Abstract
The role of the myc gene family in the biology of normal and cancer cells has been intensively studied since the early 1980s. myc genes, responding to diverse external and internal signals, express transcription factors (c-, N-, and L-Myc) that heterodimerize with Max, bind DNA, and modulate expression of a specific set of target genes. Over the last few years, expression profiling, genomic binding studies, and genetic analyses in mammals and Drosophila have led to an expanded view of Myc function. This review is focused on two major aspects of Myc: the nature of the genes and pathways that are targeted by Myc, and the role of Myc in stem cell and cancer biology.
Keywords: Myc, cancer, growth, stem cells, target genes, transcription
“Nature was not designed to make life easy for biologists” (Tudge 2007).
The myc proto-oncogene family (comprising c-myc, N-myc, and L-myc) ranks among the most exhaustively studied group of genes in biology. The profound involvement of deregulated myc genes in a wide range of human, and other animal, cancers has propelled much of this research. In normal cells, Myc proteins appear to integrate environmental signals in order to modulate a diverse, and sometimes opposing, group of cellular processes, including proliferation, growth, apoptosis, energy metabolism, and differentiation. This has made Myc fair game for researchers in different fields and has led to a staggering number of publications. Paradoxically, while a great deal of information has emerged, a unified view of Myc function has proven elusive. The difficulty stems from an apparent disconnect between Myc's dramatic effects on multiple cellular functions and its molecular characterization as a relatively weak transcriptional activator with a poorly defined set of target genes. Over the last several years, however, there has been a significant shift in how Myc function is perceived. This review focuses on two aspects of this shift: first, the growing realization that Myc possesses a considerably larger number of gene targets than anticipated and may have broad effects on chromatin modifications and genomic transcription; and second, the emergence of a more nuanced view of the interplay between Myc and cell differentiation, as well as the recognition of a critical role for Myc in the generation and maintenance of stem cells. In what follows, we summarize recent evidence that has both refined and broadened our perception of Myc's functions.
Transcriptional regulation by Myc
Myc is a basic helix–loop–helix zipper (bHLHZ) protein that heterodimerizes with the small bHLHZ protein Max and binds the E-box sequence CACGTG. E-box binding by Myc-Max has been predominantly associated with gene activation—a finding consistent with Myc's ability to recruit multiple coactivator complexes (for reviews, see Grandori et al. 2000; Adhikary and Eilers 2005; Cole and Nikiforov 2006). Yet Myc has also been associated with transcriptional repression. While there are likely to be several modes of Myc repression, at least one mechanism involves specific binding and inhibition of the transcriptional activator Miz-1 (Kleine-Kohlbrecher et al. 2006).
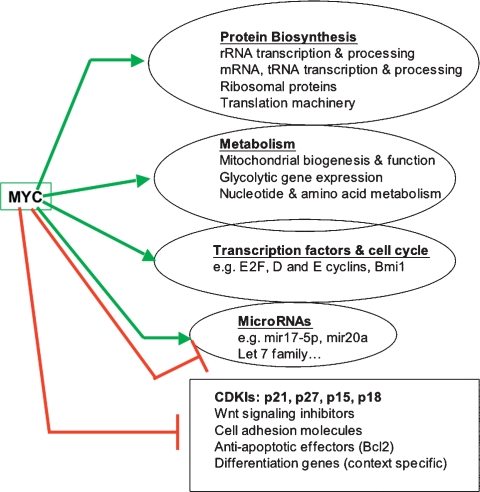
Given Myc's role in gene activation and repression, it would seem that the problem of understanding Myc function should simply reduce to identifying the subset of genes regulated by Myc. To this end, gene expression profiling has been widely employed to examine the effects of Myc overexpression in a range of biological settings. The conclusion is that hundreds to thousands of genes are modulated by augmenting Myc expression (for example, see O'Connell et al. 2003; Li et al. 2005; Schlosser et al. 2005; Lawlor et al. 2006; see also http://www.myc-cancer-gene.org/index.asp). Differences in array platforms, biological systems, time points, and data analysis make comparison between these gene lists perilous. Notwithstanding some puzzling gaps in overlap between studies, the trends that emerge indicate that Myc overexpression influences the regulation of genes that, in aggregate, encompass a broad range of biological functions (Fig. 1; Patel et al. 2004b). Predominant are up-regulated genes directed toward cell growth (defined as an increase in cell mass): ribosome biogenesis, protein synthesis, and metabolism. Modulation of growth-related gene expression was also found in Drosophila upon dMyc overexpression (Orian et al. 2005) or siRNA knockdown (Hulf et al. 2005). Such functional trends are generally consistent with biological studies indicating roles for Drosophila and mammalian Myc in cell growth (Mateyak et al. 1997; Iritani and Eisenman 1999; Johnston et al. 1999; Beier et al. 2000; Pierce et al. 2004; Hulf et al. 2005; Teleman et al. 2008). Most of the genes whose expression is induced by Myc are transcribed by RNA polymerase II. However recent studies demonstrating that Myc additionally stimulates genes transcribed by RNA polymerase III (e.g., tRNA and 5S rRNA genes) (Gomez-Roman et al. 2003; Steiger et al. 2008) and RNA polymerase I (genes encoding ribosomal RNA) (Arabi et al. 2005; Grandori et al. 2005; Grewal et al. 2005) provide further support for the idea that Myc activity is directed toward cell growth at least in part through production of components important in protein translation. Energy metabolism is also an essential aspect of cell growth and proliferation. Myc function had earlier been linked to metabolism with the finding that Myc stimulates transcription of the lactate dehydrogenase A (LDH-A) gene (Shim et al. 1997) as well as a group of nuclear genes involved in mitochondrial function (Morrish et al. 2003). Recently Myc has been shown to promote oxidative phosphorylation as well as glycolysis through coordinate transcriptional control of the mitochondrial metabolic network (Zhang et al. 2007; Morrish et al. 2008). Moreover, Myc appears to be an important regulator of the cellular response to hypoxia through its functional interactions with the hypoxia-inducible factors HIF1α and HIF2α (for review, see Gordan et al. 2007b).
Figure 1.
Summary of the functional categories of genes whose expression is modulated by augmented Myc expression. This information is derived primarily from gene expression profiling studies employing microarrays (see the text for details and references).
While many growth-related genes are activated following Myc expression, fewer genes are down-regulated by Myc; however, those that are appear to be of considerable importance. Most frequent among Myc down-regulated genes are those involved in cell cycle arrest, cell adhesion, and cell–cell communication. This also makes biological sense in that mammalian Myc has been shown to cancel expression of genes that attenuate cell cycle progression (e.g., cyclin-dependent kinase inhibitors [CDKIs]) (Staller et al. 2001; Herold et al. 2002; Knoepfler et al. 2002; Obaya et al. 2002), inhibit signal transduction pathways (Berwanger et al. 2002; Cowling et al. 2007), and reduce cell adhesion and contact (Frye et al. 2003; Wilson et al. 2004; Gebhardt et al. 2006a). The ability of mammalian Myc to abrogate the influence of proliferation arrest genes is a crucial aspect of Myc function in normal development (Zindy et al. 2006) and in tumorigenesis (Seoane et al. 2002; Oskarsson et al. 2006). Taken together, the expression profiling data indicate that Myc has the capacity to both stimulate cell growth and abrogate cell cycle inhibitors, a powerful combination of functions that, when deregulated, may drive the limitless replicative potential characteristic of nearly all tumors (Hanahan and Weinberg 2000).
