or greater than 0.5 (a value distinguishing relatively ordered from relatively disordered sites).
Figure 3.
FHSQC spectra of NRX1Δ (A, C) and NRX1+4 (B, D). Dashed boxes indicate aliased resonances from arginine Nε-Hε groups. A, B: Resonances common to the spectra of both isoforms are shown in black, resonances new to either spectrum are shown in red, and resonances that could not be clearly assigned to the first two categories are shown in green. C, D: Resonances with a heteronuclear {1H}-15N NOE value smaller (larger) than 0.5 are shown in red (black).
The FHSQC spectrum of β-NRX1Δ contains 214 backbone crosspeaks, more than the expected number of 166 based on the protein sequence. Gel filtration chromatography, native gel electrophoresis, and mass spectrometry indicate that the β-NRX1Δ sample is chemically homogeneous (data not shown), strongly suggesting a dynamic interchange between two conformations in equilibrium. A 15N ZZ-exchange experiment (Farrow et al., 1994b) with a mixing time of 1 s did not show any exchange crosspeaks, demonstrating that the kinetic rate constants for exchange between conformations is < 1 s–1 (data not shown). Of 214 crosspeaks, 148 crosspeaks could be associated with crosspeaks in the spectrum of β-NRX1+4 and 49 crosspeaks were unique to the β-NRX1Δ spectrum. The 49 unique peaks had a lower average intensity (34%) compared to the remainder of the crosspeaks, but had similar NOEs (0.77 versus 0.78) and 1H resonance linewidths (21.5 s–1 for both). These results suggest that the resonances in common between β-NRX1Δ and β-NRX1+4 represent a major species present in the sample of β-NRX1Δ with a similar conformation as β-NRX1+4. Furthermore, the unique set of resonances reflect a minor species with similar dynamic properties as the major species, which is hence also likely to be a globular conformational state.
The FHSQC spectrum for β-NRX1+4 contains 181 crosspeaks; 148 of these, as noted above are associated with corresponding major peaks in the β-NRX1Δ spectrum. Few crosspeaks in the spectrum of β-NRX1+4 are associated with the resonance shifts of the minor form of β-NRX1Δ. Instead, a total of 24 crosspeaks unique to the β-NRX1+4 spectrum appear mostly at the resonance frequencies expected for a disordered random peptide chain. Furthermore, one of these unique crosspeaks is found in the region of the spectrum characteristic of Trp side chain indole 15Nε1−1Hε1 resonances, likely corresponding to the indole of Trp226 in the splice insertion. The majority (20/24) of the unique peaks, including the putative indole of Trp226, have NOEs < 0.5 compared to an average of 0.70. These data suggest that inclusion of the splice insertion sequence shifts the conformational equilibrium that leads to supernumerary peaks for β-NRX1Δ to favor the major species, and that the splice insertion itself gives rise to the group of peaks, not present in the Δ isoform, which appear to be disordered.
Taken together, these data strongly imply that (i) the major species of β-NRX1Δ and β-NRX1+4 have similar structures; (ii) a minor species is populated in β-NRX1Δ, but not β-NRX1+4; and (iii) in the absence of a binding partner, the splice insertion sequences of β-neurexins are disordered. The very slow conformational equilibrium observed in β-NRX1Δ may reflect slow-timescale dynamics of the long β9-β10 loop that contains the splice insertion site. Insertion of 30 additional residues into this loop might be responsible for shifting the equilibrium towards the major conformation. The position of the splice insertion into a long loop combined with the flexibility of the splice insertion found by NMR likely explains our inability to produce crystals of lone neurexins containing these sequences.
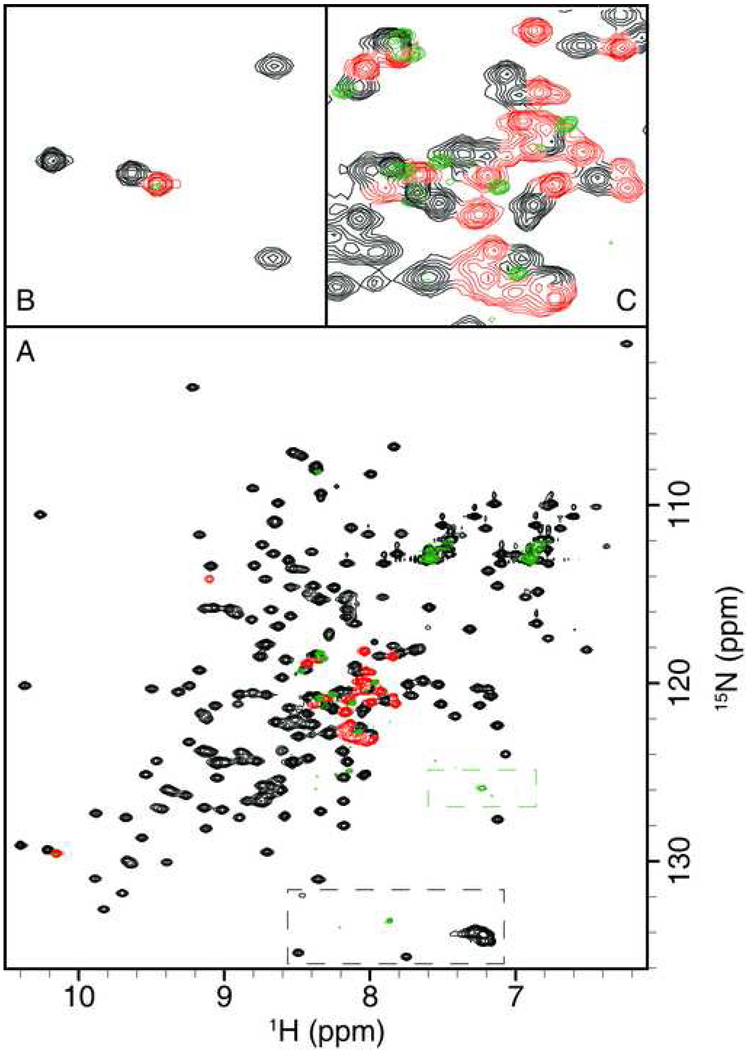
An FHSQC spectrum also was recorded for a complex between [U-99% 15N]- β-NRX1+4 and unlabeled NL1A (NL1 containing splice insertion A). The complex is 2:2 β-NRX1+4:NL1A with a total molecular mass of ∼190kDa. As shown in Figure 4, resonances for well-ordered amide groups are nearly completely absent from the spectrum owing to fast transverse relaxation. In contrast, a set of resonances is observed at resonance frequencies corresponding to amide groups with NOEs < 0.5 in the free β-NRX1+4 protein, most of which are unique to β-NRX1+4. Observation of these resonances suggests that the splice insertion in β-NRX1+4 remains largely disordered in the complex with NL1A. Importantly, the putative Trp226 indole resonance also is observed in the spectrum of the complex (see Fig. 4B); the large hydrophobic side chain of Trp226 would be expected to be part of an intermolecular interface, and consequently well-ordered, if the splice insertion sequence contributed significantly to binding between β-NRX1+4 and NL1A.
