Abstract
Particular DNA sequences have long been known to have exceptional structures and biological properties. Famous in the medical world are the trinucleotide repeat sequences, such as (CTG)n, and their association with more than a dozen neurodegenerative diseases. Numerous meetings have been held to discuss these repeats and the diseases they cause. Now, a much-needed meeting has been held to discuss other noncanonical (non-B-form) DNA structures, their properties, and their biological consequences. Although the meeting was titled “DNA palindromes: roles, consequences, and implications of structurally ambivalent DNA,” the participants discussed and debated a range of additional structures—dubbed “Z,” “HJ,” “G4,” and “H” DNA—as well as trinucleotide repeats. These remarkable structures can have profound effects on chromosomes and organisms, ranging from mutational hotspots in bacteria to causes of intellectual disability in humans. Bringing together four dozen researchers prominent in the field focused attention on these controversial DNA structures in a way that promises to spur greater understanding of DNA elements critical to life and health.
Keywords: DNA palindromes, G4 DNA, chromosomal rearrangements, cruciforms, noncanoncal DNA structures, trinucleotide repeats
Usually thought of as a linear double helix, DNA can in reality assume many different structures, some of which have profound influences on DNA’s biological functions. For example, palindromic DNA sequences, which read the same in opposite directions (but on different strands), can extrude to form a cruciform, with both strands involved, or a hairpin, with only one strand involved (Fig. 1). These structures are acted upon by enzymes that have only slight activity on linear double-helical DNA. There is extensive evidence that these and other noncanonical DNA structures can have dire consequences such as initiating chromosomal translocations, which can result in cancer or developmental defects. Only recently was a meeting held with emphasis on noncanonical DNA structures other than trinucleotide repeats. Organized by David Leach (University of Edinburgh), Susanna Lewis (University of Toronto), and Alison Rattray (National Cancer Institute), this meeting was sponsored by FASEB and held at Saxtons River, VT, July 6–11, 2008. The structures discussed included palindromes, Holliday junctions (HJs), G4 DNA, Z DNA, and trinucleotide repeats in organisms as diverse as poxvirus, bacteria, yeasts, Drosophila, mice, and humans. One was left with the feeling that “standard” (i.e., B-form) linear DNA is inert relative to the dynamic structures discussed, and that these alternative structures deserve their own meeting, which, it was agreed, should be continued on a biennial basis.
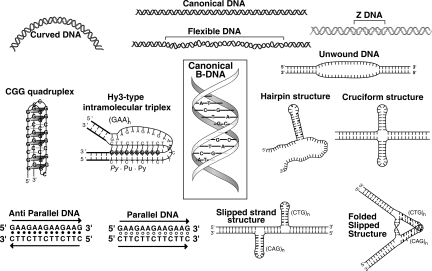
Figure 1.
Noncanonical DNA structures. Canonical B-form DNA (center, boxed) is a right-handed double helix. DNA with special nucleotide sequences can adopt other structures, many of which were discussed at the FASEB meeting reported here. Additional structures are in Figure 4. Figure courtesy of Richard Sinden.
DNA palindromes and inverted repeats—sliding into cruciforms and hairpins
A palindrome, such as the famous “A man, a plan, a canal, Panama,” reads the same in both directions. In the DNA and RNA worlds the term means that one strand reads the same in the 5′ → 3′ direction as the complementary strand reads in the 5′ → 3′ direction. An example is 5′-GTTAG|CTAAC-3′, where | indicates the center of the palindrome. If sufficiently long, these sequences can extrude to form a cruciform, with a few unpaired nucleotides at the center flanked by dsDNA (Fig. 1). When present in ssDNA, such as during replication or transcription, the palindrome can fold into a hairpin, equivalent to half of a cruciform. In both cases the capped end is a substrate for two known classes of enzymes: (1) the MRN complex of eukaryotes, and its archeal equivalent MR and bacterial equivalent SbcCD; and (2) Artemis of vertebrates, involved in cutting hairpins made by the RAG complex during V(D)J recombination, which produces active immunoglobulin genes. [MRN derives from Mre11, Rad50, and Nbs1, the polypeptides in the human complex; the Saccharomyces cerevisiae homolog of Nbs1 is Xrs2, and the Schizosaccharomyces pombe homolog of Mre11 is Rad32. MRN is used here for all species. Escherichia coli SbcC and SbcD are homologs of Rad50 and Mre11.]
Closely related to a palindrome is an inverted repeat (IR), in which there are additional, unique base pairs at the center (| in the example above). In this case, pairing between the repeats leaves extensive single-stranded loops of the unique base pairs at the tips of the cruciform (Fig. 1). The energetic cost to form this structure with single-stranded loops is large in dsDNA but not in ssDNA. Craig Benham (University of California at Davis) discussed the energetics of formation of hairpins and cruciforms, which indicate that even imperfect repeats can form these structures under the right conditions. In many cases IRs can have as profound effects as palindromes, presumably by folding during replication when the DNA is partly single-stranded.
