Abstract
The development of stomach cancer is closely associated with chronic inflammation, and the Wnt/β-catenin signaling pathway is activated in most cases of this cancer. High-mobility group A (HMGA) proteins are oncogenic chromatin factors that are primarily expressed not only in undifferentiated tissues but also in various tumors. Here we report that HMGA1 is induced by the Wnt/β-catenin pathway and maintains proliferation of gastric cancer cells. Specific knockdown of HMGA1 resulted in marked reduction of cell growth. The loss of β-catenin or its downstream c-myc decreased HMGA1 expression, whereas Wnt3a treatment increased HMGA1 and c-myc transcripts. Furthermore, Wnt3a-induced expression of HMGA1 was inhibited by c-myc knockdown, suggesting that HMGA1 is a downstream target of the Wnt/β-catenin pathway. Enhanced expression of HMGA1 coexisted with the nuclear accumulation of β-catenin in about 30% of gastric cancer tissues. To visualize the expression of HMGA1 in vivo, transgenic mice expressing endogenous HMGA1 fused to enhanced green fluorescent protein were generated and then crossed with K19-Wnt1/C2mE mice, which develop gastric tumors through activation of both the Wnt and prostaglandin E2 pathways. Expression of HMGA1-enhanced green fluorescent protein was normally detected in the forestomach, along the upper border of the glandular stomach, but its expression was also up-regulated in cancerous glandular stomach. These data suggest that HMGA1 is involved in proliferation and gastric tumor formation via the Wnt/β-catenin pathway.
Gastric cancer is the second leading cause of human cancer deaths worldwide, and it is known to be closely associated with chronic inflammation caused by Helicobacter pylori infection.1,2 This disease is an example of human oncogenesis that is etiologically induced by extrinsic or environmental factors. Despite preventive therapies and numerous efforts to identify premalignant lesions, gastric cancer is often diagnosed at the advanced stages.3,4 It is therefore crucial to understand the molecular basis of gastric tumorigenesis to identify diagnostic and therapeutic targets in this cancer.
High-mobility group A proteins (HMGA1 and HMGA2, formerly HMGI/Y and HMGI/C, respectively) are non-histone, architectural chromatin proteins that participate in various cell regulation activities, including cell growth and proliferation.5,6 HMGA1 and HMGA2 are encoded by two distinct genes, and are characterized by the presence of three DNA-binding motifs, named AT hooks, which preferentially bind stretches of AT-rich DNA sequences.7 HMGA genes are highly expressed during embryonic development, whereas their expression is down-regulated in differentiated cells in adults,8,9 though both HMGA1 and HMGA2 can be induced by mitogenic stimuli.7,10 Notably, HMGA genes are frequently reactivated in many types of human cancer, and the overexpression of HMGA proteins is linked to malignant transformation and progression in human cancers, including gastric cancer.11,12,13,14 In addition to the above reports, our recent study determined that HMGA2 maintains epithelial–mesenchymal transition in human pancreatic adenocarcinomas.15 However, the biological roles of the different HMGA proteins in different cancer phenotypes, and the induction mechanism of oncogenic HMGA genes are largely unknown.
Among the cancer-related signaling pathways, the canonical Wnt pathway, also known as the Wnt/β-catenin pathway, is involved in gastrointestinal carcinogenesis. Wnt ligands engage their receptor complex, stabilize intracellular levels of β-catenin, and allow the nuclear accumulation of β-catenin, together with the transcription factor lymphoid enhancer-binding factor 1/ T cell-specific factor, followed by transcriptional activation of the Wnt/β-catenin target genes such as c-myc and cyclin D.16 In the absence of Wnt, destruction complexes consisting of glycogen synthase kinase-3β, the adenomatous polyposis coli protein, and axin, bind and phosphorylate β-catenin, which is thus targeted for ubiquitination and proteolytic degradation. Constitutive activation of the Wnt/β-catenin pathway can occur due to mutations in the adenomatous polyposis coli, β-catenin, and axin genes during cancer development.17,18,19,20,21,22 The nuclear localization of β-catenin is a hallmark of gastric cancer tissues.23 It has been recently reported that K19-Wnt1/C2mE transgenic mice expressing Wnt1, cyclooxygenase-2 (COX2), and microsomal prostaglandin E synthase-1 in gastric epithelial cells, under the control of the cytokeratin 19 (K19) gene promoter. They develop dysplastic stomach tumors, so providing an animal model of human gastric adenocarcinoma.24 Interestingly, the activation of both Wnt and inflammation pathways was required for cancer development, since either altered pathway alone did not lead to tumor formation. Collectively, these observations suggest that the Wnt/β-catenin pathway is involved in gastric tumorigenesis, although the precise mechanisms remain undetermined.