Figure 4.
A: Overlay of the FHSQC spectrum of [U-99% 15N]-NRX1+4 in complex with unlabeled NL1A (green) with the FHSQC spectrum of [U-99% 15N]-NRX1+4 alone (black and red). In the FHSQC spectrum of the uncomplexed NRX1+4, resonances not observed for NRX1Δ are shown in red, and all remaining resonances in black. Dashed boxes (black for the spectrum of the uncomplexed NRX1+4, green for the spectrum of the complex) indicate aliased resonances from arginine Nε-Hε groups. B: Magnification of the region between 126.2 ppm and 132.4 ppm (F1) and 9.77 ppm and 10.51 ppm (F2), which is characteristic of Trp indole Nε1-Hε1 resonances. C: Magnification of the region between 117.8 ppm and 123.7 ppm (F1) and 7.78 ppm and 8.51 ppm (F2), which is characteristic of disordered backbone amide resonances.
Structures of β-Neurexin 2Δ and β-Neurexin 1Δ Ca2+ complex
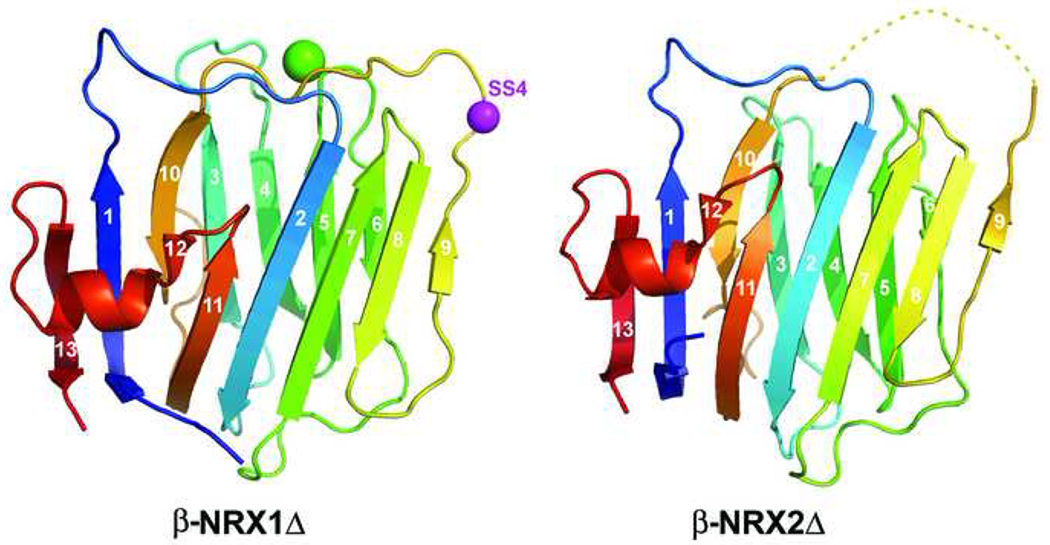
The β-NRX1Δ structure was determined by molecular replacement using PDB 1C4R as a search model (Table 2). The final structure, refined at 1.7Å resolution (R=0.176, Rfree=0.220) had 1 molecule per asymmetric unit, 1481 protein atoms, 188 water molecules, and one Ca2+ ion bound per neurexin protomer. Using this structure as search model, we determined the structure of β-NRX2Δ at 3.0Å resolution (R=0.211, Rfree=0.289; because of the limited resolution, this model includes no water molecules). In contrast with the β-NRX1Δ structure, in β-NRX2Δ the Ca2+ binding region is disordered, and no electron density is seen for a calcium ion or the expected calcium binding ligands (Fig. 5).
Table 2.