One of the most dramatic examples of palindromes causing human disease was presented by Beverly Emanuel (University of Pennsylvania) and Hiroki Kurahashi (Fujita Health University). Palindromes of ∼500 base pairs (bp) or greater on human chromosomes 11 and 22 are the sites of the most frequent recurrent human translocation (other than Robertsonian translocations between centromeres). Carriers of the Chr. 11:22 reciprocal translocation are asymptomatic except for reduced fertility, but their chromosomally unbalanced progeny suffer from severe developmental defects, including intellectual disability and cardiac defects. The hotspots for these translocations contain AT-rich palindromes at each chromosomal break point. Designated PATRR for palindromic AT-rich regions, the repeats on Chr. 11 are 99% identical to each other but share no significant homology with the PATRR on Chr. 22. The break points of the translocations are all located within 20 bp of the centers of the PATRRs. A few nucleotides are lost during translocation, suggesting that nonhomologous end-joining is involved. Presumably, at each palindrome a cruciform is cut diagonally to make a stable hairpin, or a hairpin is formed in ssDNA (see Fig. 3, below). A cut at each hairpin tip yields uncapped dsDNA ends, which after loss of a few nucleotides are joined to form the translocation.
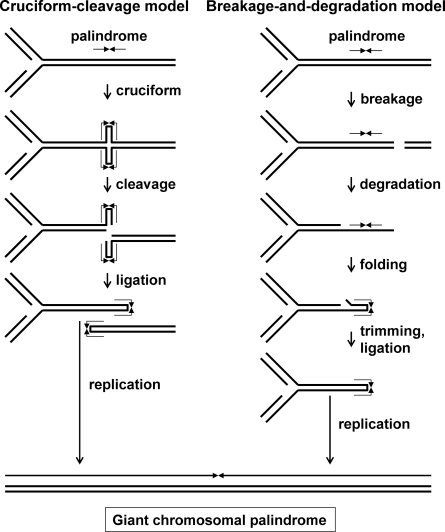
Figure 3.
Two models of palindrome-mediated gross chromosomal rearrangement. (Left) A palindrome extrudes into a cruciform, which is cleaved by an HJ resolvase. The resultant nicks are ligated to form large hairpins. Replication of a hairpin can produce a giant (chromosome-size) palindrome. (Right) A DSB near a palindrome leads, by unwinding or degradation, to ssDNA, which can fold into a large hairpin and be converted into a giant palindrome as on the left. Other rearrangements of the large hairpin can give translocations or other gross chromosomal rearrangements. Figure courtesy of David Leach.
Support for this mechanism comes from studies of plasmids with insertions of the PATRRs and variants of them. Such plasmids readily adopt the cruciform structure, and stability of the cruciform directly correlates with the rate of plasmid fusion at the PATRRs when introduced into human cells. Furthermore, the PATRR on Chr. 11 is polymorphic, and shorter PATRRs, which presumably form less stable cruciforms, translocate less frequently.
The frequency of the Chr. 11:22 translocation is so high—approximately 2 × 10−5 per DNA molecule—that it can be detected by PCR of sperm or testis biopsies. Assay of highly diluted samples shows that the two reciprocal translocation types occur in a single sperm, supporting the view that the palindromes on each chromosome are cut at about the same time, and the ends are swapped and rejoined. These translocations are not detectable by PCR in lymphoblasts or fibroblasts, and thus appear to be germline-specific, but the basis of this specificity is unclear. Emanuel noted, however, that the relevant points on Chrs. 11 and 22 are closer to each other in meiotic cells than in mitotic cells and closer than other chromosome pairs. Kurahashi suggested that the translocation occurs after meiotic replication, when the chromosomes become highly compacted for packaging into sperm heads. Consistent with this view, the three analyzed cases of the translocation arising de novo occurred in the father of the translocation carrier.
Although the molecular basis of these debilitating translocations is clearly palindromic DNA, the mechanism by which palindromes react has been most thoroughly studied in model organisms, especially bacteria and yeasts. Half of the talks at this conference dealt with microbes. Multiple strong parallels suggest that the mechanisms deduced in microbes readily pertain to multicellular species, including humans.
David Lilley (Dundee University) led off the conference with a description of the structure and branch migration of cruciforms and closely related HJs discussed below. The four arms of both junctions are arranged in a “stacked X” form, in which the arms are in two pairs of coaxially aligned, stacked helices with an angle of ∼60° between the axes (Fig. 2). The choice of partners in this coaxial alignment depends on the nucleotides at the base of the junction (and to a lesser degree the next base pairs). During branch migration, the arms must lose their coaxial stacking to adopt an open square form (Fig. 2). This open form is more stable in the absence of Mg2+ ions, which counter the repulsive negative charges of the DNA phosphate groups; these negative charges splay the arms out in the absence of Mg2+.
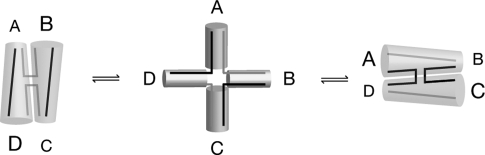
Figure 2.
