Abstract
Bloom's syndrome is caused by mutations in the BLM gene. The BLM gene product, BLM helicase, forms a complex with two other proteins, DNA topoisomerase IIIα and RMI1. In this issue of Genes & Development, Wang and colleagues (2843–2855) and Meetei and colleagues (2856–2868) report the discovery of a fourth component of this complex called RMI2. RMI2 may be a representative of a new family of OB-fold-containing proteins that are important for complex stabilization and checkpoint response.
Keywords: Bloom's syndrome, BLM, RMI1/2, RECQ, double Holliday junction, topoisomerase IIIα
In humans, mutations in the BLM gene give rise to Bloom's syndrome (BS) (Ellis et al. 1995), a rare genetic disorder that is most prevalent in the eastern European Ashkenaki Jewish population (incidence 1/50,000). The prominent physical features associated with BS include sun-sensitive skin lesions, small stature, male infertility, and a susceptibility to infections and diabetes (German 1993). Most importantly, BS patients develop various types of cancers often at a young age (for more information about the disease, visit the Bloom's Syndrome Foundation Web site at http://www.bloomssyndrome.org). Cells derived from individuals with BS exhibit genome instability, the hallmark feature of which is an elevated frequency of sister chromatid exchange (SCE). Since this debilitating genetic disorder was first reported more than five decades ago (Bloom 1954), significant effort has been exerted to try to understand the precise molecular defects associated with BS. As a consequence, we now have a good understanding of the cellular actions and activities of the BLM protein (Wu and Hickson 2006).
The human RECQ family helicases
BLM is a member of the RecQ family of DNA helicases (Ellis et al. 1995). The family is named after the Escherichia coli ortholog RecQ, which is required for the efficient induction of the DNA damage (SOS) response and suppression of illegitimate recombination leading to DNA rearrangements (Hanada et al. 1997). RecQ homologs are also present in Saccharomyces cerevisiae and Schizosaccharomyces pombe, where they are known as Sgs1 and Rqh1, respectively. Like RecQ, Sgs1 is required for the suppression of illegitimate recombination and SCEs, or crossovers, that arise by homologous recombination (Watt et al. 1996). Mutants of sgs1 show an increased sensitivity to agents that cause DNA damage or promote S-phase arrest, such as UV, hydroxyurea, and methylmethanesulfonate, indicating a role in DNA replication, recombination, and repair.
Perhaps surprisingly, although E. coli and yeast contain only one RecQ protein, there are no less than five members of this protein family in humans. They are known as RECQ1, BLM, WRN, RECQ4, and RECQ5, and all possess a domain structure to suggest that they are likely to function as DNA helicases. Indeed, RECQ1, BLM, WRN, and RECQ5 have been shown to unwind a variety of DNA substrates including replication forks, 3′ or 5′ flaps, and Holliday junctions (HJs), consistent with a role in DNA metabolism (Bachrati and Hickson 2003; Opresko et al. 2004). The one exception to this rule is RECQ4, for which it has not been possible to demonstrate helicase activity in vitro (Macris et al. 2006). In addition to the helicase activities, all five human RECQ proteins promote the reannealing of complementary ssDNAs (Bachrati and Hickson 2008).
Because the human RECQ proteins exhibit similar biochemical properties and are implicated in common DNA metabolic processes, it might be reasonable to think that these proteins have overlapping or redundant cellular functions. However, this is clearly not the case, as mutations in different RECQ proteins lead to distinct clinical syndromes (Hickson 2003). While mutations in BLM cause BS, mutations in WRN are linked to Werner Syndrome (WS), and RECQ4 is defective in Rothmund-Thomson Syndrome (RTS). Moreover, while individuals with BS or RTS exhibit various physical and mental developmental abnormalities, the most striking phenotype associated with WS is premature aging. Most importantly, all these patients, in particular those with BS, have a high risk of cancer predisposition. As a consequence, cancer is the primary cause of death for BS patients before the age of 30.
