Abstract
The transition from pachytene to Meiosis I is a key regulatory point in yeast meiosis. This transition requires Ndt80, a transcription factor that commits cells to complete meiosis by expression of a diverse set of genes. In this issue of Genes & Development, Sourirajan and Lichten (2627-2632) report that CDC5, an NDT80-regulated gene encoding yeast polo-like kinase, is sufficient for Holliday junction resolution and exit from pachytene. Determining the meiotic targets of Cdc5 therefore provides a new approach for identifying a eukaryotic Holliday resolvase.
Keywords: Saccharomyces cerevisiae, synaptonemal complex, joint molecules, crossover, cyclin-dependent kinase, Holliday junction
A major challenge for sexually reproducing organisms is the need to keep the chromosome number of a species constant from generation to generation. This challenge has been met by the evolution of a specialized cell division called meiosis, which divides the chromosome number of a cell in half to produce gametes. The fusion of two gametes at fertilization then results in offspring with the parental number of chromosomes. Meiosis has important similarities to mitosis, where sister chromatids created by chromosome replication are segregated to opposite poles of the spindle to create genetically identical daughter cells. However, to reduce the number of chromosomes in half, meiosis exhibits a fundamental difference with mitosis in that it consists of two rounds of chromosome segregation following a single round of chromosome duplication. The first division is unique to meiosis in that homologous pairs of sister chromatids disjoin to opposite poles. The second meiotic division resembles mitosis in that sister chromatids segregate into daughter cells. Meiosis I requires a series of specialized, coordinated steps during prophase I to allow paternal and maternal homologs to segregate in a reductional division. These steps include creating physical connections between homologous pairs of sister chromatids by a combination of crossovers and sister chromatid cohesion, the mono-orientation of sister kinetochores, and the loss of arm, but not centromere, cohesion at anaphase I. Execution of these steps involves the interplay between meiosis-specific factors and mitotic regulators (Marston and Amon 2004; Wan et al. 2008).
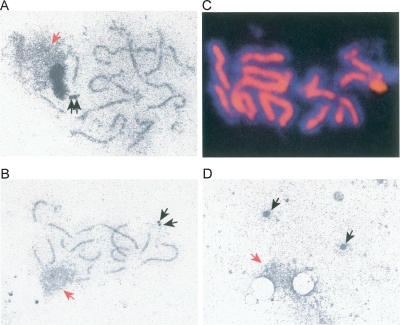
Chromosome behavior during meiotic prophase I has been analyzed both at the cytological and molecular levels. A hallmark of meiosis is the synapsis of homologous pairs of sister chromatids to form synaptonemal complexes (SCs) (Zickler and Kleckner 1999). Cytologists have divided prophase into discrete stages based on the degree of synapsis. In leptotene, sister chromatids condense along protein cores to form axial elements. This stage is followed by zygotene, where the axial elements of homologous chromosomes begin to be joined together by the insertion of a central region to form stretches of SC. Pachytene is the stage when synapsis of all of the chromosomes is complete. In budding yeast, synaptonemal formation has been analyzed both by electron microscopy as well as by immunofluorescence studies using Zip1, a meiosis-specific protein that forms part of the central region of the SC (Fig. 1A–C; Sym et al. 1993). Besides fully synapsed chromosomes, another feature of budding yeast pachytene cells is that the spindle pole bodies are duplicated but unseparated (Fig. 1A,B). The disassembly of the SCs occurs at diplotene, followed by metaphase I and anaphase I (Fig. 1D). Although SCs are found in the germ cells of many sexually reproducing organisms, the requirements for synapsis vary between species. In budding yeast and mice, for example, recombination is required to bring homologous chromosomes together, whereas synapsis is independent of recombination in fruit flies and worms (Villeneuve and Hillers 2001). The exact function of the SC is not well understood.
Figure 1.
