Abstract
One of the great paradoxes in cellular differentiation is how cells with identical DNA sequences differentiate into so many different cell types. The mechanisms underlying this process involve epigenetic regulation mediated by alterations in DNA methylation, histone posttranslational modifications, and nucleosome remodeling. It is becoming increasingly clear that disruption of the “epigenome” as a result of alterations in epigenetic regulators is a fundamental mechanism in cancer. This has major implications for the future of both molecular diagnostics as well as cancer chemotherapy.
Epigenetics is a rapidly evolving field focused on explaining how heritable changes in gene expression occur that do not involve changes in nucleotide sequence.1 Epigenetic regulation of transcription can be mediated through DNA methylation, histone modifications including histone acetylation, phosphorylation, methylation, ubiquitination, and proteolysis, and alterations in chromatin remodeling. Importantly, increasing evidence shows that epigenetic deregulation is a common mechanism in cancer. The role of DNA methylation in cancer has been extensively studied, and a number of excellent reviews have been published on this topic.2,3,4 More recently, it has become clear that histone modifications, as well as disruption of chromatin remodeling machinery, play a fundamental role in cancer, and this is our primary focus in this review. It is important to recognize that these three types of epigenetic regulation are highly interdependent. For example, patterns of histone methylation are important for establishing patterns of DNA methylation. Chromatin remodeling, in turn, is programmed in part by changes in DNA methylation and histone modifications.3
DNA Methylation
CpG-rich sequences are generally rare in the mammalian genome except for in so-called CpG islands, which are associated with centromeres, microsatellite sequences, and the proximal promoter regions of approximately half of all genes. CpG-containing sequences are cytosine methylated by a family of DNA methyltransferases or DNMTs (to date, unequivocal evidence for DNA demethylases is lacking). These methyltransferases generally exempt promoter-associated CpG islands, where <20% are CpG methylated.5
It was recognized over 25 years ago in studies of colon cancer that patterns of DNA methylation in tumor cells differed considerably from normal cells.6 These early studies, which detected DNA methylation using methylation-sensitive restriction enzymes and Southern blotting, revealed global hypomethylation of DNA sequences compared with normal cellular counterparts (Figure 1). Subsequently, many high-throughput techniques have been developed for CpG methylation profiling, including restriction landmark genomic screening, bisulfite sequencing, differential methylation hybridization, DNA immunoprecipitation using antibodies directed against 5-methylcytosine, and array or sequence-based detection methods, which have confirmed this finding of global hypomethylation in a variety of cancers.7 This hypomethylation, whose mechanism remains poorly understood, may play several roles in oncogenesis, including increasing genomic instability as well as contributing to the over expression of genes, such as MAGE, CAGE, CYCLIND2, S100A4, CD30, as well as loss of imprinting of genes such as IGF2.4 In one well-studied example, loss of imprinting of the maternally inherited IGF2 allele has been implicated in the pathogenesis of colorectal cancer and Wilm’s tumor.8 Changes in DNA methylation have also been associated with chemotherapy resistance. For example, methylation of the MLH1 gene is associated with increased resistance to cis-platinum and alkylating agents.9
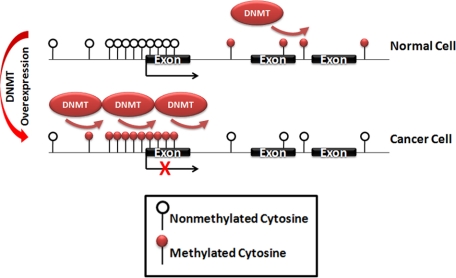
Figure 1.
Aberrant DNA methylation patterns in cancer cells as a result of DNA methyltransferase overexpression. CpG dinucleotides are methylated in normal cells, whereas CpG islands, consisting of overrepresented CpG clusters near gene regulatory regions, are unmethylated. In contrast, cancer cells generally show global hypomethylation with hypermethylation at CpG islands, resulting in gene silencing at a subset of genes, including tumor suppressor genes.