During our investigations into chromatin factors, we found that HMGA1 is induced by the Wnt/β-catenin pathway and maintains proliferation of gastric cancer cells. Depletion of HMGA1 resulted in reduced cell proliferation. Wnt3a treatment increased HMGA1, as well as c-myc transcripts, and the Wnt3a-induced expression of HMGA1 was inhibited by c-myc knockdown. Overexpression of HMGA1 was consistently correlated with the nuclear accumulation of β-catenin in human gastric cancer tissues. To visualize the Hmga1 protein in vivo, transgenic mice expressing endogenous Hmga1 fused to enhanced green fluorescent protein (EGFP) were generated and crossed with K19-Wnt1/C2mE mice. Expression of Hmga1-EGFP was normally found in the forestomach, along the upper border of the glandular stomach. In contrast, Hmga1-EGFP was up-regulated in cancerously proliferative glandular stomach. Based on the results of the present study, we discuss the role of HMGA1 in gastric tumor formation via the Wnt/β-catenin pathway.
Materials and Methods
Cell Culture and Treatment
AGS, KATO-III, and Panc1 cells (American Type Culture Collection, Manassas, VA), as well as HEK293 cells (Health Science Research Resources, Osaka, Japan) were used. Two gastric cancer cell lines, HSC39 and HSC57, were a gift from Dr. K. Yanagihara and Dr. T. Ushijima (National Cancer Center Research Institute, Tokyo, Japan). The culture conditions were: RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% (v/v) heat-inactivated fetal bovine serum for AGS, HSC39, HSC57, and KATO-III cells; 1:1 mixture of Dulbecco’s modified Eagle’s minimum essential medium and Ham’s F-12 nutrient medium supplemented with 10% fetal bovine serum for Panc1 cells; and low glucose Dulbecco’s modified Eagle’s minimum essential medium supplemented with 10% fetal bovine serum for HEK293 cells. AGS cells (1 × 105/well) were grown in 6-well plates and treated with 100 μmol/L NS-398 (Wako Pure Chemical Industries, Ltd., Osaka, Japan) or 100 μmol/L indomethacin (Wako Pure Chemical Industries, Ltd.) for 48 hours. Secreted Wnt3a was prepared from culture medium of L9 cells stably expressing Wnt3a, which were a gift from Dr. S. Takada (National Institutes of Natural Sciences, Okazaki, Japan). HEK293 cells were treated with 50% Wnt3a-condition medium for 48 hours.
Small Interfering RNA Mediated Knockdown
Small interfering (si)RNA duplexes were designed for targeting mRNAs encoding human HMGA1, HMGA2, β-catenin, and c-myc (Japan Bio Services Co., Ltd., Saitama, Japan), and are listed in Table 1. The selected siRNA sequences were submitted to human genome and Expressed Sequence Tags databases to ensure their target specificities. Validated stealth RNA interference against c-myc and its negative control was obtained from Invitrogen (Carsbad, CA). The siRNAs were transfected into the cells using Oligofectamine RNAiMAX (Invitrogen, Carsbad, CA).
Table 1.
Small Interfering RNAs Used in this Study
| Name | siRNA sequence |
|---|---|
| HMGA1 si-S | 5′-GUGCCAACACCUAAGAGACCUTT-3′ |
| HMGA1 si-AS | 5′-AGGUCUCUUAGGUGUUGGCACTT-3′ |
| HMGA1 si-S2 | 5′-GCAGGAAAAGGACGGCACUTT-3′ |
| HMGA1 si-AS2 | 5′-AGUGCCGUCCUUUUCCUGCTT-3′ |
| HMGA2 si-S | 5′-CCGGUGAGCCCUCUCCUAATT-3′ |
| HMGA2 si-AS | 5′-UUAGGAGAGGGCUCACCGGTT-3′ |
| c-myc si-S | 5′-CUAUGACCUCGACUACGACTT-3′ |
| c-myc si-AS | 5′-GUCGUAGUCGAGGUCAUAGTT-3′ |
| stealth c-myc-S | 5′-UUUCAACUGUUCUCGUCGUUUCCGC-3′ |
| stealth c-myc-AS | 5′-GCGGAAACGACGAGAACAGUUGAAA-3′ |
| β-catenin si-S | 5′-CAGUCUUACCUGGACUCUGTT-3′ |
| β-catenin si-AS | 5′-CAGAGUCCAGGUAAGACUGTT-3′ |
| GL3 si-S | 5′-CUUACGCUGAGUACUUCGATT-3′ |
| GL3 si-AS | 5′-UCGAAGUACUCAGCGUAAGTT-3′ |
Cell Proliferation Analysis
Cell proliferation was assessed by seeding AGS, HSC57, and KATO-III cells (1 × 105/well) into 6-well plates. The cells were transfected with HMGA1, HMGA2, or control siRNAs (50 pmol) on day 0, using oligofectamine RNAiMAX, according to the manufacturer’s protocols. The number of viable cells was counted using a hemocytometer. Data were obtained from three independent experiments.
