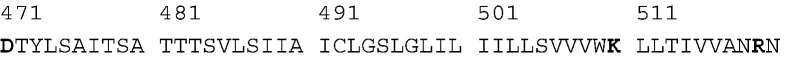Abstract
To enter cells, enveloped viruses use fusion-mediating glycoproteins to facilitate the merger of the viral and host cell membranes. These glycoproteins undergo large-scale irreversible refolding during membrane fusion. The paramyxovirus parainfluenza virus 5 mediates membrane merger through its fusion protein (F). The transmembrane (TM) domains of viral fusion proteins are typically required for fusion. The TM domain of F is particularly interesting in that it is potentially unusually long; multiple calculations suggest a TM helix length between 25 and 48 residues. Oxidative cross-linking of single-cysteine substitutions indicates the F TM trimer forms a helical bundle within the membrane. To assess the functional role of the paramyxovirus parainfluenza virus 5 F protein TM domain, alanine scanning mutagenesis was performed. Two residues located in the outer leaflet of the bilayer are critical for fusion. Multiple amino acid substitutions at these positions indicate the physical properties of the side chain play a critical role in supporting or blocking fusion. Analysis of intermediate steps in F protein refolding indicated that the mutants were not trapped at the open stalk intermediate or the prehairpin intermediate. Incorporation of a known F protein destabilizing mutation that causes a hyperfusogenic phenotype restored fusion activity to the mutants. Further, altering the curvature of the lipid bilayer by addition of oleic acid promoted fusion of the F protein mutants. In aggregate, these data indicate that the TM domain plays a functional role in fusion beyond merely anchoring the protein in the viral envelope and that it can affect the structures and steady-state concentrations of the various conformational intermediates en route to the final postfusion state. We suggest that the unusual length of this TM helix might allow it to serve as a template for formation of or specifically stabilize the lipid stalk intermediate in fusion.
Abbreviations: F, fusion protein; TM, transmembrane; PIV5, paramyxovirus parainfluenza virus 5; HN, hemagglutinin neuraminidase; HA, hemagglutinin; FP, fusion peptide; HR, heptad repeat; 6-HB, six-helix bundle; VSV, vesicular stomatitis virus; cryoEM, cryoelectron microscopy; CuP, Cu(II)(1,10-phenanthroline)3; 6-CF, 6-carboxyfluorescein; RBC, red blood cell; PAb, polyclonal antibody; LTR, long terminal repeat; LPC, lysophosphatidylcholine; OA, oleic acid; CPZ, chlorpromazine; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; p.t., posttransfection; PBS, phosphate-buffered saline; RIPA, radioimmunoprecipitation assay
Keywords: viral membrane fusion, transmembrane domain function, protein refolding intermediates, oxidative cross-linking, modeling a transmembrane domain
Introduction
Membrane fusion is a fundamental biological process that occurs in intracellular trafficking, exocytosis, resealing of plasma membranes, protein trafficking, and in the entry of enveloped viruses. This ubiquitous process is mediated and tightly controlled by a combination of specific protein machinery and lipid composition.1 Lipids spontaneously assemble into bilayer structures such as liposomes; however, lipid bilayer membranes do not spontaneously fuse.2 The spontaneous negative or positive curvature of a lipid can enhance or diminish fusion, respectively, but energy must be expended to overcome hydration repulsion between membranes and to disrupt the bilayer structure. The energy for this remodeling may be derived from the thermal fluctuations of the membrane or from specialized fusion proteins.2 Many biological processes, such as neuronal synaptic vesicle fusion, endosomal fusion, and exocytosis, employ the SNARE superfamily proteins.3 SNAREs are found in all eukaryotic organisms. All SNARE proteins have a common heptad repeat that forms four-helix coiled-coil structures, and this coiled-coil SNARE complex forms in trans to promote fusion of the two membranes in which the SNARE proteins are anchored.4 Enveloped viruses use an analogous strategy and mediate fusion with target cells through specialized fusion proteins. The paramyxovirus parainfluenza virus 5 (PIV5) requires two surface glycoproteins for this process: the attachment protein hemagglutinin neuraminidase (HN) that binds sialic acid and the fusion protein (F) that physically merges the two membranes. Paramyxovirus fusion occurs at the plasma membrane and does not require the low pH of the endosome to trigger fusion.5
The paramyxovirus F protein is a class I fusion glycoprotein that is synthesized as a type I integral membrane protein and it folds into homotrimers, is posttranslationally modified by the addition of carbohydrate chains, and then is proteolytically cleaved to become biologically active. Similar processing occurs for other class I viral fusion proteins, such as influenza virus hemagglutinin (HA), HIV gp160, retrovirus Env, Ebola GP, and SARS CoV S.5 The paramyxovirus F precursor protein (F0) is cleaved into the membrane-anchored F1 subunit and the smaller N-terminal F2 fragment. F1 contains two hydrophobic regions, the N-terminal fusion peptide (FP), located at the new N-terminus after cleavage, and the transmembrane (TM) domain, and two heptad repeat regions, HRA and HRB. HRA is located immediately C-terminal to the FP, and HRB is proximal to the TM domain.5
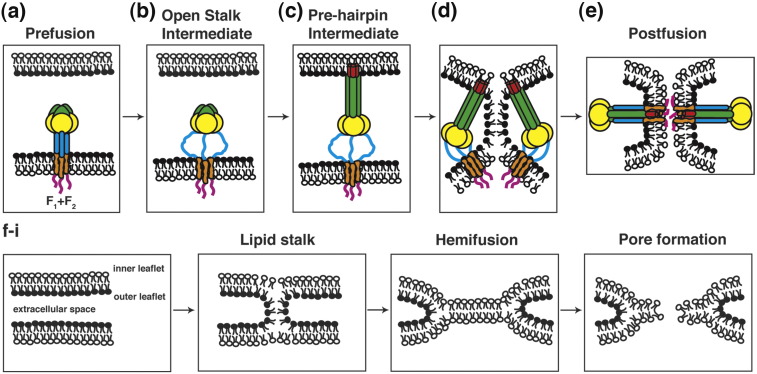
The paramyxovirus F protein folds initially into a metastable prefusion form (Fig. 1a) that upon triggering undergoes a series of large scale conformational rearrangements, proceeding down an energy gradient to form a final irreversible postfusion form (Fig. 1b–e). Recently, the crystal structures of both the uncleaved prefusion conformation of the paramyxovirus F protein and the uncleaved postfusion conformation were solved.6, 7 The prefusion form contains a globular head containing three domains (DI–DIII) attached to a trimeric coiled-coil stalk formed by the HRB region. The HRA region in the prefusion form is composed of 11 distinct segments that wrap around the DIII core in the globular head.7 This is in contrast to the postfusion form of F where HRA is extended into a long α-helix as part of the six-helix bundle (6-HB). For the postfusion structure, an unanticipated finding emerged as the available data indicate the F TM domain and/or cytoplasmic tail are important for the folding of F into the metastable prefusion form of F:6 secreted F lacking a TM domain converts to the postfusion form.
Fig. 1.
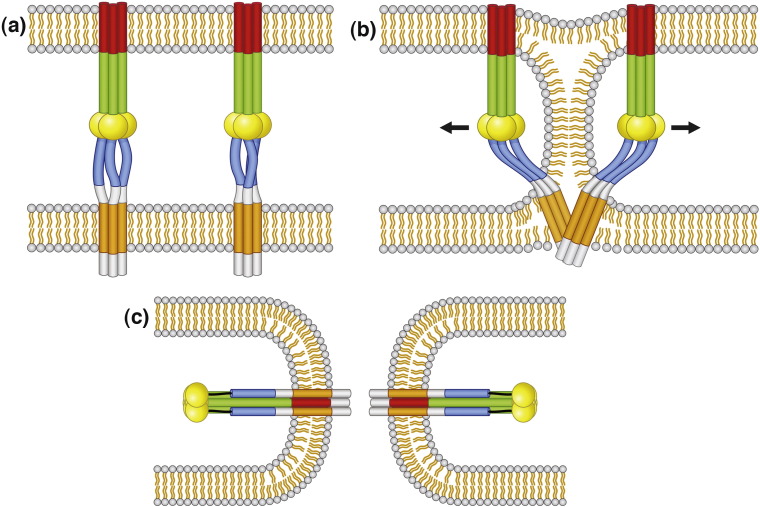
Model of membrane fusion for paramyxovirus F protein. (a) The prefusion form of F contains a globular head with the HRA region in 11 distinct sections and the HRB region is in a three-helix bundle. The F TM domain is also represented as a three-helix bundle, consistent with the oxidative cross-linking data (Fig. 3). (b) Upon HN binding to target cells (HN not shown for clarity), F is activated for fusion, and the HRB region separates, forming the open-stalk conformation where N-1 peptide can bind to HRB. At this open-stalk stage, the TM domain is still thought to be in a three-helix bundle because N-1-HAt can still bind to HRB after the addition of the oxidative cross-linker (see Fig. 3). (c) After formation of the open-stalk conformation, HRA rearranges to form the extended α-helical bundle, and the FP is inserted into the target cell membrane (the prehairpin intermediate). (d–e) Finally, the postfusion state occurs with the formation of the 6-HB. (d–e and f–i) Lipid intermediates in fusion with the F protein removed for clarity. The two bilayers contain an inner and outer leaflet and are separated by the extracellular space. During the process of F refolding to form the postfusion form, water is excluded from the extracellular space and the outer leaflets initially merge to form the lipid stalk intermediate. The lipids of the bilayers mix, forming the hemifusion intermediate, and then the fusion pore forms. F domains: FP (red), HRA (green), globular head (yellow), HRB (blue), TM domain (orange), cytoplasmic tail (pink).
Upon receptor binding, biochemical studies indicate HN induces a conformational change in F and the HRB three-helix stalk separates.8 It is hypothesized that following the melting of the HRB helices and destabilization of the head (Fig. 1b), HRA refolds to form an extended α-helical coiled coil, which enables the insertion of the fusion peptide into the target cell membrane and forms the prehairpin intermediate7 (Fig. 1c). The F protein then refolds where HRB binds into the grooves between the HRA monomers and forms the six-helix bundle (6-HB), bringing the FP and TM domain into juxtaposition within the same membrane7, 9, 10, 11 (Fig. 1e). A conceptually related final postfusion structure is formed for all enveloped virus fusion proteins. The postfusion structure consists of either an α-helical coiled-coil structure like the 6-HB in PIV5 F or β-strand structures in other fusion proteins, such as dengue virus E, Semliki Forest virus E1, and vesicular stomatitis virus (VSV) G.12 In all cases the fusion peptide and TM domain are together in the same membrane. The formation of the 6-HB and the associated free-energy change is tightly linked to the merger of the target and viral membranes.8, 13 The collapse of the prehairpin intermediate distorts the bilayers to possibly form an initial pointlike protrusion.14 Whether the insertion of the fusion peptide perturbs the lipid bilayer and lowers the distortion energy is unknown.14 Following the initial bilayer contact, membrane merger proceeds to the lipid stalk intermediate, where the outer leaflets of each bilayer merges but the inner leaflets remain separate. This stalk then expands to form the hemifusion diaphragm. Continuation of this expansion leads to formation of the fusion pore that allows for the transfer of aqueous contents (Fig. 1f–i).15
In addition to aiding in protein folding and stability, the TM domains of many viral envelope fusion proteins have been shown to have a role in fusion.16, 17, 18, 19, 20, 21, 22, 23, 24 When the TM domain of influenza virus HA was replaced with a glycosylphosphatidylinositol anchor, it resulted in a fusion protein that could mediate hemifusion by allowing the transfer of a lipid but not aqueous fluorescent probe, suggesting a role of the TM domain in pore formation and enlargement. This glycosylphosphatidylinositol-anchored HA was embedded only in the outer leaflet of the membrane, and it was proposed that the TM domain, which spans the entire membrane, affects the positive curvature on the inner leaflet that would allow for pore formation.17, 19 In addition, specific residues in the TM domain have been shown to be important for fusion of influenza virus HA,18 VSV G,20 HIV gp41,23, 24, 25 and baculovirus GP64.21
Although little is known about the role of the TM domain in paramyxoviruses, the TM domain of PIV5 F shows several features that might be relevant to function.
It has a 25-residue hydrophobic segment (spanning from 485 to 509) that is somewhat longer than the typical hydrophobic length of 20 residues for a vertically inserted TM helix. The N-terminus of the predicted TM helix is devoid of side chains such as Trp, Tyr, Arg, or Lys that typically anchor the end of the helix in the head-group region of the bilayer, and the side chains of the 15 preceding residues are uncharged and can partition easily between bilayer and aqueous phases.26, 27 This region forms the end of the coiled-coil HRB stalk and a short linker segment in the prefusion form of F. Finally, the C-terminal end of the TM helix is predicted to occur at the positively charged K510, but the seven residues following this residue are predominantly hydrophobic amino acids. Thus, if K510 snorkels into the head-group region, the TM helix might extend beyond this point. This finding is reminiscent of the TM helix of HIV gp41,23 which contains an absolutely conserved and functionally essential positively charged residue within the hydrophobic region of the TM helix. In summary, the TM helix of F appears to be longer than usual and has an unusually large number of potential orientations in membranes; these features piqued our interest and in part motivated the present study.