Data collection and refinement statistics
| β-NRX1 | β-NRX2 | |
|---|---|---|
| Data collectiona | ||
| Space group | P212121 | C2 |
| a, b, c (Å) | 45.0, 49.1, 63.6 | 95.5, 97.0, 62.6 |
| α,β,γ(°) | 90, 90, 90 | 90, 106.0, 90 |
| Resolution (Å) | 1.70 (1.76–1.70) | 3.00 (3.11–3.00) |
| Rsym | 0.058 (0.193) | 0.116 (0.430) |
| I / σI | 27.3 (6.4) | 6.5 (2.0) |
| Completeness (%) | 99.5 (95.5) | 98.7 (99.6) |
| Redundancy | 6.5 (4.7) | 3.3 (3.4) |
| Refinement | ||
| Resolution (Å) | 20–1.7 | 20–3.0 |
| No. reflections | 15154 | 10170 |
| Rwork / Rfree | 0.176/0.220 | 0.211/0.289 |
| B-factors (Å2) | ||
| All atoms | 16.3 | 41.1 |
| Protein | 15.0 | 41.2 |
| Water | 26.7 | 35.3 |
| Ca2+ | 13.9 | N/A |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.012 | 0.008 |
| Bond angles (°) | 1.42 | 1.29 |
| Ramachandran angles | ||
| % favored | 87.8 | 83.8 |
| % allowed | 11.5 | 15.3 |
| %genereously allowed | 0.7 | 1.0 |
| %disallowed) | 0.0 | 0.0 |
Rfree = Rwork calculated using 5% of the reflection data chosen randomly and omitted from the start of refinement.
Figure 5.
Structure of Δ-isoform LNS domains from β-NRX1 and β-NRX2. The ribbon diagrams are colored in a rainbow from blue to red from N- to C-terminus. β-strands are numbered, and the bound Ca2+ ion in the NRX1 structure is shown as a green sphere. The α-carbon of residue 200, the position of splice insertion 4, is shown as a magenta sphere. An eight residue stretch in the β9-β10 loop of NRX2 is disordered and indicated as a dashed yellow line.
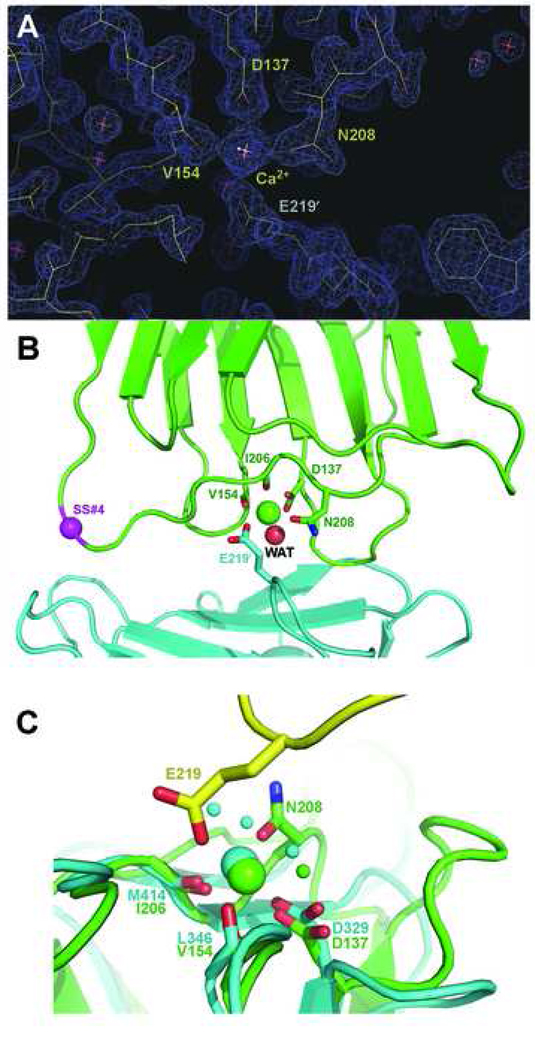
The neurexins adopt an all-β topology related to that of many lectins. The 1.7Å structure of β-NRX1Δ reveals clear electron density for the Ca2+ ion and its protein ligands (Fig. 6A). Ca2+ is ligated by residues in multiple loops at the membrane-distal end of the neurexin ectodomain, including side chain oxygen atoms of Asn 208 and Asp137, and backbone carbonyl groups from Val 154 and Ile 206, and is additionally coordinated by the side chain of Glu 219 from a symmetry-related protomer. Site-directed mutation of both residues whose side chains are involved in calcium binding, Asn 208 and Asp137, result in loss of NL binding (Graf et al., 2006), indicating that metal coordination in the LNS domain plays an important role in molecular recognition. The β-NRX1Δ structure reported here reveals an “extra” Ca2+ coordination position that is fortuitously occupied by a Glu side chain. This suggests the possibility that binding of NLs may exploit the β-NRX Ca2+ site in a similar way.
Figure 6.
Structure of the Ca2+ binding region from β-NRX1, and comparison with LNS2 from α-NRX1. (A) 1.7Å 2Fo-Fc electron density contoured at 1.0σ in the region of the Ca2+ binding site. (B) The Ca2+ ion is ligated by four ligands in the Ca2+ binding site, one water molecule, and a glutamic acid side chain from a symmetry mate. (C) Superposition of LNS2 from α-NRX1 (blue) on the β-NRX1 structure (green and yellow) shows that the Ca2+ binding sites are positioned identically in each LNS domain. Ligation by the N208 side chain appears to be common to all the β-NRX, but absent from many other LNS domains of α-NRX.
Splice insertion 4 of β-NRX1 is at position 200 (magenta sphere in Fig 5 and Fig 6B), where the 30 residue insertion sequence is located in β-NRX1+4. This site is only 12.4 Å from the bound Ca2+ ion measured from the Cα of Arg 200 in the β-NRX1Δ structure. Thus both the Ca2+ binding site implicated in direct neuroligin interactions and the splice insertion site appear to compose parts of a localized site that could provide a diversified surface for ligand interaction.