Reverse Transcription and Quantitative Real-Time PCR
Two micrograms of the total RNAs were treated with DNase I (Roche Diagnostics, Mannheim, Germany) and reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). PCR amplification was then performed using specific primers for the indicated transcripts (Tables 2 and 3). For quantification, real-time PCR analysis was performed using Power SYBR Green PCR Master Mix on an ABI Prism 7500 Sequence Detector (Applied Biosystems). PCR amplification was repeated at least three times from more than three independent experiments. The relative fold induction was quantified using the comparative threshold cycle method, and β-actin was used as a normalization control. Primer sets are listed in Table 3.
Table 2.
Oligonucleotides Used for the PCR
| Name | Primer sequence |
|---|---|
| Human | |
| HMGA1 S | 5′-TGAGTCCCGGGACAGCACTGGTAG-3′ |
| HMGA1 AS | 5′-GCGGCTAAGTGGGATGTTAGCCTTG-3′ |
| HMGA2 S | 5′-CAGGATGAGCGCACGCGGTGAGGGC-3′ |
| HMGA2 AS | 5′-CCATTTCCTAGGTCTGCCTCTTGGC-3′ |
| c-myc S | 5′-TCGTCTCAGAGAAGCTGGCCT-3′ |
| c-myc AS | 5′-CTTTTCCACAGAAACAACATCG-3′ |
| β-catenin S | 5′-CAGTTGCTTGTTCGTGCACAT-3′ |
| β-catenin AS | 5′-CAAGTCCAAGATCAGCAGTCTC-3′ |
| GAPDH S | 5′-GATGCCCCCATGTTCGT-3′ |
| GAPDH AS | 5′-CAGGGGTCTTACTCCTTGGA-3′ |
| Mouse | |
| Hmga1 S | 5′-ATGAGCGAGTCGGGCTCAAAG-3′ |
| Hmga1 AS | 5′-TCACTGCTCCTCCTCAGAG-3′ |
| Gapdh S | 5′-ATCACCATCTTCCAGGAGCGAG-3′ |
| Gapdh AS | 5′-GTTGTCATGGATGACCTTGGCC-3′ |
Table 3.
Oligonucleotides Used for the Quantitative Real-time PCR
| Name | Primer sequence |
|---|---|
| Human | |
| HMGA1 S | 5′-TCCAGGAAGGAAACCAAGG-3′ |
| HMGA1 AS | 5′-AGGACTCCTGCGAGATGC-3′ |
| c-myc S | 5′-TGCTCCATGAGGAGACACC-3′ |
| c-myc AS | 5′-CTTTTCCACAGAAACAACATCG-3′ |
| β-catenin S | 5′-GCTTTCAGTTGAGCTGACCA-3′ |
| β-catenin AS | 5′-CAAGTCCAAGATCAGCAGTCTC-3′ |
| Cyclin D1 S | 5′-GAAGATCGTCGCCACCTG-3′ |
| Cyclin D1 AS | 5′-GACCTCCTCCTCGCACTTCT-3′ |
| YWHAZ S | 5′-AGACGGAAGGTGCTGAGAAA-3′ |
| YWHAZ AS | 5′-TCAAGAACTTTTCCAAAAGAGACA-3′ |
| β-actin S | 5′-CCAACCGCGAGAAGATGA-3′ |
| β-actin AS | 5′-CCAGAGGCGTACAGGGATAG-3′ |
| Mouse | |
| Hmga1 S | 5′-CTCCAGGGAGGAAACCAAG-3′ |
| Hmga1 AS | 5′-CAGAGGACTCCTGGGAGATG-3′ |
| Wnt1 S | 5′-ACAGTAGTGGCCGATGGTG-3′ |
| Wnt1 AS | 5′-CTTGGAATCCGTCAACAGGT-3′ |
| K19 S | 5′-ATGAGATCATGGCCGAGAAG-3′ |
| K19 AS | 5′-GGTGTTCAGCTCCTCAATCC-3′ |
| Ki67 S | 5′-AGGGTAACTCGTGGAACCAA-3′ |
| Ki67 AS | 5′-TTAACTTCTTGGTGCATACAATGTC-3′ |
| β-actin S | 5′-CCAACCGTGAAAAGATGACC-3′ |
| β-actin AS | 5′-CCAGAGGCATACAGGGACAG-3′ |
Plasmids and Luciferase Assay
The human HMGA1 promoter-luciferase construct (a generous gift from Dr. K. Peeters, University of Leuven, Belgium25) was introduced into HEK293 cells, together with phRL-SV40 (1 ng) (Promega, Madison, WI) using Fugene6 (Roche Diagnosics). Luciferase activities were checked 48 hours after transfection using the dual luciferase reporter assay system (Promega). Firefly luciferase activities were normalized to Renilla luciferase activities. Luciferase activities were determined from more than three independent assays.