Despite the apparent agreement between expression profiling and functional studies, there are some concerns about interpretation of the gene expression data. Foremost is the uneasy sense that so many genes are modulated upon Myc overexpression that they could be shoehorned into nearly any functional model. Nonetheless, more precisely tailored expression profiling studies can tell us much about the broad transcriptional effects of Myc. In an elegant recent study (Lawlor et al. 2006) a Myc-estrogen receptor (ER) ligand-binding domain (Myc-ER) transgene was specifically induced in vivo in murine pancreatic β cells coexpressing anti-apoptotic Bcl-XL, thereby permitting survival, myc-driven cell cycle entry, and tumor progression. Expression profiling of cells isolated using laser capture dissection microscopy following acute Myc activation showed a wave of activation and repression ranging from several hundred genes over 24 h, to several thousand genes over a 21-d period. While many of these Myc-regulated genes are, as expected, involved in growth, cell cycle progression, and metabolism, a significant subpopulation of the affected genes had not been identified previously as regulated by Myc in other profiling experiments. Moreover, the presence of genes specific for pancreatic islets further indicates that Myc regulation can impact genes in a context-specific manner. Interestingly, the repression of many islet-specific genes is consistent with the notion that Myc overexpression suppresses the differentiated phenotype. Indeed, acute deactivation of Myc in the Myc-induced β-cell tumors results in tumor regression and re-expression of differentiation-related β-cell genes (Lawlor et al. 2006). This redifferentiation/regression effect has been previously observed in other myc-induced tumor types (Karlsson et al. 2003; Shachaf et al. 2004) and is consistent with the long held view that a failure to down-regulate Myc inhibits terminal differentiation. It is important to note, however, that in situations where commitment to a specific lineage is closely linked to an increase in proliferation, Myc can promote differentiation (see below; Gandarillas and Watt 1997; Frye et al. 2003; Wilson et al. 2004; Habib et al. 2007).
Widespread DNA binding by Myc
While the expression profiling studies provide a basis for understanding Myc's broad effects on the cell, they nonetheless leave wide open the question of what genes are regulated through direct Myc binding (i.e., primary targets) and what genes are regulated indirectly (i.e., secondary or downstream targets) as a consequence of the activities of primary targets or as part of a more general cellular response. As recently as a few years ago, most Myc-ologists would probably have predicted a small number of primary targets and a large number of downstream targets. However experiments employing chromatin immunoprecipitation (ChIP) or genomic marking methods on a promoter-wide or genome-wide scale have shown that Myc binding is more prevalent than anticipated. Moreover, the genomic binding analyses have revealed what are likely to be several novel aspects of Myc function.
Initial genomic binding studies employed arrays comprised of E-box-containing sequences (Fernandez et al. 2003), annotated promoters (Li et al. 2003), or CpG island-enriched DNA (Mao et al. 2003) to screen Myc-bound DNA fragments isolated by ChIP from several human cell types. In addition, a methylation marking procedure was employed in Drosophila tissue culture cells to identify dMyc-, dMax-, and dMnt-associated DNA sequences proximal to coding regions (Orian et al. 2003). More recently, high-throughput sequencing of ChIP DNA (ChIP-seq) has been used to locate DNA regions bound by Myc (Zeller et al. 2006). In aggregate these studies provide substantial support for the prevailing view that (1) Myc associates with Max on DNA, (2) Myc:Max complexes bind to E-box-containing DNA, and (3) E-box binding generally (but not always) corresponds to activation of gene expression. These studies also indicated that 10%–15% of genomic loci are bound by c-Myc in mammals (Fernandez et al. 2003; Li et al. 2003; Zeller et al. 2006) and by dMyc/dMax/dMnt in flies (Orian et al. 2003). As expected, the bound loci represent an extremely broad range of functions, with genes involved in biosynthesis, metabolism, signal transduction, and cell cycle predominating. In the studies that compared binding with expression arrays, it was found that a significant percentage of bound loci are also modulated at the transcriptional level when Myc abundance is altered. For example, in the P493 human B-cell line, ∼20% of 3465 putative c-Myc target genes were up-regulated or down-regulated as a consequence of c-Myc expression (Zeller et al. 2006). Moreover, when a 453 gene subset of c-Myc-bound loci was retrospectively analyzed for expression across 46 human tissues and cells, a majority showed a positive correlation with c-myc expression levels, while a smaller group displayed an inverse correlation (Li et al. 2003). Similarly, in Drosophila nearly 50% of binding sites occupied by dMyc/dMax/dMnt in tissue culture cells were mapped to genes whose expression is modulated in larvae overexpressing dMyc (Orian et al. 2003).
Although Myc binding is clearly coupled to transcription, there appear to be a significant number of binding sites that are not directly associated with gene expression. These latter sites may actually reflect nonproductive binding by Myc. However they may also represent sites that are regulated contingently, dependent on a specific cell or developmental context; as, for example, the cell type-specific differentiation genes induced by Myc in pancreatic β cells (Lawlor et al. 2006). In addition, the apparently nonproductive binding sites for Myc may regulate transcription of genes not normally included on expression arrays, such as those encoding miRNAs (He et al. 2005; O'Donnell et al. 2005; Chang et al. 2008; Schulte et al. 2008) or RNA polymerase III transcripts (Gomez-Roman et al. 2003). Finally, the recent demonstration that Myc can bind to and stimulate origins of DNA replication, independent of Myc transcriptional function (Dominguez-Sola et al. 2007), would suggest that many of the presumed nonproductive Myc-binding sites are replication origins.
Determinants of Myc genomic binding
Chromatin context for Myc binding: consequences for gene regulation
Given that on the order of 10%–15% of genomic loci can be found associated with Myc in different cell types, it is apparent that Myc binding, while widespread, must nonetheless be restricted. This idea is reinforced when one considers that a canonical E-box is expected at a frequency of once per 4 kb on average and that not all Myc binding is E-box-dependent (see below). While CpG methylation could inhibit binding to some sites (James and Eisenman 2002; Perini et al. 2005; Guccione et al. 2006), there are likely to be other determinants of Myc binding. An important recent analysis asked whether specific histone modifications correlate with genomic Myc binding and found that elevated H3-K4 and H3-K79 methylation (active marks), but not H3-K27 methylation (a repressive mark), are characteristic of Myc-binding sites (Guccione et al. 2006). Similar histone methylation determinants of Myc binding have also been shown in a genome-wide binding study of embryonic stem (ES) cells (Kim et al. 2008). These regions, located near the 5′ ends of active genes, constitute euchromatic islands. Because the H3-K4 and H3-K27 modifications were identified in Myc target genes, but in the absence of actual Myc binding, the data imply that the general transcriptional machinery is already engaged in the vicinity of these sites prior to Myc binding. This suggests that Myc prefers to bind to genes that have been accessed previously by other transcription factors. Perhaps Myc preferentially associates with genes that are already actively transcribed and functions to modulate the rate of ongoing transcriptional or post-transcriptional processes. Indeed, there is evidence in support of a role for Myc in elongation, at least at some target genes where Myc, through recruitment of P-TEFb mediates phosphorylation of the C-terminal domain of RNA polymerase II (Eberhardy and Farnham 2002; Cowling and Cole 2007). Furthermore, Myc has been known for some time to selectively influence the rate of translation of a subset of mRNAs (Shiio et al. 2002), and recent data indicate that Myc can facilitate mRNA cap methylation and translation independent of its DNA-binding activity (Cowling and Cole 2007).