The most striking feature in cell lines derived from BS patients is the high levels of SCE accompanied with the formation of multi-radial chromosomes, which is a potential key factor leading to cell transformation and tumorigenesis. However, recent studies with recq5−/− embryonic stem cells and primary embryonic fibroblasts derived from recq5−/− knockout mice demonstrated similar phenotypes to those found associated with BS. The recq5−/− blm−/− double mutants exhibited an even greater frequency of SCEs than either the recq5−/− or the blm−/− single mutants, suggesting that these two proteins suppress crossover formation using different mechanisms. Thus, BLM and RECQ5 play nonredundant roles in cancer prevention by regulating SCE formation and preventing chromosome aberration (Hu et al. 2005, 2007).
Given that BLM and RECQ5 exhibit very similar biochemical properties, why do mutations in both of these proteins contribute to an elevated SCE frequency? To gain insight into the nonredundant roles of the RECQ family proteins, recent efforts have focused on identifying the specific protein–protein interactions of each of the RECQ helicases. Importantly, it was shown that RECQ5, but not the other RECQs, forms a stable complex with chromatin-associated RNA polymerase II (RNAPII) (Aygun et al. 2008; Izumikawa et al. 2008). The interaction of RECQ5 with RNAPII might indicate that RECQ5 suppresses SCE formation by specifically preventing recombination events associated with transcription. Similarly, the architecture of the BLM protein complex has been gradually established during the past 5 years. In this regard, the discovery of an additional component of the BLM complex, as described in this issue of Genes & Development (Singh et al. 2008; Xu et al. 2008), may turn out to be significant. It is likely that further analysis of such specific protein–protein interactions will reveal the complex functions and regulatory aspects of BLM in human cells.
The BLM core complex
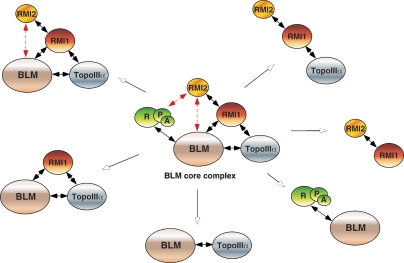
Over the past decade, many proteins have been described as potential BLM-interacting proteins, but it is now thought likely that several of them interact either transiently or indirectly with BLM. Only a few polypeptides are now considered as part of the true BLM core complex (Fig. 1), and these include topoisomerase IIIα (TopoIIIα), Replication Protein A (RPA), and RMI1. Functional interactions take place between BLM and TopoIIIα (Wu et al. 2000; Wu and Hickson 2003) and are mimicked by similar associations that take place between their yeast homologs, Sgs1 and TopoIII (Gangloff et al. 1994; Bennett et al. 2000). The combined actions of BLM with TopoIIIα leads to a remarkable biochemical reaction that results in the dissociation of DNA structures containing double HJs (dHJs), structures that represent intermediates in the process of homologous recombination (Wu and Hickson 2003). The product of the dissolution reaction is genetically silent in that the HJs are removed without any exchange of DNA between the interacting molecules (they are noncrossover products). Given that cells contain alternative pathways to resolve these intermediates, into both crossover and noncrossover products, it is now clear why the absence of BLM leads to an elevated frequency of SCEs. In essence, BLM–TopoIIIα complex acts as a suppressor of crossing over and SCE formation.
Figure 1.
Schematic diagram indicating the BLM core complex and a variety of possible subcomplexes that are thought to exist in human cells. Direct interactions between two proteins are indicated by solid black double arrows, whereas potential interactions are shown with dashed red double arrows.