Generation of Hmga1-EGFP Knock-In Mice
To generate Hmga-1-EGFP knock-in mice, 2.5- and 4-kb fragments containing the Hmga1 gene were amplified by genomic PCR from mouse bacterial artificial chromosome clone RP23-189L19, derived from C57BL/6J mice. The EGFP gene was fused in-frame to the last open reading frame before the Hmga1 translation stop codon in the 5′ homologous arm. A 2.5-kb 5′ arm of homology (EcoRI to BamHI) including exons 3, 4, and 5 before the stop codon fused EGFP gene, and a 4-kb 3′ arm of homology (XbaI/SpeI to MluI) including exon 5 after the stop codon, were cloned into 5′ and 3′ multiple-cloning sites of the pIRES-neo3 vector (Clontech Laboratories, Inc., Mountain View, CA) that lacked a synthetic intron. After sequence confirmation, the construct was linearized using MluI and introduced into wild-type TT2-KTPU8 F1 mouse embryonic stem (ES) cells by electroporation. The transfected ES cells were then cultured in selection medium containing 0.2 mg/ml G418. Southern blot analysis using a probe 5′ to the BamHI site was performed on G418 resistant colonies to identify the ES cells with correct incorporation of the targeting construct into the genome. The gene targeted ES cells were then aggregated with morulae of ICR mice. The aggregated embryos were transferred to pseudopregnant females and allowed to develop to term. The chimeric mice were bred with C57BL/6 wild-type mice, and the resulting pups were screened for the presence of the heterozygous targeted allele. The genotype of the mice was determined by Southern blot analysis and PCR of genomic DNA isolated from the tail or ear. Heterozygous mice were intercrossed to obtain homozygous mice. Hmga1-EGFP/Hmga1-EGFP mice were also crossed with K19-Wnt1/C2mE mice24 or C57BL/6 mice (as a control), to analyze the expression of Hmga1-EGFP in normal tissues and gastric tumors.
Immunohistochemistry
Mouse tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Histological sections were cut at 3 μm. Human stomach tumor tissue arrays (BioChain Institute, Inc., Hayward, CA) or mouse tissue samples were deparaffinized, and antigens were retrieved by autoclaving at 120°C for 15 minutes for β-catenin and HMGA1, in a buffer solution (0.01 M/L sodium citrate [pH 6.0] for β-catenin, 1 mmol/L EDTA/PBS [pH 9.0] for HMGA1). The slides were then incubated in methanol with 0.3% hydrogen peroxide for 30 minutes to block endogenous peroxidase activity. Thereafter, tissue sections were immersed in 0.5% BlockAce (Dainippon Sumitomo Pharma Co., Ltd., Osaka, Japan) in PBS for 30 minutes, covered with primary antibodies, and incubated overnight at 4°C. To detect nuclear β-catenin, mouse monoclonal antibodies for the stabilized (active) form of β-catenin that is dephosphorylated on Ser-37 or Thr-41 (Clone 8E7; Upstate, Charlottesville, VA) were used as the primary antibodies.24. Goat polyclonal HMG-I(Y) antibodies (N-19; Santa Cruz Biotechnology, Inc., CA) were used to detect HMGA1, and rabbit polyclonal GFP antibodies (FL; Santa Cruz Biotechnology, Inc., CA) were used to detect GFP. As the internal positive control, anti-Sp1 antibodies were used (data not shown). Visualization of the immunoreactions was performed using Histofine Simple Stain MAX-PO (Nichirei Bioscience Inc., Tokyo, Japan) and 3,3-diaminobenzidine tetrahydrochloride (Dako, Glostrup, Denmark). The slides were counterstained with hematoxylin and mounted with Malinol (Muto Pure Chemicals Co., Ltd., Tokyo, Japan).
Carcinoma cells with moderate or strong nuclear HMGA1 staining were counted as HMGA1-positive, while cells with weak nuclear staining and/or diffuse cytoplasmic staining were counted as negative. Cells with nuclear β-catenin staining were judged as β-catenin-positive, and those with membrane-associated β-catenin or no β-catenin staining were counted as negative. Positive nuclear staining for HMGA1 or β-catenin was exemplified in adenocarcinoma. HMGA1-positive cells and β-catenin-positive cells were quantitatively assessed by counting carcinoma cells (mean, 233; range, 110 to 450) in the same tissue samples.
To observe fluorescent images, mouse tissues were fixed in 4% paraformaldehyde for 3 hours, incubated in 20% sucrose overnight, and frozen in Tissue-Tek optimal cutting temperature embedding compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan). Embedded frozen tissues were sectioned at 5 μm.
Statistical Analysis
Statistical analyses were performed using JMP 7.0.1 for Windows software (SAS Institute Inc., Cary, NC). Significant differences in real-time PCR quantification were evaluated using two-tailed paired t-tests. The association between HMGA1-positive cells and β-catenin-positive cells was analyzed using Pearson’s correlation coefficient, which varies from a perfect negative correlation (−1) to a perfect positive correlation (+1). Statistical significance was considered at a probability level of 0.05 or less.