We first asked whether the TM domain formed a well-defined helical bundle in the prefusion form of the protein. This is a particularly important question, because TM helices of fusogenic proteins often have sequences known to mediate helix–helix association, such as the GXXXG motif, and mutation of these sequences sometimes, but not always, leads to proteins that are properly expressed but have impaired function.20, 28, 29 Cryoelectron microscopy (cryoEM) studies on the human immunodeficiency virus fusion protein (Env) differ, as some reports indicate the TM domains of the fusion protein are apart, like the legs of a tripod,30, 31 whereas other studies indicate the TM domains are together and form one helical bundle.32 By singly substituting each residue of the TM domain with cysteines, we have examined the structure of the TM domain in the lipid membrane. The addition of an oxidative cross-linker and formation of disulfide bonds clearly showed that one face of the TM helix mediates the formation of a homomeric TM helical bundle, most likely a trimer. To address the role of specific residues within the TM helix we performed alanine-scanning mutagenesis, and determined that two residues, 486 and 488, are critical for PIV5 F fusion. The aggregate of analysis of the steps of fusion indicate these mutants do not support hemifusion and are trapped at the lipid stalk stage just prior to formation of the hemifusion diaphragm. Our results indicate a specific amino acid sequence of the TM domain is necessary for completion of fusion. The amino acid side chains at residues 486 and 488, which are predicted to be in the outer leaflet of the F TM domain, are critical for the merger of the two lipid membranes and fusion completion. The block in fusion observed with mutation of F residues 486 and 488 could be overcome by addition of compounds that affect the curvature of the membrane bilayer.
Results
The F protein TM domains are in close proximity and likely form a three-helix bundle
The TM domain is not part of the solved crystal structure of either the prefusion form of PIV5 F7 or the postfusion form of hPIV3 F.6 Although it is thought likely that the TM domain of F would be α-helical because the hydrophobic packing of an α-helix would aid in spanning a lipid bilayer, this has not been determined. It is also unknown if the TM domains of the PIV5 F monomers interact to form a trimer. It has been shown recently that TM peptides of influenza virus HA associate with each other in model membranes.33 In the prefusion form of F, HRB forms a three-helix bundle (3-HB) and HRB is separated from the TM domain by only seven residues. Therefore, it seems likely that the F TM domain monomers would be in close proximity in the membrane if not in a 3-HB. We used oxidative disulfide cross-linking to examine the structure of the F TM domain. This method has been used to investigate the arrangement of TM domains for several membrane proteins, such as the Escherichia coli chemoreceptor,34, 35, 36 the aspartate receptor,37 the influenza virus M2 ion channel,38 and CD39.39, 40
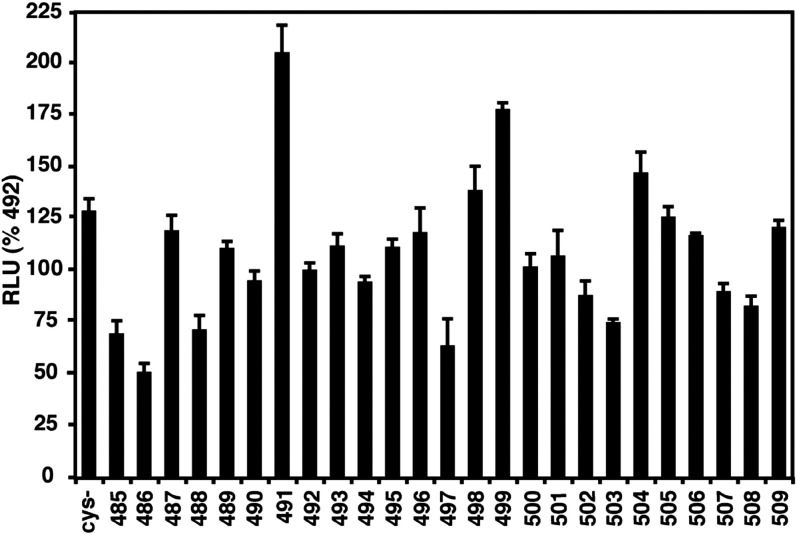
The PIV5 F protein contains 10 disulfide-bonded cysteine residues in the ectodomain and one free cysteine residue in the TM domain. This latter cysteine at residue 492 was mutated to serine, and single-cysteine substitutions were made in the TM domain in this pseudo wild-type (wt) Cys− background. All mutants in the Cys− background were expressed at the cell surface equivalently to pseudo wt F (data not shown). The fusion activity of these mutants was determined by using the luciferase reporter gene assay (Fig. 2 ). Most mutants showed fusion activity similar to that of pseudo wt (Cys−, C492S) and wt F protein (denoted as 492). Mutants 485, 486, 488, 497, and 503 exhibited some decrease in fusion activity, with the maximum reduction being ∼ 50% for mutant 486 (see Fig. 2). Mutations in the TM domain of influenza virus HA have been shown to affect raft association.41 However, none of the cysteine substitutions affected raft association of the F protein (data not shown).
Fig. 2.
Fusion activity of F TM domain cysteine substitution mutants. Each residue of the F protein TM domain was substituted with cysteine in a background in which the naturally occurring cysteine residue 492 was mutated to serine. The luciferase reporter gene assay was used to measure cell–cell fusion mediated by the single cysteine mutants in the Cys− background (pseudo wt). Vero cells were cotransfected to express HN, pseudo wt F or mutant F protein, and a luciferase reporter construct. Shown is the average of three experiments each done in triplicate and normalized to pseudo wt Cys− F.
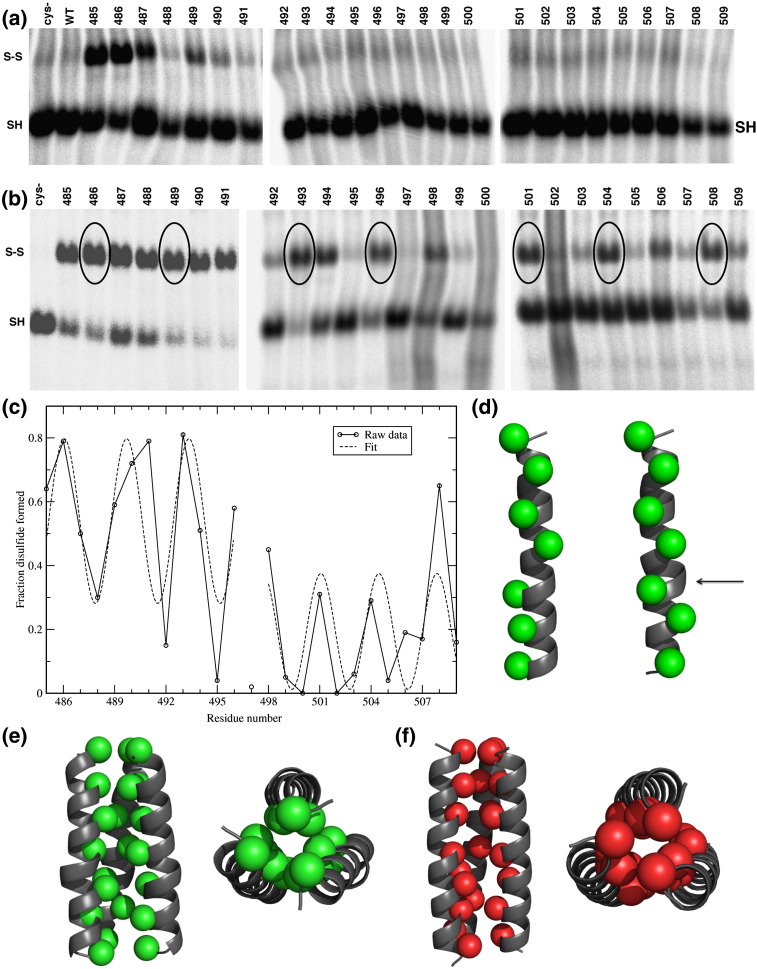
We examined the effect of oxidative cross-linking of these F TM domain single-cysteine substitutions. For each mutant, there are three available cysteines in each trimer. If these cysteines are oriented toward each other in the membrane and are within disulfide bond-forming distance, two will form a disulfide bond and leave the third cysteine unbonded. On the other hand, if the helices are not specifically interacting, one might expect that high concentrations of oxidants would be required to induce any disulfide formation in the membrane and that there would be no clear pattern for forming disulfides between residues along any one face of the helix. In Fig. 3a are shown untreated F mutants analyzed under nonreducing conditions on a 3.5% SDS-PAGE gel. Disulfide bonds formed by air oxidation should occur only in the extracellular aqueous phase and the head-group region of the outer leaflet of the bilayer.38 Air oxidation is much slower in the hydrophobic region of the bilayer, and the reducing intracellular environment minimizes air oxidation on the cytoplasmic side of the membrane. Cysteine residues in the outer leaflet of the bilayer at positions 485, 486, 487, and 489 did form disulfide bonds, indicating the individual F TM domains are in close proximity to themselves and that these positions occur near the head-group region of the outer leaflet of the bilayer.
Fig. 3.
F protein TM domain cysteine substitution and disulfide bond formation on oxidation. (a) F protein TM domain cysteine mutants in a pseudo wt background (Cys−, C492S) were expressed in HeLa CD4 LTR βgal cells, and 18 h p.t. cells were labeled with 50 μCi of 35S-Promix, Dounce homogenized, solubilized, and immunoprecipitated with an anti-F2 PAb. Polypeptides were analyzed by SDS-PAGE on a 3.5% acrylamide gel under nonreducing conditions. (b) Cells were transfected and labeled as above, Dounce homogenized, and treated with 3 mM CuP for 10 min at 37 °C. Samples were then solubilized, immunoprecipitated with an anti-F2 PAb, and polypeptides were analyzed by SDS-PAGE on a 3.5% acrylamide gel under nonreducing conditions. Circled residues indicate interacting residues in the predicted helix. (c) Periodicity of the predicted TM domain helix. The raw CuP cross-linking data in (b) were normalized by dividing the amount of disulfide formed by the total amount of protein. Also shown is the sine wave fit of the data. The data obtained with oxidation occurring at 4 °C were used for residue positions 485–491, and the data obtained with oxidation occurring at 37 °C data were used for the remaining positions (492–509). This was done to compensate for the greater disulfide bond formation that takes place in the outer leaflet and to maintain the signal on the image plate used to detect radioactivity within the linear range. (d) Ideal helix (left) and pi bulge (right) models of the TM helices. Interface positions predicted from disulfide cross-linking are shown in green spheres. For the first model, the TM domain is predicted to form a helix with a pi bulge at residues 497–500 (arrow). (e) Model of the TM helix trimer containing a pi bulge (in green). On the left is a view from within the membrane and on the right is a view from above. As in (d), interface positions are shown in green spheres. (f) Model of the TM trimer containing a stutter (in red). On the left is a view from within the membrane and on the right is a view from above. Interface Cβ positions for the model are shown in red spheres (Cα shown for G497).
The data are in very good agreement with the expected orientations of the TM helices, as inferred from deletion mutagenesis and crystallographic analysis of the protein. In the prefusion structure of F residues 446 to 476 form a three-stranded coiled coil. Not in the atomic structure is a short linker that connects to the TM domain near residues 485. Four residues (481–484) can be deleted with full retention of function, while a two-residue deletion (483–484) caused a decrease in fusogenic activity.10, 42 More strikingly, deletion of eight residues (477–484), which compose the end of the coiled coil and the entire linker, leads to nearly full retention of fusogenic activity, while deletion of only six residues (479–484) leads to a protein that is less well-expressed on the cell surface and highly inactive in fusion.10 Interestingly, if the HRB coiled coil continues into the TM region, the four-residue and eight-residue mutants would position the side chains of residues 485 and 486 in close proximity, consistent with their high level of cross-linking in Fig. 3a (see also Supplemental Fig. 2). On the other hand, there would be a strong mismatch between the axial rotation of the helical axes in the HRB coiled coil versus the TM bundle in the six-residue deletion mutant, explaining its lack of stability and function.
Cu(II)(1,10-phenanthroline)3 (CuP) was used to induce disulfide cross-linking within the lipid interior,38, 43 which is otherwise resistant to oxidation. When the F TM mutants were treated with 3 mM (final concentration) of CuP for 10 min at 37 °C, more extensive disulfide cross-linking was observed (Fig. 3b). Under conditions required to induce oxidation of the membrane-spanning regions, much more extensive cross-linking is observed in the aqueous-accessible region (residues 485–491). Within the TM region, residues 493, 494, 496, 498, 501, 504, 506, and 508 all showed disulfide bond formation. CuP (3 mM) was also added at 4, 10, and 22 °C for 10 min (data not shown). All residues near the ectodomain (485–491, 493, 494) formed disulfide bonds at all temperatures, even at 4 °C. Mutants that formed disulfide bonds after treatment with 3 mM CuP for 10 min at 37 °C also formed disulfide bonds when treatment with 3 mM CuP was for 1 or 5 min (data not shown). The effect of cross-linking on fusion could not be determined after oxidation due to the toxicity of the CuP on the live cells that are required for the fusion assays (data not shown).
Cross-linking occurs in a pattern suggestive of that of a helical bundle. Even at a qualitative level, it is clear that the extent of cross-linking follows the same periodicity as that of an α-helix (approximately 3.6 residues per turn), suggesting that one face of the helix interacts in the core of a bundle. Furthermore, the extent of cross-linking under these conditions is similar to that observed with previously characterized membrane proteins of known three-dimensional structure, suggesting that the TM domain folds into a well-defined structure.38, 43 One unusual feature, however, is that three consecutive residues near the outer leaflet of the bilayer (489–491) form cross-links quite efficiently, suggesting some malleability in this region of the protein. To better determine the nature of the interface, the extent of disulfide cross-linking was quantified and fit to a sine wave. The raw CuP cross-linking data were normalized by dividing the amount of disulfide formed by the total amount of protein (disulfide linked plus not linked). The CuP data at 37 °C were used for most of the TM domain, positions 492–509. Because cross-linking was stronger in the outer leaflet of the bilayer, low-temperature (4 °C) CuP data were used for residue positions 485–491 so that data from the two regions would be on the same scale (and to maintain linearity of the image plate response to radioactivity). This type of approach was previously used to model the structure of the TM domain of the influenza virus M2 protein,38 where a similar level of cross-linking was observed.