In contrast to TopoIIIα, which interacts specifically with BLM, RPA interacts with several RECQ family members (Bachrati and Hickson 2008). RPA is an oligonucleotide-binding, or OB-fold, containing protein and is functionally similar to the E. coli single-stranded binding protein SSB. However, whereas SSB consists of a single polypeptide, RPA is comprised of three polypeptides associated together via their OB-fold domains. The functional association of RPA with all RECQ helicases results in a stimulation of DNA unwinding, which occurs by two distinct mechanisms: (1) stabilization of the ssDNA that results from DNA unwinding, and (2) direct protein–protein interactions. Since RECQ helicases exhibit a strand-annealing activity in addition to their ATP-dependent DNA helicase activities, RPA appears capable of counteracting DNA annealing by binding and stabilizing the ssDNA products. These reactions, however, are thought to require specific protein–protein interactions since E. coli SSB cannot substitute for RPA to efficiently stimulate human RECQ helicases (Bachrati and Hickson 2008).
In 2005, another member of the BLM core complex, RMI1 (originally called BLAP75), was identified as a component of the BLM complex purified from human cells (Yin et al. 2005) and also as part of the yeast Sgs1–TopoIII complex (Chang et al. 2005; Mullen et al. 2005). Interestingly, RMI1's association with BLM appears to be accomplished by independent direct physical interactions with both BLM and TopoIIIα (Raynard et al. 2006; Chen and Brill 2007), although it is not clear whether the interactions are mutually exclusive as the BLM and TopoIIIα interaction domains on RMI1 map to the same N-terminal region (Raynard et al. 2008). Nevertheless, it has been shown that RMI1 plays an important role in stimulating dHJ dissolution catalyzed by BLM–TopoIIIα (Wu et al. 2006; Raynard et al. 2008). Recent results show that TopoIIIα–RMI1 complex also stimulates the movement or branch migration activity of BLM on single HJs, and it is thought likely that RMI1 interacts primarily with TopoIIIα within this complex (Raynard et al. 2008).
RMI1 contains two OB-fold domains and is capable of binding to ssDNA, dsDNA, or more complex DNA structures such as dHJs (Wu et al. 2006; Raynard et al. 2008). Intriguingly, unlike RPA, the DNA-binding activity of RMI1 is dispensable for the stimulation of BLM-dependent helicase activity. Thus, in contrast to the RPA-dependent stimulation that occurs through both DNA–protein and protein–protein interactions, RMI1 affects BLM function only through specific protein–protein contacts. This is an interesting difference as it suggests that the OB-fold domain proteins RPA and RMI1 exhibit functionally distinct mechanisms of stimulation.
RMI2, a new OB domain protein
The association of two OB-fold proteins, RPA and RMI1, with BLM suggests that other OB-fold proteins might interact with BLM using this common domain. This possibility leads us to the two papers in this issue (Singh et al. 2008; Xu et al. 2008) that report the identification of a novel BLM complex-associated protein that has been called RMI2. The protein was found to be present in BLM or RMI1 immunoprecipitates and was subsequently identified as a 15.8-kDa polypeptide by mass spectrometry (Singh et al. 2008; Xu et al. 2008). RMI1 was found to interact with RMI2 via the C-terminal OB-fold domain of RMI1 and the OB-fold domain of RMI2, reminiscent of the interactions that occur between the largest subunits of RPA, RPA1, and RPA2. Purified recombinant RMI1 and RMI2 stably associate to form a heterodimeric complex that has greatly improved solubility compared with either of the two individual subunits. Indeed, it is possible that the RMI1/RMI2 heterodimer is a functional protein, reminiscent of the RPA heterotrimer (Fig. 1). This observation raises the question of whether RMI might actually be a paralog of RPA. However, this is clearly not the case because—in contrast to RPA, which exhibits a high affinity for DNA—the RMI complex actually fails to display DNA-binding activity. Since RMI1 alone has been shown to bind to DNA (Wu et al. 2006; Raynard et al. 2008), it seems likely that the interaction of RMI1 with RMI2 inhibits its ability to bind DNA. Interestingly, the DNA-binding domain of RMI1 has been mapped to the same region as the RMI2-interaction domain, suggesting that inhibition occurs by competition between DNA binding versus protein–protein interaction (Raynard et al. 2008; Xu et al. 2008).