The structure of β-NRX2 was determined from crystals grown in conditions that include Ca2+ ions, however no density is observed for a Ca2+ ion or for the β9-β10 loop of the Ca2+ binding region. The apparent mobility of this region in β-NRX2 raises the possibility that the well-ordered structure observed for β-NRX1 might arise from the fortuitous ligation of the bound Ca2+ ion by a side chain from a crystal mate. However, it also remains possible that the low pH of these crystals (pH 4.9) impaired Ca2+ binding, which could also underlie the observed lack of defined structure. The disordered region of the β-NRX2 β9-β10 loop also includes residue 200, the position of splice insertion site 4. The possibility that this region is mobile agrees with the NMR data presented above: First, supernumerary cross peaks in the HSQC fingerprint region of β-NRX1Δ could indicate exchange between conformational isomers. Second, this conformational exchange likely takes place in regions near splice site 4 because inclusion of a splice insertion at this position collapses the spectrum to that of an apparent single molecular species.
Comparison of calcium binding in β-NRX1 and LNS domain 2 from α-NRX1
The structure of LNS domain 2 from α-NRX1 (Sheckler et al., 2006) revealed a Ca2+ ion bound in a position nearly identical to that reported here for β-NRX1. However, Ca2+ is coordinated differently in these domains (Fig. 6C). Each structure reveals 6 oxygen-containing ligands, including water molecules, providing octahedral coordination. In β-NRX1, four ligands are from the protein itself, one from a Glu side chain of a symmetry mate, and one water molecule. In contrast, in α-NRX1 LNS domain 2, Ca2+ is coordinated by three protein ligands and three water molecules.
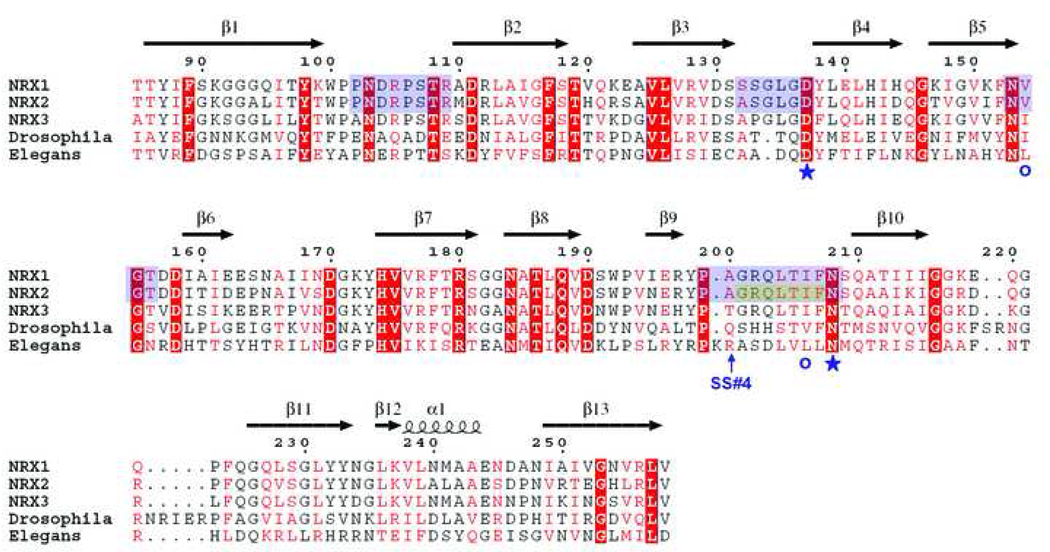
In β-NRX1, two main chain carbonyl groups and side chains from two residues, Asp 137 and Asn 208, coordinate the calcium ion. Structural alignment with the α-NRX1 LNS domain 2 structure shows that the positions of main chain carbonyl ligands and an Asp side chain are almost identical between the two structures. However, the side chain from Asn 208 found in the β-NRX1 structure has no counterpart in α-NRX1 LNS domain 2. Sequence alignments show that Asn 208 is conserved in the LNS domains of β-NRX from Drosophila to man, but this residue is not conserved in the other LNS domains of neurexins.
Comparison of β-NRX1 and β-NRX2 structures
The amino acid sequences of the β-NRX1 and β-NRX2 LNS domains are 78% identical, and their overall structures are very similar. The RMS difference of Cα coordinate positions is 0.98 Å for 157 atom pairs. RMS Cα coordinate for β-NRX1 and β-NRX2 with LNS domain 2 from α-NRX1 is larger, 1.15 and 1.18 Å for 151 and 140 atom pairs, respectively.