Myc's preference for association with promoters marked by methylated H3-K4 is also open to another interpretation—namely, that such genes may not be actively transcribed but rather retain a memory of an earlier transcriptionally active state. Importantly, H3-K4 and H3-K79 methylation marks are highly enriched in the “replacement” histone H3.3, which is assembled into nucleosomes at all phases of the cell cycle. While methylated H3.3-K4 is associated with actively transcribed loci, recent experiments in both Drosophila and vertebrate cells show that a methylatable H3.3-K4 also “marks” silent loci that were previously active but are developmentally repressed (Mito et al. 2007; Henikoff 2008; Ng and Gurdon 2008). Perhaps Myc, by recognizing H3-K4 or H3.3-K4 methylation participates in the reactivation of silenced loci; for example, during transition from mitosis into the G1 phase of the cell cycle or during genetic reprogramming to induce pluripotency (see below).
Mxd proteins can restrict Myc binding and antagonize Myc function
The Mxd family (formerly known as the Mad family) of transcriptional repressors contain Myc-related bHLHZ domains, heterodimerize with Max, and bind E-box sequences. Mxd proteins antagonize Myc function by competing with Myc for available Max, competing with Myc-Max heterodimers for available binding sites, and causing repression at bound sites (Grinberg et al. 2004; for reviews, see Hooker and Hurlin 2006; Rottmann and Luscher 2006). In fact loss-of-function mutations in Mxd proteins, particularly the ubiquitously expressed Mnt protein, can generate phenotypes resembling Myc up-regulation (Hurlin et al. 2003; Nilsson et al. 2004; Toyo-oka et al. 2006). In Drosophila, dmnt-null mutation partially rescues the early arrest of larval development characteristic of dmyc mutant larvae, by attenuating the extent of repression of a large number of growth-promoting genes upon Myc loss of function (Pierce et al. 2007). In mammalian cells, the interplay between Myc and Mxd proteins is important in growth, cell cycle progression, and differentiation (Iritani et al. 2002; McArthur et al. 2002; Popov et al. 2005; Zhu et al. 2008). These considerations suggest that the broadly expressed and highly regulated Mxd proteins normally act to restrict Myc's functional access to DNA.
In and out of the E-box?
While genomic binding experiments indicate that c-Myc protein associates with Max and activates expression of a wide range of genes through E-box binding, the data also suggest that this is not the whole story. Promoter-proximal E-boxes within CpG-rich, H3-K4-methylated, euchromatic islands indeed appear to constitute conserved high-affinity Myc-binding sites in that Myc expressed at either endogenous or at higher levels can be found E-box-associated in several different cell lines (constituting ∼11% of all promoters) (Fernandez et al. 2003; Guccione et al. 2006). However, upon Myc overexpression, previously unoccupied E-boxes were found associated with Myc protein (Fernandez et al. 2003). Depending on the cell type and the abundance of Myc, binding could encompass nearly 60% of all E-box-containing promoters (∼15% of all promoters). Furthermore, at very high Myc levels, binding to promoters lacking canonical CACGTG E-boxes and E-boxes not situated within annotated promoters is detected (Fernandez et al. 2003; Zeller et al. 2006). In another study, E-boxes within a 1-kb region of the TSS were only found associated with a quarter of c-Myc-bound promoters in a Burkitt's lymphoma cell line (Li et al. 2003). Myc has been shown to bind to noncanonical E-boxes (e.g., CATGTG, CA CATG) both in vitro (Blackwell et al. 1993) and in vivo (Grandori et al. 1996; Boyd et al. 1998; Morrish et al. 2003), and it is possible that association with such variant sequences may account in part for the non-E-box sites occupied upon Myc overexpression. Nonetheless, a recent ChIP-seq analysis of relatively high abundance Myc binding showed that while 62% of the most reliable sites possess canonical or noncanonical E-boxes within a 10-kb region, nearly 40% of binding loci did not contain any E-box (Zeller et al. 2006). Perhaps this relates to the observation that about half of the euchromatic islands bound by Myc did not contain E-boxes, suggesting that H3-K4 methylation may be an even better determinant of Myc binding than the presence of an E-box (Guccione et al. 2006).
While we know very little about putative non-E-box targets, there is circumstantial evidence suggesting that Myc's activation of RNA polymerase III-transcribed genes (Gomez-Roman et al. 2003) results from a non-E-box-dependent association with DNA. In Drosophila, null mutations in dmax, although lethal during the larval stages of development, were found to be surprisingly less severe than deletion of dmyc and/or dmnt (Steiger et al. 2008). In the absence of dMax, it was observed that dMyc is still capable of stimulating RNA polymerase III transcription, suggesting that this process may be Max-independent, and therefore E-box-independent. Myc associates with the TFIIIB subunit of the RNA polymerase III complex and mediates recruitment of GCN5 and the TRRAP complex (Gomez-Roman et al. 2003; Kenneth et al. 2007). Therefore in its stimulation of polymerase III transcription, Myc may be simply functioning as a coactivator independent of its cognate DNA-binding site. The report that the interaction of dMyc with the Groucho corepressor complex during embryonic neurogenesis is independent of dMax (Orian et al. 2007) provides another example of a possible E-box-independent activity of Myc. The notion that Myc possesses functions independent of Max had been indicated earlier by the identification of a cell line lacking the max gene (Hopewell and Ziff 1995). Moreover Max protein levels have been shown to be limiting in at least certain biological settings (Walker et al. 2005). Therefore, while Max is indisputably an essential gene in both mammals and flies (Shen-Li et al. 2000; Steiger et al. 2008), there may be a subset of important Myc functions that are not dependent on Max and therefore are likely to be independent of E-boxes for binding. Figuring out what Myc does outside of the E-box may provide a handle on these functions.