Budding yeast meiotic chromosomes at pachytene and beyond. (A,B) Electron micrographs of silver-stained SCs in nuclear spreads from cells in pachytene. Note that the spindle pole bodies indicated by black arrows are duplicated, but unseparated. The nucleolus is indicated with a red arrow. (C) Pachytene chromosomes visualized by immunostaining of Zip1 (red) and 4′,6-diamidino-2-phenylindole (DAPI) to stain the DNA (blue). (D) Electron micrograph of silver-stained nuclear spread after SC disassembly and spindle pole body separation. Images courtesy of Josef Loidl, University of Vienna.
The molecular details of recombination have been most thoroughly delineated in budding yeast where assays for many of the steps of recombination are available (Hollingsworth and Brill 2004). The mechanism for initiating recombination is highly conserved through evolution, and involves the introduction of double-strand breaks (DSBs) catalyzed by a topoisomerase-like protein called Spo11 (Keeney 2001). The 5′ ends of the breaks are resected to produce 3′ single-stranded tails. These tails are bound by the recombinases, Dmc1 and/or Rad51, RecA orthologs that mediate strand invasion of homologous chromosomes. The preference for recombination to occur between homologs instead of between sister chromatids is another difference in meiosis and is due in part to the influence of meiosis-specific axial element components (Schwacha and Kleckner 1997; Couteau et al. 2004; Niu et al. 2005). Further processing creates recombination intermediates containing two Holliday junctions (HJs) that are then preferentially resolved as crossovers (Allers and Lichten 2001). The enzyme that resolves these meiotic HJs has long been sought. While a structure-specific endonuclease, Mus81–Mms4, appears to resolve single HJs formed during fission yeast meiotic recombination, it is not responsible for the bulk of HJ resolution in budding yeast and is unlikely to be the resolvase during mammalian meiosis (Hollingsworth and Brill 2004; McPherson et al. 2004; Cromie et al. 2006). Therefore, this Holy Grail of eukaryotic recombination still remains to be found.
Budding yeast has played a major role in our understanding of meiosis. One reason is the relative ease with which cells can be induced to undergo meiosis. It would be disastrous for haploid cells to enter meiosis, and this catastrophe is prevented by demanding cells be heterozygous for mating type. In addition, a lack of nitrogen in combination with a nonfermentable carbon source is required for cells to enter the meiotic program. A standard experimental protocol to study meiotic events is to take cells at various times after transfer to sporulation (Spo) medium and use genetic, biochemical and cytological assays to compare wild-type and mutant strains. This approach has shown that DSB formation corresponds with leptotene, strand invasion with zygotene, and double HJ intermediates with pachytene (Hunter and Kleckner 2001). Resolution of double HJs to form crossovers occurs around the time of pachytene exit.
Another advantage of using budding yeast for meiotic studies is the ability of yeast cells that have entered meiosis to “return to growth” when provided with rich medium (Esposito and Klapholz 1981). Under these conditions, cells that have proceeded through premeiotic S-phase exit meiosis and chromosomes segregate equationally as in a mitotic division. Cells that are returned to growth after meiotic recombination is initiated exhibit high levels of both gene conversion and crossovers. Return to growth experiments revealed a phenomenon in budding yeast called the “commitment to meiosis.” It was observed that once cells enter Meiosis I, they no longer leave the meiotic program and proceed to make spores even when exposed to glucose (Esposito and Klapholz 1981). Cells in pachytene can return to growth, whereas cells that have begun Meiosis I cannot. Therefore a reasonable candidate for the point at which cells become committed to completing the meiotic divisions is exit from pachytene.