Results
HMGA1 Maintains Proliferation of Gastric Cancer Cells in Association with β-Catenin
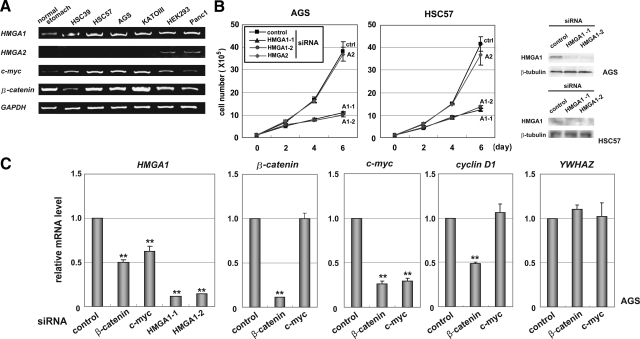
Several reports have shown that HMGA1 is overexpressed in gastric cancer,13,26,27 but the precise role of HMGA1 in the malignant phenotype remains undetermined. To examine the expression status of HMGA genes in human gastric cancer cells, we performed reverse transcription (RT)-PCR (Figure 1A). HMGA1 was expressed in all four gastric cancer cell lines (HSC39, HSC57, AGS, and KATO-III), whereas HMGA2 expression was not detected in any of the gastric cancer cells studied. In normal stomach tissue, HMGA1 was expressed at low levels, while HMGA2 was not detected. As a control, both HMGA1 and HMGA2 transcripts were found in HEK293 and Panc1 cells. To test the effect of HMGA1 on cell proliferation, we used siRNAs against HMGA1 or HMGA2 transcripts, whose knockdown effects have been previously demonstrated at both the RNA and protein levels.15 Western blot analysis showed that HMGA1 was expressed and depleted by the specific knockdown in AGS and HSC57 cells (Figure 1B). Quantitative RT-PCR analysis showed that HMGA1 was equally down-regulated by two distinct siRNAs in AGS cells (Figure 1C), and in HSC57 and KATO-III cells (data not shown). Notably, the knockdown of HMGA1 significantly reduced the growth rate of the gastric cancer cells studied, compared with the use of control and HMGA2 siRNAs. Cell death was assessed by fluorescence activated cell sorting analysis and was not increased under knockdown conditions (data not shown). These results indicate that HMGA1 is involved in maintaining the proliferation of the gastric cancer cells.
Figure 1.
HMGA1 maintains proliferation of gastric cancer cells in association with β-catenin. A: Expression status of transcripts of HMGA1, HMGA2, c-myc, and β-catenin in human gastric cancer cells. RT-PCR was performed using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control. Gastric cancer cell lines are HSC39, HSC57, AGS, and KATO-III. Normal stomach tissue, HEK293 cells, and Panc1 pancreatic cancer cells are used as controls. B: Effect of HMGA1 and HMGA2 knockdown on cell proliferation. The cell numbers were determined on days 0, 2, 4, and 6 after the small interfering (si)RNA-mediated knockdown. The knockdown efficiencies with individual siRNAs are shown in the right panel and in C (left panel). Results were obtained from three independent experiments. Error bars indicate SD. C: Effect of β-catenin and c-myc knockdown on HMGA1 expression. Quantitative RT-PCR was performed in β-catenin and c-myc-knockdown AGS cells. The relative mRNA levels with the use of control siRNAs were normalized to 1. cyclin D1 and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) genes were used as controls. Values are given as means and standard deviations from more than three independent experiments. **P < 0.01 when compared with control cells.
Nuclear localization of β-catenin, a hallmark of Wnt/β-catenin signaling activation, is found in approximately 30% to 50% of gastric cancer tissues and in many kinds of gastric cancer cell lines.23,28 Our expression studies showed that transcripts for β-catenin and c-myc, known as key factors in the Wnt/β-catenin pathway, were expressed in the gastric cancer cells studied (Figure 1A). To assess the effect of the Wnt/β-catenin pathway on HMGA1 expression, quantitative RT-PCR was performed following the selective knockdown of β-catenin or c-myc (Figure 1C). The depletion of either β-catenin or c-myc significantly reduced the expression of HMGA1 (P < 0.01), as did HMGA1 knockdown. In addition, the loss of β-catenin also decreased c-myc and cyclin D1 transcripts. Since the knockdown of β-catenin or c-myc reduced HMGA1 expression by approximately 50%, we examined whether other factors may mediate the transcriptional up-regulation of HMGA1. The use of COX2 inhibitors, NS-398 and indomethacin, decreased their proliferation but did not affect HMGA1 expression in AGS cells, suggesting that COX2 pathway unlikely influences on the expression of HMGA1 (data not shown). These results suggest that HMGA1 is involved in maintaining the growth activities of the gastric cancer cells, by acting as a downstream target of the Wnt/β-catenin pathway.
