Abstract
JNK signaling functions to induce defense mechanisms that protect organisms against acute oxidative and xenobiotic insults. Using Drosophila as a model system, we investigated the role of autophagy as such a JNK-regulated protective mechanism. We show that oxidative stress can induce autophagy in the intestinal epithelium by a mechanism that requires JNK signaling. Consistently, artificial activation of JNK in the gut gives rise to an autophagy phenotype. JNK signaling can induce the expression of several autophagy-related (ATG) genes, and the integrity of these genes is required for the stress protective function of the JNK pathway. In contrast to autophagy induced by oxidative stress, non-stress related autophagy, as it occurs for example in starving adipose or intestinal tissue, or during metamorphosis, proceeds independently of JNK signaling. Autophagy thus emerges as a multifunctional process that organisms employ in a variety of different situations using separate regulatory mechanisms.
Keywords: JNK, autophagy, oxidative stress, signal transduction, Drosophila
Introduction
Autophagy is a catabolic process by which cells degrade and recycle their own constituents through a tightly controlled lysosomal mechanism. In macro-autophagy, the process that we consider here and which we will refer to as autophagy for simplicity, cytoplasmic components, including long-lived proteins and whole organelles, become engulfed in double membrane vesicles dubbed “autophagosomes”. These vesicles then fuse with lysosomes to form autolysosomes in which degradation of the cargo commences. Proteins encoded by so-called autophagy-related genes (ATG), orchestrate different steps of the process that ultimately results in autophagic protein and organelle breakdown.
Autophagy has been implicated in many aspects of homeostasis and development (Levine, 2005; Lum et al., 2005; Rubinsztein et al., 2007; Shintani and Klionsky, 2004), and has been extensively studied in the context of metabolic regulation under conditions of fluctuating nutrient availability. In times of food deprivation, organisms activate autophagy to meet their energy requirements (Levine and Klionsky, 2004; Lum et al., 2005). The mechanisms that control autophagy in response to nutrient deprivation are relatively well understood. In higher metazoans starvation-induced autophagy is chiefly governed by changes in insulin/Tor/PI3K signaling (Arsham and Neufeld, 2006; Shintani and Klionsky, 2004).
Autophagy also makes essential contributions to animal development. During Drosophila metamorphosis, autophagy is required for the large-scale histolysis that breaks down larval tissues and makes their components available for the formation of adult structures (Butterworth et al., 1988; Lee and Baehrecke, 2001; Lee et al., 2002). This process relies on hormonal regulation by ecdysone (Rusten et al., 2004). Similarly, autophagic processes have been implicated in the normal development and morphogenesis of vertebrates (Cecconi and Levine, 2008; Melendez and Neufeld, 2008). For example, autophagy is required for embryonic cavitation (Qu et al., 2007; Yue et al., 2003), and at later stages contributes to the formation of the nervous system and the retina (Fimia et al., 2007; Mellen et al., 2008).
To be beneficial for cells and organisms autophagy has to be tightly controlled. Indeed, misregulated or excessive autophagy has been associated with a number of pathologies, including cancer (Hoyer-Hansen and Jaattela, 2008; Mathew et al., 2007; Yue et al., 2003), neurodegeneration (Hara et al., 2006; Komatsu et al., 2006) and muscle atrophy (Mammucari et al., 2007; Zhao et al., 2007).
Recently, much interest has focused on autophagy as a mechanism by which cells defend themselves against environmental stresses. The notion that autophagy can have cell protective functions first emerged based on the finding that adaptations of several organisms to unfavorable environmental conditions require ATG genes and autophagy. Examples range from sporulation in yeast to the formation of fruiting bodies in Dictyostelium and dauer larvae in C. elegans (Deutschbauer et al., 2002; Melendez et al., 2003; Otto et al., 2004). Autophagy can also confer resistance to oxidative stress. Mutations that compromise the autophagy system result in increased stress sensitivity. Drosophila loss-of-function mutants for ATG7 or ATG8a, for example, are hypersensitive to H2O2 (Juhasz et al., 2007a; Simonsen et al., 2008). Cells can respond to a variety of insults, including oxidative stress with increased autophagic activity (Girardot et al., 2004; Xiong et al., 2007). However, how the autophagy machinery senses and responds to stress is not thoroughly understood. Such regulation could occur at several levels, as autophagy can be regulated by transcriptional, as well as post-transcriptional mechanisms. Consistent with a function of gene regulation in this context, multiple reports show that ATG gene expression can be stimulated in response to stresses (Girardot et al., 2004; Thorpe et al., 2004; Xiong et al., 2007)
The work described here explores the control of autophagy by JNK signaling in Drosophila melanogaster. The JNK pathway is an evolutionarily conserved signal transduction system that can be triggered by several types of external insults, including oxidative stress (Davis, 2000; Weston and Davis, 2007). Stress signals are conveyed by a MAP kinase cascade, which in Drosophila, consists of one of several JNKKKs (Jun kinase kinase kinases), a JNKK, the MKK7 ortholog Hemipterous (Hep) and the JNK, Basket (Bsk). The duration and extent of JNK responses is tightly controlled and restricted in time and space by a number of negative feedback mechanisms. One of these mechanisms relies on the transcriptional activation of the JNK specific MAP kinase phosphatase puckered (puc) (Martin-Blanco et al., 1998).
JNK signaling has been implicated in the regulation of a range of cellular stress responses. These include the induction of antioxidant and repair programs, and, depending on the nature of the inducing signal and the cell type, apoptosis. Such responses can be orchestrated by changes in gene expression mediated by several transcription factors that are regulated by JNK signaling. In addition, JNK can directly regulate cell responses such as apoptosis by phosphorylating effector molecules.
We have shown previously that activation of the JNK pathway can protect fruit flies against oxidative toxicity. For example, flies in which JNK signaling is elevated due to the loss of one copy of the gene encoding the JNK phosphatase Puckered, or by over expressing the JNK kinase Hep, gain resistance to the free radical inducing drug paraquat (Wang et al., 2003; Wang et al., 2005).
JNK signaling emerges as a regulator of multiple mechanisms that cells can engage to increase resistance against external stresses. Recent studies in cell culture models indicate that autophagy is one of these responses (Li et al., 2006; Ogata et al., 2006; Pattingre et al., 2008; Wei et al., 2008). Here we show that autophagy is part of a JNK-induced stress defense program in an intact organism, Drosophila melanogaster, and explore the relationship between stress-induced autophagy and autophagy that is regulated in response to metabolic or developmental signals.