Alternative conformations of DNA at the junctions of arms in cruciforms and HJs. In the center panel, the four arms are splayed apart into a square, forming a roughly planar structure. On the left, arms A and D are coaxially stacked to form a nearly continuous duplex, and arms B and C are similarly stacked. Note that the two DNA strands indicated by black lines (denoted “continuous”) are each in one stack, whereas the gray strands (denoted “discontinuous”) span different stacks. On the right, arms A and B are in one stack, and the gray strands are continuous. Figure courtesy of David Lilley.
These structures of the four-arm junction were initially inferred from the mobilities of branched DNA during electrophoresis through agarose under various ionic conditions and by fluorescence resonance energy transfer experiments. More than a decade later, the stacked X structure was directly verified by X-ray crystallography of DNA alone and bound by HJ resolvases. Now, Yuri Lyubchenko (University of Nebraska) showed dramatic pictures, captured by atomic force microscopy (AFM), of HJs undergoing branch migration. In the presence of Mg2+ ions the junction arms remain in a stable conformation, with two different, approximately equally frequent arm angles observed (Fig. 2). By adding EDTA to the buffer during imaging (to lower the free Mg2+ ion concentration), he showed that only those HJs in the open square form undergo branch migration. Thus, the textbook picture of branch migration by “rotary diffusion” of parallel or anti-parallel arms is not tenable. Lyubchenko’s AFM results confirm earlier inferences, but “seeing is believing” or, better, knowing with certainty.
Even more exciting were the single-molecule studies of HJs with two fluorescent tags that reveal by light microscopy the time that the arms spend in one or another orientation. These studies, illustrated for immobile HJs by Lilley, show that the dwell times of the alternative stacking conformers can vary considerably when the central sequence is altered. The kinetic data allow calculation of the conformational equilibrium constants, which, thankfully, agree well with previous inferences. Lyubchenko applied the same technology to follow branch migration in individual mobile HJs. His studies indicated that branch migration consists of consecutive migration and folding steps and suggested that one “hop” can be >1 bp, perhaps over the entire region of available homology (5 bp). Thus, at this stage the structure of HJs and their spontaneous branch migration are well established at the level of whole molecules. The current single-molecule experiments promise to reveal the structure of HJs in solution and their branch migration with much greater resolution—at the level of individual base pairs.
Less well established is the occurrence of cruciforms and the enzymes that cleave them and HJs in eukaryotic cells. Extensive evidence indicates that palindromes can extrude into a cruciform in isolated supercoiled circular DNA and in circular plasmids in Escherichia coli cells. More controversial has been the occurrence of cruciforms in eukaryotic cells. Atina Coté (in Lewis’s group at the University of Toronto) showed that in Saccharomyces cerevisiae circular DNA containing a palindrome gives rise to linear DNA with both ends capped with a hairpin. The simplest way to generate this structure is to extrude the palindrome into a cruciform, cut the base of the cruciform diagonally, and seal the resulting nicks with a DNA ligase (Fig. 3, left). Two observations support this view: The linear structure is detectable only if the cells contain Mus81 (a good candidate for an HJ resolvase; see below) but also lack Sae2 or the MRN complex (both of which cleave hairpins; see below). Additional evidence for cruciforms in eukaryotes is also discussed below. Although many of the observed DNA rearrangements arising at palindromes could occur during replication, via single-stranded hairpins, others are more readily accounted for by cruciforms.
Working with palindromes and inverted repeats in E. coli, Leach showed that a double-strand break (DSB) arises during or shortly after replication of the repeats. Contrary to the prevailing view, he showed that the DSB does not result from “collapse” of the replication fork, since that would produce only one double-stranded end. Rather, linear dsDNA from both sides of the repeats is detected by Southern blot hybridizations. These ends are potent substrates for RecBCD enzyme, which avidly binds and acts on dsDNA ends; coupled with RecA strand exchange protein, RecBCD enzyme promotes Chi hotspot-stimulated recombination at and near the palindrome, as expected. In the absence of RecA and SbcCD, which cuts dsDNA with an end “capped” by a hairpin or a protein, giant palindromes are observed by Southern blot analysis. These giant palindromes likely arise when the IR folds back on itself and primes self-templated DNA synthesis, giving rise to a replication fork (Fig. 3) that proceeds around the entire chromosome. This scheme of palindrome elongation was a theme of several subsequent talks on both prokaryotes and eukaryotes.