It was considered that the disulfide cross-linking could occur between two trimers. However, when more than 100-fold different amounts of F protein were expressed, there was no change in the level of cross-linking as would be expected to occur for intertrimer cross-links (data not shown). In addition, higher molecular weight species of trimer–trimer formation, in addition to cross-links within a trimer, were not observed on gels (data not shown). Further, the extensive nature of the cross-links for some mutants and not others argues for cross-links within the TM domains within a trimer.
Examination of the F TM domain data showed that the cross-linking efficiency oscillated as a function of the sequence position with approximately the same periodicity as the α-helix (3.6 residues per turn). However, G497 deviated from this trend, showing a lower degree of cross-linking than expected. Given the unusual characteristics of glycine residues, residue 497 was removed from the calculation and separate fits were made to the data before and after this residue. The periodicity was also indicative of a helical conformation (residues 485–496: 3.65 ± 0.18 residues per turn; residues 498–509: 3.40 ± 0.21 residues per turn; Fig. 3c). The pattern is interrupted by the insertion of one residue near residue 497. Thus, the fit parameters suggest a packed helical TM interface made up of residues 486, 489, 493, 501, 504, and 508 (circled in Fig. 3b) with a potential distortion to the helix around residue 497.
To create models for the TM region, we assume it forms a trimeric bundle and that efficient cross-linking occurred only at residues close together in the three-dimensional structure. One way to allow insertion of a single residue into α-helices is by inserting a pi bulge.44, 45 Figure 3d shows an example of a 3-fold symmetric bundle created by using the backbone of a TM helix containing a pi bulge from the Na-dependent aspartate transporter structure (Protein Data Bank code 2nwl, 2.96 Å).46 The sequence of the TM region of F was threaded onto this backbone, and Gly497 was positioned within the pi bulge. The effect of a pi bulge on the positioning of the residues that cross-link efficiently (when mutated to Cys) can be seen by comparing it to the result expected for an ideal helix and without a pi bulge (Fig. 3d). The pi bulge allows both the interfaces above and below the bulge to occur on the same face of the helix. A bundle was created from this helix by applying C3 symmetry followed by energy minimization (Fig. 3e).
A second way insertions are accommodated in coiled coils is by variation of the helix packing in a common structural motif known as a “stutter.”47 A geometric model incorporating this insertion was created by correlating the cross-linking data with the Cα distances of the trimeric coiled coil from influenza virus HA. As the stutters in HA result in different local distortions,48 pre- and postfusion HA structures49, 50 were searched for one that matched the cross-linking data as described in Supplemental Methods. Threading the sequence of the TM helix onto this backbone template gave a model in good qualitative agreement with the cross-linking data (Fig. 3f). In summary, the cross-linking data show good evidence to support the formation of a 3-HB, and stereochemically reasonable bundles can be constructed that are consistent with the data.
F. protein TM domain residues bordering the ectodomain have a key role in fusion
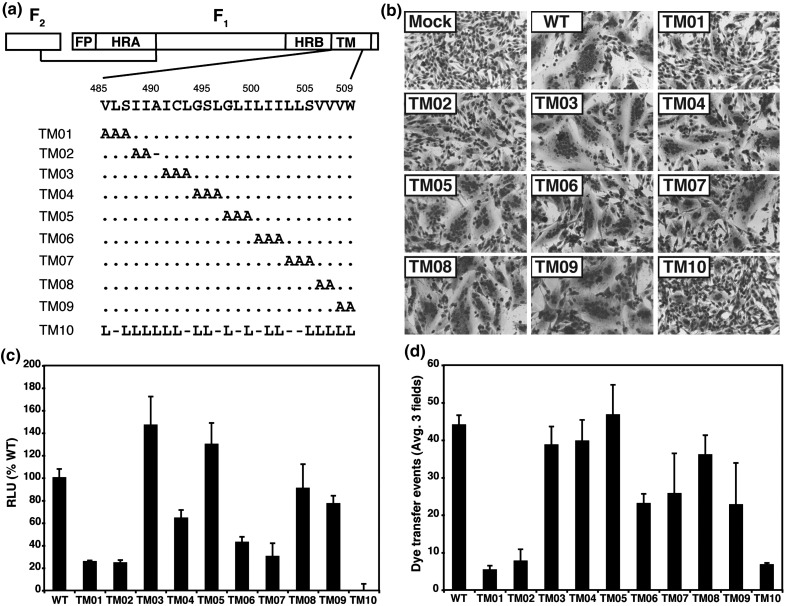
To determine the role of the F protein TM domain in fusion, alanine-scanning mutagenesis was performed in groups of two or three amino acids for the 25 residues of the PIV5 F protein TM domain. Further, the entire TM domain was replaced en bloc with 25 leucine residues (Fig. 4a). Cell surface expression of the F protein mutants TM01–TM09 was equivalent to wt PIV5 F (Table 1 ). However, mutant TM10 was not expressed at the cell surface; thus, F TM10 is likely to be a malfolded protein that is not transported through the exocytic pathway to the cell surface.51 Previously, when the TM domain of PIV5 HN was replaced with leucine residues, there was no discernable effect on the cell surface expression of HN and no effect on its biological function.52 The ability of the F TM domain alanine scanning mutants to cause cell–cell fusion was determined by three assays: (1) syncytia formation, (2) a luciferase reporter assay, and (3) a dye transfer assay. F TM domain mutants TM03–TM09 formed similar-sized syncytia as compared to wt F protein, but mutant F proteins TM01 and TM02 did not cause syncytia formation, although these proteins were well expressed at the cell surface (Fig. 4b). Whereas several of the TM mutants showed some decrease in fusion in the quantitative luciferase reporter and dye transfer assays, TM01 and TM02 caused a consistent and major reduction in fusion in all assays used (Fig. 4c and d).
Fig. 4.
Analysis of fusion activity of alanine mutations in the F protein TM domain. (a) Schematic diagram of the PIV5 F protein. The positions of the fusion peptide (FP), heptad repeat A (HRA), heptad repeat B (HRB), and TM domain are shown. The TM domain is taken as beginning at V485 and ending at W509. The alanine substitutions in the alanine scan for mutants TM01 through TM10 are shown. For mutant TM10, all TM domain residues were replaced with leucine. (b) Representative micrographs of syncytia formed at 20 h p.t. in BHK-21F cells expressing PIV5 HN and either wt F or F containing a TM domain mutation. Mock, expression of HN alone. (c) Luciferase reporter gene assay of cell–cell fusion mediated by the F protein TM domain mutants. Vero cells were cotransfected to express HN, wt F or mutant F protein, and a luciferase reporter construct. Shown is the average of three experiments each performed in triplicate and the data normalized to wt F. (d) Quantification of cell–cell fusion obtained from the dye transfer assay. Effector CV-1 cells were infected with vaccinia virus vTF7-3 and transfected with DNA encoding HN and wt F or F TM domain mutant. RBCs were labeled with the lipidic probe R18 and the aqueous probe 6-CF. Labeled RBCs were bound to CV-1 cells for 1 h at 4 °C and then incubated at 37 °C for 15 min before visualization by confocal microscopy. Cell–cell fusion was observed as the transfer of red R18 and green 6-CF from the target RBCs to the effector CV-1 cells. Shown is the quantification of 6-CF. The means and error bars are from three microscopic fields.
Table 1.
Relative surface expression of mutant F proteins
| RMFI (% wt) | |
|---|---|
| wt | 100 |
| TM01 | 102 |
| TM02 | 99 |
| TM03 | 103 |
| TM04 | 98 |
| TM05 | 94 |
| TM06 | 87 |
| TM07 | 90 |
| TM08 | 111 |
| TM09 | 97 |
| TM10 | 9 |
| V485A | 102 |
| L486A | 81 |
| S487A | 99 |
| I488A | 89 |
| I489A | 99 |
| S443P | 113 |
| S443P/L486A | 67 |
| S443P/I488A | 81 |
| G105A | 97 |
| G105A/L486A | 78 |
| G105A/I488A | 85 |
| G109A | 10 |
| G109A/L486A | 11 |
| G109A/I488A | 9 |
Surface expression levels were determined by flow cytometry using F-specific mAb F1a.
F. TM domain residues L486 and I488 are key residues involved in fusion
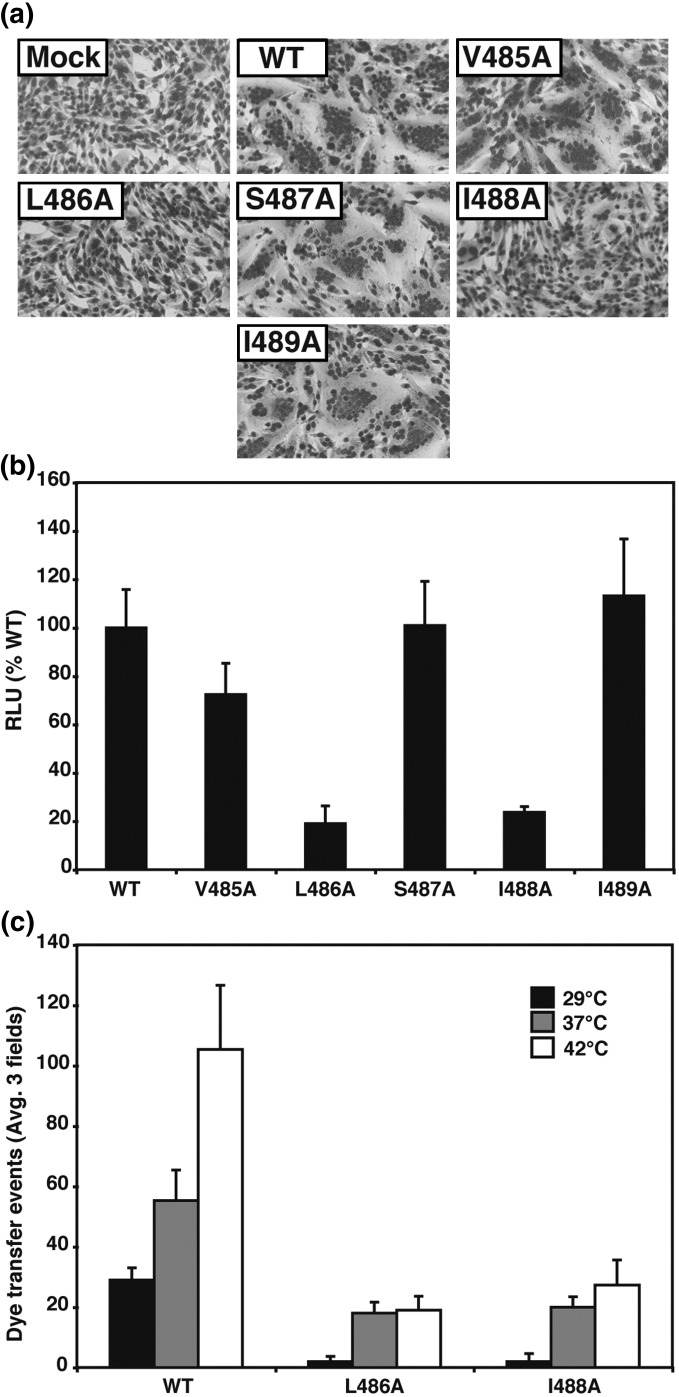
To determine further which residues in mutants TM01 and TM02 are responsible for the greatly reduced fusion activity, the first five residues, 485–489, of the F protein TM domain were changed individually to alanine. All these mutants were expressed at the cell surface at levels similar to that of wt F (Table 1). F TM domain mutants V485A, S487A, and I489A formed syncytia at levels similar to that of wt F, but F TM domain mutants L486A and I488A did not cause syncytia formation (Fig. 5a). These mutants also showed a substantial decrease in fusion in the luciferase reporter and dye transfer assays (Fig. 5b and c). In all cases where fusion occurred, both the lipidic dye, R18 (data not shown), and the aqueous content mixing dye, 6-carboxyfluorescein (6-CF), were transferred to the CV-1 cells, whereas F TM domain mutants L486A and I488A did not cause the transfer of R18 or 6-CF. Thus, these mutants do not cause either lipid mixing or content mixing. Previously, we determined that an increase in temperature can be a surrogate for HN triggering fusion.8, 53 To examine temperature-dependent triggering of the F TM domain mutants, red blood cells (RBCs) were labeled with 6-CF and bound to effector CV-1 cells expressing HN and F proteins and labeled with SYTO-17 at 4 °C. The target–effector complexes were then incubated at 29, 37, or 42 °C for 15 min, and the number of fusion events was measured by confocal microscopy (Fig. 5c). For wt F, the number of dye transfer events increased with increasing temperature. The amount of fusion mediated by F TM domain mutants L486A and I488A increased between 29 and 37 °C, but increasing the temperature beyond 37 °C did not enhance fusion. The mutants L486A and I488A did not exhibit a hemifusion phenotype, i.e., transfer of the lipidic dye R18 to target cells in the absence of transfer of the aqueous dye 6-CF. It is possible that these mutations either stabilize the F protein and prevent F protein from attaining its lowest energy postfusion conformation or affect protein–membrane interactions that stabilize an intermediate in the fusion pathway. For example these mutant F proteins may be trapped at a folding/fusion intermediate that cannot be overcome by an increase in temperature.
Fig. 5.
Analysis of fusion activity of alanine point mutants for F TM domain residues 485–489. (a) Representative micrographs of syncytia formed at 20 h p.t. in BHK-21F cells expressing PIV5 HN and either wt F or F containing TM domain point mutations or HN alone (mock). (b) Luciferase reporter gene assay to measure cell–cell fusion mediated by the F point mutants V485A-I489A. Vero cells were cotransfected to express HN, wt F or F point mutant proteins, and a luciferase reporter construct. Shown is the average of three experiments each done in triplicate and normalized to wt F. (c) Cell–cell fusion of F point mutants at different temperatures. After RBCs labeled with 6-CF were bound to CV-1 cells labeled with SYTO-17, the temperature was raised to 29, 37, or 42 °C for 15 min to activate fusion and dye transfer. Shown is the quantification of green 6-CF dye transfer events from three microscopic fields.