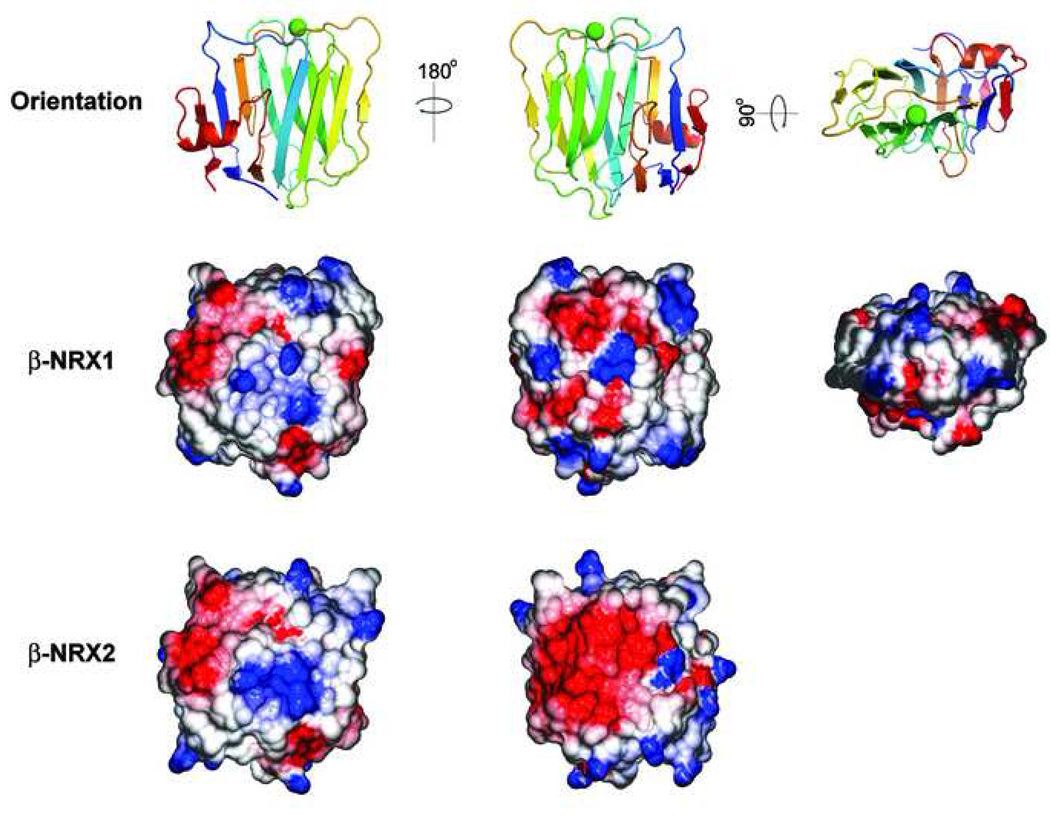
The residues that contribute Ca2+ binding side chain ligands are conserved (as they are in the LNS domains of all β-neurexins). Despite the disordered state of the Ca2+ binding region in the β-NRX2 structure, the surface residues on this face of the molecule are remarkably similar. The Ca2+ binding site is formed from residues donated from the β3-β4, β5-β6, and β9-β10 loops, and is situated on an edge of the β-sandwich that also contains elements from the β1-β2 loop. Since neurexin-neuroligin interaction is strictly Ca2+-dependent, this edge of the molecule is most likely to interact with neuroligins. Remarkably, the residues in each of these loops – including those disordered in β-NRX2 – are identical between β-NRX1 and β-NRX2, and there is only a single substitution in β-NRX3.
To find possible regions of functional difference we examined the molecular surface charge potentials of the β-NRX1 and β-NRX2 structures (Fig. 7) using the program GRASP2 (Petrey and Honig, 2003). The surface potentials are very similar, except that in β-NRX2 the surface of the β1-containing sheet has a large area of negative charge potential. This is not due to the presence of additional negatively charged side chains, but to the absence of two positively charged side chains found in β-NRX1, Lys 147 an Lys 151.
Figure 7.
Comparison of electrostatic surface potential in β-NRX1 and β-NRX2. Surface potentials calculated using the program GRASP (Petrey and Honig, 2003) are displayed in blue (positive) and red (negative) for the potential range +5 to −5 kT. The “top” surface is not shown for β-NRX2 because eight residues in the β9-β10 loop which contribute the top surface are disordered.
Discussion
The data presented here adds to prior studies which suggest a mechanism for Ca2+-dependent ligand recognition for neurexins. Prior mutagenesis data implicates the Ca2+ binding site of neurexin LNS domains in binding their ligands. β-NRX1 loses its ability to bind neuroligin (Graf et al., 2006), and α-NRX1 LNS domain 2 cannot bind dystroglycan when the residue corresponding to Asp 137 is mutated (Sugita et al., 2001). The β-NRX1 ectododmain crystal structure presented here shows that Ca2+ is coordinated incompletely by β-NRX1, leaving open liganding positions that can be occupied by functional groups donated from a partner molecule. In the β-NRX1 structure such an “extra” ligand is fortuitously donated by a Glu side chain from a symmetry-related β-NRX1 protomer. This suggests the possibility that neuroligins, and other NRX ligands may bind similarly, by sharing Ca2+ coordination with NRX. This mode of interaction is reminiscent of adhesive binding in integrins through their MIDAS motif (Lee et al., 1995). The magnesium ions bound at the MIDAS sites of integrins exhibit octahedral coordination, which is common for magnesium centers. In contrast, although Ca2+ ions are sometimes found in octahedral geometry, it is more common that they adopt pentagonal bipyramid coordination (Nakayama and Kretsinger, 1994). How octahedral calcium coordination is important for NRX ligand binding characteristics is not yet clear. Other than their common coordination of an “extra” Glu ligand to complete the metal coordination sphere, the NRX and integrin MIDAS sites are structurally dissimilar.
Ca2+ binding in the neuroligin-binding β-NRX LNS domains differs from that found for the dystroglycan-binding LNS domain 2 from α-NRX. Although the Ca2+ ions are bound at essentially identical positions within each domain, the coordination chemistry differs. Interestingly, in both structures, Ca2+ is coordinated by NRX in an octahedral geometry rather than the more common pentagonal bipyramid coordination found for many proteins including those of the EF-hand and cadherin families (Nakayama and Kretsinger, 1994). β-NRX donates 4 ligands: 2 carbonyl oxygen atoms, and side chain oxygen atoms from the β-NRX conserved side chains Asp 137 and Asn 208. In contrast, LNS domain 2 from α-NRX donates only three ligands because Asn 208 is replaced by a Ser in this protein, and does not partake in Ca2+ ligation. The more complete coordination of Ca2+ in β-NRX may imbue greater binding affinity for the ion, and also may alter binding specificity for partner proteins.
Since the β-NRX Ca2+ binding surface is strongly implicated in binding to neuroligins, it is remarkable that the residues that make up this edge of the LNS domain, which includes contributions from the β1-β2, β3-β4, β5-β6, and β9-β10 loops, are essentially identical in neurexins 1, 2, and 3 (Fig. 8). Promiscuous binding between the various β-NRX and NLs can thus be understood through the near identity of the β-NRX at their likely NL binding surface. Whether other β-NRX regions participate in binding is now unclear and must await determination of complex structures. Although clear differences in the β-NRX1 and β-NRX2 ectodomain structures are apparent, like the large negative surface on the β1-containing sheet unique to β-NRX2, the functional significance of these differences is not yet clear.