Results
JNK signaling regulates ATG gene expression
Autophagy is a pleiotropically acting process that is regulated at multiple levels including gene expression. In many organisms the transcription of ATG genes, which encode proteins required for the regulation and execution of autophagy, is increased in adverse conditions, such as starvation or stress (Gorski et al., 2003; Juhasz et al., 2007a; Lee et al., 2003; Zinke et al., 2002). To determine whether the expression of ATG genes might be responsive to oxidative stress in Drosophila, we exposed young adult flies (3-5 days old) to paraquat-laced food and measured the mRNA levels of two representative autophagy genes, ATG1 and ATG18.2/CG8678, by quantitative real time RT PCR analysis (Fig. 1A). The abundance of both mRNA species rose transiently under these conditions, and, interestingly, this rise was paralleled by a similar response of the prototype JNK target gene puckered (Fig. 1A). JNK signaling is a key regulator of stress inducible gene expression, and we asked whether ATG genes might, like puckered, be regulated by the JNK pathway. To address this question, we ubiquitously raised JNK activity in larvae by expressing a constitutively active form of the Drosophila JNKK Hemipterous, Hepact, under the control of the temperature inducible Gal4/Gal80ts TARGET system (McGuire et al., 2003; Weber et al., 2000). This system relies on the transcription factor Gal4 under the control of a temperature sensitive version of its selective inhibitor Gal80 (Gal80ts). At the permissive temperature (20°C), Gal80ts blocks Gal4 activity thereby preventing the expression of Hepact from a UAS driven transgene. At the restrictive temperature (29°C), Gal80ts is inactive and Gal4 is free to activate the transcription of Hepact resulting in strong activation of JNK. Using real time RT PCR, we assessed the response of 13 ATG mRNAs to JNK activation in this system. Equivalent measurements were conducted on a control strain, which was isogenic to the experimental flies, except that it lacks the UAS Hepact transgene and therefore cannot induce the expression of the JNK gain-of-function mutant. Initially, the levels of ATG mRNAs were normalized to Gal4 mRNA, which served as an internal control for RNA recovery and the real time PCR process. Fig. 1B and supplemental table 1 display the ratio of ATG mRNA levels in the experimental flies in which Hepact was induced for 6 hours and the controls. Among 13 ATG mRNAs that were assessed by real time RT PCR, six, including the one encoding the key regulator ATG1, were moderately, but significantly, elevated in response to JNK stimulation (Fig. 1B, suppl. Table 1). It should be pointed out that RNA levels were measured in total fly lysates and that the relatively modest increase represents an average of all cell types thus sampled which might mask higher induction levels in specific tissues. A similar 2 to 5-fold transcriptional response had been reported for several ATG genes in genomic profiling experiments that tracked gene expression changes in conditions of LPS exposure or starvation (Boutros et al., 2002; Girardot et al., 2004; Gorski et al., 2003; Jasper et al., 2001; Lee et al., 2003; Zinke et al., 2002). We conclude that JNK signaling is sufficient to direct transcriptional induction of multiple ATG genes in a similar manner as physiological changes or stress exposure.
Figure 1. Oxidative stress and JNK signaling stimulate the expression of ATG genes.
A) ATG1, ATG18.2, and puc mRNA levels rise transiently after adult flies are exposed to food containing paraquat. Total RNAs of 3-5 day old flies treated with paraquat, or mock-treated, for the time points indicated were prepared, and the mRNA level of the above-mentioned three genes were quantitated by real time RT PCR. For each time point, six independent biological repeats were carried out in triplicate each. The ratio between paraquat-treated and control samples is shown as fold induction (mean ± standard error). The confidence in the changes of ATG1, ATG18.2, and puc mRNA levels of upon paraquat-treatment was determined by Student's t-test. The p values for the comparison of the paraquat-treated group at the 10.5 hour time point to control is: puc: p<0.00001; ATG1: p<0.00001. ATG18.2: p<0.00001.
B) Elevated expression of multiple ATG genes in response to JNK activation by the constitutively active JNKK, Hepact, expressed in early 3rd instar larvae using the inducible Gal80ts system (McGuire et al., 2003). Larvae of the genotype T80Gal4, UAS-eGFP/UAS-Hepact; Tub Gal80ts/+ express Hepact at 29°C, but not at 21°C. The reference strain is T80Gal4, UAS-eGFP/+; tub-Gal80ts/+. The mRNA levels of six ATG genes and puc, a known JNK target gene, were measured by real time RT PCR before and after induction of Hepact expression for six hours or mock induction in the control strain. The histogram shows that ratio of RNA levels between the control and the Hepact expressing strain. Before induction (0 hrs Hepact), this ratio is close to 1 as depicted in the left histogram. After transgene activation ATG and puc mRNA levels in the UAS-Hepact strain exceeded those in the reference strain by a factor of 2 to 5 (right panel, 6 hrs Hepact). The differential activation of the ATG genes was statistically evaluated by Student's t-test. Single asterisks indicate that the expression level of ATG genes in the JNK activation group was significantly up-regulated (p<0.05), and double asterisks signify p<0.01. p values are ATG1: p=0.006, ATG4: p=0.006, ATG6: p=0.003; ATG8a (CG32672): p=0.019; ATG18.1 (CG7986): p=0.002; ATG18.2 (CG8678): p=0.005. The fold induction is shown as mean ± standard deviation of three independent experiments, each in triplicate.
Autophagy as a protective mechanism against oxidative stress
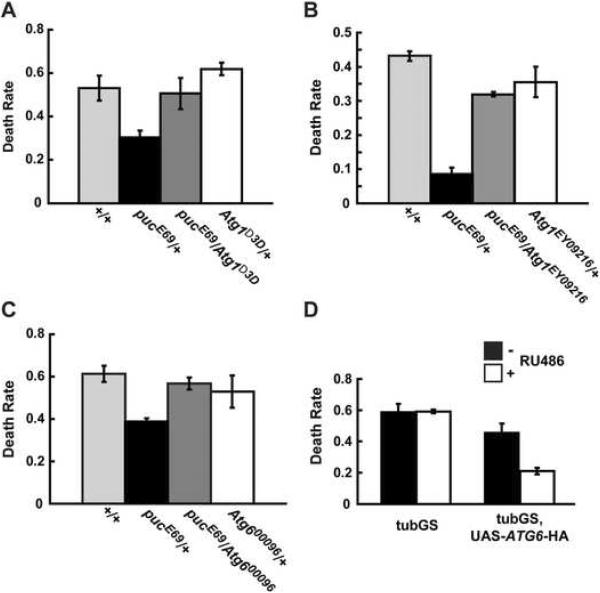
JNK regulates mechanisms that defend organisms against environmental insults and counteract the detrimental effects of oxidative damage on cell functionality (Wang et al., 2003). This protective role is illustrated by the stress resistant phenotype of flies in which JNK activity is artificially elevated (Wang et al., 2003). Our finding that JNK can activate ATG gene expression in Drosophila suggested that autophagy might contribute to JNK-dependent stress defenses. To explore such a potential functional relationship between JNK signaling and autophagy, we conducted genetic interaction experiments. Fig. 2 shows that, as published previously (Wang et al., 2003), flies in which JNK signaling is elevated due to the loss of one copy of the gene encoding the JNK-phosphatase Puckered (genotype pucE69/+), are more resistant against paraquat toxicity than otherwise isogenic wild type animals. This resistance was lost and mortality reverted to that of wild type controls when ATG1 or ATG6 levels were reduced in the pucE69/+ strain by introducing the respective loss-of-function alleles (Fig. 2 A-C). We conclude that the ATG1 and ATG6 genes, and thus presumably autophagy, contribute to the protective function of JNK in acutely stressed flies. Heterozygosity for ATG1 or ATG6 loss-of-function alleles in an otherwise wild type background does not affect paraquat resistance significantly, suggesting that their gene dose is only limiting under conditions of elevated JNK signaling.