Continuing the discussion of E. coli, Sue Lovett (Brandeis University) unraveled the molecular basis of a mutational hyper-hotspot—a quasi-palindrome (one in which the repeats have extensive but incomplete complementarity). She found that spontaneous mutations in the thyA gene most frequently change the same base pair, A131 → T131, and occur at ∼200 times the genome average base-pair mutation rate. Inspection of the nucleotide sequence revealed that the A131T mutation increases the extent of complementarity within a 17-bp quasi-palindrome. Quasipalindromes had previously been observed at mutational hotspots in S. cerevisiae by Fred Sherman and in E. coli and phage T4 by Lynn Ripley and Richard Sinden, who postulated intermolecular switching of templates during DNA replication. Lovett proposed an alternative: The nascent strand folds back on itself to provide a new (intramolecular) template. She supported this model by showing that increasing the extent of complementarity in the quasi-palindrome increases the mutation rate, presumably by making the “fold-back” more likely during normal replication; conversely, decreasing complementarity decreases the mutation rate. Furthermore, elimination of three error-prone polymerases (PolII, PolIV, and PolV) increases the mutation rate, presumably by eliminating, surprisingly, a competing error-free repair mechanism. Each step of increased complementarity propels the mutation rate ever higher, and one might expect the entire genome to become a palindrome. Presumably, the countervailing instability of large palindromes prevents this fate of the genome.
Stalling of the replication fork at a palindrome or IR is likely involved in the rearrangements described here. But direct evidence for such stalling was lacking until the work of Sergei Mirkin (Tufts University), who reported that DNA replication in E. coli, S. cerevisiae, and monkey cells is stalled at IRs, as assayed by two-dimensional gel electrophoresis. Spacers of 12–86 bp have little effect on stalling, implying that hairpins, not cruciforms, cause stalling. CGG repeats, expansions of which are responsible for chromosomal fragility and hereditary neurological disorders in humans (see below), also induce stalling. In S. cerevisiae, stalling at IRs and CGG repeats is diminished by both Tof1 and Mrc1, proteins that stabilize replication forks. These proteins facilitate stalling at the rDNA pause site, which depends on the Fob1 protein. Thus, Tof1 andMrc1 have opposite roles in structure-mediated and protein-mediated stalling, indicating that the mechanisms, and perhaps the consequences, of the two types of stalling are different.
In S. pombe Antony Carr (University of Sussex) found that a palindrome or IR at a stalled replication fork gives rise to translocations and giant palindromes. Here, stalling is dependent on a protein bound to a special DNA sequence, which together make a replication fork barrier. Individually, the palindrome, IR, and barrier are innocuous. But the palindrome or IR, combined with the barrier, results in gross chromosomal rearrangements. These likely include dicentric chromosomes, since chromosomes lagging behind others at mitosis are observed in approximately one out of four of the cells. Physical analysis of DNA reveals recombinant rearranged chromosomes, but no DSBs are detectable. Carr postulated that template switching at the folded hairpin, without an intervening DSB, gives rise to a giant palindrome.
A mechanism for de novo palindrome creation involving aberrant replication was discussed by Rattray. Working with S. cerevisiae, she showed that an HO endonuclease-induced DSB that has homology on only one side of the break can give rise to a large palindrome (6–8 kb). This mechanism is similar to one of the reactions described by Leach (see above) and to the amplification of rDNA in Tetrahymena described ∼15 years ago, both of which involve a DSB near an initial small palindrome or IR (Fig. 3, right). Remarkably, in S. cerevisiae, the reaction appears to require only tiny IRs, in the range of 4–6 bp, with only a few base pairs separating them. In other words, almost any stretch of DNA is likely to contain a sequence capable of seeding the formation of much larger palindromes. These large palindromes are recovered only if the cells lack the MRN and Sae2 proteins, which can cleave hairpins, presumed intermediates in the reaction.
Rattray also noted that a scheme of this type could account for the symmetric fusion of chromosome fragments in McClintock’s breakage–fusion–bridge (BFB) cycle observed 75 years ago, if fold-back priming occurred on the centromeric side of a break. The essence of other BFB models is that replication of a broken chromosome produces two sister chromatids whose ends then fuse to produce dicentric symmetric chromosome fragments. Rattray’s data suggest an alternative mechanism whereby the symmetric, fused chromosome fragments arise through replication. A broken chromosome end provides the primer for fold-back replication, rather than serving as a substrate for fusion after replication. Additional experiments with modern genetic tools and physical assays may settle how these fused chromosomes form.
Equally stunning effects of IRs on genome rearrangements were reported by Kirill Lobachev (Georgia Institute of Technology). In S. cerevisiae, an IR of the ∼300-bp Alu sequence from human DNA increases ∼25,000-fold the rate of loss of the chromosome arm distal to the IR. Analysis of DNA by two-dimensional gel electrophoresis showed that replication pauses in a 1- to 2-kb region around the IR, showing that these elements are inherently difficult to replicate, as also reported by Mirkin. As anticipated, crippling replication, by mutation in one or another DNA polymerase gene, further augments the rate of chromosome arm loss. Comparative genome hybridization (CGH) reveals that chromosome fragmentation at the IR is often accompanied by translocations and amplifications. DSBs, observed by Southern blot analysis, arise at the IR but are not dependent on MRN, Sae2, or Mus81–Mms4, enzymes that can cut hairpins and cruciforms. The IR presumably folds into a hairpin or cruciform during replication, but the cutting enzyme in this case remains elusive.