Figure 8.
Sequence alignment of the LNS domains from rat β-NRX1, 2, and 3, and the ectodomains from Drosophila and C. Elegans neurexins. The secondary structure of β-NRX1 is shown above the alignment. Residues that ligate Ca2+ are denoted as blue stars (side chain ligation) or circles (main chain carbonyl ligation). The position of splice site 4 is indicated. The loop regions that contribute to the Ca2+ binding face of the β-NRX are highlighted in blue. The disordered region of the β-NRX2 structure is highlighted in gray.
The SPR binding data presented here show that the presence of splice insertion sequence 4 in β-NRX1 has a moderate (∼3-fold) inhibitory effect on interactions with both NL1A and NL2A. This moderate weakening of interaction is consistent with the idea that splice insertion sequence 4 may introduce steric hindrance to the β-NRX/NL interaction which would not necessarily be revealed in the semi-quantitative assay systems employed in previous studies (Boucard et al., 2005; Chih et al., 2006; Graf et al., 2006). Although quantitative binding measurements have not yet been performed, prior reports suggest that binding between β-NRX1+4 and NL1B is much weaker than binding between β-NRX1Δ and NL1B or binding between β-NRX1+4 and other neuroligins (Boucard et al., 2005; Chih et al., 2006; Graf et al., 2006). This more severe reduction in binding affinity depends on the presence of glycosylation in the NL1 B insertion sequence. The carbohydrate-dependence suggests that, the primary mechanism that limits interactions with NL1B involves steric hindrance, similar as for β-NRX1+4 interactions with NL1A and NL2A. Notably, the NRX splice insertion 4 does not contain glycosylation sites but the amino acid sequence is highly conserved between species. This high sequence conservation seems at odds with a role solely in steric hindrance, which would not be expected to provide strong evolutionary constraints on sequence. Moreover, the NMR data presented here show that the splice site 4 insertion sequence fails to adopt an ordered structure even in the β-NRX1+4/NL1A complex. This disordered conformation and the high level of sequence conservation suggest the possibility that splice insertion 4 may play another functional role, perhaps in recruiting binding partners, other than neuroligins, to the synapse.
Methods
Protein expression and purification
Based on the crystal structure from Rudenko et al. (1999), we produced expression constructs corresponding to the ordered part of the extra cellular domains for rat β-NRX1 (residues 86–288) and β-NRX2 (residues 87–290). These regions were cloned into the pGEX6p-1 vector (GE Healthcare) to produce an N-terminal GST fusion. Protein was produced by over expression in E.coli BL21 cells grown at 37°C and induced with IPTG. Cells were sonicated in lysis buffer (150mM NaCl, 20mM Tris pH8, 2mM EDTA, 1mM DTT, 1mM TCEP and 100ng/mL Leupeptin), followed by centrifugation at 25,000×g for 1h at 4°C. After centrifugation, the supernatant was passed over a glutathione Sepharose affinity column. Fusion proteins were eluted from the column with 20mM reduced glutathione in lysis buffer. The GST tags were removed by incubation with PreScission protease during dialysis at 4°C over night (150mM NaCl, 10mM Tris pH8, 3mM CaCl2, 1mM DTT). Subsequently, the samples were passed over a MonoQ column equilibrated in 150mM NaCl, 10mM Tris pH8, 3mM CaCl2. The flow-through was applied to a Superdex S75 column. For the purification of β-neurexin2Δ the lysis buffer contained 0.1% (v/v) Triton X-100. Samples for NMR studies were produced as above except that E. coli were grown in M9 minimal medium (Sambrook et al., 2001) prepared with 15NH4Cl (Cambridge Isotope Laboratories).
Neuroligin 1A was produced using a mammalian cell expression system based on the pCEP4 vector from Invitrogen. pCEP4 contains the gene coding for the Epstein Barr nuclear antigen (EBNA) as well as the EBNA replication start site. This combination allows for the episomal replication of the plasmid. We produced pCEP4-based expression constructs encoding the NL1A ectodomain fused to a C-terminal 8xHistidine affinity tag. The expression vector encodes the cDNA for residues 1–612, corresponding to the AChE-like region of the maturely spliced ectodomain including the native NL1signal sequence. Stably transformed neuroligin expressing HEK-293 cells were cultured in 10-layer cell farms (each of which yields 2 liters of conditioned medium). Secreted neuroligin ectodomains were purified by Ni2+ affinity chromatography followed by ion exchange and gel filtration steps. Briefly, conditioned medium was collected and brought to final concentrations of 500mM NaCl, 20mM Tris pH 8.0, 20mM imidazole and 3mM CaCl2. The medium was filtered through a 0.22µm filter and passed over HisTrap HP columns (GE Healthcare). The columns were washed with 500mM NaCl, 20mM Tris pH 8.0, 20mM imidazole and 3mM CaCl2 and the protein eluted with 500mM NaCl, 20mM Tris pH 8.0, 250mM imidazole and 3mM CaCl2. The eluate was dialyzed against 100mM NaCl, 20mM Bis-Tris pH 6.0 and 3mM CaCl2 at 4°C over night and then applied to a Mono S column equilibrated with the same buffer. The flow-through was passed over a Mono Q column equilibrated with the same buffer and eluted with a linear gradient at about 275mM NaCl. Finally, the protein was subjected to size exclusion chromatography (Superdex 200) in 150mM NaCl, 10mM Tris pH 8.0 and 3mM CaCl2.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed using a Beckman XL-I ultracentrifuge in the absorption mode with an An60 Ti rotor equipped with two-channel cells with sapphire windows. All measurements were performed in a buffer containing 10 mM Tris-Cl, pH 8.0, 150mM NaCl, and 3mM CaCl2. Three independent experiments were performed: β-NRX1+4 in isolation at 35µM; NL1A in isolation at 11µM; and a NRX1+4:NL1A mixture composed of 18µM β-NRX1+4 and 6µM NL1A, giving a 3:1 NRX:NL molar ratio. Since each NL dimer is expected to bind two NRX molecules, this constitutes a molar excess of NRX. Samples were spun at 5,000 rpm for 16h at 25 °C, and scans were collected every two minutes. Concentration profile time-derivative analysis was used to determine molecular masses using SedAnal v4.78 (Walter Stafford and Peter Sherwood, http://www.bbri.org/AUCRL.html).