Figure 2. ATG genes are required for JNK-induced oxidative stress tolerance in adult flies.
A) Heterozygosity for the gene encoding the JNK phosphatase Puckered (pucE69/+), which causes mildly elevated levels of JNK activity, confers increased resistance against paraquat. Loss of one copy of ATG1 due to heterozygosity for the ATG1Δ3D null allele, in ATG1Δ3D/pucE69 flies, significantly decreases JNK-dependent paraquat resistance to a level approaching that of isogenic control flies (+/+). Loss of one copy of ATG1 in an otherwise wild type background has no significant effect on paraquat resistance in comparison to wild type flies. Similar effects were seen with an independent allele of ATG1, ATG1EY09216(B) and with a loss of function allele of ATG6, ATG600096(C). All stocks were isogenized by extensive backcrossing (at least 10 generations) into the cognate genetic backgrounds (w1118 and ry506). The exact genotypes are listed below. Paraquat exposure times and levels were calibrated to achieve approximately 50% lethality in the wild type controls (15 to 20 mM paraquat for 24 to 48 hours). Data are shown for female flies. Panels A), B), and C) show independently conducted experiments. While the mortality of dietary paraquat is reproducible from vial to vial within one experiment, there is a considerable degree of variability if the experiments are done on different days. This is probably caused by batch variations of food, paraquat or other parameters such slight fluctuations in temperature or humidity. Importantly, the direct comparisons as depicted in Fig 2 are between mutants in isogenic conditions and treated in parallel. The results for the different allele combinations are qualitatively similar between males and females (data not shown).
D) Ubiquitous overexpression of ATG6 from a UAS-ATG6-HA transgene driven by the RU486-inducible tubGSGal4 promotes the survival of flies challenged by paraquat. Induction of ATG6 expression by feeding flies with RU486 for 4 days decreased mortality of a subsequent 20 mM paraquat exposure compared to a mock-treated group. Data for male flies are shown, females display a comparable effect. p values between the death rate of different genotypes were obtained via Student's t-test. The histograms shown above depict a representative experiment out of 2-4 independent repeats for each set of genotypes. Sample size and p-values used in this figure are as follows:
A) +/+, n=120; pucE69/+, n=98; pucE69/ATG1Δ3D, n=69; ATG1Δ3D/+, n=79; p values: (+/+)/(pucE69/+), p=0.009; (+/+)/(pucE69/ATG1Δ3D), p=0.784; (+/+)/(ATG1Δ3D/+), p=0.114; (pucE69/+)/(pucE69/ATG1Δ3D), p=0.011; (pucE69/+)/(ATG1Δ3D/+), p=0.004; (pucE69/ATG1Δ3D)/(ATG1Δ3D/+), p=0.246; B) ATG1EY09216/+, n=58; +/+, n=120; pucE69/ATG1EY09216, n=59; pucE69/+, n=104; p values: (+/+)/(pucE69/+), p= 0.002; (+/+)/(pucE69/ATG1EY09216), p=0.002; (+/+)/(ATG1EY09216/+), p=0.235; (pucE69/+)/(pucE69/ATG1EY09216), p= 0.012; (pucE69/+)/(ATG1EY09216/+), p=0.047; (pucE69/ATG1EY09216)/(ATG1EY09216/+), p=0.455; C) ATG600096/+, n=88; +/+, n=82; pucE69/ATG600096, n=77; pucE69/+, n=125; p values: (+/+)/(pucE69/+), p=0.005; (+/+)/(pucE69/ATG600096), p=0.190; (+/+)/(ATG600096/+), p=0.058; (pucE69/+)/(pucE69/ATG600096), p= 0.002; (pucE69/+)/(ATG600096/+), p=0.011; (pucE69/ATG600096)/(ATG600096/+), p=0.395; D) tubGS>w1118: +RU486, n=152, -RU486, n=144, p-value= 0.930; tubGS>UAS-ATG6-HA: +RU486, n=118, -RU486, n=107, p-value=0.012.
If induction of ATG gene transcription is required for JNK-mediated stress resistance, as suggested by the above experiments, we may ask if expression of ATG genes by itself might be sufficient to protect flies against stress. To address this question we ectopically expressed the ATG6 gene in adult flies. Forced expression of ATG6, also referred to as beclin1, has previously been shown to be sufficient to drive yeast as well as mammalian cells into autophagy (Liang et al., 1999). Using the RU486-inducible tubulin-GeneSwitch-Gal4 driver (referred to as tubGSGal4 below), we globally overexpressed ATG6 from a UAS-transgene by feeding two to three day old flies with food containing RU486. Four days later the animals were exposed to semi lethal concentrations of paraquat as in the previous experiments. Fig. 2D shows that ATG6 overexpression can significantly increase the resistance to paraquat. Thus, transcriptional activation of autophagy per se appears to promote survival of flies under conditions of oxidative stress. Similar findings were recently reported by Simonsen et al. who showed that overexpression of ATG8a (which, like ATG6, is encoded by a JNK responsive gene according to our experiments, Fig. 1B) in Drosophila neuronal tissues is sufficient to confer stress resistance (Simonsen et al., 2008).
Oxidative stress and autophagy in the Drosophila midgut
Next, we asked whether JNK might regulate autophagy in vivo under conditions of oxidative stress. We chose the Drosophila larval intestinal epithelium to study oxidative stress responses and their potential relation to autophagy in an intact tissue. The gut is an excellent organ for these experiments. Intestinal epithelia directly encounter, and react to, environmental insults, for example in the form of toxicants and pro-oxidants contaminating food. The larval gut is amenable to genetic manipulations and can be readily dissected for cell level analysis by confocal microscopy. Exposure of early third instar larvae (at a stage when metamorphosis associated autophagy has not yet commenced) to paraquat-containing food or to food supplemented with 1.5 % H2O2 induces the prominent appearance of autophagosomes in intestinal cells as visualized by Lysotracker Red staining (Fig. 3A). Lysotracker Red, a fluorescent acidotropic probe, labels acidic organelles, including lysosomes and autolysosomes, in living cells and can provide an indication of autophagic activity (Munafo and Colombo, 2001; Rusten et al., 2004; Scott et al., 2004). A second GFP-based marker was employed to validate the identification of autophagosomes and autolysosomes. This fluorescent probe (Rusten et al., 2004) consists of the human ATG8 homolog, LC3B, fused to GFP. GFP-LC3 specifically accumulates in and labels autophagosomes when expressed in Drosophila cells. Consistent with the results of the Lysotracker red stain, the GFP-LC3 assay confirms the stress-dependent accumulation of autophagosomes in the intestinal epithelium. The guts of control larvae that were fed with regular food showed little or no autophagosomes.