F TM domain mutants L486A and I488A form the open-stalk and prehairpin intermediates of fusion
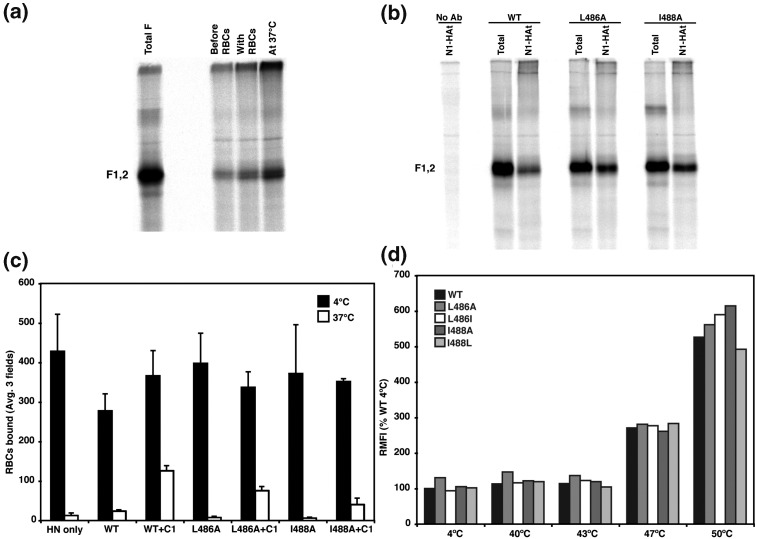
For PIV5 entry into cells by fusion, HN binds to its ligand, sialic acid, and by an unknown process begins the activation of F. The earliest stage of fusion that has been determined was inferred from functional properties of the N-1 peptide (which is derived from the HRA region of F). N-1 peptide can bind to the HRB region of F after HN has bound to target cells at 4 °C and inhibits fusion.8 Based on the atomic structure of prefusion F this step in fusion has been called the open-stalk conformation with the HRB helices melting and breaking the interactions at the base of the head but leaving the head domain largely intact.7 To determine if F TM domain mutants L486A and I488A can attain the F open-stalk conformation, a modified N-1 peptide, N-1-HAt, which contains an 11-residue HA tag (YPYDVPDYASL) at the C-terminus of N-1, was synthesized. Peptide binding was determined by the ability of the HA tag monoclonal antibody (mAb) to immunoprecipitate wt F protein. It was found that the N-1-HAt bound to F at 4 °C when target RBCs containing the HN receptor sialic acid were present but N-1-HAt did not bind to F as well in the absence of target RBCs. (Fig. 6a). To test if F TM domain mutants L486A and I488A reached the open-stalk stage of fusion, N-1-HAt peptide was incubated with cells expressing HN and one of the mutant F proteins at 4 °C in the presence of 0.5% hematocrit target RBCs (Fig. 6b). The HA tag mAb 12CA5 was used to immunoprecipitate the F protein containing bound peptide, and the total F in the lysate was immunoprecipitated with a polyclonal antibody (PAb) specific for F protein. N-1-HAt bound to wt F and F TM domain mutants L486A, and I488A, and the band for these mutants was generally more intense than that of the wt. This observation indicates the HRB region was accessible in all F proteins, and that the alanine substitutions in the TM domain might stabilize somewhat the formation of the open-stalk conformation of F protein.
Fig. 6.
Analysis of the protein conformation of F mutants L486A and I488A and examination of the steps of fusion attained by these mutants. (a) HA-tagged N-1 peptide binds to the open stalk intermediate of wt F in the presence of HN and target cells (RBCs). HeLa CD4 LTR βgal cells expressing HN and wt F were metabolically labeled with 400 μCi of 35S-Promix. Lane 1, wt F was immunoprecipitated with a PAb specific for F (PAb vacF) to indicate the total amount of wt F. Cells were incubated with 0.5% of RBCs. HA-tagged N-1 peptide was added either before the RBC incubation, during the RBC incubation, or during the 37 °C incubation used to initiate fusion. The F protein was then immunoprecipitated using the HA tag specific antibody 12CA5. Only a significant amount of F was immunoprecipitated when the N-1-HAt peptide was added with RBCs or during the 37 °C incubation when the open-stalk intermediate has formed. (b) Immunoprecipitation of the open-stalk intermediate for F mutants L486A and I488A. HeLa CD4 LTR βgal cells expressing HN and wt F, F L486A, or F I488A were metabolically labeled with 400 μCi of 35S-Promix. Lane 1, wt F plus N-1-HAt peptide with no antibody added; lanes 2, 4, and 6, HN- and wt F-, L486A- or I488A-expressing cells were incubated with 0.5% of RBCs at 4 °C in the absence of N-1-HAt peptide and were immunoprecipitated using PAb vac F (represents total F); lanes 3, 5, and 7, the N-1-HAt peptide was added during the RBC incubation at 4 °C. The F protein was co-immunoprecipitated with mAb 12CA5. The polypeptides were analyzed by SDS-PAGE under nonreducing conditions on 15% acrylamide gels. (c) Quantification of RBC binding for HN only or HN plus wt F-, F L486A-, and F I488A-expressing CV-1 cells. CV-1 cells labeled with SYTO 17 were incubated with RBCs labeled with 6-CF for 1 h at 4 °C. C-1 peptide was also added to some samples expressing wt F, F L486A, and F I488A during the 37 °C incubation for 15 min to capture the prehairpin intermediate. Black bars represent number of RBCs bound at 4 °C, and white bars represent the number of RBCs bound after the 15 min 37 °C incubation. Means and error bars shown are from three microscopic fields. (d) The conformation of wt F, F L486A, F L486I, F I488A, and F I488L mutant F on the surface of cells. This was determined by reactivity with postfusion specific mAb 6–7 at 4, 40, 43, 47, or 50 °C. HeLa CD4 LTR βgal cells expressing wt F, F L486A, F L486I, F I488A, or F I488L F protein were heated to 4, 40, 43, 47, or 50 °C for 10 min before binding mAb 6–7 at 4 °C for 30 min. Antibody reactivity was measured by flow cytometry. All data are normalized to wt F at 4 °C.
The second known step in the F protein refolding event after formation of the open stalk intermediate is thought to be that HRA refolds and the fusion peptide is inserted into the target cell membrane to form the prehairpin intermediate.7, 8 At this stage of fusion, the C-1 peptide, derived from the HRB region of F, specifically inhibits PIV5 F-mediated fusion54 by binding in the grooves on the outside of the HRA coiled coil.10 To capture the prehairpin conformation, the C-1 peptide was used in an RBC retention assay.8 To examine the F TM domain mutants for prehairpin formation, target RBCs labeled with 6-CF were bound at 4 °C to CV-1 cells that co-expressed F and HN and were labeled with STYO 17. When the RBCs that have bound at 4 °C to the CV-1 cell are warmed to 37 °C, the majority of RBCs either fuse or are released due to the neuraminidase activity of HN that is active at 37 °C but not at 4 °C.8, 55 In the presence of C-1 peptide, the F protein forms the prehairpin intermediate and the RBCs remain bound to the CV-1 cells because the fusion peptide has inserted into the RBC membrane, but further refolding is blocked and fusion is inhibited.8 Addition of C-1 peptide did not affect the number of RBCs bound at 4 °C (Fig. 6c). However, when cells expressing F TM domain mutants L486A and I488A with bound RBCs were warmed to 37 °C in the absence of C-1 peptide, there was a near-complete loss of bound RBCs, suggesting that these mutants are not trapped at the prehairpin intermediate (Fig. 6c). To determine if these mutants reached the prehairpin intermediate or are unable to transition between the open-stalk conformation and the prehairpin intermediate, 40 μM C-1 peptide was added. At 37 °C there was an increase in the number of RBCs retained as compared to L486A and I488A at 37 °C without the addition of peptide, although the number was not as large as for wt F (Fig. 6c). These data indicate that the F TM domain mutants form the prehairpin intermediate, but the reproducible lower amount of C-1 binding for mutants L486A and I486A could indicate some impairment in prehairpin formation for these two mutants.
F TM domain mutants 486 and 488 have closely related postfusion conformations as wt F protein
MAb 6–7 recognizes only the postfusion conformation of the F protein and not the prefusion conformation of the F protein.8, 53, 56 Thus, mAb 6–7 can be used to determine if F TM mutants L486A and I488A undergo the F protein refolding event that accompanies membrane fusion. It was found that although F TM domain mutants L486A and I488A cause greatly decreased fusion compared to wt F and TM domain mutants L486I and I488L, wt F and the four mutant F proteins exhibited the same mAb 6–7 reactivity at 4, 40, 43, 47, and 50 °C, as determined by flow cytometry, supporting the notion that the F protein TM domain mutants can proceed through the known intermediates of fusion and that they have a conformation closely related to postfusion F, even though L486A and L488A are essentially fusion inactive (Fig. 6d). Thus, although the steady-state concentrations of the prehairpin intermediate might be affected by these mutations, the extent and rate of conversion to the postfusion form recognized by mAB 6–7 would appear to be essentially the same as that of wt.
The addition of a hyperfusogenic mutation can rescue fusion for F TM mutants L486A and I488A
It has been shown previously that a mutant of the W3A isolate of PIV5, S443P, demonstrates a lower temperature requirement for fusion activation, faster fusion kinetics, and independence of HN activation.53 Because for F S443P extensive syncytia formation occurs at room temperature in the absence of HN co-expression, F S443P and related mutants57 have been termed hyperfusogenic. It is thought that F mutation S443P destabilizes the interactions between the top of the HRB three-helix bundle and the linker to the IgG-like domain II,7 hence lowering the energy barrier for conversion to the open-stalk conformation. Two other hyperfusogenic mutants that lower the temperature requirement for fusion and enable HN-independent fusion are Gly to Ala mutations in the fusion peptide G105A (previously referred to as G3A) and G109A (G7A).58, 59 Both mutations are highly destabilizing: the G109A not only destabilizes the F protein on cell surface expression but also inactivates the F protein relatively quickly and inactivates F for fusion if target cells are not present.59
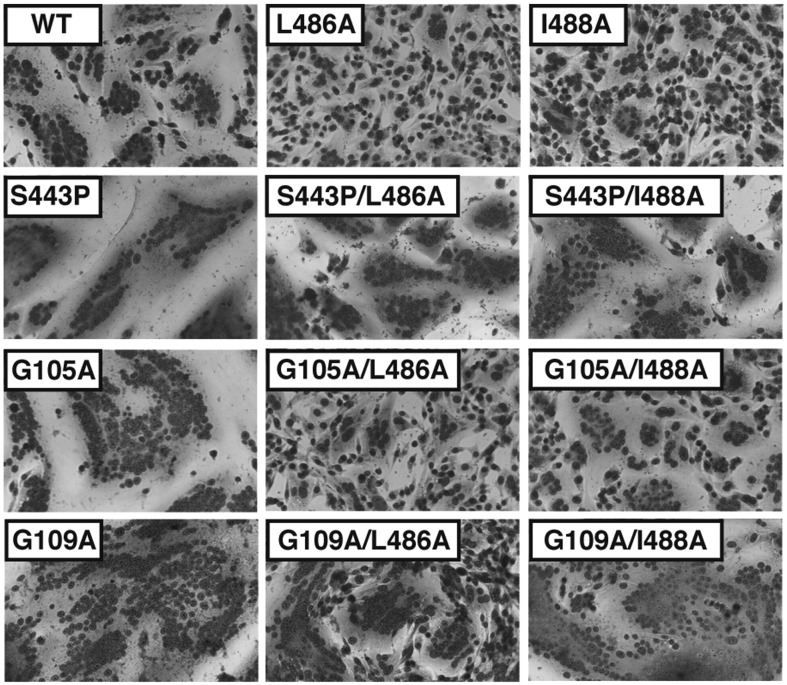
It seemed possible that if F TM domain mutants are blocked in causing fusion at a stage beyond the prehairpin intermediate, then incorporation of a hyperfusogenic mutation into the TM domain mutants might overcome the fusion block, probably due to the increased kinetics of fusion that is thought to be caused by triggering a greater number of F trimers at any one time. The L486A and I488A mutations were introduced into three hyperfusogenic backgrounds, S443P, G105A, and G109A F, to create the double mutants S443P/L486A, S443P/I488A, G105A/L486A, G105A/I488A, G109A/L486A, and G109A/I488A (Fig. 7 ). The F double mutants were expressed in HeLa CD4 long terminal repeat (LTR) β-galactosidase (βgal) cells and cell surface abundance was determined by flow cytometry. S443P/L486A, S443P/I488A, G105A/L486A, and G105A/I488A were surface expressed similarly to wt F, whereas the surface expression of G109A, G109A/L486A, and G109A/I488A was only 10% of that of wt F (Table 1). Expression in BHK-21F cells showed that F mutants S443P/L486A, S443P/I488A, G109A/L486A, and G109A/I488A exhibited extensive syncytia formation (hyperfusogenic) and syncytia formation was independent of HN expression (data not shown). F G105A/L486A and F G105A/I488A showed higher levels of syncytia formation than F L486A and F I488A but less than that of F G109A protein, consistent with earlier data for F mutant G105A.59 Thus, whereas F TM mutants L486A and I488A cannot cause fusion, the addition of the hyperfusogenic mutants to create double mutants lowers the energy barrier to fusion and overcomes the block to fusion caused by the L486A and I488A mutations. These data indicate that the F TM domain mutations L486A and I488A have not caused the F proteins to convert to an inactive “spent” conformation and the F TM mutant proteins have not veered off the fusion pathway. It may be that increasing the number of active F molecules increases the probability of a successful fusion event—perhaps the membrane state and the number of active F molecules are parameters that interact in a manner unknown to determine a successful outcome. Alternatively, the “lifetime” of the prehairpin intermediate may be different between mutant and wt, and the presence of a greater number of active F molecules could enhance the probability of successful fusion. This second explanation is also consistent with the peptide pull-down results (Fig. 6).