The enzyme that cuts at a palindrome during replication is, however, known for S. pombe, as discussed by Gerry Smith (Fred Hutchinson Cancer Research Center). During meiotic replication, DSBs arise at a 160-bp palindrome and are dependent on the MRN complex, which presumably cleaves the palindromic hairpin structure formed on ssDNA of the lagging strand. The palindrome, likely via the DSB, is a recombination hotspot and can generate crossovers, which are almost entirely dependent on the Mus81 HJ resolvase, as discussed below. Note that DSB formation at the repeated sequence is MRN-dependent in S. pombe but MRNindependent in S. cerevisiae. It is not clear whether this difference reflects a fundamental difference in species, in palindrome versus IR, or in chromosome behavior with and without replication or during meiosis versus mitosis. Nevertheless, these observations show that there is more than one way to make a DSB at repeated sequences.
A remarkably high-frequency “revision” of palindromes in mice was described by Lewis. Fortuitously, a 15.6-kb perfect palindrome was obtained as a “transgene” in a mouse chromosome. This palindrome is inherited as a single Mendelian trait (i.e., in half of the progeny), indicating that it is not lethal or significantly deleterious. But ∼50% of the palindromes in the progeny have a rearrangement, often deletions ranging from ∼20 bp to several kilobases at the center (the type of rearrangement that allows a palindrome to be stably inherited in wild-type E. coli). In a transformed cell line derived from the founder mouse the palindrome is rearranged at a rate of ∼0.5% per cell division. These mitotic rearrangements can be as simple as deletion of a single GC base pair at the center of the palindrome, or deletion of 4 bp or alteration of 3 bp, for example. It is remarkable that so few base pairs out of >15,000 bp can make the difference between survival or not.
Palindromes may be in constant flux during evolution, too. Lewis compared the available genome sequences of humans, chimpanzees, and macaques, and concluded that large palindromes often arise but are quickly purged from genomes. This conclusion is well supported by experimental evidence, such as that reported here. But they are so quickly purged from E. coli that these especially intriguing sequences of the human genome and other genomes are missing from the “reference sequences” in general use, because these are almost universally based on DNA propagated in E. coli. Lewis pointed out that “personal genomes” may have limited utility until these sequences are included.
Perhaps the medically most important role of palindrome elongation was described by Hisashi Tanaka (Cleveland Clinic Foundation), who provided evidence that this process underlies gene amplification in the development of cancer. As a model for gene amplification, he inserted into Chinese hamster ovary cells a cassette containing three elements—the site for dsDNA cleavage by I-SceI endonuclease, a 497-bp IR, and the gene for dihydrofolate reductase (DHFR). Cleavage was induced, and several days later cells resistant to a low level of methotrexate were selected (i.e., for increased expression of DHFR, which commonly arises by gene amplification). As described in cases above, the small palindrome was converted into a giant palindrome, with two copies of DHFR. This process mimics the natural amplification of a single rDNA gene via conversion of a small IR into a large palindrome in macronuclear development of the ciliate Tetrahymena, as shown 15 years ago. Using this knowledge, Tanaka then developed a genome-wide assay for small (or large) palindromic DNA based on “snap-back” DNA, which rapidly renatures and is resistant to the ssDNA-specific nuclease S1. Microarray analyses with snapback DNA as probe reveals three to 20 IRs per chromosome in the reference human genome. (Here, IR is defined as >1-kb-long arms with >90% identity and separated by <10 kb of unique DNA.) Strikingly, he identified a 27-kb IR at the boundary of a 1-Mb region amplified in colon cancer cells. Based on all the evidence cited above and other work, it seems likely that the 27-kb IR was responsible for the amplification that made the cells cancerous. Even more “cancer-prone” palindromes may be lurking in our cells, because, as noted above by Lewis, the human genome sequence is particularly deficient in palindromic DNA. We all await methods to get these sequences.
HJs—slippery intermediates of recombination
Closely related to the cruciforms made by palindromes and IRs are HJs, DNA intermediates in genetic recombination. The center of a cruciform is identical to the center of an HJ (Figs. 1, 2). Both structures can migrate by unpairing the bases on one side of the center and reforming base pairs with their complements, an event made possible by the nucleotide sequence identities of the dsDNAs. Enzymes that cleave HJs also cleave cruciforms, which in fact were the substrates for first detecting HJ cleaving enzymes, called HJ resolvases, >25 years ago.