Figure 3. Oxidative stress increases autophagy markers in Drosophila larval midgut.
A) The pictures in the left hand panel (control) show midgut epithelial cells of mock-treated 3rd instar larvae. GFP-LC3-marked autophagosomes (green) and Lysotracker Red-labeled lysosomes (red) are rare and do not coincide. 10 hours exposure to food containing 100 mM paraquat (middle panel, paraquat) or 7 hours exposure to food with 1.5% H2O2 (right hand panel, H2O2) induced the appearance of numerous LC3-GFP and Lysotracker Red double positive large autolysosomes. The bars below each panel show quantitative analyses of GFP-LC3 foci observed in each condition. Autophagy activity was defined as follows: high: >10 GFP-LC3 foci/cell; medium: 6-10 GFP-LC3 foci/cell; low: <6 GFP-LC3 foci/cell. In each group, more than 80 cells were randomly picked for GFP-LC3 focus counting. Quantification was done by Pearson Chi-square test in SPSS 13.0. p-values: in paraquat treatment, treated/control< 0.00001; in H2O2 treatment: treated/control< 0.00001.
B)ATG1 is required for starvation- and paraquat-induced autophagy. Midgut preparations from wild type or ATG1 homozygous mutant third instar larvae are shown. The larvae had either been starved for 1 hour in the presence of 5% sucrose only, exposed to food containing 100 mM paraquat or maintained on control food. The two left hand panels show that, as expected, starvation-induced autophagy activity is largely suppressed (fewer Lysotracker Red-positive foci) in an ATG1 mutant background. The right hand panels show that paraquat-induced autophagy is similarly decreased in ATG1 deficient intestinal cells. The bars below each panel show a quantification of Lysotracker Red-positive foci observed in each condition. Autophagy activity was defined as follows: high: >10 Lysotracker Red foci/cell; medium: 6-10 Lysotracker Red foci/cell; low: <6 Lysotracker Red foci/cell. Lysotracker Red-positive foci in 35-61 randomly chosen cells were counted for quantification using Pearson Chi-square test in SPSS 13.0. p-values: in starvation, ATG1-/-/control< 0.0001; in paraquat treatment: ATG1-/-/control= 0.0007.
C) Larval midgut treated with 100 mM paraquat for 8 hours (right panel) shows elevated JNK activity as measured by the JNK reporter puc-LacZ. The untreated control shown in the left panel displays basal activity of the reporter in the connection between proventriculus, midgut (peritrophic mill), and foregut ring.
Genotypes: A) UAS-GFP-LC3/+; DaGal4/+; B) ATG1-/-: ATG1Δ3D/ ATG1Δ3D; Control: ATG1Δ3D /TM3; C) pucE69, ry/TM3, ry
Scale bars: in A) 10 μm and B) 18.75 μm.
To confirm that the Lysotracker red positive foci appearing in oxidatively stressed intestine are genuine autolysosomes, we repeated the experiment in larvae that are homozygous for the ATG1 mutant allele ATG1Δ3D (Scott et al., 2004). Even though such animals are deficient for autophagy, they are viable until pupal stages of development. Under these ATG1 mutant conditions the abundance of Lysotracker positive foci in paraquat-exposed gut is strongly reduced (Fig. 3B). In further support of the validity of the Lysotracker assay for monitoring autophagy in the gut, we found that starvation which is accepted as a bona fide autophagy-inducing condition causes a similar phenotype as paraquat treatment. Control experiments using food coloring showed that the larvae ingest the paraquat and H2O2 containing foods, suggesting that the autophagy response is not caused by food avoidance and starvation (data not shown).
Coincident with increased autophagy in paraquat-treated gut, JNK signaling is activated as visualized by the activity of a JNK responsive reporter, pucLacZ (Fig. 3C). To investigate the correlation between JNK signaling and autophagy in the intestine further, we asked whether the induction of autophagy markers after oxidative stress might be mediated by JNK. To this end, we fed paraquat to larvae in which JNK signaling was suppressed by the expression of dominant negative Drosophila JNK (BskDN). BskDN has been established as an effective and specific inhibitor of JNK activity in transgenic flies and cells (Weber et al., 2000). Figure 4 shows a marked suppression in the size and number of paraquat-induced autophagosomes in clones in which JNK activation is precluded by BskDN expression. Similar effects were observed in clones that were homozygous for the bsk170B null allele (data not shown). These results demonstrate that JNK is necessary for the autophagic stress response in the larval gut.
Figure 4. Oxidative stress increases autophagosome abundance in a JNK-dependent manner.
Expression of dominant negative Drosophila JNK (BskDN) in GFP-labeled clones of midgut epithelium does not affect unstressed intestinal cells (compare top two panels). However, paraquat-induced autophagy, as visualized by GFP-LC3, is strongly suppressed in BskDN expressing clones (compare bottom two panels). The histograms below to the micrographs show the quantitative analysis of autophagosome abundance per cell. The number of GFP-LC3 labeled autophagosomes or autolysomes per cell was counted and is expressed as follows: high: >10 GFP-LC3 foci/cell; medium: 6-10 GFP-LC3 foci/cell; low: <6 GFP-LC3 foci/cell. At least 30 clones were analyzed per condition. Statistical analysis was done in SPSS 13.0 by Pearson Chi test. p-values: BskDN,GFP-LC3 group: paraquat/control, p=0.038; GFP-LC3 group: paraquat/control, p<0.00001; control: BskDN,GFP-LC3/GFP-LC3, p=0.230; paraquat: BskDN,GFP-LC3/GFP-LC3, p<0.00001.
Genotypes: Control clones: hsFLP/+; UAS-GFP-LC3/+; Act>y>Gal4/+
BskDN clones: hsFLP/+; UAS-GFP-LC3/+; Act>y>Gal4/UAS-BskDN
Scale bars: 18.75 μm.
Next, we tested whether JNK activation might not only be necessary but also sufficient to trigger autophagy. As organism-wide activation of Hepact is lethal, we conducted this experiment using clonal expression of Hepact. Employing a Flp-based expression system (see Materials and Methods), Hepact along with GFP-LC3 was expressed in a subset of early third instar larval intestinal epithelial cells to induce high levels of JNK activity. Fig. 5A shows a preponderance of autolysosomes in such clones identified by double labeling with Lysotracker Red and GFP-LC3. GFP-LC3-negative cells in which JNK is not over-activated (outlined in white in Fig. 5A) show only basal levels of Lysotracker Red foci. To gauge whether JNK-activated autophagy might be a widespread phenomenon, we also tested whether JNK activation might have the same stimulatory effect in the fat body, an organ that is well known for its prominent autophagy activity (Rusten et al., 2004; Scott et al., 2004). In Hepact expressing cells, autophagic activity as measured by Lysotracker Red foci is more prominent than in surrounding wild type cells (Fig. 5A). Thus, JNK appears to function as a cell-autonomously acting trigger for autophagy in multiple tissues.