Interactions with other transcriptional regulators influence Myc binding
Association of Myc or Myc-Max heterodimers with other DNA-binding factors or complexes also appears to contribute to establishment, or inhibition, of Myc binding at E-boxes or other sites. As mentioned earlier, Myc directly interacts with and inhibits the activity of the BTB-POZ domain protein Miz1 (Seoane et al. 2001; Staller et al. 2001). Myc has also been reported to inhibit multiple factors, including Sp1, NF-Y, and Smad2/Smad3 (Gartel et al. 2001; Izumi et al. 2001; Feng et al. 2002). Other interactions of Myc with transcription factors may not necessarily be direct. Hence association of Myc and the ER, detected in situations where an E-box is proximal to an estrogen response element, are enhanced in a ligand-dependent manner by interaction with the TRRAP coactivator complex. TRRAP apparently acts as a bridge between the proteins at their proximal binding sites (Cheng et al. 2006). In addition, in response to hypoxia HIF-2α has been reported to bind to Max and thereby augment Myc-Max DNA-binding and transcriptional activity (Gordan et al. 2007a). How many other factors Myc interacts with, the extent to which these associations affect Myc binding, the mechanisms underlying Myc's inhibition or activation of these factors, and the degree to which these interactions are important for Myc's biological functions all remain to be determined.
Myc degradation and the dynamics of DNA binding
The widespread binding of Myc to DNA, its recruitment of multiple coregulatory molecules, its interactions with other DNA-binding transcription factors, and its additional involvement in processes such as DNA replication would at first blush imply that Myc is expressed at relatively high levels. This is unlikely to be the case. Previous estimates of endogenous c-Myc proteins levels by radioimmunoassay reported on the order of several thousand molecules per cell in proliferating normal fibroblasts, probably fewer than the number of genomic binding sites (Waters et al. 1991). A possible solution to this apparent paradox comes from consideration of the short half-life of Myc proteins (t1/2 = ∼20 min). During the last several years, it has been demonstrated that the proteasome-mediated degradation of c-Myc is directed by at least two ubiquitin ligases—Fbw7, which recognizes the Myc Box I phosphodegron (Welcker et al. 2004a, b; Yada et al. 2004), and Skp2, which interacts with a non-phospho-dependent binding site on Myc (Kim et al. 2003; von der Lehr et al. 2003). Recent evidence also links the HectH9/Huwe1 ubiquitin-ligase to degradation of N-Myc during differentiation (Zhao et al. 2008). Mutations in Fbw7 are associated with several types of tumors and lead to increased abundance of Myc and other Fbw7 targets critical for cell proliferation (for review, see Welcker and Clurman 2008). Moreover, the ubiquitin-specific protease USP28 associates with, and stabilizes, Myc by interacting with Fbw7 (Popov et al. 2007). USP28 is detected at high levels in colon and breast carcinomas and contributes to proliferation of these tumors. Although stabilization of Myc has been shown to be important in both normal and neoplastic settings (Kenney et al. 2003; Welcker et al. 2004a; Yeh et al. 2004; Cartwright et al. 2005; Hemann et al. 2005), the “stabilized” Myc protein nonetheless still possesses a measurable, and often quite rapid, rate of turnover. Several lines of evidence also suggest a link between Myc turnover and its transcriptional activity. Degradation of c-Myc through the Skp2 complex has been reported to stimulate Myc-dependent transcription (Kim et al. 2003; von der Lehr et al. 2003). In addition, chemical inhibitors of the proteasome or knockdown or the Fbw7 ligase induce accumulation of Myc in nucleoli, an organelle in which Myc is normally present at only very low levels (Arabi et al. 2003; Welcker et al. 2004a). However Myc has a nucleolar function in that it directly stimulates RNA polymerase I transcription of rDNA (Arabi et al. 2005; Grandori et al. 2005). This suggests that Myc is normally turned over very rapidly in the nucleolus in concert with the high rate of rRNA production.
The continuous degradation of Myc protein implies that Myc binding at a given site is transient. Yet the rapid rate of synthesis of Myc, required to balance its rate of degradation and maintain steady-state levels, would act to continuously generate newly synthesized Myc proteins with the potential to occupy previously unbound target sites. In other words, not all Myc targets would be bound within a single cell at a given moment, but over time, all targets will come to be transiently occupied. This idea is supported by quantitative measurements of binding site occupancy by other transcription factors indicating dwell times of seconds to minutes for many factors (Phair et al. 2004; Meshorer et al. 2006). Having the full complement of Myc targets dependent on continual synthesis of limiting amounts of Myc protein would tend to make these targets highly sensitive to even small changes in abundance of Myc protein, providing a rationale for the multiple levels at which myc gene expression is regulated transcriptionally and post-transcriptionally.
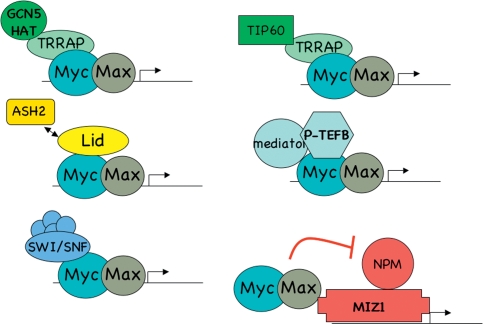
The rapid turnover of Myc proteins may also impact Myc transcriptional regulation in another way: by permitting rapid exchange of Myc recruited coregulators. Myc has been reported to recruit a bewildering number of transcriptional coregulators (Fig. 2). These include the TRRAP coactivator, which in turn binds the HAT GCN5 and other subunits involved in histone modification; the Tip60 complex (Cole and Nikiforov 2006); the Pim1 kinase (Zippo et al. 2007); the Lid/Rbp2 H3-K4 demethylase (Secombe et al. 2007); the USP22 H2A deubiquitinating enzyme (Zhang et al. 2008); and the HectH9 ubiquitin ligase (Adhikary et al. 2005) among others. These coregulators are thought to mediate histone modifications or remodeling but may also modify other associated factors or Myc itself (Vervoorts et al. 2003; Patel et al. 2004a; Adhikary et al. 2005). While a single Myc polypeptide could in principle interact with several of these proteins at once, an alternative possibility is that Myc recruits distinct complexes to specific subsets of target genes (Fig. 2). Moreover, the continuous degradation and replacement of Myc proteins, bearing distinct sets of coregulators at genomic binding sites, could result in a sequential change in coregulator composition at gene targets. Exchange of coactivators and corepressors has been previously shown to be critical in transcriptional regulation by NFκB and nuclear hormone receptors (Baek et al. 2002; Perissi et al. 2004). Myc's recruitment of multiple coregulators could potentially lead to spreading of chromatin modifications and to nucleosome instability, perhaps explaining the widespread changes in histone modifications and DNA accessibility observed in cells with myc deletions (Knoepfler et al. 2006). It is conceivable that Myc's role in the induction of pluripotency (see below) is to broadly induce the open chromatin structure characteristic of stem cells (Niwa 2007).
Figure 2.
Transcriptional complexes associated with Myc. Depicted are individual coregulator complexes known to interact with Myc. In principle, several complexes could simultaneously associate with single Myc proteins. The binding of a Myc-Max heterodimer to the Miz-1 is shown, resulting in repression through inhibition of nucleophosmin (NPM). Many other protein interactions have been described for Myc (see the text).