Paradoxically, local hypermethylation of specific genes also appears to play an important role in cancer (Figure 1). The first hypermethylated gene identified was calcitonin, which is hypermethylated in a subset of small cell carcinoma cases.10 This was followed by identification of a number of bona fide tumor suppressors, including RB, VHL, and BRCA1.4 Recently, expression of potentially important micro-RNAs has also been shown to be regulated by DNA.11 Interestingly, many genes that are mutated in familial cancers, such as BRCA1, are also hypermethylated or deleted in sporadic cancer cases. In addition, hypermethylation of some genes can promote genomic instability. For example, the mismatch repair gene MLH1 is hypermethylated in colorectal cancers associated with microsatellite instability.12
DNA methylation is intimately associated with histone modifications. Methyl CpG proteins such as MECP2, MBD1, and MBD2, which specifically bind to CpG methylated DNA, are associated with histone deacetylase (HDAC)-containing complexes so that they “erase” the transcription-activating histone acetyl marks. Increasing evidence indicates that patterns of repressive histone methylation, specifically histone H3 lysine 27 methylation established in stem cells, correlates with genes that are commonly hypermethylated in cancer.13
Histone Modifications
Histone Acetylation
Various mechanisms of histone modification may contribute to epigenetic gene regulation. Histone tail acetylation at lysine residues on histones H3 and H4 is associated with transcriptional activation. Acetylation neutralizes the negative charge of DNA and generally renders DNA more accessible to transcription factors. In addition, histone acetylation creates marks that are “read” by chromatin-associated proteins, many of which have evolutionarily conserved domains, termed bromodomains, that selectively interact with acetylated lysines. Experimentally, histone acetylation (and other modifications such as methylation) are readily detected by chromatin immunoprecipitation.
A wide variety of histone acetyltransferases (HATs) have been identified, a number of which have been implicated with aberrant transcriptional activation in cancer (Table 1). The HATs are, in turn, modulated by a number of HDACs, which fall into three general classes: class I HDACs, which are homologous to yeast Rpd3, class II HDACs, which are homologous to yeast Hda1, and class III HDACs, which are distinguished by their dependence on NAD+. HDAC inhibitors are finding increasing clinical applications. As is a general theme with histone-modifying enzymes, a number of nonhistone substrates have been identified, including proteins important for carcinogenesis, such as p53, GATA-1, and E2F1.14,15,16
Table 1.
Epigenetic Regulators Altered in Cancer
| Histone regulator class | Epigenetic regulator | Function | Histone modification | Associated cancer | Alteration in cancer |
|---|---|---|---|---|---|
| Writer | DNMT1 | DNMT | Methyl CpG | Various types | Overexpressed |
| Writer | DNMT3a | DNMT | Methyl CpG | Various types | Overexpressed |
| Writer | DNMT3b | DNMT | Methyl CpG | Various types | Overexpressed |
| Writer | p300 | HAT | Multiple lysines | Leukemia, myelodysplasia | Translocation/inactivating mutation |
| Writer | CBP | HAT | Multiple lysines | Leukemia, myelodysplasia | Translocation/inactivating mutation |
| Writer | MOZ | HAT | Multiple lysines | Leukemia | Translocation |
| Writer | MORF | HAT | Multiple lysines | Leukemia | Translocation |
| Writer | HDAC1-3, 6 | HDAC | General | Various types | Overexpressed |
| Writer | RIZ1 | HMT | H3K9 | Various types | Down-regulation/mutation |
| Writer | EZH2 | HMT | H3K27 | Various types | Overexpressed |
| Writer | MLL1 | HMT | H3K4 | Leukemia, lymphoma | Translocation |
| Writer | SMYD3 | HMT | H3K4 | Colorectal, hepatocellular carcinoma | Overexpressed |
| Writer | DOT1L | HMT | H3K79 | Leukemia | Deregulated recruitment |
| Writer | NSD1 | HMT | H3K36 | Leukemia, hepatocellular carcinoma | Translocation/inactivating mutations |
| Writer | NSD2/MMSET | HMT | H3K36 | Multiple myeloma | Translocation |
| Writer | NSD3 | HMT | H3K36 | Breast cancer | Translocation/overexpressed |
| Eraser | JMJD2C/GASC1 | Histone demethylase | H3K9 | Various types | Overexpressed |
| Reader | HP1 | Methylated histone-binding protein | H3K9 | Breast cancer, melanoma | Down-regulation |
| Reader | ING1-5 | Methylated histone-binding protein | H3K4 | Various types | Down-regulation/mutation |
| Reader | MBD1-4 | Methyl-CpG-binding protein | Methyl CpG | Various types | Overexpressed |
| Reader | MeCP2 | Methyl-CpG-binding protein | Methyl CpG | Various types | Overexpressed |
| Remodeler | INI1 | SWI/SNF complex | Malignant rhabdoid tumor | Inactivating mutations | |
| Remodeler | BRM | SWI/SNF complex | Various types | Inactivating mutations | |
| Remodeler | BRG1 | SWI/SNF complex | Various types | Inactivating mutations | |
| Remodeler | ATRX | SWI/SNF complex | Myelodysplasia | Inactivating mutations |
Histone Methylation
Whereas histone acetylation is highly dynamic, modification of histones by mono-, di-, and trimethylation of lysine residues, both in the histone tail as well as core nucleosomes, is thought to be a more lasting modification that comprises a form of “cellular memory.” Studies of Drosophila have identified two groups of proteins that are associated with either transcriptional maintenance or repression that are termed Trithorax or Polycomb group proteins, respectively. Several Trithorax proteins, most notably the mixed lineage leukemia protein MLL, are histone H3 lysine 4 methyltransferases. In contrast, Polycomb group proteins such as EZH2 have histone H3 lysine 27 methyltransferase activity, which plays important roles in silencing at euchromatic regions as well as in maintenance of heterochromatic regions in association with histone H3 lysine 9 methylation.