Figure 5. A) JNK signaling is sufficient to activate autophagy markers in the larval gut and fat body.
Ectopic expression of Hepact in clones of larval gut cells (marked with GFP-LC3) strongly increases the number of Lysotracker/GFP-LC3 marked foci (arrowheads), indicating increased autophagic activity. Out of 71 Hepact-expressing clones, 43 (61%) showed enhanced autophagy. Non-clonal wild type cells (non-GFP stained cells, outlined by dashed line) show only sporadic Lysotracker Red foci.
The same experiment was also conducted in larval fat body cells. Note that in this case the clones are marked by expression of regular GFP (not fused to LC3) hence the autophagosomes are not labeled in the green fat body panel. Out of 24 fat bodies that clonally expressed Hepact, 19 (79%) showed enhanced numbers of Lysotracker Red stained foci.
B) Transmission Electron Microscopy of midgut cells undergoing autophagy in response to JNK signaling or starvation. No obvious autophagosome structures were detected in the control midgut cell shown in the upper panel. Conversely, similar cells in which JNK has been ectopically activated by Hepact expression (middle panel), or midgut cells from starved larvae (lower panel) display frequent autolysosomes. These organelles are discernable as membranous vesicles containing whole organelles (green arrowhead) or electron-dense material, presumably representing degradation products (red arrowhead). “nu” indicates nucleus. Insets show the autophagosomes at higher magnification.
Genotypes:
A) control clones in midgut: hsFLP/+; UAS-GFP-LC3/+, UAS-Hepact/+; Act>y>Gal4/+ Hepact clones in midgut: hsFLP/+; UAS-GFP-LC3, UAS-Hepact/+; Act-Gal4/+
B) control clones in fat body: hsFLP/+; UAS-GFP, Act>y>Gal4/UAS-Hepact Hepact clones in fat body: hsFLP/+; UAS-GFP, Act-Gal4/UAS-Hepact
C) Control and starved: w; T80Gal4, UAS-eGFP/+; tub-Gal80ts/+ Hepact : w; T80Gal4,UAS-eGFP/UAS-Hepact; tub-Gal80ts/+ Scale bars: A) midgut: 18.75 μm; fat body: 15.87 μm; C) 1μm
To further characterize JNK-induced autophagy, we inspected thin sections of intestinal epithelium by Transmission Electron Microscopy. Ectopic activation of JNK by Hepact expression in this tissue caused the frequent appearance of structures resembling autophagosomes and autolysosomes at different stages of the autophagy process. In some cases, remnants of organelles, such as mitochondria, engulfed by membranes are discernible (Fig. 5B, green arrowheads). Electron-dense material in other vesicles presumably represents protein breakdown products (Fig. 5B, red arrowheads) as they are frequently observed in mature autolysosomes (Ghadially, 1988; Juhasz et al., 2007a; Lee et al., 2002). The vesicles observed in Hepact expressing gut epithelium closely resemble the autophagosomes induced by starvation in the same tissue (Fig. 5B). Similar structures are only very rarely seen in sections of wild type tissue prepared from well-fed larvae. The electron microscopic analysis therefore supports the conclusion that JNK signaling can induce autophagy in the Drosophila gut and fat body.
JNK is not required for developmentally controlled and starvation-induced autophagy
The data so far show that autophagy can be regarded as a JNK-regulated stress response that helps the organism to cope with oxidative attack, possibly by removing damaged macromolecular components that might otherwise interfere with vital cellular functions. Autophagy has, however, also been implicated into processes that are not obviously related to oxidative stress response, including development and metabolism. This raises the question of whether JNK is universally required for the control of autophagy or whether its role is restricted to autophagic responses to cell stress.
To address this question, we modeled starvation-induced autophagy by transferring larvae to 5% sucrose as the sole energy source. After three to four hours on sucrose, the expected increase of autophagy in intestinal cells became evident by the appearance of abundant Lysotracker Red-positive foci (Fig. 6A). This type of starvation response results from a loss of PI3K/Akt/Tor signaling (Scott et al., 2004). Consistently, clones of intestinal epithelial cells in which PI3K activity is artificially maintained at high levels during starvation by the expression of the large subunit of PI3K (Dp110), are refractory to the induction of autophagy (Fig. 6A). Similar to the cells of the intestinal epithelium, the cells in the fat body undergo autophagy in our starvation regime (Fig. 6B). To test whether JNK signaling might be involved in the induction of autophagy by starvation, we generated cell clones in which BskDN is expressed (labeled by GFP in Fig. 6B). Whereas PI3K activation drastically diminishes the appearance of Lysotracker red positive autophagosomes after starvation (Figure 6B), suppression of JNK activity affects neither the autophagosomes count in starving adipose tissue nor in starving gut (Fig. 6A). In agreement with these data we found that starvation-induced autophagy also proceeds unimpeded in bsk170B JNK null clones in Drosophila midgut and fat body (data not shown). We conclude that JNK is not required for starvation-induced autophagy in either tissue.
Figure 6. Inhibition of JNK signaling does not affect autophagy elicited by starvation or during development.
A) In starved larval midgut cells autophagy, as monitored by the frequency of Lysotracker Red stained autolysomes, is prevalent. Suppression of JNK activity in BskDN-overexpressing clones (labeled with GFP, green, and outlined in white on the bottom panel) displays a similar number of autophagic vesicles (arrowheads) as surrounding control cells. In contrast, autophagy is suppressed in clones overexpressing the catalytic subunit of PI3K, Dp110 (green cells).
B) In fat body of early third instar larvae, before the onset of metamorphosis, autophagy can be induced by starvation and the resultant loss of PI3K signaling (Scott et al., 2004). Overexpression of Dp110, but not of UAS-BskDN (GFP positive cell clones) inhibits this starvation-induced autophagy.
C) The prominent autophagy in late third instar fat body that is associated with metamorphosis is not affected in UAS-BskDN-overexpressing clones (marked by GFP expression, green) or in clones homozygous for the JNK null allele bsk170B (marked by the absence of GFP). The clones, outlined with a white line in the bottom panel, show a similar pattern of Lysotracker red stained autophagosomes (arrowheads) to the surrounding non-clonal cells.
The histograms below to the micrographs show the quantitative analysis of autophagosome number per cell in clones expressing the UAS constructs indicated, or in the case of the right hand panel of C, clones that are homozygous for the JNK loss of function allele bsk170B. The autophagosome count in non clonal control cells is represented in the same manner. The number of Lysotracker Red foci per cell was counted and is categorized as follows: high: >10 Lysotracker Red foci /cell; medium: 6-10 Lysotracker Red foci /cell; low: <6 Lysotracker Red foci/cell. At least 16 clonal cells were analyzed per genotype per condition. Statistical quantification was performed in SPSS 13.0 using Pearson Chi test. p-values are: in A) BskDN: control/starvation, p=0.947, Dp110: control/starvation, p<0.00001; in B) BskDN: control/starvation, p=0.310, Dp110: control/starvation, p=0.0095; in C) control/BskDN, p=0.252; control/bsk170B, p=0.157;.