c-myc Induces HMGA1 Expression in the Wnt/β-Catenin Pathway
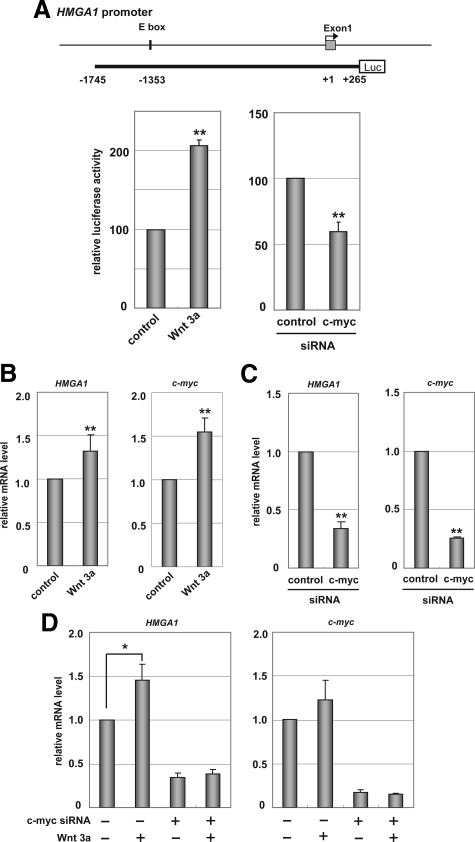
We used HEK293 cells that have no constitutive Wnt/β-catenin activation but accumulate nuclear β-catenin after treatment with Wnt3a29,30 to investigate how the Wnt/β-catenin pathway induced the expression of HMGA1. We used a luciferase reporter assay in HEK293 cells (Figure 2A) to examine the transcriptional role of Wnt/β-catenin in the human HMGA1 gene promoter. An E-box motif in the HMGA1 gene promoter (at position −1353 from the transcriptional start site) has been reported to bind c-myc,10,31 and we therefore used a reporter plasmid containing the promoter region (nucleotides −1745 to + 265) upstream of the luciferase gene. Treatment of the cells with Wnt3a increased the HMGA1 promoter activity by about twofold, while the depletion of c-myc reduced the luciferase activity relative to the control. These data suggest that the HMGA1 promoter can be induced by Wnt3a and c-myc. Under Wnt3a treatment, we then measured the mRNA levels of endogenous HMGA1 in HEK293 cells, using quantitative RT-PCR analysis (Figure 2B). Wnt3a up-regulated the expression of the HMGA1 and c-myc genes (P < 0.01). In addition, c-myc knockdown reduced the expression of HMGA1 (P < 0.01) (Figure 2C), suggesting that c-myc mediates HMGA1 expression. We treated the c-myc knockdown cells with Wnt3a to determine whether c-myc is required for Wnt3a-induced HMGA1 expression (Figure 2D). The depletion of c-myc reduced HMGA1 expression in the control cells and inhibited HMGA1 induction in Wnt3a-treated cells. The reduction of HMGA1 by the knockdown of c-myc was found in Wnt3a-untreated cells as well as Wnt3a-treated cells, suggesting that c-myc maintains the basal levels of HMGA1 expression. Collectively, these data suggest that the expression of HMGA1 is positively controlled at least in part via the Wnt/β-catenin/c-myc pathway.
Figure 2.
The wnt/β-catenin/c-myc pathway induces expression of HMGA1. A: Effect of Wnt3a and c-myc knockdown on human HMGA1 gene promoter. The human HMGA1 promoter (the region from −1745 to + 265 from the transcriptional start site, containing the E box that binds c-myc-luciferase construct25 was introduced into HEK293 cells, together with phRL-SV40 (1 ng). Luciferase activities were checked 48 hours after transfection using the dual luciferase reporter assay. Luciferase activities of the control were normalized to 100. **P < 0.01 when compared with control cells. B–D: Expression of endogenous HMGA1 and c-myc genes. Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis of HMGA1 and c-myc transcripts in HEK293 cells was performed using Wnt3a-treatment (B), c-myc knockdown (C), and a combination of Wnt3a and c-myc knockdown (D). The depletion of c-myc inhibited Wnt3a-induced HMGA1 expression. The relative control mRNA levels were normalized to 1. Values are given as means and standard deviations from more than three independent experiments. Asterisks indicate statistically significant differences compared with control cells (*P < 0.05, **P < 0.01).