Recently, a number of “readers” of methylated histone tails have been identified. One group of these proteins contains chromodomains, which recognize histones methylated on lysine 9 or 27. One possible role for these chromodomain- containing proteins is to target Polycomb repression complexes to sites of transcriptional regulation and DNA replication. The latter has been implicated as a possible means of perpetuating the lysine 27 methyl mark during DNA replication.17
Although it is clear that histone methylation is involved in establishing long-term patterns of gene expression, it is increasingly apparent that this modification is also dynamic. Indeed, a variety of histone demethylases, including LSD1 and the jumonji family of proteins,18,19 have been identified.
Other Histone Modifications
Replacement of modified histones with unmodified or variant histones is another possible mechanism of changing expression patterns.20 Interestingly, proteolytic cleavage of histone tails has also been recognized as a mechanism for “erasing” histone modifications.21 Like HATs, histone methyltransferases (HMTs) have activity on a variety of nonhistone substrates, such as p53, which may have important cancer implications.22
Further adding to the complexity of posttranslational modifications, histones undergo a variety of other modifications, including phosphorylation, sumoylation, and ubiquitination. Histone monoubiquitination (as opposed to polyubiquitination, which is associated with proteosomal degradation) occurs not on tails but on the core histones H2A at lysine 119 (mediated by RING1A) and H2B on lysine 123 (mediated by BRE1/RAD6). These modifications are required for the subsequent methylation at lysine 27 and lysines 4 and 79, respectively.23
Disruption of “Writers,” “Readers,” and “Erasers” of the Histone Code in Cancer
Ample evidence suggests that the “histone code” is deranged in cancer. For example, cancer cells commonly show loss of lysine 16 acetylation and lysine 20 methylation.24 Furthermore, global changes in histone acetylation and methylation are seen in cancer cells when compared with normal cells, and these changes can be used to predict disease outcome in tumors such as prostate cancer.25 For example, differentiation of embryonic stem cells is accompanied by the appearance of large regions of H3K9-dimethylated chromatin (>4 Mb) termed LOCKs. LOCKs are common in differentiated tissues such as liver and brain and have been found to be dramatically reduced in cancer cell lines.26
Heterochomatin Protein 1
In recent years, a number of readers, writers and erasers of the histone code have been implicated in carcinogenesis. Heterochromatin protein 1 (HP1) is a good example of a histone reader that is disrupted in cancer. The HP1 family, which is composed of HP1α (CBX5), HP1β, (CBX1) and HP1γ (CBX3), plays an integral role in maintaining silenced heterochromatin. All three family members specifically bind histone H3 that is methylated on lysine 9 (H3K9), a transcriptionally repressive modification associated with heterochromatin and silenced euchromatin.27 HP1 is targeted to methylated H3K9 via the chromodomain.28 This mark is deposited by several proteins, including the Suppressor of variegation, Enhancer of Zeste and Trithorax (SET) domain HMT SUV39H1.29 HP1 dimerization results in recruitment of other H3K9 HMTs, leading to additional HP1 recruitment.30 HP1 recruitment of Suv4-20 HMTs and the Dnmt3a/3b DNA methyltransferases establishes a complete transcriptionally repressed state.27
HP1γ is localized to silenced euchromatic sites, and HP1α and HP1β are localized to pericentric chromatin. Loss of HP1 function results in kinetochore defects, defective chromosome condensation and segregation, and impaired telomere function. HP1 down-regulation has been noted in metastatic breast cancer, papillary thyroid carcinoma, and medulloblastoma.31,32 Strikingly, overexpression of HP1 in metastatic breast cancer cells decreased invasiveness, whereas knockdown of HP1 in nonmetastatic cells increased invasiveness, suggesting HP1 functions as a metastasis suppressor.33 Frame shift, missense mutations, and epigenetic silencing also contribute to HP1 down-regulation, allowing for cancer progression.31,32 HP1 is also recruited to the cell cycle control gene, cyclin E, indicating a direct link to cell proliferation following HP1 inhibition.34 As noted above, chromosome instability, including aneuploidy and telomere fusion, results from reduction or overexpression of HP1, respectively.31 Thus, down-regulation of HP1 in cancer likely contributes to tumor progression by leading to aneuploidy of other chromosomal abnormalities.