Myc at the root of stem cells—implications for tumorigenesis
The ability of Myc to bind to a staggeringly large number of genomic loci has also been demonstrated in ES cells (Kim et al. 2008) and is likely to underlie its activity as part of the “magic quartet” of transcription factors that can reprogram somatic cells to induced pluripotent stem (iPS) cells. Several independent studies found that—while not absolutely required—ectopic expression of Myc augments the ability of Oct4, Sox2, and Klf4 to induce the formation of pluripotent cells from mouse and human fibroblasts, liver cells, and mature B cells by a factor of twofold to 10-fold, depending on the cell type (Okita et al. 2007; Takahashi et al. 2007; Wernig et al. 2007; Hanna et al. 2008). Expression of ectopic Myc is silenced in the resulting iPS cells, arguing that high levels of Myc are only transiently required in the establishment of pluripotency and/or the expansion of a critical cell pool. Perhaps pharmacological strategies or delivery vectors (e.g., adenovirus) that transiently elevate levels of Myc will eventually be able to substitute for the ectopic expression of Myc and thereby escape the tumorigenic potential of the iPS cells in which Myc has been expressed.
A role for endogenous Myc proteins in the proliferation of stem and progenitor cells has been documented in several studies. For example, Drosophila neuroblasts undergo an asymmetric division that leads to the exit of one of the daughter cells from the cell cycle and its subsequent terminal differentiation. This exit is mediated by the asymmetric distribution of a cell fate determinant, Brat (Betschinger et al. 2006). Brat is a negative regulator of cell growth that has properties of a tumor suppressor protein since in its absence both daughter cells continue to proliferate as neuroblasts. Brat exerts its function as a negative regulator of Drosophila Myc (dMyc), arguing that expression of dMyc is critical for maintaining proliferation and inhibiting differentiation of neuronal progenitor cells. Similarly, the conditional deletion of mammalian N-Myc, a gene closely related to c-Myc that is highly expressed in neuronal progenitor cells, induces the premature differentiation of neuronal progenitor cells in the cerebral cortex and in neurospheres in culture, arguing that N-Myc is critical for preventing the premature differentiation of neuronal precursor cells in mammals (Knoepfler et al. 2002). N-Myc has also been shown to be required in a dose-dependent manner in developing lung to maintain undifferentiated proliferating distal progenitor cells (Okubo et al. 2005). Very recently, placental rescue experiments in mice have demonstrated a primary requirement for c-myc specifically in hematopoietic stem and progenitor cells—indeed, the loss of these populations appears to be the major defect responsible for embryonic lethality at embryonic day 12.5 (E12.5) (Dubois et al. 2008).
Conversely, ectopic expression of Myc inhibits the differentiation of murine ES cells. The self-renewal capacity of ES cells depends on the presence of leukemia-inhibitory factor, LIF. By binding to its cognate receptor, LIF activates the transcription factor STAT3, which in turn activates expression of c-myc. Constitutive expression of either STAT3 or a mutant allele of c-Myc (T58A), which is not recognized by the Fbw7 ubiquitin ligase and is therefore more stable in the absence of growth factors than the wild-type protein, maintains ES cells in a self-renewing and pluripotent state even when LIF is removed from the medium (Cartwright et al. 2005). In contrast, inhibition of Myc using a dominant interfering form induces differentiation of ES cells even in the presence of LIF, demonstrating that Myc is critical for the maintenance of ES cell self-renewal. Furthermore, numerous experiments document the ability of Myc to inhibit terminal differentiation of multiple cell types in culture and to promote tumorigenesis in vivo when expressed in differentiated cells. In a number of cases, the resulting tumors have been transplanted, formally demonstrating the presence of tumor cells that are capable of self-renewal; examples are Myc-induced lymphomas and liver tumors (Shachaf et al. 2004; Giuriato et al. 2006).
While the notion that Myc simply functions to inhibit differentiation and promote proliferation is satisfying, it is becoming increasingly clear that Myc's role in differentiation is more nuanced and complex. In skin, ectopic expression of Myc under the control of the keratin14 promoter, which is active in stem and progenitor cells, induces a long-term depletion of stem cells (Arnold and Watt 2001; Waikel et al. 2001). As a consequence, one phenotype observed in such mice is a severe defect in wound healing, which requires the mobilization of stem cells from their niche. At the same time, ectopic expression of Myc promotes both proliferation and an accumulation of differentiated cell layers in the epidermis. In vitro, Myc promotes the proliferation of keratinocytes in short-term assays, but leads to a loss of clonogenic potential and promotes differentiation of keratinocytes in long-term culture (Gandarillas and Watt 1997; Watt et al. 2008). Similarly, ectopic expression of c-Myc in hematopoietic cells leads to a depletion of stem cells, a finding consistent with the observation that targeted deletion of c-Myc leads to an expansion of stem cell pools and a concomitant loss of all differentiated cell lineages (Wilson et al. 2004). These data do not address the issue of whether the proliferation of hematopoietic stem cells (HSCs) per se requires Myc activity. This is because HSCs express the functionally redundant paralog N-Myc that could in principle drive self-renewal in the absence of c-Myc. However, the data demonstrate that levels of Myc have a role in controlling exit from the stem cell niche.
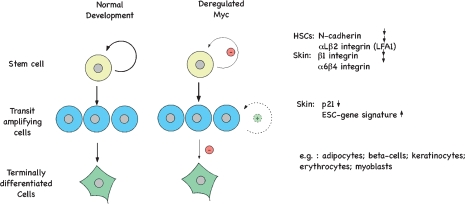
In both keratinocytes and HSCs, at least part of Myc's role in driving differentiation involves its ability to reduce adhesive interactions of stem cells with their niche (Fig. 3; Frye et al. 2003; Wilson et al. 2004). In keratinocytes, Myc represses, through binding to Miz1, a wide spectrum of genes encoding cell–cell and cell substratum adhesion proteins, including the integrins β1 and α6β4 (Gebhardt et al. 2006b). Re-expression of integrin β1 normalizes the premature differentiation phenotypes in mice expressing ectopic Myc, demonstrating that integrin β1 is an important effector of Myc in this pathway. HSCs lacking c-Myc express enhanced levels of the adhesion receptor LFA-1, N-cadherin, and several integrins (Wilson et al. 2004). Myc represses the promoter of the CD11a gene, which encodes a subunit of LFA-1, suggesting that some of these changes in expression reflect a direct repression of the respective genes by Myc (Lopez-Rodriguez et al. 2000). Notably, other and less direct mechanisms may also be at play: For example, expression of tert, a target gene of Myc that encodes the catalytic subunit of telomerase, promotes stem cell mobilization (Flores et al. 2005). Furthermore, maintaining the relative quiescence of stem cells requires p21cip1 and repression of p21cip1 expression may be another mechanism through which Myc promotes exit from the stem cell compartment (Cheng et al. 2000).
Figure 3.
Myc influences self-renewal and differentiation of stem cells. The effects of Myc on expression of genes considered critical for differentiation and proliferation are indicated. See the text for details.