Genotypes:
A), B), and C) BskDN clones: hsFLP/+; Act>CD2>Gal4,UAS-eGFP/+; UAS-BskDN/+ and Dp110 clones: hsFLP/+; Act>CD2>Gal4, UAS-eGFP/UAS-DP110
C) bsk170B clones: hsFLP/+; ubi-GFP, FRT40A/ bsk170B, FRT40A, and FRT40A clones: hsFLP/+; ubi-GFP, FRT40A/FRT40A.
Scale bars: in C) bsk170B clone, 25.4 μm, and all other clones: 18.75 μm.
In addition to the metabolic regulation of autophagy, developmental signals can activate the process. Ecdysone-induced histolysis of larval fat body during metamorphosis, for example, relies on sustained autophagy on a large scale (Juhasz et al., 2007a; Lee et al., 2002; Rusten et al., 2004). At the late third instar larval stage when the animals prepare to pupate, sustained autophagy can be readily observed in the fat body by Lysotracker Red staining (Rusten et al., 2004) and Fig. 6C. To investigate whether such developmentally programmed autophagy might require JNK signaling, we generated clones of fat body cells in which JNK activity is abrogated either by expression of BskDN or by homozygosity for a null allele of basked (bsk170B, marked by the absence of GFP in Fig. 6C). In spite of the loss of JNK signaling in these clones, the abundance of autophagosomes remains unchanged as visualized by Lysotracker Red staining. These data indicate that, developmentally controlled autophagy during metamorphosis, is regulated by mechanisms that act independently of JNK. Consistent with this conclusion, no metamorphosis-related phenotypes have been reported for JNK pathway mutants.
Discussion
Autophagy is now recognized as a process that has multiple functions in addition to balancing energy homeostasis. It has become increasingly apparent that cells also rely on autophagy for protecting themselves against a battery of potentially harmful insults. For example, autophagy can contribute to cellular defenses against pathogens, bacterial toxins, oxidative stress and ER stress (Bernales et al., 2006; Gutierrez et al., 2004; Juhasz et al., 2007a; Levine, 2005; Nakagawa et al., 2004; Ogata et al., 2006; Ogawa et al., 2005; Simonsen et al., 2008; Xiong et al., 2007; Yorimitsu et al., 2006).
To explore the function and regulation in organismic stress responses we studied the Drosophila intestinal epithelium. This tissue is directly exposed and potentially vulnerable to oxidative stress in the form of dietary toxicants or of H2O2 that can be internally generated as part of pathogen defenses (Ha et al., 2005a; Ha et al., 2005b). It can therefore be expected that the gut employs potent defense and regeneration systems. Here we show that exposure of the Drosophila intestine to oxidative stress, or deliberate activation of JNK signaling in the gut epithelium, results in a prominent rise of autophagosome density as monitored by Lysotracker red or GFP-LC3 staining. This effect resembles the well-established induction of autophagy in this and other tissues in response to starvation. Ultrastructure analysis by transmission electron microscopy confirms that JNK can effectively induce the formation of bona fide autophagosomes. The combined evidence from histology and microscopic analyses, the genetic interactions between JNK and ATG genes, and the induction of ATG gene expression by JNK, support the conclusion that JNK and oxidative stress can induce autophagy in the Drosophila gut.
Our data suggest that JNK signaling induces autophagy, at least in part, by transcriptional activation of ATG genes. Such a mechanism would be consistent with several previous reports indicating that conditions that stimulate autophagy, such as starvation and stress, also lead to increased expression levels of ATG genes (Girardot et al., 2004; Xiong et al., 2007). Furthermore, the deliberate expression of ATG1, ATG6 or ATG8a by itself is sufficient to drive cells into autophagy (Liang et al., 1999; Scott et al., 2007; Simonsen et al., 2008). It is therefore plausible that the JNK-induced increases in ATG gene expression levels observed here might drive and/or sustain autophagy in stressed organs. However, it is also clear that autophagy can be controlled by mechanisms other than gene expression. For instance, protein phosphorylation, lipidation, and processing events have been shown to regulate the process (Kabeya et al., 2004; Klionsky et al., 2008; Mizushima et al., 1998; Scott et al., 2007). Two recent studies conducted in mammalian cell lines indicate that JNK can induce autophagy by phosphorylating Bcl2, thereby relieving its inhibitory effect on Beclin 1, the ATG6 homolog (Pattingre et al., 2008; Wei et al., 2008). It thus emerges that JNK may impinge on autophagy at multiple regulatory levels. Such a scenario bears an interesting resemblance of the better-understood role of JNK in the regulation of apoptosis, a process that it can control by transcriptional, as well as non-transcriptional mechanisms (Tournier et al., 2000; Weston and Davis, 2007). It is at present a matter of speculation how these different layers of regulation are integrated and how they may have evolved. In this regard it is interesting that the only anti-death Bcl2 family member in Drosophila, the buffy gene product, does not contain JNK phosphorylation sites (Quinn et al., 2003), suggesting that Bcl2-dependent mechanisms do not contribute to JNK induced autophagy in Drosophila. It is possible that in flies JNK acts predominantly at the transcription level, and that the role of Bcl2 in this context has evolved later.
The oxidative stress hypothesis of aging predicts that bolstering resistance against oxidative damage can extend longevity of organisms, and we have shown previously that JNK signaling can control aging by deploying cellular oxidative stress defenses (Wang et al., 2003). The data presented here, which indicate that JNK-mediated induction of autophagy can increase oxidative stress resistance, therefore raise the question of whether such a mechanism might be also relevant for the regulation of longevity. Interestingly, several published studies support the conclusion that ATG genes, and by extension the process of autophagy, are required for the lifespan extending effects of caloric restriction or reduced Tor signaling (Hansen et al., 2008; Juhasz et al., 2007a; Melendez et al., 2003). A recent study even finds that over expression of ATG8a under the control of one particular neuron specific promoter is sufficient to extend lifespan and confer stress resistance in Drosophila (Simonsen et al., 2008).
The downstream transcription factor(s) that mediate the activation of ATG genes in response to JNK signaling are not known at this point. However, recent experiments by Juhasz et al. indicate that the transcription factor FoxO is required for the induction of autophagy in flies that have been deprived of food (Juhasz et al., 2007b). In mammals it has been shown that the FoxO can induce ATG gene expression (Mammucari et al., 2007; Zhao et al., 2007; Zhao et al., 2008). We have reported Drosophila FoxO to be critical for JNK-mediated stress resistance (Luo et al., 2007; Wang et al., 2005). FoxO is therefore a good candidate to execute the transcriptional activation of ATG gene expression in response to JNK signaling. Further experiments are required to determine the mechanisms by which the transcriptional regulation of autophagy proceeds.