Studies of meiotic gene expression show that groups of genes are transcribed in successive waves. Based on the timing of their expression, these waves have been broken down into classes described as early, middle, and late (Chu et al. 1998; Primig et al. 2000). Early meiotic genes include many of the genes required for recombination and synapsis such as SPO11, DMC1, and ZIP1. Early meiotic gene transcription is induced by the activator Ime1, which is expressed in response to both genetic and nutritional signals (Vershon and Pierce 2000). The second wave of gene expression contains middle sporulation genes required for meiotic progression such as CLB1, CLB3, and CLB4, as well as spore formation (e.g., GIP1 and SPS1) (Neiman 2005; Carlile and Amon 2008). This wave of gene expression is dependent upon a transcription factor called NDT80 (Chu and Herskowitz 1998). NDT80 itself is a meiosis-specific gene whose transcription is regulated in two steps. The first step of NDT80 transcription has been described to have “delayed early” timing, in that it occurs after early meiotic gene transcription but before the middle sporulation genes are expressed (Chu and Herskowitz 1998). This stage requires both Ime1 and the kinase activity of an early meiotic gene product, Ime2 (Pak and Segall 2002). Ndt80 protein resulting from this delayed early transcription binds to cis-acting sequences in the NDT80 promoter called middle sporulation elements and then acts as a potent transcriptional activator of itself (Chu and Herskowitz 1998). The resulting Ndt80 protein is more abundant, activated by phosphorylation, and binds to middle sporulation elements upstream of many middle and late sporulation genes to induce their transcription (Chu and Herskowitz 1998; Benjamin et al. 2003).
NDT80 is a key regulator that controls exit from pachytene. ndt80Δ mutants exhibit a classic pachytene arrest with fully synapsed chromosomes and duplicated, but unseparated, spindle pole bodies (Xu et al. 1995). The meiotic recombination checkpoint responds to unrepaired recombination intermediates by arresting cells through inhibition of Ndt80 activity (Chu and Herskowitz 1998; Tung et al. 2000). Although DSB formation and noncrossovers are normal in the absence of NDT80, double HJ resolution is decreased, raising the interesting possibility that the meiotic HJ resolvase is either directly or indirectly regulated by NDT80 (Allers and Lichten 2001). Benjamin et al. (2003) developed a way of conditionally expressing NDT80 during meiosis by placing NDT80 under the control of the GAL promoter (NDT80-IN). The GAL4 activator was then fused to the estradiol receptor (Gal4-ER). In the absence of estradiol, the Gal4-ER fusion is present in the cytoplasm, NDT80 is not expressed and cells arrest in pachytene. Addition of estradiol to the Spo medium results in the movement of Gal4-ER to the nucleus and transcription of NDT80. Induction of NDT80 with estradiol is sufficient for cells to synchronously complete meiosis and form spores (Benjamin et al. 2003; Carlile and Amon 2008).
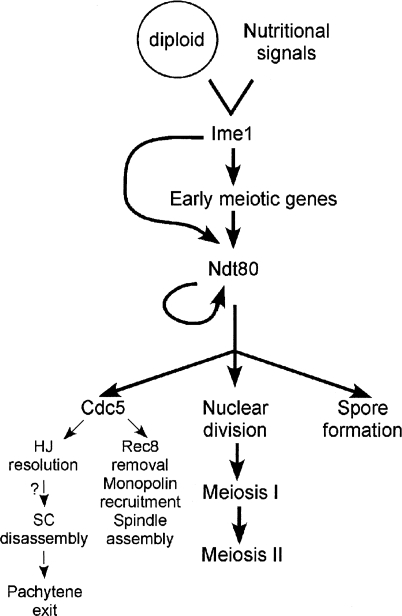
It was initially thought that commitment to meiosis could not be due to a single event because “there appears to be a series of individual commitments to different landmark events, rather than one early commitment event for the whole process” and because it is possible “to separate commitment to the two primary genetic events of meiosis, genetic recombination and chromosome segregation…” (Esposito and Klapholz 1981). It now appears as though commitment to meiosis does result from a single event—the induction of NDT80, which then proceeds to transcribe genes that control a diverse set of coordinated, but independent, processes such as exit from pachytene, the meiotic divisions and the packaging of haploid nuclei into spores (Fig. 2).
Figure 2.