The stable association of RMI1 with RMI2 prompted the Wang group (Xu et al. 2008) and the Meetei group (Singh et al. 2008) to analyze the effects of the heterodimer on the ability of BLM–TopoIIIα to promote dHJ dissolution. If RMI2 was expected to greatly stimulate dHJ dissolution, then the authors would have been very disappointed, as little significant effect was observed. Indeed, the small amount of stimulation seen by the Meetei group (Singh et al. 2008) may well be due simply to the enhanced stability of the RMI complex compared with RMI1 alone.
RMI2 may be required for a subset of BLM reactions
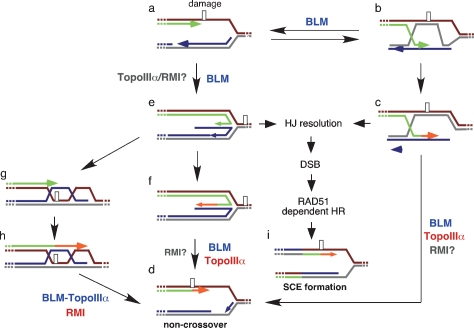
Homologous recombination provides an important cellular mechanism to repair double-strand breaks caused by DNA-damaging agents. Normally SCE formation by homologous recombination is low, most probably due to the dHJ dissolution activity of the BLM complex (Wu and Hickson 2003). Similarly, spontaneous SCE formation, which could occur during the repair of stalled replication forks, is also suppressed (Kuzminov 2001; Cox 2002). In a normal cell, there are several alternative mechanisms to resume replication at the stalled replication fork, and it is thought that BLM complex may act in multiple pathways that ultimately lead to fork recovery without crossover formation (Fig. 2). In some pathways of fork repair, the DNA helicase activity of BLM alone might be sufficient for recovery, possibly by the promotion of fork regression, while other pathways such as those involving dHJs require the presence of a more complete BLM complex containing TopoIIIα and possibly the RMI proteins.
Figure 2.
Potential roles of BLM and its interaction partners in human cells Schematic diagram indicating alternative pathways for the repair of stalled replication forks that arise as a result of DNA damage. Leading and lagging strand synthesis is shown in green and blue, respectively. (a–c) Following fork blockage, RAD51 recombinase can promote strand exchange between paired sisters, such that the DNA synthesis can bypass the damage in the leading strand by use of the sister chromatid. (c,d) The resulting recombination intermediate may then be dissociated by BLM–TopoIIIα to give rise to a noncrossover product. (e) Alternatively, BLM could promote fork regression to produce a chicken foot structure, or HJ, with the newly synthesized strands annealed with each other. (f) The newly synthesized strand from the undamaged parental strand can then serve as a template for lesion bypass. (d) The HJ may then be dissociated back to a replication fork allowing replication to continue. (g) A further possibility is that the 5′-end of the newly synthesized single-strand of DNA could fold back and reanneal to its original parental strand. DNA synthesis and branch migration could also give rise to dHJs (h), which in turn can be dissociated by BLM complex back to a replication fork structure without crossover formation (d). On the other hand, in the absence of a functional BLM complex, HJs formed by either fork regression (e) or by RAD51-dependent strand exchange reactions (c) could be resolved by HJ resolvases leading to DSB formation, which in turn can be repaired by homologous recombination to restore the replication fork with crossover (i). The involvement of BLM in each pathway is indicated, as are the potential roles of BLM interacting proteins (red). In some cases, the roles of BLM-interacting proteins are unknown (gray).