The findings presented here indicate that the diverse cues that can cause a cell to undergo autophagy, including metabolic, hormonal and stress signals, are transmitted by distinct signaling systems. While the JNK pathway induces autophagy in response to oxidative stress, changes in PI3K and Tor pathways stimulate autophagy in response to food deprivation and ecdysone in a JNK-independent manner. Consistent with the conclusion that the induction of autophagy in conditions of limited food supply does not involve the JNK signaling pathway, we do not detect activation of a JNK reporter gene in gut cells under the starvation conditions employed here, which nevertheless effectively induce autophagy (data not shown). However, protracted or extreme starvation may derail vital cell functions causing stress and a JNK response. For example, mammalian cells that are cultured in nutrient-deprived media activate JNK and consequently autophagy (Pattingre et al., 2008; Wei et al., 2008).
The induction of autophagy by oxidative stress so far discussed contrasts with developmentally programmed autophagy as it prominently occurs during metamorphosis in the Drosophila fat body. This mechanism is hormonally regulated by the ecdysone system and does not appear to require JNK.
Further genetic and molecular studies on the complex regulation of autophagy in different biological settings and organ systems are a high priority. Insight into the pleiotropic functions of this process will be valuable not only for developmental and cell biology, but also for the understanding of pathologies that are correlated with oxidative damage to cells and tissues, such as aging related and degenerative diseases.
Experimental Procedures
Fly stocks
The following stocks were obtained from the Bloomington Drosophila stock center: OreR, w1118, ry506, Ubi-GFP, FRT40A, UAS-GFP-LC3, ATG1EY09216, ATG600096, and Act>y>Gal4. ATG1Δ3Dwas a gift from Thomas Neufeld (Scott et al., 2004). pucE69 was a gift from Enrique Martin-Blanco. bsk170B , FRT40A/Cyo, UAS-Hepact, and UAS-BskDN were gifts from Marek Mlodzik. hsFLP; Act>CD2>Gal4, UAS-eGFP is described in (Britton et al., 2002) and T80Gal4, UAS-EGFP; tubGal80ts in (Hyun et al., 2006). UAS-Dp110 was a gift from Sally Leevers (Leevers et al., 1996). Tub-GeneSwitch-Gal4 (tubGSGal4) was a gift from Scott Pletcher.
The pUAST-ATG6-HA transgene was generated by PCR amplification of the ATG6 coding sequence from larval cDNA (forward primer: GAGAATTCGTGGCAACTCTGGTAGCAGT; reverse primer: GATCTAGACGGTGACACAAACTGTGAAG, EcoR I and Xba I cloning sites are underlined). The ATG6 coding sequence is C-terminally fused to an HA epitope tag.
All flies were maintained at 25°C and 60% humidity unless otherwise stated.
Oxidative stress treatments
The pucE69 and ATG600096 stocks used for oxidative stress resistance experiments (Fig. 2) were backcrossed into the ry506 background for at least 10 generations, and ATG1EY09216was backcrossed to w1118 flies for at least 10 generations. Accordingly, w1118 flies were used as isogenic wild type controls for experiments with ATG1Δ3D and ATG1EY09216flies, and ry506 flies for ATG600096 flies. All crosses were set up at 25°C and 60% humidity. Newly eclosed flies were collected daily and mated for one day before male and female flies were separated. Males and females collected for three consecutive days were pooled. Prior to stress exposure the flies were starved for 6 hours at 25°C by maintaining them in vials only in the presence of water-soaked filter paper, to avoid desiccation. After the pre-starvation the animals were transferred to 50 ml vials containing food with paraquat or control food and incubated in the dark, at a density of 30-40 flies per vial. The food used in these experiments consisted of 1% low melting agarose (Amresco), 5% sucrose (J.T. Baker), and 15 mM paraquat (Arcos). Food was made freshly and the stressed flies were flipped to fresh food every 2 days. Dead flies were counted every 12 hours. When the cumulative death rate of the control flies reached 50%, the cumulative death rate of all other genotypes was calculated. The statistical significance of the difference in cumulative death rates among the various genotypes was examined by Student's t-test.
For paraquat or H2O2 treatment larvae were staged to early 3rd instar by molting and the morphology of the mouth hooks. Prior to stress exposure these early 3rd instar larvae were starved in the presence of water-soaked filter paper for 45 min to 1 hour at 25°C unless otherwise stated. Subsequently, larvae were treated in the dark with food containing either paraquat or H2O2 (Sigma) prepared as follows: paraquat or H2O2 were added to low melting agarose solution after it had cooled down to 37°C to avoid decomposition of the reagents. The final concentration of the ingredients was: 4% autoclaved yeast, 0.5% low melting agarose, 5% sucrose, and 100 mM paraquat or 1.5% H2O2 in double distilled H2O.
For experiments using the gene switch driver tubGSGal4, 2-3 days old adult flies mated for 1 day were split into two groups per sex: one was fed with food containing 200 μM RU486 (Cayman) to induce UAS-driven transgene expression, the other one was fed with control food. All groups were flipped to fresh food every 2 days for a total of 4 days. Subsequently, flies were transferred to paraquat or control food, in the continued presence or absence of 200 μM RU486.
Starvation
For starvation experiments early 3rd instar larvae of the desired genotypes were transferred from standard yeast, molasses food to 5% sucrose in 0.5% low melting agarose for 3 to 4 hours before dissection. Control larvae were kept on regular food until dissection.
RNA purification and Quantitative Real-Time PCR
Larvae in which the expression of Hepact is inducible by heat shock (T80Gal4, UAS-eGFP/UAS-Hepact; tub-Gal80ts/+), or control larvae (T80Gal4, UAS-eGFP/+; tub-Gal80ts/+) were generated. The temperature sensitive mutant form of Gal80, Gal80ts, inhibits Gal4 and thus keeps the UAS-controlled Hepact inactive at the permissive temperature 20°C (McGuire et al., 2003). Upon incubation at 37°C and maintenance at the restrictive temperature of 29°C, Gal80ts is inactivated and Gal4 is liberated to activate UAS-Hepact expression and consequently JNK signaling. Larvae of the desired genotype were raised at 20°C until they reached early third instar larval stage, and then heat-treated at 37°C for 1.5 hours to induce Hepact expression. The heat-treated larvae were maintained at 29°C for an additional 6 hours. All larvae were collected and washed three times in PBS. Total RNAs were prepared by Trizol (Invitrogen) purification following the manufacturer's protocol. 5 μg total RNA from each genotype was reverse-transcribed into cDNA using SuperScript II reverse-transcriptase (Invitrogen). The resultant cDNA was used as the template for quantitative real-time PCR on an iCycler iQ™ system (BioRad). The PCR data were collected and analyzed using the iCycler 3.0 software (BioRad). In these experiments, Gal4 mRNA was used as internal control to normalize the mRNA levels of the analyzed genes. Their fold induction after JNK activation, as shown in Fig. 1 was calculated as follows: first, the raw mRNA levels of the analyzed genes were normalized relative to Gal4 mRNA; second, the ratio between these normalized values for the Hepact expressing and control stocks were calculated.