SPR binding analysis
Binding assays were performed using a Biacore T100 biosensor, equipped with a Series S NTA sensor chip (GE Healthcare, Piscataway NJ). Neuroligins were coupled over independent flow cells using a covalent immobilization approach (Willard et al., 2006) at 25ºC in HBS running buffer (10mM HEPES pH 7.4, 150mM NaCl). The flow cell of interest was charged using a 60-second pulse of 500µM of NiSO4 at 20µL/min. The surface was activated using a 7-minute injection of 50mM N-hydroxysuccinimide (NHS) / 200mM 1-ethyl-3(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) (Sigma-Aldrich, St. Louis) at the same flow rate. Nrlg 1A at 16µg/mL, was injected over the activated surface at 20µL/min. Any remaining activated groups were blocked using a 4-minute injection of 1.0 M ethanolamine pH 8.5 (Sigma-Aldrich, St. Louis) at 20µL/min. NiSO4 was removed from the surface using five 10-second injections of 350 mM EDTA at 100µL/min. The levels of immobilized NL1A and NL2A ranged between 500–700 RU. Flow cell 2 remained unmodified to serve as a reference flow cell.
Equilibrium binding analysis was performed at 25ºC in a running buffer of 10mM Tris-HCl pH 7.4, 150mM NaCl, 3mM CaCl2, 0.005% (v/v) P20 supplemented with 0.5 mg/mL BSA for β-NRX1Δ4 and 1 mg/mL BSA for β-NRX1+4 binding experiments. The concentrations of each neurexin tested for binding are shown in Figure 2. Neurexin concentrations were prepared in running buffer using a two-fold dilution series. Samples were injected in order of increasing concentration with each concentration tested for binding in triplicate. Each sample was flowed over the NL-immobilized surfaces for 60s at a flow rate of 50µL/min, followed by a 60s dissociation phase. Running buffer was injected for 60s at 50µL/min at the end of each cycle to minimize sample carryover into the next injection. Buffer blanks were injected throughout the experiment for double referencing (Myszka, 1999).
Data analysis was performed using Scrubber 2.0 (BioLogic Software,Pty., Australia). At equilibrium, the responses were plotted against the neurexin concentrations and fit to a 1:1 binding model to calculate the KD.
Crystallographic analysis
We used a Mosquito crystallization robot to screen through commercially prepared sets of crystallization reagents. We found crystallization “hits” for each of the neurexins lacking splice insertion sequences: NRX1Δ, NRX2Δ, and NRX3Δ. Despite extensive screening (at least 1000 conditions for each protein at each 4°C and 20°C) no crystals could be obtained for any of the neurexins that included a splice insertion sequence. Optimized crystals of NRX1Δ were obtained in 0.1 M Tris-Cl, pH 9.0, 18% PEG 8000. These crystals belonged to space group P212121, with a=45.0, b=49.1, c=63.6Å, had one molecule per asymmetric unit, and diffracted x-rays to 1.7Å resolution. The crystals were cryo-protected for freezing in liquid nitrogen in mother liquor supplemented with 35% sucrose. Optimized crystals of NRX2Δ were obtained in 15% PEG 4000, 0.2M (NH4)2SO4, 0.1M sodium acetate, pH 4.6. These crystals were in space group C2, with a=95.5, b=97.0, c=62.6, β=106.0°, have three molecules per asymmetric unit, and diffract x-rays to 3.0Å resolution. The crystals were cryo-protected for freezing in liquid nitrogen in mother liquor supplemented with 35% glycerol.
The β-NRX1Δ structure was determined by molecular replacement using PDB 1C4R as a search model. The final structure, refined at 1.7Å using CNS (Brunger et al., 1998) and Refmac (Consortium, 1994) resolution (R=0.176, Rfree=0.220) had 1 molecule per asymmetric unit, 1481 protein atoms, 188 water molecules, and one Ca2+ ion bound per neurexin protomer. Using this structure as search model, we determined the structure of β-NRX2Δ at 3.0Å resolution (R=0.211, Rfree=0.289; because of the limited resolution, this model includes no water molecules). NCS restraints were used in the initial stages of refinement for β-NRX2Δ, but were removed for the final few rounds of refinement. Crystallographic statistics are shown in Table 1.