Fig. 7.
Syncytia formation mediated by F mutants L486A and I488A containing a second destabilizing mutation. Representative micrographs of syncytia formed in BHK-21F cells 20 h p.t. Cells were cotransfected with pCAGGS HN DNA and pCAGGS DNA encoding wt F, F L486A, or F I488A, the hyperfusogenic mutants F S443P, F G105A, or F G109A, or the double mutants containing a hyperfusogenic mutation and also F L486A or F I488A.
Changing the curvature of the membrane rescues fusion of F TM domain mutants L486A and I488A
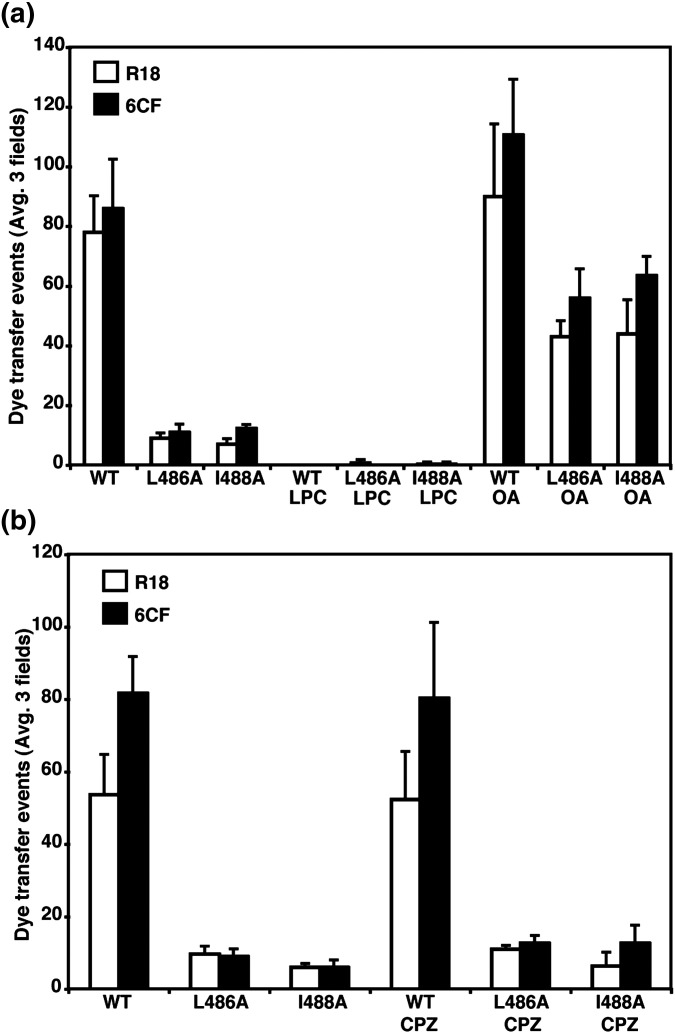
In addition to the F protein conformational intermediates of fusion, the changes occurring in the lipid bilayers can be broken down into several intermediates. It is thought that formation of the F protein 6-HB brings the membranes of the virus and the target cell together8, 10 and exclusion of water molecules permits formation of the hemifusion stalk. This stalk is an initial lipid connection between the proximal membrane leaflets.60 The stalk then expands to form the hemifusion diaphragm, which is a single bilayer segment, before the fusion pore forms.60 Lipids can affect membrane fusion based on their molecular shape. The cone-shaped lipid stearoyl lysophosphatidylcholine (LPC) has a positive spontaneous curvature, which is predicted to hinder the transition to hemifusion when present in the outer bilayer. The cone-shaped lipid oleic acid (OA) has a negative spontaneous curvature, which favors hemifusion when present in the outer bilayer.61 The addition of LPC and OA to cells expressing HN and F proteins has been shown previously to inhibit and favor an increase in fusion, respectively.8 To examine whether addition of cone-shaped lipids would overcome the block in fusion of F TM domain mutants, LPC was added to CV-1 cells expressing HN and wt F, F L486A, or F I488A before and during binding of R18- and 6-CF-labeled RBCs at 4 °C. On warming to 37 °C, neither lipid mixing (R18 transfer) nor cytoplasmic content mixing (6-CF transfer) occurred between the RBCs and the CV-1 effector cells (Fig. 8a). In contrast, addition of OA to the effector cells not only increased wt F fusion, but also caused a significant increase in lipid and content fusion for F TM mutants L486A and I488A (Fig. 8a). Chlorpromazine (CPZ), a drug that causes positive curvature of the lipid bilayer, ruptures the hemifusion diaphragm and causes pore formation.61 The addition of CPZ to effector cells did not cause an increase in lipid or contents fusion for F TM domain mutants L486A or I488A (Fig. 8b). Thus, these data suggest that the block in fusion of F TM domain mutants L486A and I488A occurs during the lipid intermediate stages of fusion. It is not known at which stage of the lipid intermediates the F protein attains its final postfusion form, although it is usually considered that refolding of the prehairpin intermediate to the postfusion form occurs across the stages of the lipid intermediates. The augmenting of fusion by OA that confers negative curvature to the membrane suggests that the F TM domain mutants are delayed/arrested at the lipid stalk intermediate and do not reach the hemifusion diaphragm or fusion pore stages of fusion.
Fig. 8.
Altering the curvature of the lipid membrane can rescue fusion for the F L486A and I488A mutants. (a) Quantification of cell–cell fusion in the dye transfer assay. Effector CV-1 cells were infected with vaccinia virus vTF7-3 and transfected with HN and wt F or mutant F DNA. RBCs were dually labeled with R18 (open bars) and the 6-CF (black bars). Labeled RBCs were bound to CV-1 cells for 1 h at 4 °C and then incubated at 37 °C for 15 min before visualization by confocal microscopy. Prior to incubation at 37 °C, 10 μM LPC or 10 μM OA was added for 15 min at 4 °C. 10 μM LPC or 10 μM OA was also present when the temperature was raised to 37 °C. The means and error bars are from three microscopic fields. (b) Quantification of dye transfer with dually labeled RBCs as above, but with 0.5 mM CPZ was added for 1 min at room temperature and then washed out before the temperature was raised to 37 °C to trigger fusion.
Evidence that the physical properties of the side chains of F TM domain residues 486 and 488 control fusion activity
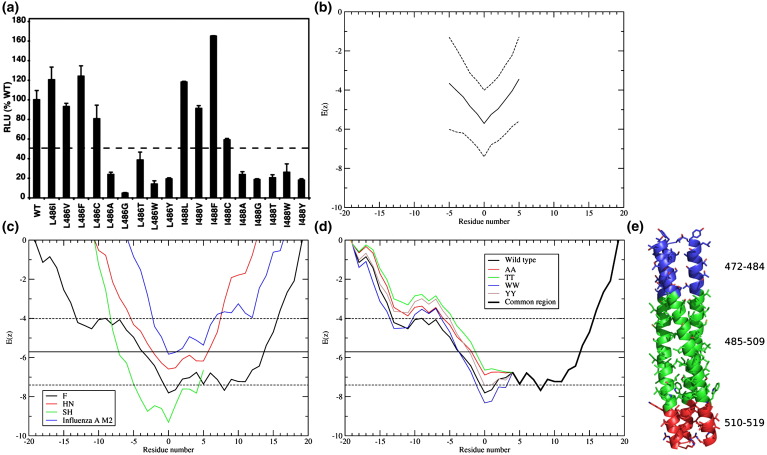
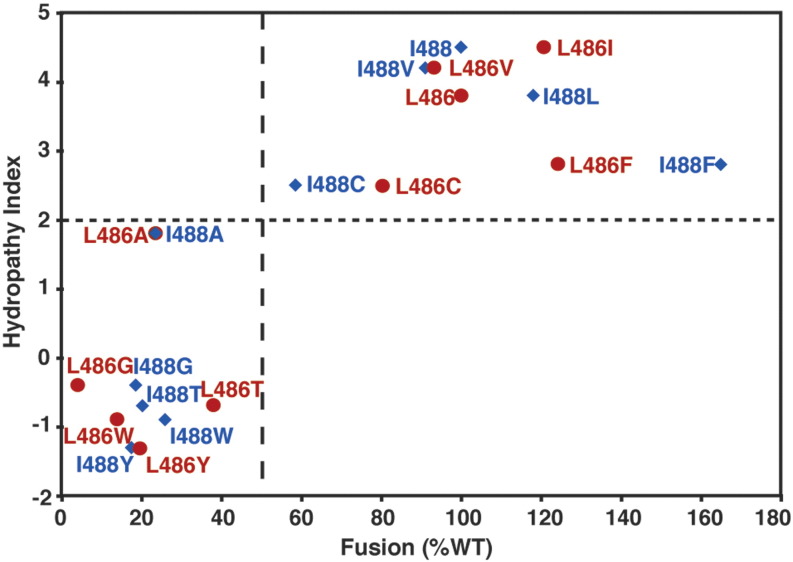
If the TM region of F plays an active role in stabilizing the lipid stalk or other lipidic intermediates, then one might expect the nature of the residues near the head-group region to have a profound effect on fusion. Therefore, further substitutions were made at F TM domain residues 486 and 488 to test how the distinct potentials of different amino acid side chains to partition into different regions of the membrane26, 62, 63 would affect fusion activity. L486 and I488 were each substituted with I/L, V, F, C, A, G, T, W, or Y (Fig. 9 ). I, L, V, and F are residues that prefer to be buried deep in a bilayer, experiencing a favorable free energy of transfer from water to the bilayer center. By contrast, C, A, G, and T, show relatively flat potential profiles, indicating that transfer to the bilayer is neither strongly favored nor disfavored. Finally, W and Y have a strong tendency to localize in the head-group region of the bilayer, relative to either the bilayer center or the aqueous phase. The fusion activity of each of the mutants expressed in Vero cells was determined using a luciferase gene reporter assay. Fusion activity was severely decreased (less than 50% of wt F) for residue 486 or 488 substitutions A, G, T, W, and Y. Substitution for C yielded ∼ 60% to 80% fusion, and substitutions for I/L, V, and F showed no difference in fusion activity compared to wt F protein (Fig. 9a).
Fig. 9.
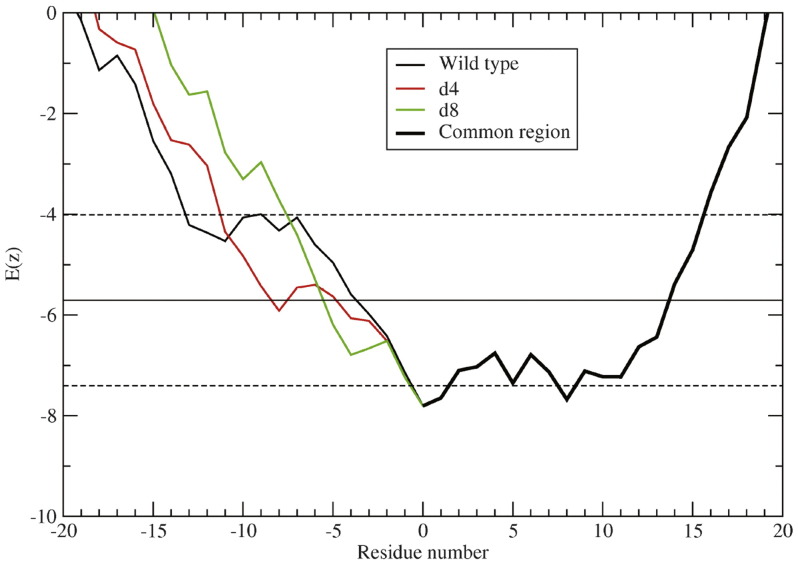
Fusion activity of F proteins depends on the hydrophobicity of substituted residues at TM residues L486 and I488. (a) Cell–cell fusion of substitutions made at residues 486 and 488 were quantified using the luciferase reporter gene assay. Vero cells were cotransfected to express HN and wt F or mutant F proteins, and a luciferase reporter construct. At 16 h p.t. BSR T7/5 cells were overlaid on the Vero cells, and 6 h post overlay, the luciferase activity of each sample was read. Dashed line indicates 50% of wt F fusion. Shown is the average of three experiments each done in triplicate and normalized to wt F. The next three panels show sequence suitability of protein sequences in the membrane quantified by the E(z) scale. (b) The mean (continuous line) and one standard deviation above and below the average (dashed line) E(z) score for 153 TM helices of known structure containing a minimum of 20 residues.64 Twenty-residue sliding windows were used for each helix sequence, with the residues renumbered such that the lowest-scoring (most membrane suitable) window occurs at residue number 0. The results for all 153 proteins were averaged after the renumbering. (c) E(z) results for four viral TM proteins: PIV5 F, PIV5 HN, PIV5 SH, and influenza (flu) A M2. Horizontal lines show the mean (continuous) and one standard deviation above and below (dashed) values for the 153 TM helices in (a). (d) E(z) results for four double mutants of PIV5 F. Residue positions L486 and I488 were mutated to alanine (A), threonine (T), tryptophan (W), and tyrosine (Y). Windows that do not include either mutation are shown in thick black (common region). (e) Atomistic model of the proposed extended TM region of PIV5 F. The canonical TM is shown in green, while the N-terminal (blue) and C-terminal (red) regions that are more suitable for the membrane than one standard deviation above the mean are also shown. This region is expected to be more suitable for the membrane than any window in 24 of the 153 TM domain sequences.