HJ resolvases are well established in bacteria and their phages; examples are E. coli RuvC and phage T7 endonuclease I. These enzymes make symmetrically placed cuts at the base of the HJ, such that the nicks in the cleavage products can be ligated without further processing to produce intact recombinant DNA. Symmetrically cleaving HJ resolvases have been detected in mitochondria of eukaryotes, but nuclear forms have been elusive, even though it is widely thought that crossovers (reciprocal recombinants) can arise only by HJ cleavage. Smith reported that during S. pombe meiosis HJs appear and accumulate in mus81 mutants, which are deficient in crossing over but not in nonreciprocal recombination (gene conversion), as expected for loss of an HJ resolvase. The S. pombe Mus81–Eme1 complex more rapidly cleaves nicked HJs than intact HJs. This feature has led some investigators to doubt that Mus81–Eme1 is an HJ resolvase, but as Lilley pointed out this property is shared by well-accepted bacterial and phage HJ resolvases. Mutant analyses have shown that Mus81 (with a partner protein) is not the only HJ resolvase in S. cerevisiae and mice, but Mus81 may well be a widespread HJ resolvase sharing the role with other proteins in some species. Remarkably, the S. pombe HJs are almost exclusively single, not the double HJs that predominate in S. cerevisiae meiosis and in most current models of recombination and DSB repair. These observations invite re-examination of the enzymes and mechanisms of meiotic recombination in other species.
An identified HJ resolvase plays an essential role for pox virus multiplication in animals, as Frederic Bushman (University of Pennsylvania) discussed. The virions of pox viruses contain 130- to 300-kb linear dsDNA whose ends are capped by hairpins formed at terminal IRs of ∼60 bp. Replication generates long DNA concatemers, which must be cut into unit-size pieces for packaging. This apparently occurs by the extrusion of a cruciform at the IRs and cleavage by the viral A22 HJ resolvase. Deletion mutants lacking A22 accumulate concatemers, and the purified A22 protein cleaves HJs. A22 also cleaves other branched structures, much as does the phage T7 endonuclease I HJ resolvase, suggesting that the two proteins play similar roles in removing branched structures from DNA during viral packaging. Nevertheless, the requirement for A22 in converting IRs into linear DNA, coupled with the enzymatic data, strongly suggests that A22 is a bona fide eukaryotic virus HJ resolvase. In this case a palindrome is beneficial, perhaps even essential, to life, at least for this virus. There may be other benefits of palindromes that have escaped notice, due to the emphasis on their deleterious effects.
Z DNA—a zigzag from left-handed DNA to a role in innate immunity
The first crystal structure of DNA (dCGCGCG), obtained in 1979, showed that this DNA was in a left-handed form, dramatically different from the right-handed DNA inferred from X-ray diffraction of DNA fibers in 1953. The phosphate-sugar backbone of the crystallized DNA had a zig-zag contour, so the structure was designated Z DNA (Fig. 1). Alex Rich (Massachusetts Institute of Technology), in whose laboratory Z DNA was discovered, reviewed its history and recent evidence that Z DNA plays an important role in cells. For years, the presence of Z DNA in cells has been elusive and highly controversial, partly because it is unstable and persists only under seemingly nonphysiological conditions, such as 4N NaCl.
To support the existence and importance of Z DNA in cells, Rich discussed proteins that bind Z DNA and their roles in viral infections. For example, the E3L protein of vaccinia virus has a Z DNA-binding domain, as does the editing enzyme dsRNA adenosine deaminase (ADAR-1). Deletion of the E3L Z DNA-binding domain renders the virus nonpathogenic, but swapping in part of the ADAR-1 Z DNA-binding domain restores pathogenicity. This swap of ∼60 amino acids leaves unchanged only ∼12 amino acids, most of which make direct contact with the DNA in cocrystals with the Z DNA-binding part of human ADAR-1. Additional single amino acid changes in the Z DNA-binding domain can abolish or restore Z DNA binding and pathogenicity in parallel. Most recent is the finding that a human protein DLM, with two Z DNA-binding domains, is the DNA-dependent activator of interferon regulatory factors. This cytosolic factor is an important part of the innate immune response, which is immediately available upon first infection (as opposed to the delay in acquired immunity). Because these proteins can bind both right-handed (B-form) and left-handed (Z-form) dsDNA, albeit with different domains, some uncertainty about their physiological ligands persists. But the abundance of potential Z DNA-forming sequences in more than three out of four of human gene promoters strengthens the argument that Z DNA is an important regulatory factor. Rich also pointed out that the density of potential Z DNA-forming sequences is markedly higher in sequences of viruses that provoke the interferon response than in those that do not. He proposed that these sequences comprise a “pathogen associated molecular pattern,” which may extend to some bacteria as well as to viruses.
Additional strong evidence for intracellular Z DNA was described by Karen Vasquez (M.D. Anderson Cancer Center), who pointed out that mammalian chromosomal deletions and translocations often have potential Z DNA-forming sequences at the novel joints. Potential Z DNA-forming sequences inserted into plasmid DNA increases mutation rates as much as 20-fold when the plasmids are incubated in mammalian cell-free extracts. High mutation rates are observed with or without overt replication, suggesting that DNA repair activity, rather than replication, is impeded by Z DNA and induces mutation. The deletions often arise between short identical sequences, suggesting that nonhomologous end-joining is involved. Most telling is the behavior of Z DNA with an adjacent lacZ reporter gene inserted into a mouse chromosome. This DNA is stable in the initial “founder” mouse, but in as many as 20% of its F1 progeny lacZ is lost or altered, as assayed by PCR. As in the examples of palindrome-associated human translocations discussed by Emanuel and Kurahashi, instability may be highest in the germline, perhaps during meiosis itself. As the evidence mounts, it is hard to escape the conclusion that DNA with potential Z structure behaves dramatically differently than “ordinary” DNA.