These findings have implications for the oncogenic effects of deregulated MYC genes. For instance, the ability of Myc to reduce the interactions of stem cells with their niche suggests that tumors that express high levels of Myc are unlikely to be derived from stem cells—at least in tissues where such niche interactions are critical for stem cell identity. Skin tumors provide one example of this idea: Undifferentiated basal cell carcinomas express very low levels of Myc, whereas amplifications of myc are frequent in squamous cell carcinoma, a tumor derived from more differentiated cells (Boukamp 2005). Indeed, the ability of Myc to promote exit of stem cells from the niche may constitute a fail-safe mechanism—similar to Myc-induced apoptosis or Ras-induced senescence—that protects organisms from stem cell-derived carcinomas.
Moreover, because high levels of Myc perturb terminal differentiation and enhance self-renewal of committed and differentiated cells, one might expect that during tumor progression Myc would promote the formation of cancer-initiating cells that retain developmental plasticity but differ from the normal stem cells of the tissue from which the tumor has been derived. Several pieces of evidence support this view. For example, expression of Myc in HSCs leads to the rapid formation of pre B-cell lymphomas if apoptosis is blocked by coexpression of Bcl-2. Cells derived from these lymphomas can be differentiated into either B-lymphocytes or macrophages in vitro (Strasser et al. 1996). Similarly, cells derived from liver tumors that have been generated using a tetracycline-inducible Myc transgene differentiate into hepatocytes and biliary cells that go on to form bile ducts when the transgene is turned off; some of these cells also express a marker of liver stem cells, cytokeratin 19 (Shachaf et al. 2004). Clearly, therefore, cells in both tumor types retain the potential to differentiate along different lineages. At the molecular level, this is reflected in the ability of Myc to induce the expression, in differentiated cells, of a gene subset that resembles the expression signatures of ES cells (Wong et al. 2008). As mentioned before, both the liver tumors and the lymphomas are transplantable, documenting that they contain tumor-initiating cells; indeed, limiting dilution analysis shows that the introduction of Myc into primary keratinocytes that have been transformed by an oncogenic allele of Ras and IKBα strongly increases the frequency of tumor-initiating cells (Wong et al. 2008). Similarly, the percentage of Myc-induced lymphoma cells capable of initiating lymphoma formation upon transplantation is very high (Kelly et al. 2007). In contrast, such cells fail to provide long-term reconstitution of the hematopoietic system in lethally irradiated mice, arguing that they do not contain normal HSCs. Further support for this view also comes from recent elegant knockout studies of the c-Myc gene. Two studies have demonstrated that c-Myc is required for the formation of intestinal tumors that are initiated by conditional deletion of the APC gene (Sansom et al. 2007) and of skin tumors in the classic DMBA/TPA driven model of Ras-dependent skin carcinogenesis (Oskarsson et al. 2006). Importantly, the latter study showed that the effect of Myc deletion had no effect on the normal development and tissue homeostasis in untreated skin but specifically abrogated skin tumorigenesis.
Conclusion
Myc proteins act as master regulators of two very distinct genetic programs: On the one hand, they stimulate virtually all nuclear processes leading to enhanced cell growth (the accumulation of cell mass); on the other hand, Myc proteins cancel the cell cycle arrest induced by multiple growth-inhibitory pathways. Both these activities of Myc appear to be tightly coupled with critical developmental decisions. Notably, induction of Myc often prompts exit from the stem cell compartment, while conversely, down-regulation of Myc is required for terminal differentiation in multiple lineages. Together, the observations suggest that a key function of Myc during normal development is to drive expansion of transit amplifying cells. Due to this unique combination of properties, oncogenic deregulation of Myc expression generates cells possessing a tumor phenotype that has no counterpart during normal development. We suggest that, at least in some tissues, targeting Myc may provide a strategy that selectively hits tumor initiating cells but spares tissue homeostasis. The identification of druggable Myc targets, as well as enzymes that are critical for the stability and function of Myc itself, may provide therapeutic inroads for the many cancers in which Myc deregulation is critical.
Acknowledgments
We are grateful to members of both our laboratories for continuing discussions on the Myc enigma. Work described in this review from the laboratory of R.N.E. was supported by the NCI grants RO1-CA20525 and RO1-CA57138.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1712408.
References
- Adhikary S., Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Arabi A., Rustum C., Hallberg E., Wright A.P. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J. Cell Sci. 2003;116:1707–1717. doi: 10.1242/jcs.00370. [DOI] [PubMed] [Google Scholar]
- Arabi A., Wu S., Shiue C., Ridderstrale K., Larsson L.-G., Wright A.P.H. c-Myc associates with ribosomal DNA in the nucleolus and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Arnold I., Watt F.M. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Baek S.H., Ohgi K.A., Rose D.W., Koo E.H., Glass C.K., Rosenfeld M.G. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Beier R., Burgin A., Kiermaier A., Fero M., Karsunky H., Saffrich R., Moroy T., Ansorge W., Roberts J., Eilers M. Induction of cyclin E–cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. EMBO J. 2000;19:5813–5823. doi: 10.1093/emboj/19.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwanger B., Hartmann O., Bergmann E., Bernard S., Nielsen D., Krause M., Kartal A., Flynn D., Wiedemeyer R., Schwab M., et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2:377–386. doi: 10.1016/s1535-6108(02)00179-4. [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K., Huang J., Ma A., Kretzner L., Alt F.W., Eisenman R.N., Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P. Non-melanoma skin cancer: What drives tumor development and progression? Carcinogenesis. 2005;26:1657–1667. doi: 10.1093/carcin/bgi123. [DOI] [PubMed] [Google Scholar]
- Boyd K.E., Wells J., Gutman J., Bartley S.M., Farnham P.J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc. Natl. Acad. Sci. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., Scadden D.T. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science (New York, NY) 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Cheng A.S., Jin V.X., Fan M., Smith L.T., Liyanarachchi S., Yan P.S., Leu Y.W., Chan M.W., Plass C., Nephew K.P., et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-α responsive promoters. Mol. Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Cole M.D., Nikiforov M.A. Transcriptional activation by the Myc oncoprotein. Curr. Top. Microbiol Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- Cowling V.H., Cole M.D. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling V.H., D'Cruz C.M., Chodosh L.A., Cole M.D. c-myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol. Cell. Biol. 2007;27:5135–5146. doi: 10.1128/MCB.02282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Ying C.Y., Grandori C., Ruggiero L., Chen B., Li M., Galloway D.A., Gu W., Gautier J., Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Dubois N.C., Adolphe C., Ehninger A., Wang R.A., Robertson E.J., Trumpp A. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development. 2008;135:2455–2465. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- Eberhardy S.R., Farnham P.J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- Feng X.H., Liang Y.Y., Liang M., Zhai W., Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15(Ink4B) Mol. Cell. 2002;9:133–143. doi: 10.1016/s1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- Fernandez P.C., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. Genomic targets of the human c-Myc protein. Genes & Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I., Cayuela M.L., Blasco M.A. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- Frye M., Gardner C., Li E.R., Arnold I., Watt F.M. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- Gandarillas A., Watt F.M. c-Myc promotes differentiation of human epidermal stem cells. Genes & Dev. 1997;11:2869–2882. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel A.L., Ye X., Goufman E., Shianov P., Hay N., Najmabadi F., Tyner A.L. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A., Frye M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsasser H.-P., Eilers M. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J. Cell Biol. 2006a;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A., Frye M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsasser H.P., Eilers M. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J. Cell Biol. 2006b;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuriato S., Ryeom S., Fan A.C., Bachireddy P., Lynch R.C., Rioth M.J., van Riggelen J., Kopelman A.M., Passegue E., Tang F., et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc. Natl. Acad. Sci. 2006;103:16266–16271. doi: 10.1073/pnas.0608017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N., Grandori C., Eisenman R.N., White R.J. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- Gordan J.D., Bertout J.A., Hu C.J., Diehl J.A., Simon M.C. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007a;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan J.D., Thompson C.B., Simon M.C. HIF and c-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007b;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Mac J., Siebelt F., Ayer D.E., Eisenman R.N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N. The MYC/MAX/MAD network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Grewal S.S., Li L., Orian A., Eisenman R.N., Edgar B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- Grinberg A.V., Hu C.D., Kerppola T.K. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol. Cell. Biol. 2004;24:4294–4308. doi: 10.1128/MCB.24.10.4294-4308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E., Martinato F., Finocchiaro G., Luzi L., Tizzoni L., Dall' Olio V., Zardo G., Nervi C., Bernard L., Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Habib T., Park H., Tsang M., de Alboran I.M., Nicks A., Wilson L., Knoepfler P.S., Andrews S., Rawlings D.J., Eisenman R.N., et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J. Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Schorderet P., Carey B.W., Beard C., Wernig M., Creyghton M.P., Steine E.J., Cassady J.P., Foreman R., et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann M.T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J.A., Cordon-Cardo C., Cleveland J.L., Tansey W.P., Lowe S.W. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Herold S., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- Hooker C.W., Hurlin P.J. Of Myc and Mnt. J. Cell Sci. 2006;119:208–216. doi: 10.1242/jcs.02815. [DOI] [PubMed] [Google Scholar]
- Hopewell R., Ziff E.B. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol. Cell. Biol. 1995;15:3470–3478. doi: 10.1128/mcb.15.7.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulf T., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol. Cell. Biol. 2005;25:3401–3410. doi: 10.1128/MCB.25.9.3401-3410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin P.J., Zhou Z.Q., Toyo-Oka K., Ota S., Walker W.L., Hirotsune S., Wynshaw-Boris A. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J. 2003;22:4584–4596. doi: 10.1093/emboj/cdg442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani B.M., Eisenman R.N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani B.M., Delrow J., Grandori C., Gomez I., Klacking M., Carlos L.S., Eisenman R.N. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 2002;21:4820–4830. doi: 10.1093/emboj/cdf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H., Molander C., Penn L.Z., Ishisaki A., Kohno K., Funa K. Mechanism for the transcriptional repression by c-Myc on PDGF β-receptor. J. Cell Sci. 2001;114:1533–1544. doi: 10.1242/jcs.114.8.1533. [DOI] [PubMed] [Google Scholar]
- James L., Eisenman R.N. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc. Natl. Acad. Sci. 2002;99:10429–10434. doi: 10.1073/pnas.162369299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.A., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P. Drosophila myc regulates growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A., Giuriato S., Tang F., Fung-Weier J., Levan G., Felsher D.W. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood. 2003;101:2797–2803. doi: 10.1182/blood-2002-10-3091. [DOI] [PubMed] [Google Scholar]
- Kelly P.N., Dakic A., Adams J.M., Nutt S.L., Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Kenneth N.S., Ramsbottom B.A., Gomez-Roman N., Marshall L., Cole P.A., White R.J. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. 2007;104:14917–14922. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.M., Widlund H.R., Rowitch D.H. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2003;131:217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Herbst A., Tworkowski K.A., Salghetti S.E., Tansey W.P. Skp2 regulates myc protein stability and activity. Mol. Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D., Adhikary S., Eilers M. Mechanisms of transcriptional repression by Myc. Curr. Top. Microbiol. Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S., Cheng P.F., Eisenman R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes & Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler P.S., Zhang X.-y., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor E.R., Soucek L., Brown-Swigart L., Shchors K., Bialucha C.U., Evan G.I. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006;66:4591–4601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- Li Z., Van Calcar S., Qu C., Cavenee W.K., Zhang M.Q., Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O'Donnell K.A., Kim J.-w., Yustein J.T., Lee L.A., Dang C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez C., Delgado M.D., Puig-Kroger A., Nueda A., Munoz E., Leon J., Bernabeu C., Corbi A.L. c-Myc inhibits CD11a and CD11c leukocyte integrin promoters. Eur. J. Immunol. 2000;30:2465–2471. doi: 10.1002/1521-4141(200009)30:9<2465::AID-IMMU2465>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Mao D.Y., Watson J.D., Yan P.S., Barsyte-Lovejoy D., Khosravi F., Wong W.W., Farnham P.J., Huang T.H., Penn L.Z. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Mateyak M.K., Obaya A.J., Adachi S., Sedivy J.M. Phenotypes of c-myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- McArthur G.A., Foley K.P., Fero M.L., Walkley C.R., Deans A.J., Roberts J.M., Eisenman R.N. MAD1 and p27(KIP1) cooperate to promote terminal differentiation of granulocytes and to inhibit Myc expression and cyclin E-CDK2 activity. Mol. Cell. Biol. 2002;22:3014–3023. doi: 10.1128/MCB.22.9.3014-3023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J.G., Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Morrish F., Giedt C., Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes & Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F., Neretti N., Sedivy J.M., Hockenbery D.M. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R.K., Gurdon J.B. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- Nilsson J.A., Maclean K.H., Keller U.B., Pendeville H., Baudino T.A., Cleveland J.L. Mnt loss triggers myc transcription targets, proliferation, apoptosis, and transformation. Mol. Cell. Biol. 2004;24:1560–1569. doi: 10.1128/MCB.24.4.1560-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. Open conformation chromatin and pluripotency. Genes & Dev. 2007;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- Obaya A.J., Kotenko I., Cole M.D., Sedivy J.M. The proto-oncogene c-myc acts through the cyclin-dependent kinase (Cdk) inhibitor p27(Kip1) to facilitate the activation of Cdk4/6 and early G(1) phase progression. J. Biol. Chem. 2002;277:31263–31269. doi: 10.1074/jbc.M202528200. [DOI] [PubMed] [Google Scholar]