Correlation between HMGA1 and β-Catenin Expression in Human Gastric Cancer Tissues
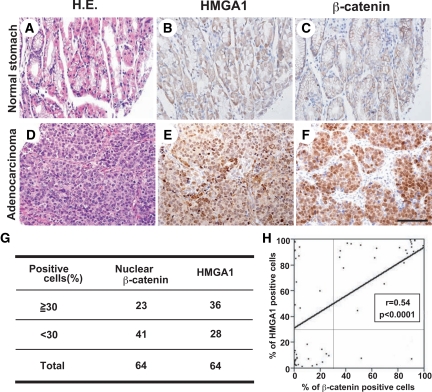
To investigate the involvement of HMGA1 in Wnt/β-catenin signaling in vivo, we examined 64 primary gastric carcinoma tissues, using immunohistochemical techniques (Figure 3). Representative images are shown in Figure 3A, and the data for each tissue are summarized in Supplemental Table 132 at http://ajp.amjpathol.org. Nuclear accumulation of HMGA1 and β-catenin was found in gastric adenocarcinomas (Figure 3D–F), but not in normal stomach tissues (Figure 3A, A–C). High HMGA1 expression was found in 36 out of the 64 gastric carcinomas studied (56.3%), where HMGA1 was densely stained in the nuclei of more than 30% of the cancer cells (Figure 3G). Similarly, β-catenin was highly expressed in 23 out of 64 cancer tissues (35.9%), where nuclear β-catenin was detected in more than 30% of the cancer cells. As in Figure 3D–F, high expression of both HMGA1 and β-catenin was observed in nine carcinoma tissue samples. To assess the correlation between HMGA1 and β-catenin expression, we compared the percentages of HMGA1-positive cells and nuclear β-catenin-positive cells in the same samples (Figure 3H) and analyzed the results using Pearson’s correlation coefficient analysis. There was a positive correlation between HMGA1-positive and β-catenin-positive cells in the gastric cancer tissues (r = 0.54, P < 0.0001). In addition, there was no significant correlation of the expression status of HMGA1 or β-catenin with the histological type of gastric cancers (Supplemental Table 132 at http://ajp.amjpathol.org). These results suggest that enhanced expression of HMGA1 is correlated with Wnt/β-catenin signaling in naturally occurring gastric cancer.
Figure 3.
Correlation between HMGA1 and β-catenin expression in human gastric cancer tissues. Serial sections of normal stomach (A–C) and gastric adenocarcinoma (D–F). Normal and noncancerous tissues showed very weak staining for HMGA1 and nuclear β-catenin. In contrast, HMGA1 and β-catenin were highly accumulated in the nuclei of cancer cells. H&E staining (A, D), immunostaining for HMGA1 (B, E), and nuclear β-catenin (C, F). Scale bar = 100 μm. G: Percentage of HMGA1-positive cells and β-catenin-positive cells in gastric cancer tissues. High expression of HMGA1 was found in 36 out of 64 (56.3%) human primary gastric carcinomas studied, where HMGA1 was densely stained in the nuclei of more than 30% of the cancer cells. Nuclear expression of β-catenin was detected in 23 out of 64 cancer tissues (35.9%), where nuclear β-catenin was stained in more than 30% of the cancer cells. H: Percentage correlations between HMGA1-positive cells and β-catenin-positive cells in gastric cancer tissues. HMGA1-positive cells and β-catenin-positive cells were counted in the same samples (mean, 233; range, 110 to 450). Data were analyzed using Pearson’s correlation coefficient. Supplemental Table S132 at http://ajp.amjpathol.org shows a summary of tumor grade and stage and the immunohistochemical data.
Generation of Hmga1-EGFP Knock-In Mice
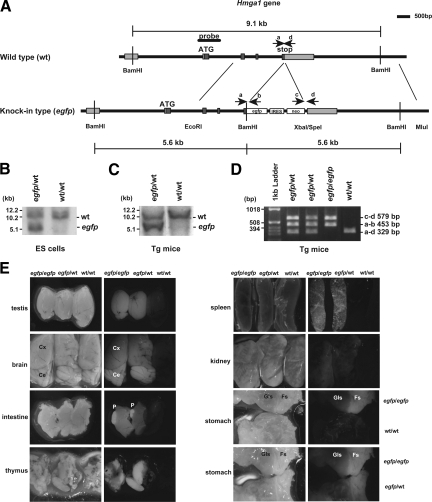
To visualize the expression of Hmga1 in vivo, we generated knock-in mice harboring the Hmga1 gene fused to the EGFP gene (Hmga1-EGFP). The mouse Hmga1 gene has five exons (Figure 4A), and EGFP-IRES-neo was inserted into exon 5 of the Hmga1 gene in-frame, together with a deletion of the stop codon of the gene, through homologous recombination. The targeted knock-in allele resulted in expression of the Hmga1-EGFP fusion gene, driven by the endogenous promoter. After mouse ES cells were transfected with the targeting vector and selected by EGFP-positive and neomycin selection, we confirmed the occurrence of the expected homologous recombination in the cells by Southern blot analysis (Figure 4B). We obtained heterozygous mice (Hmga1-EGFP/wild-type), and then homozygous mice (Hmga1-EGFP/Hmga1-EGFP), as indicated in Figures 4C and 4D. A fluorescent macroscopic analysis of adult mice revealed that Hmga1-EGFP was markedly expressed in testis, cerebrum (Cx), cerebellum (Ce), Payer’s patch (P), thymus, and spleen (Figure 4E). Low levels of Hmga1-EGFP were found in the kidney and liver (data not shown). In the stomach, it was of interest that Hmga1-EGFP was highly detected in the forestomach (Fs), but not in the glandular stomach (Gls), which is homologous to human stomach tissue. These observations not only agreed with a previous report on Hmga1 mRNA levels in tissues,33 but also determined the distribution of Hmga1 in specific parts of living tissues and organs.