RMI2, the stabilizer of the BLM core complex
While the full spectrum of the potential biochemical functions of RMI2 requires further investigation, it seems likely that RMI2 may not serve primarily as a catalytic component of the BLM core complex. Indeed, current evidence suggests that the function of RMI2 is to act as a stabilization factor for the BLM core complex in vivo. Using RNAi oligos to independently knock down each of the BLM core complex components, it was shown that the cellular stabilities of RMI1, RMI2, and TopoIIIα proteins were dependent on each other (Singh et al. 2008; Xu et al. 2008). Indeed, the loss of any one protein greatly reduced the protein levels of the other two factors. In contrast, however, the protein levels of BLM remained unchanged; nor was BLM required for the stability of the RMI1, RMI2, or TopoIIIα proteins. One possibility is that RMI1–RMI2–TopoIIIα forms a stable heterotrimer, which in turn interacts with BLM to form the BLM core complex (Fig. 1). Consistent with this proposal, Xu et al. (2008) observed the RMI1–RMI2– TopoIIIα complex in cell-free extracts. Since RMI1 interacts with both RMI2 and TopoIIIα, it is to be expected that RMI1 knockdown would affect the stability of RMI2 and TopoIIIα. At the present time, however, it is not clear whether RMI2 interacts directly with TopoIIIα, and it is perhaps more likely that TopoIIIα and RMI2 affect each other indirectly through RMI1. The presence of a RMI1–RMI2–TopoIIIα subcomplex raises the question of whether this complex might have a cellular role that is independent of BLM. It also opens the possibility that BLM might be stabilized through interactions with alternative protein factors in the absence of the RMI1–RMI2–TopoIIIα subcomplex.
In addition to its stabilization role, studies with a mutant form of RMI2K121A indicate a regulatory function for RMI2 in terms of defining the interaction of BLM with the RMI1–RMI2–TopoIIIα subcomplex (Xu et al. 2008). Indeed, the K121A mutation resulted in a complete failure to interact with BLM, and this occurred independently of either RMI1 or TopoIIIα (while both of these proteins retained the ability to interact with the mutant). However, it remains puzzling why the interaction between BLM and the RMI1–RMI2K121A–TopoIIIα subcomplex was completely abolished, since both RMI1 and TopoIIIα can interact independently with BLM. Precisely how the K121A mutation in RMI2 affects the interaction of BLM with RMI1–RMI2–TopoIIIα subcomplex clearly needs further investigation.
Beyond the BLM core complex
While the biochemical contributions of RMI2 to biochemical reactions mediated by the BLM complex remain questionable, it is clear that the presence of RMI2 is critical for the stability of the BLM core complex. It is also likely that RMI2 serves as a mediator that links BLM with other factors important for DNA damage and cell cycle checkpoint response. Firstly, RMI2 was found to be important for the recruitment or stabilization of BLM complexes to sites of DNA damage, as measured by the formation of BLM and RMI1 foci after DNA-damaging treatment. Of course, it is possible that the decrease in RMI1 focus formation in the RMI2-depleted cells is a direct result of the reduced concentration of RMI1 protein. However, given that the protein levels of BLM do not decrease after RMI2 depletion, and yet the number of cells with BLM foci were reduced significantly, this is thought to be unlikely. Interestingly, RMI2-depleted cells show an elevated frequency of SCEs (Xu et al. 2008) and an increased number of chromosome breaks (Singh et al. 2008). One possible scenario is that the RMI1–RMI2–TopoIIIα complex recognizes sites of DNA damage and relocalizes BLM to these sites through protein–protein interactions. This possibility is supported by observations indicating that RMI2 helps to target BLM to chromatin (Singh et al. 2008). However, since the RMI1–RMI2 subcomplex shows little affinity with DNA, the interactions with DNA must be achieved through TopoIIIα. Alternatively, RMI1–RMI2–TopoIIIα might act as a bridge that links BLM to other DNA damage response factors and subsequently facilitates the recruitment of BLM to the sites of repair.