Inhibitor of Growth 1
The inhibitor of growth (ING) protein ING1 is another reader of the histone code that was discovered through a candidate tumor suppressor screen using cDNA from normal and breast cancer cell lines (Figure 2A).35 Subsequent phylogenetic analysis revealed four more members of the ING family, ING2–5.36 Consistent with their role as putative tumor suppressors, ING proteins interact with p53 to induce apoptosis, cellular senescence, and growth arrest.37 ING proteins also associate with a large chromatin-remodeling complex that includes HDAC1, Sin3a, and SAP30,38,39 which functions as a general repressor of transcription.40 The domain required for interaction with Sin3-HDAC is essential for ING-dependent cell cycle arrest.38
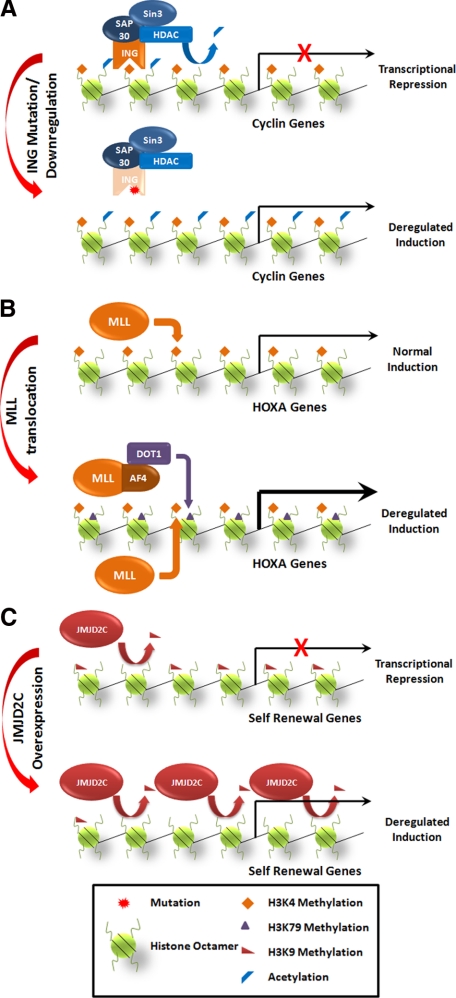
Figure 2.
Disruption of histone readers, writers, and erasers in cancer. A: ING proteins use PHD fingers to recognize the trimethylated histone H3K4. Cell cycle arrest is mediated by the ING protein binding to proliferative genes, such as cyclins, and recruitment of HDAC complexes that deacetylate histone tails, resulting in gene silencing. Inactivating mutations or down-regulation of ING proteins results in deregulated cyclin expression and proliferation. B: MLL is a histone H3K4 methyltransferase that is required for maintenance of HOX gene expression. MLL translocations interact with various partners, including AF4, leading to the aberrant recruitment of the histone H3K79 methyltransferase Dot1. Dot1-mediated methylation at H3K79 results in deregulated expression of HOX genes, which are critical for transformation. Other proteins recruited by MLL fusion proteins that may also play a role in transcriptional activity are not shown. C: JMJD2C normally functions to demethylate H3K9, leading to transcriptional activation. In cancers that overexpress JMJD2C, a global reduction in H3K9 is observed, resulting in demethylation and increased expression of target genes such as self renewal genes, which likely contribute to tumorigenesis.