G4 DNA—a quartet of guanines prominent in acquired immunity
A century ago, solutions of GMP, unlike those of other nucleotides, were noted to form gels, and from physical studies a half century ago it was proposed that four guanine bases at the corners of a planar square can basepair to form what is now called G4 DNA (Fig. 4A). DNA with three or more Gs in a row, at four or more close positions, can form multiple highly stable structures. As with Z DNA, the existence of G4 DNA in cells has been controversial, for much the same reasons. Timothy London (in Kevin Hiom’s group at the MRC Laboratory of Molecular Biology) reported work from his laboratory and others showing that G4 DNA can be unwound by a limited set of helicases. These include the human BLM and FANC-J helicases, which unwind DNA in opposite directions and are altered in patients with cancer-prone Bloom’s syndrome and Fanconi’s anemia, respectively. Caenorhabditis elegans mutants lacking Dog-1, the homolog of FANC-J, accumulate deletions with end points at runs of G; dog-1 him-6 mutants, lacking the BLM homolog in addition, have an enhanced phenotype. Human FANC-J-deficient cells, analyzed by CGH, also have deletions ending at potential G4-forming sequences. Additional evidence for intracellular G4 DNA is that chicken DT-40 cells lacking a FANC-J homolog are hypersensitive to small molecules that stabilize G4 DNA. These compounds were identified in screens for chemicals that would stabilize telomeres, which have G4-forming potential, but they may prove useful for studying G4 DNA in other contexts as well.
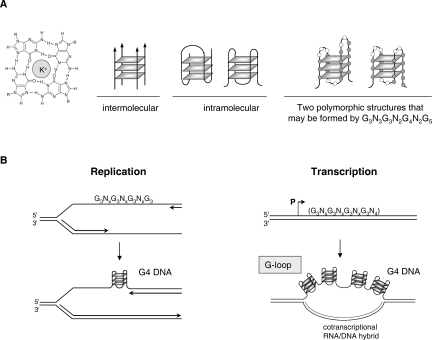
Figure 4.
G4 DNA structures. (A, left) Hydrogen bonding of four guanine bases (guanine quartet or G4). (Center) Three possible inter- and intrastrand pairings of G4. (Right) Two polymorphic structures that may be formed by G3N3G3N2G4 N2G5. Each plane segment is a G4. (B) Formation of intrastrand G4 structures during replication (left) or transcription (right). Figure courtesy of Nancy Maizels.
Runs of G with the potential to form G4 DNA are greatly enriched in the region of the human immunoglobulin (Ig) genes involved in switching from one class of Ig to another. Nancy Maizels (University of Washington) showed that G4 DNA arises at high frequency when the G-rich strand of the Ig switch region DNA is specifically on the nontemplate strand during transcription (Fig. 4B). Such G4 structures can be observed by electron microscopy of transcriptionally active plasmids extracted from E. coli cells, adding to the evidence that G4 DNA is physiologically important. She also showed that human exonuclease I (ExoI) can digest the G4 DNA strand of such transcription intermediates if there is an ExoI entry site, such as a nick provided by activation-induced deaminase (AID). These studies reveal another tool that cells have to deal with impediments to transcription and replication of DNA with noncanonical structures.
H DNA and slipped DNA—tying DNA into pseudoknots
DNA with mirror symmetry in its DNA sequence—that in which one strand reads the same in both directions, such as 5′-AATGGTAA-3′—can form a three-stranded structure called H DNA (Fig. 1). Here, one strand in a repeat loops back to form a triplex with the other repeat; its complement remains unpaired. Mirkin first described this structure in 1987. Richard Sinden (Florida Institute of Technology) described a related structure formed by DNA with the sequence (CCTG)n found at the myotonic dystrophy type 2 (DM2) locus. As an example of genetic “anticipation,” this DNA reveals an increasingly noticeable phenotype as n increases with each generation. Only when n > 104 is the disease truly debilitating. Sinden showed that this DNA can form a slipped structure when it is heated and cooled to form dsDNA with two single-stranded loops (Fig. 1), which apparently can interact with each other to form “donuts” visible by AFM. These structures are stable up to 55°C, unlike the much less stable cruciforms, Z DNA, and H DNA. The sequence (CCTG)n is the first example of slipped DNA that can arise simply from DNA supercoiling. Notably, Z DNA adjacent to this sequence diminishes its propensity to form the slipped structure. Thus, Z DNA, with its left-handed twisting, reduces the supercoiling density and can be protective in some contexts. In E. coli plasmids the slipped structure arises when n is more than ∼40; when n is ∼170, slipped DNA forms without heating and cooling but is dependent on supercoiling. These thresholds are vastly different from the threshold for human disease. Nevertheless, repeat length determines the disease phenotype, just as in the trinucleotide repeat diseases so thoroughly studied, and all of these diseases manifest anticipation.