- O'Connell B.C., Cheung A.F., Simkevich C.P., Tam W., Ren X., Mateyak M.K., Sedivy J.M. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okubo T., Knoepfler P.S., Eisenman R.N., Hogan B.L. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132:1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.M., Cowley S.M., Yost C., et al. Genomic binding by the Drosophila Myc, Max, Mad.Mnt transcription factor network. Genes & Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A., Grewal S.S., Knoepfler P.S., Edgar B.A., Parkhurst S.M., Eisenman R.N. Genomic binding and transcriptional regulation by the Drosophila myc and mnt transcription factors. Cold Spring Harb. Symp. Quant. Biol. 2005;70:1–10. doi: 10.1101/sqb.2005.70.019. [DOI] [PubMed] [Google Scholar]
- Orian A., Delrow J.J., Rosales Nieves A.E., Abed M., Metzger D., Paroush Z., Eisenman R.N., Parkhurst S.M. A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc. Natl. Acad. Sci. 2007;104:15771–15776. doi: 10.1073/pnas.0707418104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T., Essers M.A., Dubois N., Offner S., Dubey C., Roger C., Metzger D., Chambon P., Hummler E., Beard P., et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes & Dev. 2006;20:2024–2029. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.J., Rakowski C., Chatterjee C., Lieberman P.M., Lane W.S., et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 2004a;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J.H., Loboda A.P., Showe M.K., Showe L.C., McMahon S.B. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer. 2004b;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Perini G., Diolaiti D., Porro A., Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl. Acad. Sci. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Aggarwal A., Glass C.K., Rose D.W., Rosenfeld M.G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Phair R.D., Scaffidi P., Elbi C., Vecerova J., Dey A., Ozato K., Brown D.T., Hager G., Bustin M., Misteli T. Global nature of dynamic protein–chromatin interactions in vivo: Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S.B., Yost C., Britton J.S., Loo L.W., Flynn E.M., Edgar B.A., Eisenman R.N. dMyc is required for larval growth and endoreplication in Drosophila. Development. 2004;131:2317–2327. doi: 10.1242/dev.01108. [DOI] [PubMed] [Google Scholar]
- Pierce S.B., Yost C., Anderson S.A., Flynn E.M., Delrow J., Eisenman R.N. Drosophila growth and development in the absence of dMyc and dMnt. Dev. Biol. 2007;315:303–316. doi: 10.1016/j.ydbio.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N., Wahlstrom T., Hurlin P.J., Henriksson M. Mnt transcriptional repressor is functionally regulated during cell cycle progression. Oncogene. 2005;24:8326–8337. doi: 10.1038/sj.onc.1208961. [DOI] [PubMed] [Google Scholar]
- Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S.J., Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- Rottmann S., Luscher B. The Mad side of the Max network: Antagonising the function of Myc and more. Curr. Top. Microbiol. Immunol. 2006;302:63–122. doi: 10.1007/3-540-32952-8_4. [DOI] [PubMed] [Google Scholar]
- Sansom O.J., Meniel V.S., Muncan V., Phesse T.J., Wilkins J.A., Reed K.R., Vass J.K., Athineos D., Clevers H., Clarke A.R. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- Schlosser I., Holzel M., Hoffmann R., Burtscher H., Kohlhuber F., Schuhmacher M., Chapman R., Weidle U.H., Eick D. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–524. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- Schulte J.H., Horn S., Otto T., Samans B., Heukamp L.C., Eilers U.C., Krause M., Astrahantseff K., Klein-Hitpass L., Buettner R., et al. MYCN regulates oncogenic microRNAs in neuroblastoma. Int. J. Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L., Eisenman R.N. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massague J. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Seoane J., Le H.V., Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Shachaf C.M., Kopelman A.M., Arvanitis C., Karlsson A., Beer S., Mandl S., Bachmann M.H., Borowsky A.D., Ruebner B., Cardiff R.D., et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Shen-Li H., O'Hagan R.C., Hou H.J., Horner J.W., Lee H.W., DePinho R.A. Essential role for Max in early embryonic growth and development. Genes & Dev. 2000;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- Shiio Y., Donohoe S., Yi E.C., Goodlett D.R., Aebersold R., Eisenman R.N. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 2002;21:5088–5096. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Dolde C., Lewis B.C., We C.-S., Dang G., Jungmann R.A., Dalla-Favera R., Dang C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massague J., Hänel F., et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Steiger D., Furrer M., Schwinkendorf D., Gallant P. Max-independent functions of Myc in Drosophila melanogaster. Nat. Genet. 2008;40:1084–1091. doi: 10.1038/ng.178. [DOI] [PubMed] [Google Scholar]
- Strasser A., Elefanty A.G., Harris A.W., Cory S. Progenitor tumours from Emu–bcl-2–myc transgenic mice have lymphomyeloid differentiation potential and reveal developmental differences in cell survival. EMBO J. 1996;15:3823–3834. [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Teleman A.A., Hietakangas V., Sayadian A.C., Cohen S.M. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K., Bowen T.J., Hirotsune S., Li Z., Jain S., Ota S., Lozach L.E., Bassett I.G., Lozach J., Rosenfeld M.G., et al. Mnt-deficient mammary glands exhibit impaired involution and tumors with characteristics of Myc overexpression. Cancer Res. 2006;66:5565–5573. doi: 10.1158/0008-5472.CAN-05-2683. [DOI] [PubMed] [Google Scholar]
- Tudge C. The tree. Crown Publishers; New York: 2007. [Google Scholar]
- Vervoorts J., Luscher-Firzlaff J.M., Rottmann S., Lilischkis R., Walsemann G., Dohmann K., Austen M., Luscher B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K.I., et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Waikel R.L., Kawachi Y., Waikel P.A., Wang X.J., Roop D.R. Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 2001;28:165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- Walker W., Zhou Z.Q., Ota S., Wynshaw-Boris A., Hurlin P.J. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J. Cell Biol. 2005;169:405–413. doi: 10.1083/jcb.200411013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C.M., Littlewood T.D., Hancock D.C., Moore J.P., Evan G.I. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991;6:797–805. [PubMed] [Google Scholar]
- Watt F.M., Frye M., Benitah S.A. MYC in mammalian epidermis: How can an oncogene stimulate differentiation? Nat. Rev. Cancer. 2008;8:234–242. doi: 10.1038/nrc2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Clurman B.E. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian A., Grim J.A., Eisenman R.N., Clurman B.E. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 2004a;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J., Grim J.A., Harper J.W., Eisenman R.N., Clurman B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. 2004b;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes & Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D.J., Liu H., Ridky T.W., Cassarino D., Segal E., Chang H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K.I. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- Zeller K.I., Zhao X., Lee C.W.H., Chiu K.P., Yao F., Yustein J.T., Ooi H.S., Orlov Y.L., Shahab A., Yong H.C., et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K.I., Dang C.V., Semenza G.L. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., McMahon S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Heng J.I., Guardavaccaro D., Jiang R., Pagano M., Guillemot F., Iavarone A., Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Blenis J., Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Knoepfler P.S., Xie S., Sherr C.J., Eisenman R., Roussel M. N-Myc and the cyclin-dependent kinase inhibitors, p18Ink4c and p27Kip1, coordinately regulate cerebellar development. Proc. Natl. Acad. Sci. 2006;103:11579–11583. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A., De Robertis A., Serafini R., Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat. Cell Biol. 2007;9:932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]