All ING proteins share a plant homeodomain (PHD), which preferentially binds di- and tri-methylated H3K4.41,42 The PHD finger of ING2 also interacts with phosphatidylinositol-5-phosphate, aiding in ING2 localization to chromatin.43 Mutations detected within the PHD finger result in premature stop codons or disrupted Zn2+ coordination and improper folding of the domain.44 This likely disrupts ING binding to methylated H3K4, resulting in improper regulation of target genes such as p21 and cyclin B1, which are thought to be important for tumorigenesis in cells with ING disruption.45,46 These mutations have been described in breast cancer, melanoma, head and neck and esophageal squamous cell carcinoma.47 Similarly, nuclear localization signal mutations lead to ING exclusion from the nucleus and have been found in brain and breast tumors, as well as melanoma and lymphoblastic leukemia.47 Additionally, ING proteins have been found to be down-regulated by either loss of heterozygosity or promoter hypermethylation in a variety of tumors, including breast, gastric, esophageal, blood, lung, and brain cancers.37,47,48 Reduction of expression of ING1 has been noted in >50% of cases of head and neck cancer and esophageal squamous cell carcinoma and 25% of ovarian cancers.49,50
Mixed Lineage Leukemia
The Mixed Lineage Leukemia (MLL) gene is rearranged in human lymphoid and myeloid acute leukemias (Figure 2B).51 MLL is an example of a histone code writer; MLL has HMT activity specific for histone H3K4 that is mediated by its carboxyl terminal SET domain52,53 Mll knockout mice are embryonic lethal at day E10.5 and show defects of the axial skeleton and hematopoietic system that are accompanied by defects in HOX gene expression and histone H3K4 methylation.54 Translocations involving MLL result in fusion of N-terminal sequences of MLL up to and including a DNA methyltransferase homology (CXXC) domain to one of >60 translocation partners. These translocations consistently delete the more C terminal PHD fingers, which have been shown to inhibit transformation.55,56,57
Despite deletion of the SET domain, MLL fusion proteins potently up-regulate target genes, including HOXA7, HOXA9, and the HOX cofactor MEIS1, which are essential for MLL fusion protein-mediated transformation.58 This transcriptional activation appears to be mediated via recruitment of a complex containing multiple MLL translocation partners, including AF4, AF5q31, and LAF4, in addition to two proteins with enzymatic activity that stimulate transcriptional elongation: CDK9, which together with cyclin T1 or 2 comprises the pTEFb complex, and DOT1L, a histone H3 lysine 79-specific HMT59 that has previously been shown to interact with the MLL translocation partner AF10 (Figure 2B).60 Histone H3 K79 methylation is associated with active transcription.61 Interestingly, MLL rearranged leukemias show abnormally high lysine 79 methylation that is broadly distributed across the HOXA and MEIS1 loci.62 Preliminary experiments suggest that inhibition of DOT1 methyltransferase activity inhibits HOX expression and the growth of cells with MLL rearrangements. Although DOT1 specificity for MLL-rearranged leukemias remains to be established, these data suggest that DOT1 may be a promising therapeutic target.63
Enhancer of Zeste 2
Enhancer of Zeste (EZH2), a member of the Polycomb group of proteins, is another histone code writer that is disrupted in cancer. EZH2 has intrinsic histone H3K27 methyltransferase activity and assembles into a multiprotein complex termed Polycomb repressive complex 2 (PRC2), which consists of EZH2, the WD40 repeat protein EED, and the zinc finger protein SUZ12.64,65 The methylation of histone H3 lysine 27 catalyzed by this complex is recognized by a second Polycomb complex, PRC1, which is primarily composed of HPC, HPH, RING1, and BMI1; PRC1 binds and maintains a state of transcriptional repression.66,67 Collectively, these complexes inhibit expression of a variety of proteins including the HOX genes and thereby antagonize the activity of Trithorax group proteins such as MLL.68 It has been reported that DNMTs and HDACs, such as SIRT1, are recruited by the PRC2 complex and contribute to gene silencing,69 thereby linking two major silencing pathways: histone H3 lysine 27 methylation and DNA methylation. However, the recent report70 that EZH2 down-regulation restores expression of H3K27-targeted genes without affecting DNA methylation raises questions about the significance of DNA methylation in Polycomb-mediated repression.