Cdc5 defines a new branch in the NDT80 regulon. This figure is adapted from Chu and Herskowitz (1998) with permission from Elsevier (http://www.sciencedirect.com/science/journal/10972765). The question mark indicates uncertainty about whether there is a mechanistic connection between HJ resolution and SC formation and, if so, which process comes first.
As in mitotic cells, cyclin-dependent kinase (CDK) plays a critical role in meiosis. CDKs are comprised of catalytic and cyclin subunits. In budding yeast, there is a single catalytic component called Cdc28. In vegetative yeast cells, there are both G1 cyclins (Clns) and mitotic cyclins (Clbs). In meiotic cells, the function of the Cdc28–Cln complexes in promoting DNA replication has been taken over by Ime2 (Dirick et al. 1998). CLB2, the predominant cyclin in vegetative cells, is not expressed (Grandin and Reed 1993). CLB1, CLB3, and CLB4 all contain middle sporulation elements in their promoters and are part of the NDT80 regulon (Chu and Herskowitz 1998). CDK activity during meiotic prophase is comprised of primarily of Cdc28–Clb5, 6, and this CDK complex is essential for premeiotic S-phase and DSB formation (Stuart and Wittenberg 1998; Henderson et al. 2006; Carlile and Amon 2008). Several different approaches have been used to inactivate CDK during meiosis, including deletion of CLB5 and CLB6. clb5 clb6 diploids fail to undergo premeiotic S phase but express the NDT80-regulated middle genes, CLB1, CLB3, SPS1, and SPS2, indicating that NDT80 function is not regulated by Cdc28–Clb5,6 (Stuart and Wittenberg 1998). Other approaches to inactivate CDK in meiosis include the use of temperature-sensitive (ts) cdc28 mutants and analog-sensitive versions of CDC28 (cdc28-as) whose kinase activity can be inhibited by addition of a purine analog (1-NM-PP1) to the medium. At high temperature, cdc28ts diploids accumulate in pachytene (Shuster and Byers 1989). Similarly, a low level of inhibitor (0.5 μM 1-NM-PP1) allows premeiotic S-phase but not the meiotic divisions, even though NDT80 is expressed (Benjamin et al. 2003). One interpretation of this result is that Cdc28–Clb1 is needed for pachytene exit.
A second evolutionarily conserved family with important functions in mitosis contains the polo-like kinases (Plks). Plks from yeast to mammals are important for mitotic entry, spindle assembly, chromosome segregation and cytokinesis (Barr et al. 2004). In addition to a catalytic domain, Plks contain a conserved region called a polo-box domain. The polo-box domain is a phospho-serine/threonine-binding module that acts to direct Plks to their substrates (Elia et al. 2003). The preferred sequence for the polo-box domain is serine–phosphoserine–proline (S-pS-P). This sequence overlaps with CDK consensus phosphorylation sites (T/S-P-X-K/R), suggesting that some Plk substrates may first be primed by CDK phosphorylation.
Budding yeast contains a single Plk called Cdc5 (Barr et al. 2004). In addition to mitosis, CDC5 also has important functions in meiotic cells. The CDC5 promoter contains three middle sporulation elements, exhibits middle sporulation gene transcriptional kinetics and Cdc5 protein is not induced in ndt80Δ diploids, indicating that CDC5 is an NDT80-regulated gene (Clyne et al. 2003). To create null versions of this essential gene, CDC5 was placed under the control of either the SCC1/MCD1 or CLB2 promoters (cdc5-nm) (Clyne et al. 2003; Lee and Amon 2003). These promoters are down-regulated during meiosis and immunoblot analysis showed that Cdc5 protein disappears between 2 and 3 h after transfer into Spo medium. Depletion of Cdc5 has no effect on premeiotic S-phase, transcription of early meiotic or NDT80-regulated genes. However, the bulk of cells fail to enter the meiotic divisions and there is a defect in SC disassembly. cdc5-nm prevents cohesin cleavage and removal from chromosomes. The cdc5-nm phenotype is not representative of a classic pachytene arrest, however, in that the duplicated spindle pole bodies separate to form metaphase I spindles. Intriguingly, cdc5-nm diploids fail to resolve HJs efficiently and exhibit a concomitant reduction in crossovers, while noncrossovers are unaffected (Clyne et al. 2003). This result raises the intriguing possibility that the ndt80Δ recombination defect is due to a failure to transcribe CDC5.