In addition to DNA damage-induced focus formation, RMI2 also influences the mitotic phosphorylation of BLM (Singh et al. 2008). One possibility is that RMI2, in collaboration with RMI1 and TopoIIIα, mediates interactions between the mitotic checkpoint kinases and BLM. It has been shown that RMI2 itself is phosphorylated by the mitotic checkpoint kinases, but the physiological relevance of this phosphorylation event remains to be elucidated. With these issues in mind and the fact that BLM is a highly interactive protein that is capable of transient association with a number of proteins involving in different aspects of DNA metabolism, it will be interesting to determine whether RMI2 plays a specific role in either connecting or excluding BLM from other DNA repair or replication factors beyond those present within the BLM core complex.
Acknowledgments
Y.L. is supported by grants from the NIH, American Cancer Society, American Federation for Aging Research, and Breast Cancer Alliance. S.C.W. is supported by grants from Cancer Research UK, the Louis-Jeantet Foundation, and the EU DNA Repair Consortium.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1732808.
References
- Aygun O., Svejstrup J.Q., Liu Y. A RECQ5–RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C.Z., Hickson I.D. Helicases, suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C.Z., Hickson I.D. RECQ helicases: Guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Bennett R.J., Noirot-Gros M.F., Wang J.C. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs: Probably a syndrome entity. AMA Am. J. Dis. Child. 1954;88:754–758. [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Zhang C.Y., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G.A., Boone C., Brown G.W. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RECQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.F., Brill S.J. Binding and activation of DNA topoisomerase III by the RMI1 subunit. J. Biol. Chem. 2007;282:28971–28979. doi: 10.1074/jbc.M705427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.M. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Blooms Syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type-I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog—A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom syndrome: A Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- Hanada K., Ukita T., Kohno Y., Saito K., Kato J., Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I.D. RECQ helicase: Caretakers of the genome. Nat. Rev. Mol. Cell Biol. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hu Y.D., Lu X.C., Barnes E., Yan M., Lou H., Luo G.B. RECQL5 and BLM RECQ DNA helicases have non-redundant roles in suppressing crossovers. Mol. Cell. Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., et al. RECQL5 helicase regulates homologous recombination and suppresses tumor formation via disruption of RAD51 presynaptic filaments. Genes & Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa K., Yanagida M., Hayano T., Tachikawa H., Komatsu W., Shimamoto A., Futami K., Furiuichi Y., Shinkawa T., Yamauchi Y., et al. Association of human DNA helicase RECQ5β with RNA polymerase II and its possible role in transcription. Biochem. J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. DNA replication meets genetic exchange: Chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macris M.A., Krejci L., Bussen W., Shimamoto A., Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst.) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Mullen J.R., Nallaseth F.S., Lan Y.Q., Slagle C.E., Brill S.J. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1–Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko P.L., Cheng W.H., Bohr V.A. Junction of RECQ helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- Raynard S., Bussen W., Sung P. A double Holliday junction dissolvasome comprising BLM, Topoisomerase IIIα, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Raynard S., Zhao W.M., Bussen W., Lu L., Ding Y.Y., Meetei A.R., Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIα-dependent Holliday junction processing. J. Biol. Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T.R., Ali A.M., Busygina V., Raynard S., Fan Q., Du C.-H., Andreassen P.R., Sung P., Meetei A.R. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase–double Holliday junction dissolvasome. Genes & Dev. 2008 doi: 10.1101/gad.1725108. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Hickson I.D., Borts R.H., Louis E.J. Sgs1, a homolog of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. DNA Helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies S.L., North P.S., Goulaouic H., Riou J.F., Turley H., Gatter K.C., Hickson I.D. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C.Z., Ou J., Xu C., Yin J.H., Chang M., Wang W., Li L., Brown G.W., Hickson I.D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Guo R., Sobeck A., Bachrati C.Z., Yang J., Enomoto T., Brown G.W., Hoatlin M.E., Hickson I.D., Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes & Dev. 2008 doi: 10.1101/gad.1708608. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J.H., Sobeck A., Xu C., Meetei A.R., Hoatlin M., Li L., Wang W. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]