Up-regulation of EZH2 is seen in a number of tumor types, including lymphomas, prostate cancer, and breast cancer, where the expression level appears to correlate with disease progression.68,71,72,73 EZH2 may contribute to cancer progression by maintaining a stem cell-like phenotype. Overexpression of EZH2 has been shown to prevent exhaustion of hematopoietic stem cells in serially transplanted mice, and ES cell lines cannot be established from Ezh2−/− blastocysts.74,75 Importantly, a causal link between EZH2 and cancer was established when it was shown that overexpression of EZH2 in the B-cell-derived Ramos cell line or multiple myeloma cells caused increased proliferation.71,76 Conversely, differentiation of the promyelocytic HL60 cell line results in down-regulation of EZH2. Furthermore, RNA interference-mediated knockdown of EZH2 causes growth arrest at the G2-M phase in prostate cells and suppression of DNA synthesis in HL60 cells.72,77 Importantly, the HMT activity of EZH2 and the deacetylase activity of EED-recruited HDACs are necessary for EZH2-mediated cell proliferation and target gene repression.72
EZH2 mRNA and protein levels are low in benign prostate and increase progressively from localized to metastatic tumors, suggesting EZH2 could be a useful prognostic indicator as well as a potential therapeutic target.72 Interestingly, EZH2 expression is regulated by micro-RNA-101, which is encoded by a locus that is commonly deleted in prostate cancer. The miR-101 locus is deleted at one or both loci in 37.5% of clinically localized prostate cancer and 66.7% of metastatic prostate tumors, suggesting loss of micro-RNA-101 leads to EZH2 overexpression and cancer progression mediated by deregulated epigenetic mechanisms.78
Jumonji Domain Containing 2C
Jumonji domain containing 2C (JMJD2C), also known as GASC1, is one of a family of three histone demethylases (including JMJD2A and JMJD2B) that is amplified in a variety of cancers and functions as erasers of the histone code (Figure 2C).79 The jumonji domain family is characterized by the presence of the jumonji domain, which is the catalytically active histone demethylase. The jumonji domain in JMJD2C is specific for di- and trimethylated histone H3 lysine 9.80 JMJD2C overexpression dramatically reduces histone H3 lysine 9 methylation, resulting in delocalization of HP-1 and thereby impairing heterochromatin formation.80 Considerable insights into JMJD2 function have come from studies of embryonic stem cell differentiation, which is accompanied by widespread increases in histone H3 lysine 9 methylation. Oct4, one of the key transcription factors involved in maintaining embryonic stem cell self-renewal, regulates JMJD2 expression, which in turn regulates expression of Nanog, another transcription factor critical for stem cell maintenance. Furthermore, depletion of JMJD2C or JMJD2A results in embryonic stem cell differentiation, suggesting that JMJD2 promotes stem cell self-renewal.81
JMJD2 overexpression has been reported in a number of human tumors, including esophageal squamous cell carcinoma, desmoplastic medulloblastoma, and occasional cases of MALT lymphoma, the latter as a result of chromosomal translocation with the IgH locus.82,83,84 Consistent with its role as an oncoprotein, inhibition of JMJD2C expression in esophageal carcinoma or U2OS osteosarcoma cells results in decreased proliferation.80
Chromatin Remodeling
The nucleosome presents a barrier to transcription factor binding as well as transcriptional elongation. A variety of evolutionarily conserved mechanisms exist to overcome this, most notably the SWItch/Sucrose NonFermentable (SWI/SNF) ATP-dependent chromatin remodeling complex.85 Perhaps through promoting chromatin accessibility to both coactivators and repressors through nucleosome displacement or through DNA displacement from nucleosomes, the SWI/SNF complex has both positive or repressive effects on transcription depending on chromatin context.86 Determining the precise role of chromatin remodeling in cancer has been somewhat hampered by the relatively difficult and insensitive testing methods available, which use DNase hypersensitivity and Southern blot analysis for assessing nucleosomal position (phasing) in tumor tissues.
Perhaps the best studied example of chromatin-remodeling enzyme disruptions in cancer is the loss of expression of the INI1 (SNF5/SMARCB1/BAF47), a core component of the SWI/SNF complex, in malignant rhabdoid tumors as well as other primitive undifferentiated pediatric sarcomas. This aggressive tumor of childhood appears to be the result of deregulation of multiple oncogenic pathways, including cyclin D1 up-regulation.87,88 Mutations have also been identified in the BRG1 ATPase of the SWI/SNF complex in a variety of solid tumors, including lung, prostate, pancreas, colorectal, and breast carcinoma, among others.87 BRG1 interacts with Rb, so it has been postulated that BRG1 mutations disrupt the ability of Rb to act as a tumor suppressor (Figure 3).87,89 In addition, constitutional mutations of ATRX, a SNF2 family chromatin remodeling protein, are associated with a variety of developmental abnormalities, including facial dysmorphism, mental retardation, and α thalassemia.90 Acquired mutations of ATRX are associated with the α thalassemia myelodysplastic syndrome. As further evidence of the interplay between different epigenetic modifications, ATRX mutations are also associated with abnormal patterns of DNA methylation.91
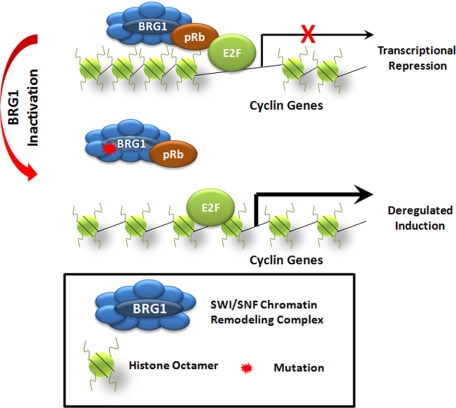
Figure 3.