When the fusion activity was plotted against the hydrophobicity index of each residue,65 those amino acid residues with a hydrophobicity index below 2 (A, G, T, W, and Y) show decreased fusion, whereas those amino acid residues with a higher hydrophobicity index exhibited wt F protein fusion activity (Supplemental Fig. 1). The E(z) potential was used to test further how the substitutions would affect the propensity of the TM sequence to insert into a bilayer.26 This potential, which is based on a statistical analysis of the crystallographic database of 153 membrane proteins,64 defines the potential for a given residue to reside at various depths in the membrane, and it is useful for determining which regions of a TM domain have the greatest preference to reside in specific regions of the bilayer.
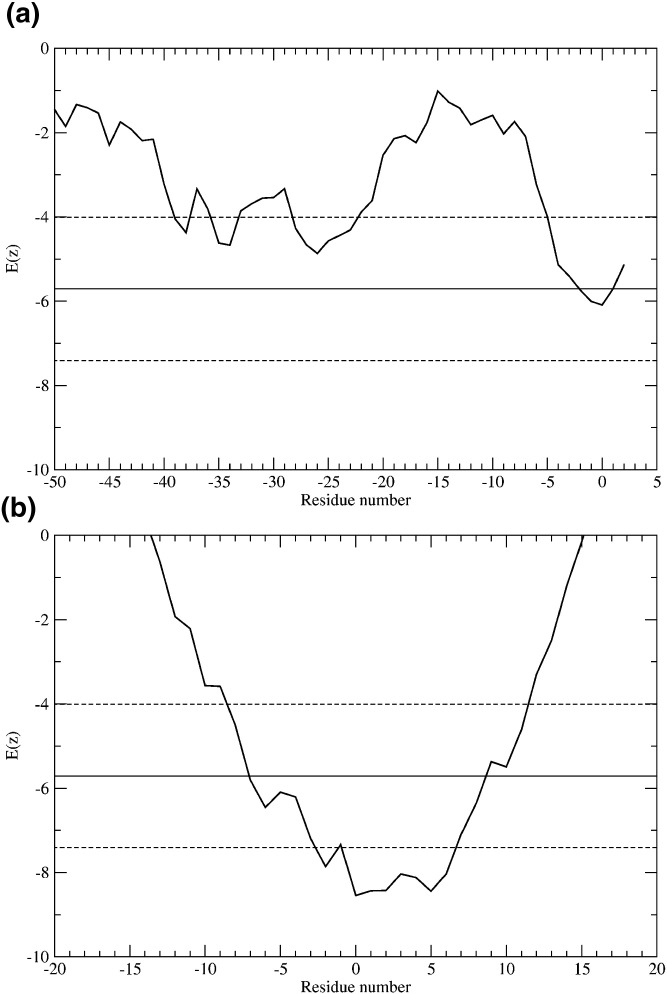
Figure 9b shows a plot of the E(z) potential scores generated by computing the mean score for successive 20-residue segments across the sequence. The results are plotted for the average, and for segments with scores plus or minus a single standard deviation from the mean. In each case the sequences funnel down to a relatively sharp minimum. Because each point reflects the score for a 20-residue sequence, the minimum identifies the 20-residue segment with the highest propensity to lie vertical to the bilayer surface. This figure also compares the profiles of the F protein with that for the other two integral membrane proteins of the PIV5 (SH and HN)66, 67 and the M2 proton channel of influenza A virus,68 the latter of which contains both a TM helix as well as a C-terminal cytoplasmic helical segment (Fig. 9c). The sequences of SH, HN, and M2 funnel in the usual manner, and 20–30 residue stretches are observed with a high propensity to span the bilayer. Interestingly, however, the F protein sequence shows an unusually broad minimum, indicative of a much more extended sequence with some propensity to lie within the bilayer. This property is conserved among the paramyxovirus F proteins tested (measles virus, mumps virus, Newcastle disease virus and human parainfluenza viruses 2 and 3), although the profile for PIV5 is the broadest (Supplemental Fig. 3). For PIV5 F, the absolute minimum in the curve suggests the TM helix begins at residue 485, in good agreement with the air-oxidation cross-linking data, showing this to be near the head-group region. However, with a very small energetic cost, the native TM helical bundle could translocate up or down through the membrane or lie at an oblique angle.
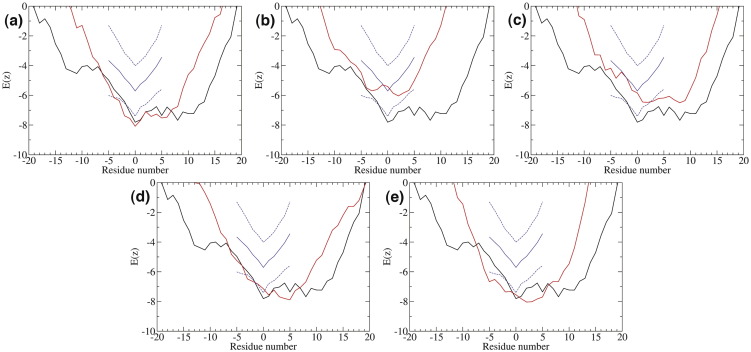
Interestingly, the scores for a 15-contiguous-point N-terminal minimum lies within one standard deviation of the mean value for the databank TM helices. This region includes a portion of the membrane-proximal end of the crystallographically defined structure of the HRB. Figure 9d illustrates the effects of the mutations on the cumulative E(z) potential score for the N-terminal region for the mutations at residues 486 and 488. To help visualize the effects, the graphs were computed for double mutants, but the same trends hold for the individual mutants. The mutations change the 20-residue window of the protein with the most membrane-favorable sequence. Instead of the 20-residue window 485–504 being the most membrane favorable, the best window becomes 493–512. Additionally, with the exception of Trp, all the mutations have the effect of moving the most membrane suitable 20-residue window from 485–504 in the wild type to 493–512 in the mutants. Clearly, other regions of the protein are important for determining the boundary of the TM helix so it is difficult to interpret this as a literal movement of the TM in the resting state. Nevertheless, if helical movements are important for the fusion mechanism, these changes would upset the energetics of fusion. Figure 9e shows an atomistic model of the proposed extended TM domain region of PIV5 F. The canonical TM is shown in green, while the N-terminal (blue) and C-terminal (red) regions that are more suitable for the membrane than one standard deviation above the mean are also shown. The extended TM domain region is expected to be more suitable for the membrane than any window in 24 of the 153 TM domain sequences.26
Discussion
For ion channel proteins that have multiple TM spanning domains, atomic structure determinations have enabled a distinction between those TM α-helices that serve architectural structural roles and those that act as the aqueous pore/ionic selectivity filter.69 For the homotetrameric influenza virus M2 proton-selective ion channel protein, each polypeptide chain only spans the membrane once and the single hydrophobic domain found in each polypeptide chain has to act as the endoplasmic reticulum membrane insertion sequence, the membrane anchoring sequence and the pore and gate of the channel.38, 68, 70, 71, 72 However, for the majority of integral membrane proteins that span a membrane once, the role of the TM domain besides being a membrane anchorage domain is largely unknown.
For many viral proteins that mediate membrane fusion, mutagenesis studies on the TM domain have yielded a wide variety of results. However, the preponderance of data indicates that the TM domain of these fusion proteins is not simply a string of hydrophobic amino acids that span a lipid bilayer but there is amino acid sequence specificity to the TM domain, implying specific roles of TM domain amino acid residues in fusion protein function. Specificity of amino acid residue implies specific structural features or interactions of these TM domain residues with the lipid bilayers and, thus, a direct role of amino acid side chains in the process of membrane fusion, further implying that membrane fusion is not simply a solely lipidic event.
To study the features of the PIV5 F protein TM domain for membrane fusion activity, alanine-scanning mutagenesis was used. The F protein TM domain was found to exhibit sequence dependence for fusion activity and it was also observed that a block of 20 leucine residues could not substitute for the entire PIV5 F TM domain even to obtain cell surface expression of F. Upon further substitution, residues L486 and I488 were found to play a key role in fusion, where altering the hydrophobicity of the side chain at these residues profoundly affected fusion activity (Fig. 9a). Although F mutants L486A and I488A were deficient for fusion, biological activity could be rescued by the addition of the destabilizing mutations F S443P, F G105A, or F G109A (Fig. 7), indicating F L486A and I488A were not trapped in an intermediate conformation nor had the mutant F proteins veered irreversibly off the fusion pathway. It is generally thought that F-mediated membrane fusion requires the action of several trimers and the hyperfusion phenotype of destabilizing mutants may be due to more synchronous F-activation events in an otherwise stochastic process. Thus, addition of a destabilizing mutation to F proteins that are blocked for fusion may simply alter the lifetimes of conformational intermediates, driving the fusion process by mass action. Detailed analysis indicated F L486A and F I488A form the prefusion conformation of F, the open-stalk intermediate, the prehairpin intermediate, and F can be converted to a conformation that is close to, or at the postfusion form (Fig. 1a–e). As discussed above, the precise timing of the conversion of the prehairpin conformation to the postfusion form with the formation of the lipid intermediates is not known, but refolding of F may occur across all the lipid intermediate stages. Evidence from studies of HIV gp120/gp41 suggest some 6-HB formation occurs after a pore has formed.13, 73
Thus, it seemed likely that the block in fusion for the F L486A and F I488A proteins corresponds to their inability to complete the lipidic stages of fusion (Fig. 1d and e). The data obtained using OA to confer negative spontaneous curvature of the membrane and concomitantly causing a large increase in fusion activity of F L486A and F I488A indicate that fusion is delayed/arrested at the lipid stalk intermediate (Fig. 1f–i). These findings suggest that the TM domain might play an active role in the lipidic steps in fusion, as discussed previously.16, 17, 18, 19 In fusion, the formation of the 6-HB and the postfusion form of F brings together the TM domain and the FP, which excludes water between the membranes, and this local dehydration allows for membrane contact.2 There is a high-energy requirement for the presumptive next stage, the lipid stalk.74
One goal of this work was to define the structural features of the TM domain and to relate these to potential roles in the mechanism of fusion. The available atomic structures of ion channel proteins indicate that the multiple TM domains are mostly α-helical69 and for the very few known viral proteins, the structures of the TM domains of integral membrane proteins are α-helical.71, 72, 75 Even less is known as to whether the TM α-helices are together in the membrane forming helical bundles or whether each α-helix is separate from the others. Clearly, for the influenza virus M2 ion channel, the TM domains form a four-helix bundle.71, 72 CryoEM studies on the human immunodeficiency virus fusion protein (Env) differ, as some reports indicate the TM domains of the fusion protein are apart, like the legs of a tripod,30, 31 whereas other studies indicate the TM domains are together and form one helical bundle.32 The substitution of F TM domain residues with cysteine and cross-linking induced by air oxidation confirmed the N-terminal boundary of the TM in the membrane, and treatment with CuP indicates the TM domains in the F trimer form a TM helical bundle. Further, the extensive cross-linking of residues in the outer leaflet of the bilayer suggests that in this region the helices have significant flexibility or are only partially folded, thereby allowing three consecutive residues to become very highly cross-linked (Fig. 3b). Quantification of the extent of disulfide bond formation and modeling studies indicate that the data fit best a model in which the F TM domain forms a 3-HB within the membrane. A break in the regular seven-residue period seen for coiled coils suggest that a residue was inserted near the center of the bilayer, resulting in either a pi bulge at residues 497–500 (Fig. 3d and e) or a “stutter” geometry (Fig. 3f). The PIV5 TM domain contains glycine at residues 494 and 497, but they do not appear to be essential to function or to mediate tight interhelical interactions. Residue L486 is predicted to be at the interface of the 3-HB, although the fusion data (in the presence of oleic acid) indicate this residue is likely to interact with the lipids of the bilayer. Presumably, the structural malleability that enables the residues close to the ectodomain to form disulfide bonds also enables residue L486 to populate different conformations that either face the interior of the predicted 3-HB or interact with the lipid bilayer.
The energy necessary to form the lipid stalk may be derived from the fusion protein, which may generate bilayer stresses that are relaxed by forming the stalk intermediate.2 For PIV5 F protein, the outer leaflet residues L486 and I488 may facilitate the negative curvature of the outer lipid leaflet that is necessary to merge the two bilayers, and residues with higher hydrophobicity may provide a more negative curvature (Fig. 1f–i).
The long potential length of the TM sequence of paramyxovirus F proteins (Fig. 9 and Supplemental Fig. 3) suggests an additional possible role for this domain in fusion. The length would allow the helix to move dynamically into different orientations and/or lie at extreme angles in the membrane. In this work, the extraviral end of the TM helix in the resting state was inferred from the Cys cross-linking as well as the E(z) potential, which were in good agreement. However, if this were the only orientation sampled by the helix during fusion, then mutation of residue I488 to Trp should not negatively impact function because it would be on the exposed face of the 3-HB located in the head-group region, where Trp is quite stabilizing. If, however, dynamic changes in the orientation within the bilayer were required, then this anchoring substituent would upset the energetic balance between the states adopted along the reaction coordinate. Given that the mutants appear to be defective in lipid stalk formation, and that the TM helix has a hydrophobic region both preceding and following it, we consider it possible that the TM region might act as a template to stabilize the lipid stalk intermediate. The N-terminal hydrophobic extension would encourage the diffusion of lipids out of the outer leaflet of the bilayer and along the stalk-like template, and initiating the formation of the lipid stalk (Fig. 10 ). Decreasing the hydrophobicity of the protein by mutating I488 or L486 to less polar residues or stabilizing the initial configuration by inclusion of Trp and Tyr would increase the energetic cost of translocation of the helix through the membrane and migration of lipids up the stalk.
Fig. 10.