In this issue of Genes & Development, Sourirajan and Lichten (2008) address the roles of CDK and Cdc5 in HJ resolution, crossover formation, and SC disassembly. To look at CDK function, strains were constructed containing a cdc28-as allele and NDT80-IN. In the absence of estradiol and 1-NM-PP1, CDK is active and the cells arrest in pachytene with unresolved HJs due to the absence of NDT80. Some crossovers are present, most likely resulting from a HJ-independent pathway mediated by Mus81–Mms4 (de los Santos et al. 2003). Induction of NDT80 transcription by addition of estradiol results in rapid disappearance of HJs, an increase in crossovers, proteolysis of Zip1, and disassembly of SCs. When estradiol is added 1 h after inactivation of Cdc28-as with a high level of inhibitor (5 μM 1-NM-PP1), these events still occur although there is a slight delay. The spindle pole bodies do not separate, nor do cells enter Meiosis I, demonstrating that Cdc28–Clb1 kinase is inactive. While this result indicates that Cdc28–Clb1 is not required for exit from pachytene, it does not rule out the possibility that phosphorylation by Cdc28–Clb5,6 prior to the ndt80 arrest is necessary, perhaps by priming proteins that will later become substrates for other kinases such as Cdc5. If true, then these phosphorylation events would have to be quite stable, as Sourirajan and Lichten (2008) inactivated Cdc28 an hour before inducing Ndt80, potentially allowing time for phosphatases to act, yet still saw no effect.
CDC5 is necessary for efficient HJ resolution and SC disassembly but is it sufficient? Sourirajan and Lichten (2008) placed CDC5 under the control of GAL4-ER in an ndt80Δ diploid to create CDC5-IN ndt80Δ. Addition of estradiol to this diploid results in the specific transcription of CDC5, leaving the remaining genes in the NDT80 regulon off. Remarkably, induction of CDC5 alone allows HJ resolution leading to an increased number of crossovers, as well as disassembly of SCs as measured by a loss of Zip1 staining from meiotic chromosomes. Induction of a kinase dead version of CDC5 does not rescue these phenotypes, indicating that Cdc5 phosphorylation of one or more target proteins at the ndt80Δ arrest is all that is needed to complete recombination and exit pachytene.
The fact that CDC5 is the only NDT80-regulated gene required for HJ resolution and SC disassembly focuses the search for proteins involved in these processes to those phosphorylated by Cdc5. This discovery is exciting because it suggests a new way to look for the budding yeast meiotic HJ resolvase. Identification of this recombination enzyme has proved intractable to genetic approaches, perhaps because the genes are essential or redundant or because the enzyme is comprised of more than one subunit. The finding that meiotic HJ activity is controlled by a single kinase indicates that either the resolvase itself, or other proteins important for controlling resolvase, are present at the ndt80Δ arrest waiting to be phosphorylated by Cdc5. Therefore, identification of Cdc5 substrates may ultimately reveal the proteins responsible for nuclear HJ activity in budding yeast. Recently developed novel approaches for the identification of kinase targets using analog-sensitive kinases make this an attainable goal (Allen et al. 2007; Snead et al. 2007; Blethrow et al. 2008). Furthermore the CDC5-IN ndt80Δ system should facilitate this search since it allows exit from pachytene but not entry into Meiosis I, thereby potentially limiting the Cdc5 substrates phosphorylated in this window to those directly involved in HJ resolution and SC disassembly.