SWI/SNF-mediated nucleosomal remodeling and transcription in cancer. BRG1 is a DNA-dependent protein that functions in a SWI/SNF complex, which can remodel histones in several ways to make transcriptional start sites more accessible to transcription machinery. For example, SWI/SNF complexes allow histones to “slide” along DNA to expose DNA sequences as well as allow DNA looping away from histones to increase accessibility. BRG1 can also recruit pRb to regulate E2F target genes. Mutations in BRG1 in cancer inhibit the function of SWI/SNF complexes resulting to deregulated transcription.
Implications for Therapy
Success with the development of kinase inhibitors such as imatinib in CML therapy raises hopes that epigenetic regulators may also be attractive therapeutic targets. Currently, two types of epigenetic-based therapies have made their way into clinical use, HDAC inhibitors (HDACi) and inhibitors of DNA methyltransferases.92 Tumor cells generally show higher sensitivity to HDACi than normal cells.93 HDACi have shown particular efficacy against cutaneous T cell lymphomas and one, suberoylanilide hydroxamic acid (vorinostat), has been Food and Drug Administration approved for this application.94 The mechanisms through which HDACi inhibit growth or kill tumor cells, however, remains unclear. Although HDACi can inhibit tumor growth through the up-regulation of the cyclin-dependent kinase inhibitor p21, many other mechanisms of HDACi action have been identified, including inhibiting DNA repair mechanisms and acetylating nonhistone proteins.92 A number of HDAC inhibitors, including valproic acid, vorinostat, depsipeptide, and many others are currently in clinical trials for solid tumors with generally mixed results.
The inhibitors of DNMT in most wide clinical use are nucleoside analogs that get converted to dNTPs and become incorporated into DNA in place of cytosine during DNA replication. Of these, 5-azacytidine received Food and Drug Administration approval for myelodysplastic disorders and leukemia in 200495 and decitabine (5-Aza-2′-deoxcytidine) in 2006.96 These inhibitors, and many others such as zebularine, 5-fluoro-2′-deoxycytidine, and 5,6-dihydro-5-azacytidine, are currently in clinical trials for a wide range of hematological malignancies and for solid tumors, where their efficacy in general has been less.92 In addition, some studies suggest the combination of HDAC and DNA methylation inhibitors for cancer therapy are more effective than either agent alone.97
One of the biggest concerns with current epigenetic regulators is their nonspecific effects. Induction of global hypomethylation, for example, has the potential to activate other oncogenes as well as induce additional genomic instability. HDAC inhibitors have many “off target” effects that contribute to their toxicity. It is likely that additional inhibitors will be developed that target specific HMTs such as EZH2 or DOT1. Alternatively, inhibitors might be developed to inhibit DNA or histone recognition modules such as CXXC or PHD domains.
Another exciting possibility is the use of epigenetic modifiers to induce tumor antigens that can then be used as targets for cancer immunotherapy. Cancer testis antigens as an example are normally expressed in male germ cells. Cancer testis antigens are generally highly immunogenic and, when reactivated by DNA demethylating agents, show promise when combined with cancer immunotherapy.98 Similar approaches are being explored to restore hormone sensitivity such as reinducing immunotherapy, thereby targeting reactivated estrogen receptor re-expression in conjunction with tamoxifen therapy.99
Footnotes
Address reprint requests to Jay L. Hess, Department of Pathology, University of Michigan Medical School, 5240 Medical Sciences 1, 1301 Catherine Road, Ann Arbor MI 48109. E-mail: jayhess@med.umich.edu.