Hypothesized mechanism of lipid stalk intermediate stabilization. (a) Prefusion. A simplified representation of two PIV5 F protein trimers in the prefusion state is shown. The canonical TM domain is shown in orange. The regions hypothesized to enter the membrane in the lipid stalk intermediate are shown as white extensions of the TM domain. The fusion peptide is shown in red, HRA in green, HRB in blue, and the globular head is shown in yellow spheres. (b) Lipid stalk. As the lipid stalk forms, the extended TM domain completely enters the membrane environment, stabilizing the lipid stalk state and lowering the energetic cost of forming this intermediate. Arrows represent the movement of the globular head away from the stalk in the postfusion state. (c) Postfusion. Two F proteins are shown after refolding of the proteins into the postfusion state.
Furthermore, the hydrophobic stretch at the cytoplasmic C-terminus of the TM helix might allow the TM 3-HB to translocate outward, extending the length of the exposed patch on the extracellular side of the bilayer (Fig. 10). Our calculations and experimental results suggest that the energy of the system is finely balanced, allowing this translocation to occur at minimal energetic cost. In the resting state, the TM bundle is stably inserted in the bilayer, but vertical translocations within the bilayer should occur at a relatively small energetic cost due to the lack of N-terminal anchoring residues and the hydrophobicity of the residues on both sides of the TM helix. More importantly, hairpin formation might provide a strong driving force for translocation of the TM helices in the bilayer. This conformational transition has the net effect of pulling the FP and the TM helix (located in opposing bilayers) toward one another, effectively applying a mechanical force favoring translocation of the N-terminus of the TM helix out of the bilayer, and the C-terminus of the TM helix deeper into the bilayer. The concomitant increase in length of the hydrophobic N-terminal extracellular region would provide a greater surface to serve as a template for nonbilayer lipidic intermediates. Finally, we note that in the prehairpin intermediate the 3-HB of HRA has a very hydrophobic surface prior to docking of the HRB helix and formation of the water-soluble postfusion 6-HB. Thus, the 3-HB of HRA could serve a similar role to encourage the initiation of a lipid-coated stalk projecting from the target cell's membrane.
It is possible that as the last stage in the F protein refolding event the TM domain interacts with the FP and forms another 6-HB. The process of converting prehairpin F to the postfusion form of F may provide the necessary energy to exclude the water between the membranes and form the lipid intermediates (Fig. 1c–e). At the first stage of fusion, the outer leaflets of the two bilayers have just merged but have not mixed (hemifusion). The TM domain and FP may still be segregated in separate bilayers, and the interaction between the TM domain and FP could drive the formation of the hemifusion intermediate. Hyperfusogenic mutants G105A and G109A are located within the FP. These two mutants rescue fusion of L486A and I488A (Fig. 7). Although hypothesized previously that these destabilizing mutants G105A and G109A may cause hyperfusion by lowering the energy necessary to drive the various fusion intermediates,59 it is also possible that these FP mutants affect the potential interaction with the TM domain and destabilize and lower the energy requirement to mix the bilayers and drive the hemifusion intermediate and form the fusion pore (Fig. 1c–e).
In summary, the results presented here demonstrate that the TM helix of F plays an active role in fusion. It forms a well-defined TM bundle, and subtle modulations to its sequence give rise to large changes in activity. We further suggest that its unusual sequence characteristics might allow it to play an active role in templating the formation of the lipid stalk intermediate. We note that other proteins such as HIV gp41 also show unusually long hydrophobic TM helices, and changing its physical properties strongly impacts fusion.23 Additionally, dengue E protein (a class II fusion protein) has two TM helices, one of which has a long membrane suitable region that has an E(z) score one deviation from the average of 153 TM domains (Supplemental Fig. 4a). VSV (class III) has a single TM helix that that also has a long membrane suitable region (Supplemental Fig. 4b). Although fusion is intrinsically a complex process with multiple transition states it would appear that viral fusogenic proteins share not only similar structures and conformational changes, but they might also specifically stabilize well-defined intermediates in the fusion pathway.
Materials and Methods
Cells, virus, and plasmids
BHK-21F, Vero, BSR T7/5, CV-1, and HeLa CD4 LTR βgal cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). BHK-21F cells were grown in DMEM supplemented with 10% tryptose phosphate broth, and HeLa CD4 LTR βgal cells were grown in DMEM supplemented with 200 μg/ml geneticin, 100 μg/ml hygromycin B, and 20 mM Hepes (pH 7.4). The recombinant vaccinia virus (vTF7-3) that expresses T7 RNA polymerase was grown in CV-1 cells as described previously.76 pCAGGS and pGEM2X plasmids encoding PIV5 F, PIV5 HN, and PIV5 S443P F have been described previously.53 pCAGGS plasmids encoding PIV5 G105A and G107A F have also been previously described.58, 59 PIV5 F proteins containing TM domain amino acid residue substitutions were made by four-primer PCR with Tgo DNA polymerase and then by cloning into pCAGGS PIV5 F and pGEM2X PIV5 F. Plasmids encoding the double substitutions were made by subcloning pCAGGS L486A and I488A F into pCAGGS plasmids encoding S443P, G105A, or G107A F. Mutations were confirmed by nucleotide sequencing using an Applied Biosystems 3100-Avant automated DNA sequencer.
Expression of F and HN glycoproteins
PIV5 F and HN cDNAs cloned in the pCAGGS vector were expressed in BHK-21F, Vero, and HeLa CD4 LTR βgal cells by transient transfection using the Lipofectamine Plus expression system (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Transfected Vero cells were incubated for 4 h at 37 °C before the addition of DMEM containing 2% FBS and incubated a further 18 h at 37 °C. PIV5 F and HN cDNAs in the pGEM2X vector were expressed using the recombinant vaccinia virus–T7 RNA polymerase transient expression system (vac T7).76 CV-1 cells in six-well dishes containing glass cover slides were infected at a multiplicity of infection of 10 plaque-forming units with vTF7-3 for 1 h at 37 °C. The cells were then transfected with 1.0 μg each of pGEM2X F and HN DNA using liposomes prepared as previously described.77 After 4 h at 37 °C, DMEM with 10% FBS was added and cells were incubated at 33 °C overnight.
Syncytia formation
Monolayers of BHK-21F cells in six-well plates were transfected with 1.0 μg each of pCAGGS PIV5 F and HN DNA as described above. At 20 h posttransfection (p.t.), cells were fixed and stained using a Hema 3 stain (Fisher Scientific, Pittsburgh, PA) according to the manufacturer's instructions, and photographs were taken with a digital camera (DCS 760, Kodak, Rochester, NY) attached to an inverted phase-contrast microscope (Diaphot, Nikon, Melville, NY).
Luciferase reporter gene assay
To quantify cell–cell fusion, a luciferase reporter gene assay was performed as previously described.57 Briefly, Vero cell monolayers in six-well plates were transfected with 1.0 μg each of three plasmids, luciferase control DNA expressing the T7 promoter (Promega, Madison, WI), pCAGGS PIV5 F, and pCAGGS PIV HN. At 16 h p.t., BSR T7/5 cells expressing the T7 RNA polymerase were overlaid onto the Vero cells and incubated at 37 °C for 6 h. The monolayers were then washed, lysed, and clarified by centrifugation per the manufacturer's instructions (Promega). For each sample, 150 μl of lysate was loaded into a 96-well plate. The luciferase activity of each lysate was quantified using 150 μl luciferase assay substrate (Promega) and an Lmax luminescence microplate reader (Molecular Devices, Sunnyvale, CA).
Dye transfer assays
Human RBCs were singly labeled with the aqueous dye 6-CF (Invitrogen) or dual labeled with 6-CF and the lipid probe octadecyl rhodamine F chloride (R18, Invitrogen). CV-1 cells were grown on glass cover slides and F and HN were expressed using the vac T7 expression system. To visualize effector cells when using singly labeled RBCs, CV-1 cells were labeled with 1 μM SYTO-17 nucleic acid dye (Invitrogen) for 1 h at 37 °C. Analysis of lipid and aqueous dye transfer was performed as previously described.57 Fusion was quantified by counting positive cells by using a scanning confocal microscope (LSM 5 Pascal, Carl Zeiss MicroImaging, Inc., Thornwood, NY) and averaging the fusion events obtained from three separate fields. For HN-independent retention of RBCs, SYTO-17 CV-1 cells were incubated with 0.1% hematocrit 6-CF-labeled RBCs. Following incubation with RBCs, cells were incubated at either 4 or 37 °C for 15 min. During this warming stage, 40 μM C-1 peptide was added to some of the samples. C-1 peptide was expressed in bacteria and purified as previously described.54 For the temperature dependence of dye transfer, samples were incubated at 29, 37, and 42 °C for 15 min after the binding of target RBCs. For dye transfer experiments using the addition of lipids, fresh solutions of 10 μM LPC (Avanti Polar Lipids, Birmingham, AL) or 10 μM OA (Sigma-Aldrich, St. Louis, MO) in phosphate-buffered saline (PBS) were made. After binding R18/6-CF dual-labeled RBCs as above, CV-1 cells were incubated in cold LPC and OA solutions for 15 min at 4 °C. The temperature was then shifted to 37 °C by changing the bathing solution with new PBS containing LPC or OA prewarmed to 37 °C and plates were incubated at 37 °C for 15 min as above. For dye transfer experiments with CPZ (Sigma), cells were prepared and R18/6-CF RBCs were bound as above. After raising the temperature to 37 °C for 15 min, 0.5 mM CPZ in PBS was added to the CV-1 cells for 1 min. Cells were extensively washed with PBS without drug.
Flow cytometry
To quantify cell surface expression and to determine the protein conformation of the F protein, monolayers of HeLa CD4 LTR βgal cells in six-well plates were transfected with 1.0 μg of pCAGGS PIV5 F DNA as described above and flow cytometry was performed as previously described using fluorescein-isothicyanate-labeled secondary antibody.78 For surface expression, mAb F1a79 was used at 1:100 dilution. To examine conformational rearrangements in the F protein, mAb 6–780 was used at 1:30 dilution. Before the addition of mAb 6–7, warmed PBS was added to the samples and the plates were incubated at 40, 43, 47, or 50 °C for 10 min and washed with cold PBS. The fluorescence intensity of 10,000 cells was measured by using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Capture of open-stalk intermediate
Monolayers of HeLa CD4 LTR βgal cells in 6-cm dishes were transfected with 2.0 μg each of pCAGGS PIV5 F and HN DNA as described above. At 18 h p.t., cells were starved with Cys- and Met-deficient DMEM for 30 min. The cells were labeled with 400 μCi of 35S-Promix (GE Healthcare Bio-Sciences, Piscataway, NJ) in 1 ml of Cys- and Met-deficient DMEM for 1 h. To allow for newly synthesized F proteins to reach the cell surface, the samples were chased with DMEM without serum for 2 h. The cells were washed with PBS and incubated with 1 ml of 0.5% hematocrit RBCs with or without 80 μg of N-1-HAt peptide for 1 h at 4 °C. N-1-HAt peptide was expressed in bacteria and purified as previously described.54 After washing at least five times with PBS to remove any unbound RBCs, the samples were incubated with 1 ml DMEM without serum containing 60 μg of anti-HA mAb 12CA5 for 3 h at 4 °C. The cells were washed another five times with PBS, then lysed with cold radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors and 50 mM iodoacetamide.81 Clarified lysates were incubated with 40 μl protein A–Sepharose beads overnight at 4 °C. The samples were washed three times with RIPA buffer containing 0.3 M NaCl, three times with RIPA buffer containing 0.15 M NaCl, once with 50 mM Tris buffer [0.25 mM EDTA (ethylenediaminetetraacetic acid), 0.15 M NaCl (pH 7.4)], and polypeptides were analyzed by SDS-PAGE on 15% acrylamide gels under nonreducing conditions in the absence of DTT.
Oxidative cross-linking
Monolayers of HeLa CD4 LTR βgal cells in six-well plates were transfected with 1.0 μg each of pCAGGS PIV5 F and HN DNA as described above. At 18 h p.t., cells were starved with Cys- and Met-deficient DMEM for 30 min. The cells were then labeled with 50 μCi of 35S-Promix (GE Healthcare Bio-Sciences) in 1 ml of Cys- and Met-deficient DMEM for 1 h. Cells were Dounce homogenized in cold RSB buffer [10 mM Tris (pH 7.4), 10 mM KCl, and 15 mM MgCl2], and 3 mM CuP (final concentration, freshly made) was added for 10 min at 37 or 4 °C. The reaction was stopped with 10 mM EDTA and 10 mM N-ethylmaleimide to chelate the copper and block free sulfhydryl groups. Samples were solubilized by adding 2× RIPA buffer plus 100 mM iodoacetamine and protease inhibitors as previously described81 and were clarified by centrifugation for 10 min at 55,000 rpm in a Beckman TLA100 rotor. Samples were incubated for 2 h at 4 °C with 10 μl of rabbit polyclonal anti-F2 peptide antiserum and then incubated with 40 μl protein A–Sepharose beads overnight at 4 °C. Samples were washed with RIPA buffer as above and polypeptides were analyzed by SDS-PAGE on 3.5% borate–acetate gels82 under nonreducing conditions in the absence of DTT.
Modeling of the TM domain
To better understand the periodic interface between the F protein TM domains, the raw radioactivity values were normalized by dividing the amount of disulfide formed by the total amount of protein (disulfide linked plus not linked). Because cross-linking is stronger toward the outside of the membrane and saturation can occur on the image plates of the Fuji Image Analyzer (Valhalla, NY), two sets of cross-linking data were combined for the analysis. First, cross-linking data using CuP at 37 °C were used for most of the TM positions, residues 492–509. To put the residues that are in the outer leaflet on the same scale, low-temperature (4 °C) CuP data were used for residue positions 485–491. Given the normalized data, a sine wave was fit according to the following formula:
where x is the residue number, y is the normalized degree of cross-linking, a is the amplitude of the sine wave, b is the phase offset, c is the α-helical periodicity, and d is the y offset of the sine wave. Values were fit by nonlinear regression. To visualize the cross-linking data on a single helix, cartoon representations of an ideal helix and a helix with a pi bulge were created with the interface residues labeled in spheres using the program PyMOL [DeLano, W. L. The PyMOL Molecular Graphics System (2002)]† . The modeling protocol for the TM trimer is described in the Supplemental Methods.