For the analysis of ATG gene expression in paraquat-treated adults (Fig. 1a), 3-5 day old OreR flies were starved with just water for 6 hours at 25°C, before transferring them to 15 mM paraquat and 5% sucrose in low melting agarose or the same food without paraquat. Total RNAs were prepared at the indicated time points and PCR reactions were carried out as described above. The internal control for mRNA level in these experiments was rp49 mRNA. The induction values shown represent the ratios between the normalized ATG mRNA levels of the paraquat-treated and control samples.
All primers used in this study are listed in supplemental table 2.
β-galactosidase analysis
Dissected midguts of early 3rd instar larvae fed with 100 mM paraquat or with 1.5% H2O2, were fixed in 1% glutaraldehyde in PBS with 2 mM MgCl2 at room temperature for 15 min, and then washed twice for 10 minutes in PBS containing 2 mM MgCl2. Then the larval guts were incubated with X-Gal -galactosidase in β staining solution (3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6, 150 mM NaCl, 1 mM MgCl2, and 0.1 M phosphate buffer with 0.27% X-Gal) overnight at room temperature. The staining reaction was stopped by washing the samples with 1x PBS twice for 10 minutes. Samples were kept in 80% sterile glycerol overnight at 4°C before mounting onto slides for microscopic analysis.
Mosaic clone analysis
Somatic clones expressing UAS-controlled transgenes, were induced employing the hsFLP; Act>y+>Gal4 system unless otherwise stated. For the analysis of clones overexpressing UAS-BskDN, UAS-GFP-LC3 after paraquat treatment, the cross and resulting progeny larvae were kept at 21°C at low density and heat-treated at 37°C for 2 hours at the early 1st instar larval stage. Heat-treated larvae were kept at 21°C until they reached early third instar stage. To generate bsk170B JNK null clones, 10 to 16 hours old embryos of experimental (hsFLP/+; bsk170B, FRT40A/ubi-GFP, FRT40A) and control groups (hsFLP/+; FRT40A/ubi-GFP, FRT40A) were moved to 37°C for 2 hours. The hatched larvae were incubated further at 25°C and analyzed at early third instar larval stage.
Microscopy and image processing
Larval fat bodies and midguts were dissected and stained without fixation in 1 μM Lysotracker Red (Molecular Probes) in 1x PBS for 60 seconds. Stained samples were immediately mounted with DABCO-Mowiol medium and analyzed on a Leica SP2 confocal microscope.
The Transmission Electron Microscopy analysis of larval gut proceeded as follows: Dissected third instar larval midguts were initially fixed in 2% glutaraldehyde in 0.1M sodium cacodylate buffer (pH 7.4) overnight, rinsed 15 minutes for 2 times in 0.1M sodium cacodylate buffer, post fixed in 1% osmium tetroxide combined with 1.5% potassium ferrocyanide in 0.1M sodium cacodylate buffer for 30 min, and rinsed in distilled water 15 min. Then the fixed guts were dehydrated in a graded ethanol series (50%, 65%, 80%, 95%, and 3 times at 100%) for 25 minutes at each concentration, transferred to (1:1) 100% ethanol : propylene oxide (P.O.), (1:2) ethanol : P.O., 100% P.O. for 2 times for 45 minutes in each mixture. The dehydrated guts were infiltrated with EPON/Araldite resin (Electron Microscopy sciences), embedded in BEEM capsules (Electron Microscopy sciences) with fresh resin to be polymerized at 70°C for 2 days. The embedded guts were thin sectioned at 70 nm onto grids with Reichert-Jung Ultracut E microtome, stained on grids with 2% uranyl acetate for 10 min, rinsed with distilled water, and stained with 0.3% lead citrate solution. The images were acquired on a Hitachi 7650 TEM with an attached 11 Megapixel Gatan digital camera.
The pictures used in this manuscript are assembled in Photoshop CS3 and Illustrator CS3.
Supplementary Material
Acknowledgements
We thank Mirka Uhlirova, Dan Li and Henri Jasper for helpful discussions and comments on the manuscript. Christine Sommers provided invaluable technical help and Karen Bentley performed expert electron microscopy. We are grateful to the following colleagues for gifts of fly stocks and reagents: Thomas Neufeld, Enrique Martin-Blanco, Marek Mlodzik, Sally Leevers, Jessica Britton, Joogyung Hyun and the Bloomington Stock Center. This work was supported by NIH grant RO1 AG 026691 to DB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–97. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3:711–22. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–49. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Butterworth FM, Emerson L, Rasch EM. Maturation and degeneration of the fat body in the Drosophila larva and pupa as revealed by morphometric analysis. Tissue Cell. 1988;20:255–68. doi: 10.1016/0040-8166(88)90047-x. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–52. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:15530–5. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007 doi: 10.1038/nature05925. advanced online publication. [DOI] [PubMed] [Google Scholar]
- Ghadially FN. Ultrastructural Pathology of the Cell and Matrix. Butterworths; London: 1988. [Google Scholar]
- Girardot F, Monnier V, Tricoire H. Genome wide analysis of common and specific stress responses in adult drosophila melanogaster. BMC Genomics. 2004;5:74. doi: 10.1186/1471-2164-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13:358–63. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005a;310:847–50. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ. An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell. 2005b;8:125–32. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Jaattela M. Autophagy - an emerging target for cancer therapy. Autophagy. 2008;4 doi: 10.4161/auto.5921. [DOI] [PubMed] [Google Scholar]
- Hyun J, Becam I, Yanicostas C, Bohmann D. Control of G2/M transition by Drosophila Fos. Mol Cell Biol. 2006;26:8293–302. doi: 10.1128/MCB.02455-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper H, Benes V, Schwager C, Sauer S, Clauder-Munster S, Ansorge W, Bohmann D. The genomic response of the Drosophila embryo to JNK signaling. Dev Cell. 2001;1:579–86. doi: 10.1016/s1534-5807(01)00045-4. [DOI] [PubMed] [Google Scholar]
- Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007a;21:3061–6. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, Sass M. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007b;14:1181–90. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–55. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13:350–7. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- Lee CY, Cooksey BA, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–11. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. Embo J. 1996;15:6584–94. [PMC free article] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–8. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Luo X, Puig O, Hyun J, Bohmann D, Jasper H. Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. Embo J. 2007;26:380–90. doi: 10.1038/sj.emboj.7601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–70. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–8. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–60. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mellen MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Munafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–29. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem. 2004;279:15621–9. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2008 doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S, Richardson H. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. Embo J. 2003;22:3568–79. doi: 10.1093/emboj/cdg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–92. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative stress-response genes. Proc Natl Acad Sci U S A. 2004;101:6564–9. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science. 2000;288:870–4. doi: 10.1126/science.288.5467.870. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5:811–6. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–25. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Weber U, Paricio N, Mlodzik M. Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development. 2000;127:3619–29. doi: 10.1242/dev.127.16.3619. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–88. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–9. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–9. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Goldberg AL. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy. 2008;4:378–80. doi: 10.4161/auto.5633. [DOI] [PubMed] [Google Scholar]
- Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. Embo J. 2002;21:6162–73. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.