References
- Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–710. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Feinberg A, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Teodoridis JM, Strathdee G, Brown R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Resist Updat. 2004;7:267–278. doi: 10.1016/j.drup.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Höppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46:2917–2922. [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc strain PCC 7120. J Bacteriol. 2008;190:6084–6096. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, Allis CD. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Pérez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- Moss TJ, Wallrath LL. Connections between epigenetic gene silencing and human disease. Mutat Res. 2007;618:163–174. doi: 10.1016/j.mrfmmm.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Allis CD, Chi Chromatin remodeling and cancer—Part I: covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Norwood LE, Moss TJ, Margaryan NV, Cook SL, Wright L, Seftor EA, Hendrix MJ, Kirschmann DA, Wallrath LL. A requirement for dimerization of HP1Hsα in suppression of breast cancer invasion. J Biol Chem. 2006;281:18668–18676. doi: 10.1074/jbc.M512454200. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol. 2005;22:104–116. doi: 10.1093/molbev/msh256. [DOI] [PubMed] [Google Scholar]
- Shi X, Gozani O. The fellowships of the INGs. J Cell Biochem. 2005;96:1127–36. doi: 10.1002/jcb.20625. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Zeremski M, Neznanov N, Li M, Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV, Qin J. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem. 2001;276:8734–8739. doi: 10.1074/jbc.M007664200. [DOI] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Côté J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Sohn H, Xue L, Firestone GL, Bjeldanes LF. 3,3′-Diindolylmethane is a novel mitochondrial H-ATP synthase inhibitor that can induce p21(Cip1/Waf1) expression by induction of oxidative stress in human breast cancer cells. Cancer Res. 2006;66:4880–4887. doi: 10.1158/0008-5472.CAN-05-4162. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Seki N, Ozaki T, Kato M, Kuno T, Nakagawa T, Watanabe K, Miyazaki K, Ohira M, Hayashi S, Hosoda M, Tokita H, Mizuguchi H, Hayakawa T, Todo S, Nakagawara A. Identification of the p33(ING1)-regulated genes that include cyclin B1 and proto-oncogene DEK by using cDNA microarray in a mouse mammary epithelial cell line, NMuMG. Cancer Res. 2002;62:2203–2209. [PubMed] [Google Scholar]
- Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R. The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer. 2008;123:1483–1490. doi: 10.1002/ijc.23790. [DOI] [PubMed] [Google Scholar]
- Coles AH, Jones SN. The ING gene family in the regulation of cell growth and tumorigenesis. J Cell Physiol. 2008;218:45–57. doi: 10.1002/jcp.21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- Shen DH, Chan KY, Khoo US, Ngan HY, Xue WC, Chiu PM, Ip P, Cheung AN. Epigenetic and genetic alterations of p33ING1b in ovarian cancer. Carcinogenesis. 2005;26:855–863. doi: 10.1093/carcin/bgi011. [DOI] [PubMed] [Google Scholar]
- Ziemin-van der Poel S, McCabe NR, Gill HJ, Espinosa R, III, Patel Y, Harden A, Rubinelli P, Smith SD, LeBeau MM, Rowley JD. Identification of a gene MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Giannola D, Udager AM, Hess JL. The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood. 2008;112:4690–4693. doi: 10.1182/blood-2008-01-134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Santillan DA, Koonce M, Wei W, Luo R, Thirman MJ, Zeleznik-Le NJ, Diaz MO. Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res. 2008;68:6199–6207. doi: 10.1158/0008-5472.CAN-07-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis[erratum appears in Cell 2005, 121:809]. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, Kutok JL, Kung AL, Armstrong SA. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multi-protein complex containing the enhancer of Zeste. Protein Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, Willemze R, Otte AP. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol. 2001;112:950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–6280. doi: 10.1038/sj.onc.1208771. [DOI] [PubMed] [Google Scholar]
- Fukuyama T, Otsuka T, Shigematsu H, Uchida N, Arima F, Ohno Y, Iwasaki H, Fukuda T, Niho Y. Proliferative involvement of ENX-1, a putative human polycomb group gene, in haematopoietic cells. Br J Haematol. 2000;108:842–847. doi: 10.1046/j.1365-2141.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of JMJD2 family genes in silico. Int J Oncol. 2004;24:1623–1628. [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrbrecht A, Müller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P, Reifenberger G, Weber RG. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- Italiano A, Attias R, Aurias A, Pérot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23–p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Vinatzer U, Gollinger M, Müllauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2. JMJD2C, and CNN3. Clin Cancer Res. 2008;14:6426–6431. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Kadonaga JT. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, Kalpana GV. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Higgs DR. Molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet. 2000;97:204–212. doi: 10.1002/1096-8628(200023)97:3<204::AID-AJMG1038>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC, Pazdur R. FDA: Approval summary—azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Letendre L. Drug therapy for myelodysplastic syndrome: building evidence for action. Cancer. 2006;106:1650–1652. doi: 10.1002/cncr.21793. [DOI] [PubMed] [Google Scholar]
- Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- Calabrò L, Fonsatti E, Altomonte M, Pezzani L, Colizzi F, Nanni P, Gattei V, Sigalotti L, Maio M. Methylation-regulated expression of cancer testis antigens in primary effusion lymphoma: immunotherapeutic implications. J Cell Physiol. 2005;202:474–477. doi: 10.1002/jcp.20133. [DOI] [PubMed] [Google Scholar]
- Glaser KB. HDAC inhibitors: clinical update and mechanism-based potential. Biochem Pharmacol. 2007;74:659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]