Trinucleotide repeats—expansions that debilitate
Nearly two dozen human neurodegenerative diseases are caused by expansion of trinucleotide repeats, such as (CTG)n. Depending on the disease, the repeats can occur in the coding sequence of a gene, 5′ or 3′ of it, or in an intron; thus, either the level of expression of the gene or the properties of the encoded protein can be altered and effect the disease. In virtually all cases examined there is a threshold value for n: When n is less than the threshold, the repeat number is relatively stable from generation to generation and there is no overt phenotype, but when n reaches the threshold, n can suddenly expand and the disease is manifest. For example, Huntington’s disease is caused when the number of CAG repeats in the coding sequence of the IT15 gene exceeds ∼36. Remarkably, if n = 32 or less, the person is unaffected. The repeated sequence can form a hairpin (Fig. 1), and the stability of the hairpin in naked DNA correlates with the risk of expansion in humans. Much attention has thus been focused on conditions and proteins that affect the stability of the hairpin form. Instability is often greater in the germline than in somatic tissues, but some instances of somatic mosaicism are known.
Bob Lahue (National University of Ireland) designed a clever way to study trinucleotide repeat stability in S. cerevisiae. He inserted (CAG)n between two elements of a promoter controlling the URA3 gene. When n = 25, the cells are Ura+, but with lesser or greater values of n the cells are Ura−; both Ura+ and Ura− cells can be readily selected. This scheme allowed him to show that there is a sharp threshold at n ≈ 15 for stability of this repeat: The rate of expansion is 10 times greater when n = 25, and 10 times less when n = 10. Thus, the rate of expansion varies >100-fold when n varies more than threefold, the hallmark of a threshold effect. Why the threshold value for this repeat is ∼15 in S. cerevisiae but ∼35 in humans is not clear. What is clear from Lahue’s study is that DNA repair functions greatly affect the rate of expansion. Mutants lacking Srs2 DNA helicase activity or Rad27 DNA flap endonuclease or altered in the Pol30 (PCNA) “sliding clamp” have 40- to 100-fold increased rates of expansion; in the rad27 pol30 double mutant the rate is ∼1000-fold increased, indicating at least two modes of maintenance of stability. Expansion, but not contraction, rates are affected by these mutations. Curiously, the Sgs1 DNA helicase has no detectable role in instability of this repeat, and only expansion, not contraction, of the repeat is affected in srs2 mutants. This specificity of DNA helicase activity or biological role or both is frequently observed and helps to explain why cells have so many DNA helicases: Human cells have dozens of putative DNA helicases. The basis of their specificity remains to be worked out in most cases, but certain helicases appear to be specialized for handling noncanonical DNA structures.
Expansion of the (CGG)n repeat in the 5′ untranslated region of the human FMR1 gene is the basis for three distinct diseases and chromosome fragility, as reported by Karen Usdin (National Institutes of Health). These repeats can form various structures, such as hairpins and tetraplexes. In unaffected people, n is in the range of ∼5–40, but if n is in the range of 50–200, Fragile X-associated tremor and an ataxia syndrome can result; female carriers are also at risk of reduced fertility. In maternally transmitted alleles, n can greatly expand to 1000 or more, and give rise to children with Fragile X syndrome, the most common cause of inherited intellectual disability.
FMR1 is one of several sites at which chromosomes break when replication is restricted by various means, such as folate deficiency or exposure to aphidicolin, a DNA polymerase inhibitor. Apparently, the repeats at FMR1 are particularly hard to replicate and the chromosomes break during segregation. As expected, checkpoint functions are activated under these restricted conditions, and checkpoint deficiency exacerbates the problem.
The mouse FMR1 homolog has only eight copies of (CGG), but when the region is altered to contain ∼120 copies the mice develop cerebellar pathology, mimicking the situation in humans. Although expansion typically occurs with the addition of a small number of repeats, rare large jumps do occur. Usdin found that the repeat number was more stable in mice with 120 copies than in those with 190. These data suggest a threshold for stability, just as with other repeats such as the (CAG) repeats discussed by Lahue. The cause of the threshold effect may well be a key to understanding how to prevent the shift from unaffected to debilitated, the “anticipation” effect.
Conclusion
This meeting highlighted features of DNA likely unanticipated at the discovery of its canonical structure. We now see that DNA can assume multiple structures, as well as multiple functions. Although some investigators may assume that the structures discussed at this meeting are rare and rightly out of mainstream consciousness, they clearly have profound effects, especially in certain human diseases. Further studies may well reveal additional structures with subtle or profound effects, likely to be reported at future, eagerly anticipated meetings.
Acknowledgments
I am deeply grateful to many speakers and, especially, Susanna Lewis, Alison Rattray, and David Lilley for many helpful suggestions and corrections and to Richard Sinden, David Lilley, David Leach, and Nancy Maizels for figures. Research in my laboratory is supported by grants GM031693 and GM032194 from the National Institutes of Health.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1724708.