NMR spectroscopy
Samples for NMR spectroscopy were 0.6 mM β-NRX1Δ or β-NRX1+4 for the spectra of the uncomplexed neurexins, and 0.25 mM β-NRX1+4 with 0.27 mM NL1A for the spectrum of the complex (10% D2O/90% H2O, 10 mM Bis-Tris, 150 mM NaCl, 3 mM CaCl2, pH 6.5). An excess of NL1A was used to ensure that all β-NRX1 molecules would be bound to NL1A partners. All spectra of the uncomplexed neurexins were recorded at 300 K using a Bruker DRX600 (Columbia University) NMR spectrometer equipped with a triple-resonance Z-axis gradient cryoprobe. The FHSQC spectrum of the complex was recorded using an Avance800 (New York Structural Biology Center) NMR spectrometer equipped with a triple-resonance Z-axis gradient cryoprobe. Two-dimensional FHSQC spectra of the uncomplexed neurexins were recorded using (256 X 512) complex (t1 X t2) points and spectral widths of (3000 Hz X 8993 Hz) in (F1 X F2), ZZ-exchange spectra of the uncomplexed neurexins were recorded using (256 X 512) complex (t1 X t2) points and spectral widths of (3041 Hz X 8993 Hz) in (F1 X F2), and {1H}-15N NOE spectra of the uncomplexed neurexins were recorded using (512 X 512) complex (t1 X t2) points and spectral widths of (3000 Hz X 8993 Hz) in (F1 X F2). The FHSQC spectrum of the β-NRX1+4:NL complex was recorded using (384 X 1024) complex (t1 X t2) points and a spectral width of (3333 Hz X 14006 Hz). FHSQC (Mori et al., 1995), ZZ-exchange (Farrow et al., 1994b), and {1H}-15N NOE (Farrow et al., 1994a) spectra were recorded using standard pulse sequences. The mixing times for the ZZ-exchange experiment were 0.01, 0.5, 1.0 s. The {1H}-15N NOE experiment used a 1H saturation time of 3 s and a recycle delay of 3 s for the control spectrum. The steady-state NOE was calculated as the ratio of the intensity of the saturated spectrum to the intensity of the control (unsaturated) spectrum. All spectra were processed using NMRpipe (Delaglio et al., 1995) and analyzed using Sparky (T. D. Goddard, and D. G. Kneller, SPARKY 3, University of California, San Francisco).
Acknowledgements
We acknowledge funding from the NIH (GM50291 to AGP, NS045014 to PS, 1U54 CA121852-01A1 to LS). BH is an investigator of the Howard Hughes Medical Institute. NT was supported by a Boehringer Ingelheim Fonds fellowship. X-ray data were acquired at the X4A and X4C beamlines of the National Synchrotron Light Source, Brookhaven National Laboratory. NMR spectrometers used here, and the X4 beamlines are operated by the New York Structural Biology Center. We thank P.D. Kwong for helpful suggestions on the manuscript.
Footnotes
Accession numbers
Coordinates and structure factors have been deposited in the protein databank; accession numbers are 3BOD (β-NRX1) and 3BOP (β-NRX2).
References
- Bolliger MF, Frei K, Winterhalter KH, Gloor SM. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochem J. 2001;356:581–588. doi: 10.1042/0264-6021:3560581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha-and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Brose N. Synaptic cell adhesion proteins and synaptogenesis in the mammalian central nervous system. Naturwissenschaften. 1999;86:516–524. doi: 10.1007/s001140050666. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Budreck EC, Scheiffele P. Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. The European journal of neuroscience. 2007;26:1738–1748. doi: 10.1111/j.1460-9568.2007.05842.x. [DOI] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Science. Vol. 307. New York, N.Y: 2005. Control of excitatory and inhibitory synapse formation by neuroligins; pp. 1324–1328. [DOI] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Sudhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Grishaev A, Whitten AE, Tsigelny I, Taylor P, Trewhella J. Synaptic arrangement of the neuroligin/beta-neurexin complex revealed by X-ray and neutron scattering. Structure. 2007;15:693–705. doi: 10.1016/j.str.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TC. The CCP4 suite: programs for protein crystallography. Acta crystallographica. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dean C, Dresbach T. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 2006;29:21–29. doi: 10.1016/j.tins.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994a;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. Journal of biomolecular NMR. 1994b;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- Ferns MJ, Campanelli JT, Hoch W, Scheller RH, Hall Z. The ability of agrin to cluster AChRs depends on alternative splicing and on cell surface proteoglycans. Neuron. 1993;11:491–502. doi: 10.1016/0896-6273(93)90153-i. [DOI] [PubMed] [Google Scholar]
- Garber K. Science. Vol. 317. New York, N.Y: 2007. Neuroscience Autism’s cause may reside in abnormalities at the synapse; pp. 190–191. [DOI] [PubMed] [Google Scholar]
- Graf ER, Kang Y, Hauner AM, Craig AM. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J Neurosci. 2006;26:4256–4265. doi: 10.1523/JNEUROSCI.1253-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- Missler M, Fernandez-Chacon R, Sudhof TC. The making of neurexins. J Neurochem. 1998;71:1339–1347. doi: 10.1046/j.1471-4159.1998.71041339.x. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. Journal of magnetic resonance. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Kretsinger RH. Evolution of the EF-hand family of proteins. Annual review of biophysics and biomolecular structure. 1994;23:473–507. doi: 10.1146/annurev.bb.23.060194.002353. [DOI] [PubMed] [Google Scholar]
- Petrenko AG, Ullrich B, Missler M, Krasnoperov V, Rosahl TW, Sudhof TC. Structure and evolution of neurexophilin. J Neurosci. 1996;16:4360–4369. doi: 10.1523/JNEUROSCI.16-14-04360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods in enzymology. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Nguyen T, Chelliah Y, Sudhof TC, Deisenhofer J. The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Sheckler LR, Henry L, Sugita S, Sudhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+ binding and the effects of alternative splicing. J Biol Chem. 2006;81:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci U S A. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. Science. Vol. 318. New York, N.Y: 2007. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice; pp. 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Sudhof TC. Neurexin III alpha: extensive alternative splicing generates membrane-bound and soluble forms. Proc Natl Acad Sci U S A. 1993;90:6410–6414. doi: 10.1073/pnas.90.14.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K, Johnson C, Shing HP. Principles of Physical Biochemistry. New York: Prentice Hall; 2005. [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- Willard FS, Siderovski DP. Covalent immobilization of histidine-tagged proteins for surface plasmon resonance. Anal Biochem. 2006;353:147–149. doi: 10.1016/j.ab.2006.02.004. [DOI] [PubMed] [Google Scholar]