Cdc5-regulated proteins now define a new, genetically separable, pathway in the Ndt80 regulon, in addition to the meiotic division and spore formation pathways noted by Chu and Herskowitz (1998) (Fig. 2). These proteins regulate not only processes directly involved in Meiosis I chromosome segregation such as monopolin recruitment and removal of cohesion, but also some that are important for pachytene exit. The fact that out of all the genes regulated by Ndt80, CDC5 alone is sufficient for HJ resolution and SC disassembly raises interesting questions. For example, is the dismantling of SCs mechanistically linked to crossover formation or are these processes independently regulated by Cdc5 (Fig. 2)? Sourirajan and Lichten (2008) show that SC disassembly does not occur simply by the degradation of Zip1, since induction of CDC5-IN in the ndt80Δ background results in SC breakdown while Zip1 protein remains intact. How does Cdc5 phosphorylation therefore allow the removal of Zip1 from meiotic chromosomes? The elegant experiments by Sourirajan and Lichten (2008) have made obtaining the answers to these important questions possible.
Acknowledgments
I am grateful to Bruce Futchter and Aaron Neiman for helpful comments on the manuscript, and to Josef Loidl who provided images and advice for Figure 1. Work from my laboratory is supported by R01 GM50717.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1727508.
References
- Allen J.A., Li M., Brinkworth C.S., Paulson J.L., Wang D., Hubner A., Chou W.-H., Davis R.J., Burlingame A.L., Messing R.O., et al. A semisynthetic epitope for kinase substrates. Nat. Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Barr F.A., Sillje H.H.W., Nigg E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Benjamin K.R., Zhang C., Shokat K.M., Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes & Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow J.D., Glavy J.S., Morgan D.O., Shokat K.M. Covalent capture of kinase-specific phosphopeptides reveals Cdk1–cyclin B substrates. Proc. Natl. Acad. Sci. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T.M., Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Clyne R.K., Katis V.L., Jessop L., Benjamin K.R., Herskowitz I., Lichten M., Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Cromie G.A., Hyppa R.W., Taylor A.F., Zakharyevich K., Hunter N., Smith G.R. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L., Goetsch L., Ammerer G., Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6 associated kinase in Saccharomyces cerevisiae. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- Elia A.E.H., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantely L.C., Smerdon S.J., Yaffe M.B. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-Box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- Esposito R.E., Klapholz S. Meiosis and ascospore development. In The molecular biology of the yeast Saccharomyces cerevisiae: Life cycle and inheritance. In: Strathern J.N., et al., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1981. pp. 211–287. [Google Scholar]
- Grandin N., Reed S.I. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K.A., Kee K., Maleki S., Santini P., Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Brill S.J. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes & Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nat. Rev. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- McPherson J.P., Lemmers B., Chahwan R., Pamidi A., Migon E., Matysiak-Zablocki E., Moynahan M.E., Essers J., Hanada K., Poonepalli A., et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- Neiman A.M. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N.M. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Segall J. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:6417–6429. doi: 10.1128/MCB.22.18.6417-6429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shuster E.O., Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead J.L., Sullivan M., Lowery D.M., Cohen M.S., Zhang C., Randle D.H., Taunton J., Yaffe M.B., Morgan D.O., Shokat K.M. A coupled chemical-genetic and bioinformatic approach to polo-like kinase pathway exploration. Chem. Biol. 2007;14:1261–1272. doi: 10.1016/j.chembiol.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A., Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes & Dev. 2008 doi: 10.1101/gad.1711408. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D., Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes & Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Engebrecht J., Roeder G.S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Tung K.S., Hong E.J., Roeder G.S. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. 2000;97:12187–12192. doi: 10.1073/pnas.220464597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A.K., Pierce M. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 2000;12:334–339. doi: 10.1016/s0955-0674(00)00104-6. [DOI] [PubMed] [Google Scholar]
- Villeneuve A.M., Hillers K.J. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K.M., Boulton S.J., Hollingsworth N.M. Cdc28–Clb5 (CDK-S) and Cdc7–Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes & Dev. 2008;22:386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura M., Padmore R., Klein C., Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]