Acknowledgements
We thank Charles Russell and Sarah Connolly for helpful discussions. The N-1 HA-tag peptide construct was kindly provided by Sarah Connolly. This work was supported in part by Research Grant R01 AI 23173 from the National Institute of Allergy and Infectious Diseases. M.L.Z.B and J.E.D. were supported by National Institutes of Health Medical Scientist Training Program Grants T32 GM08152-18 and HL107971, respectively. RAL is an investigator of the Howard Hughes Medical Institute.
Edited by J. Bowie
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2008.12.029
Appendix A.
Supplemental Fig. 1.
Fusion activity of F proteins depends on the hydrophobicity of substituted residues at TM residues L486 and I488. Plot of fusion activity from Fig. 9 versus the Kyte and Doolittle hydropathy index for each substituted residue. Substitutions at 486 are shown in red and substitutions at 488 are shown in blue. Wt F is denoted as L486 or I488. Long dashed line indicates 50% of wt F fusion, and short dashed line indicates hydropathy index of substitutions producing less than 50% of F fusion. All substitutions in lower left quadrant are deficient in fusion.
Supplemental Fig. 2.
PIV5 linker deletion mutants have similar E(z) scores. The wt F sequence (black) is shown with two deletion mutants: d4 (red) represents the deletion of residues 481–484 and d8 (green) represents the deletion of residues 477–484. These mutants showed similar fusogenic activity to the wt F protein experimentally (Baker el., 1999 ref. 10). Unlike the hypofusogenic mutations shown in Fig. 9, both deletion mutants show similar or greater membrane suitability than the wt F protein over an extended region N-terminal to the canonical TM segment. As in Fig. 9, the mean (solid line) and one standard deviation above and below the average (dashed line) E(z) score for 153 TM domain helices of known structure is shown. 20 residue sliding windows were used and the residues renumbered such that the lowest scoring (most membrane suitable) window occurs at residue number 0. Windows not affected by either mutation are shown in thick black (common region).
Supplemental Fig. 3.
Paramyxovirus F proteins contain broad minima on the E(z) scale. The E(z) score of each of five homologous fusion proteins are shown in red dash: (A) mumps virus, (B) measles virus, (C) Newcastle disease virus (NDV), (D) human parainfluenza virus 2 (HPIV2), (E) human parainfluenza virus 3 (HPIV3). For comparison, in each window the PIV5 F (black solid) is shown. In addition, the mean (blue solid) and one standard deviation from the mean (blue dash) E(z) scores for a database of 153 TM domain helices of known structure are shown. 20 residue sliding windows were used and the residues renumbered such that the lowest scoring (most membrane suitable) window of the F protein occurs at residue number 0. A multiple sequence alignment was used to renumber the homologs to match the PIV5 F protein numbering.
Supplemental Fig. 4.
Analysis of an example of the TM domains of a class 2 and a class 3 viral fusion protein on the E(z) scale. The E(z) scores of two viral fusion proteins from different fusion protein classes were considered: (A) Dengue E protein (class II) and (B) VSV G protein (class III). As in other E(z) figures, a sliding window is used such that each residue number in the figure represents a 20 residue window. Dengue E protein is predicted to have two TM helices, the leftmost being the longer. VSV G protein is predicted to have a single, long TM helix. As in Fig. 9, the mean (solid line) and one standard deviation above and below the average (dashed line) E(z) score for 153 TM domain helices of known structure is shown.
References
- 1.Tamm L.K., Crane J., Kiessling V. Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr. Opin. Struct. Biol. 2003;13:453–466. doi: 10.1016/s0959-440x(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 2.Chernomordik L.V., Kozlov M.M. Protein–lipid interplay in fusion and fusion of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 3.Martens S., McMahon H.T. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell. Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 4.Wickner W., Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb R.A., Parks G.D. Paramyxoviridae: The viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th edit. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 1449–1496. [Google Scholar]
- 6.Yin H.-S., Paterson R.G., Wen X., Lamb R.A., Jardetzky T.S. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl Acad. Sci. USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H.S., Wen X., Paterson R.G., Lamb R.A., Jardetzky T.S. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell C.J., Jardetzky T.S., Lamb R.A. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 10.Baker K.A., Dutch R.E., Lamb R.A., Jardetzky T.S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 11.Lamb R.A., Jardetzky T.S. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melikyan G.B., Markosyan R.M., Hemmati H., Delmedico M.K., Lambert D.M., Cohen F.S. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melikyan G.B., Lin S., Roth M.G., Cohen F.S. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell. 1999;10:1821–1836. doi: 10.1091/mbc.10.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melikyan G.B., Grener S.A., Ok D.C., Cohen F.S. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 1997;136:995–1005. doi: 10.1083/jcb.136.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melikyan G.B., Markosyan R.M., Roth M.G., Cohen F.S. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol. Biol. Cell. 2000;11:1143–1152. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemble G.W., Danieli T., White J.M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 20.Cleverley D.Z., Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl Acad. Sci. USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Blissard G.W. Functional analysis of the transmembrane (TM) domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: substitution of heterologous TM domains. J. Virol. 2008;82:3329–3341. doi: 10.1128/JVI.02104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West J.T., Johnston P.B., Dubay S.R., Hunter E. Mutations within the putative membrane-spanning domain in the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation, and infectivity. J. Virol. 2001;75:9601–9612. doi: 10.1128/JVI.75.20.9601-9612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang L., Yue L., Hunter E. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell–cell fusion and virus infection. J. Virol. 2008;82:5417–5428. doi: 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helseth E., Olshevsky U., Gabuzda D., Ardman B., Haseltine W., Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owens R.J., Burke C., Rose J.K. Mutations in the membrane-spanning domain of the human immunodeficiency envelope glycoprotein that affect fusion activity. J. Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senes A., Chadi D.C., Law P.B., Walters R.F., Nanda V., Degrado W.F. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J. Mol. Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Hessa T., Meindl-Beinker N.M., Bernsel A., Kim H., Sato Y., Lerch-Bader M. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong R.T., Kushnir A.S., White J.M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyauchi K., Komano J., Yokomaku Y., Sugiura W., Yamamoto N., Matsuda Z. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 2005;79:4720–4729. doi: 10.1128/JVI.79.8.4720-4729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forster F., Medalia O., Zauberman N., Baumeister W., Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl Acad. Sci. USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou T., Xu L., Dey B., Hessell A.J., Van Ryk D., Xiang S.H. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanetti G., Briggs J.A.G., Grunewald K., Sattentau Q.J., Fuller S.D. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:790–797. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D.K., Cheng S.F., Kantchev E.A.B., Lin C.H., Liu Y.T. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;6 doi: 10.1186/1741-7007-6-2. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee G.F., Burrows G.G., Lebert M.R., Dutton D.P., Hazelbauer G.L. Deducing the organization of a transmembrane domain by disulfide cross-linking. J. Biol. Chem. 1994;269:29920–29927. [PubMed] [Google Scholar]
- 35.Lee G.F., Lebert M.R., Lilly A.A., Hazelbauer G.L. Transmembrane signalling characterized in bacterials chemoreceptors by using sulfhydryl cross-linking in vivo. Proc. Natl Acad. Sci. USA. 1995;92:3391–3395. doi: 10.1073/pnas.92.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee G.F., Dutton D.P., Hazelbauer G.L. Identification of functionally important helical faces in transmembrane segments by scanning mutagenesis. Proc. Natl Acad. Sci. USA. 1995;92:5416–5420. doi: 10.1073/pnas.92.12.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falke J.J., Koshland D.E., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 38.Bauer C.M., Pinto L.H., Cross T.A., Lamb R.A. The influenza virus M2 ion channel protein: probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 39.Grinthal A., Guidotti G. Dynamic motions of CD39 transmembrane domains regulate and are regulated by the enzymatic active site. Biochemistry. 2004;43:13849–13858. doi: 10.1021/bi048644x. [DOI] [PubMed] [Google Scholar]
- 40.Grinthal A., Guidotti G. Bilayer mechanical properties regulate the transmembrane helix mobility and enzymatic state of CD39. Biochemistry. 2007;46:279–290. doi: 10.1021/bi061052p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda M., Leser G.P., Russell C.J., Lamb R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl Acad. Sci. USA. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J., Dutch R.E., Lamb R.A. Proper spacing between heptad repeat B and the transmembrane domain boundary of the paramyxovirus SV5 F protein is critical for biological activity. Virology. 1997;239:327–339. doi: 10.1006/viro.1997.8917. [DOI] [PubMed] [Google Scholar]
- 43.Bass R.B., Butler S.L., Chervitz S.A., Gloor S.L., Falke J.J. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Ende B.M., Sharom F.J., Davis J.H. The transmembrane domain of Neu in a lipid bilayer: molecular dynamics simulations. Eur. Biophys. J. 2004;33:596–610. doi: 10.1007/s00249-004-0407-2. [DOI] [PubMed] [Google Scholar]
- 45.Luecke H. Atomic resolution structures of bacteriorhodopsin photocycle intermediates: the role of discrete water molecules in the function of this light-driven ion pump. Biochim. Biophys. Acta. 2000;1460:133–156. doi: 10.1016/s0005-2728(00)00135-3. [DOI] [PubMed] [Google Scholar]
- 46.Boudker O., Ryan R.M., Yernool D., Shimamoto K., Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–393. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- 47.Brown J.H., Cohen C., Parry D.A. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 48.Strelkov S.V., Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- 49.Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–375. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 50.Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 51.Doms R.W., Lamb R.A., Rose J.K., Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 52.Leser G.P., Ector K.J., Ng D.T., Shaughnessy M.A., Lamb R.A. The signal for clathrin-mediated endocytosis of the paramyxovirus SV5 HN protein resides at the transmembrane domain–ectodomain boundary region. Virology. 1999;262:79–92. doi: 10.1006/viro.1999.9890. [DOI] [PubMed] [Google Scholar]
- 53.Paterson R.G., Russell C.J., Lamb R.A. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 54.Joshi S.B., Dutch R.E., Lamb R.A. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
- 55.Porotto M., Murrell M., Greengard O., Doctor L., Moscona A. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 2005;79:2383–2392. doi: 10.1128/JVI.79.4.2383-2392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connolly S.A., Leser G.P., Yin H.S., Jardetzky T.S., Lamb R.A. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl Acad. Sci. USA. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell C.J., Kantor K.L., Jardetzky T.S., Lamb R.A. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 2003;163:363–374. doi: 10.1083/jcb.200305130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horvath C.M., Lamb R.A. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell C.J., Jardetzky T.S., Lamb R.A. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 2004;78:13727–13742. doi: 10.1128/JVI.78.24.13727-13742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chernomordik L.V., Kozlov M.M. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 61.Chernomordik L.V., Kozlov M.M. Membranes of the world unite! J. Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 63.White S.H., von Heijne G. How translocons select transmembrane helices. Annu. Rev. Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 64.Gimpelev M., Forrest L.R., Murray D., Honig B. Helical packing patterns in membrane and soluble proteins. Biophys. J. 2004;87:4075–4086. doi: 10.1529/biophysj.104.049288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 66.Hiebert S.W., Paterson R.G., Lamb R.A. Identification and predicted sequence of a previously unrecognized small hydrophobic protein, SH, of the paramyxovirus simian virus 5. J. Virol. 1985;55:744–751. doi: 10.1128/jvi.55.3.744-751.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiebert S.W., Paterson R.G., Lamb R.A. Hemagglutinin-neuraminidase protein of the paramyxovirus simian virus 5: Nucleotide sequence of the mRNA predicts an N-terminal membrane anchor. J. Virol. 1985;54:1–6. doi: 10.1128/jvi.54.1.1-6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamb R.A., Zebedee S.L., Richardson C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 69.von Heijne G. The membrane protein universe: what's out there and why bother? J. Intern. Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 70.Pinto L.H., Dieckmann G.R., Gandhi C.S., Papworth C.G., Braman J., Shaughnessy M.A. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc. Natl Acad. Sci. USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnell J.R., Chou J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stouffer A.L., Acharya R., Salom D., Levine A.S., Di Costanzo L., Soto C.S. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Markosyan R.M., Cohen F.S., Melikyan G.B. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell. 2003;14:926–938. doi: 10.1091/mbc.E02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang L., Huang H.W. Observation of a membrane fusion intermediate structure. Science. 2002;297:1817–1818. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y., Corver J., Chipman P.R., Zhang W., Pletnev S.V., Sedlak D. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuerst T.R., Niles E.G., Studier F.W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rose J.K., Buonocore L., Whitt M.A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 78.Waning D.L., Russell C.J., Jardetzky T.S., Lamb R.A. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl Acad. Sci. USA. 2004;101:9217–9222. doi: 10.1073/pnas.0403339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Randall R.E., Young D.F., Goswami K.K.A., Russell W.C. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 80.Tsurudome M., Ito M., Nishio M., Kawano M., Komada H., Ito Y. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 2001;75:8999–9009. doi: 10.1128/JVI.75.19.8999-9009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paterson R.G., Lamb R.A. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A., Elliott R.M., editors. Molecular Virology: A Practical Approach. IRL Oxford University Press; Oxford, UK: 1993. pp. 35–73. [Google Scholar]
- 82.Gething M.-J., McCammon K., Sambrook F. Protein folding and intracellular transport. Evaluation of conformational changes in nascent exocytotic proteins. Methods Cell Biol. 1989;32:185–206. doi: 10.1016/s0091-679x(08)61171-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.