Figure 4.
Characterization of Hmga1-EGFP knock-in mice. A: Generation of Hmga1-EGFP knock-in mice. Schematic representation of the mouse Hmga1 gene locus and targeting construct are shown. In the knock-in type allele (EGFP), a 2.5-kb 5′ arm of homology (EcoRI to BamHI) containing exons 3, 4, and 5 before the stop codon was fused to the EGFP gene, IRES and neo sequences linked to a 4-kb 3′ arm of homology (XbaI/SpeI to MluI) containing exon 5 after the stop codon. B: Southern blot analysis using the indicated probe was performed to identify the embryonic stem (ES) cells that had correctly incorporated the targeting construct into the genome. C, D: The genotype of the Hmga1-EGFP knock-in mice was determined by Southern blot analysis (C) and polymerase chain reaction (D) of genomic DNAs using specific primers (a: 5′-TCATCCCCCTTTTGCAGAGG-3′, b: 5′-CGCTCCTGGACGTAGCCTTC-3′, c: 5′-TACCTGCCCATTCGACCACC-3′, d: 5′-ACAGGGACTGAGCCGAATCC-3′). E: Expression of Hmga1-EGFP protein in the knock-in mice. Fluorescent macroscopic analysis demonstrated the expression of Hmga1-EGFP in testis, cerebrum (Cx), cerebellum (Ce), Payer’s patch (P), thymus, spleen, kidney and forestomach (Fs), but not in kidney and glandular stomach (Gls). Homozygous (EGFP/EGFP), heterozygous (EGFP/wt), and wild-type (w/w) mice are indicated.
Tumor-Associated Expression of HMGA1 in K19-Wnt1/C2mE/Hmga1-EGFP Mice
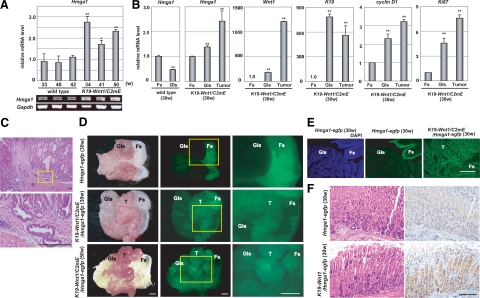
K19-Wnt1/C2mE transgenic mice have been reported to simultaneously overexpress Wnt1, COX-2, and prostaglandin E synthase-1 in gastric epithelial cells under the control of the K19 gene promoter, which is transcriptionally active in the gastric epithelium,24 resulting in the development of dysplastic gastric tumors at the upper glandular stomach, with similar histology to human gastric adenocarcinomas. We examined the expression status of Hmga1 in K19-Wnt1/C2mE mice using RT-PCR and found that it was highly expressed in stomach tumors in these mice, compared with age-matched and non-cancerous glandular stomach controls (*P < 0.05 and **P < 0.01) (Figure 5A). We also determined the levels of Hmga1 expression in the forestomach, glandular stomach, and tumors, using quantitative RT-PCR (Figure 5B). The mRNA levels of Hmga1 in the forestomach showed no significant differences between wild-type and K19-Wnt1/C2mE mice, and were normalized to 1, based on more than three independent experiments. In wild-type mice, the expression levels of Hmga1 in the glandular stomach were about 50% lower than those in the forestomach. In contrast, the expression of Hmga1 was increased in the glandular stomach in K19-Wnt1/C2mE mice (P < 0.01). Moreover, Hmga1 expression was markedly increased in the tumor tissues (P < 0.01). In addition to Hmga1 induction, Wnt1 and cytokeratin 19 (K19) were also highly expressed in the glandular stomach and the tumors (P < 0.01). The expression level of endogenous Wnt1, which was detected by the 3′ untranslated region of the mRNAs, did not increase in K19-Wnt1/C2mE mice (data not shown). Further, the expression of cyclin D1 and the proliferation marker Ki-67 significantly increased in the glandular stomach and the tumors (P < 0.01), suggesting the hyperproliferative state in these tissues. Nuclear localization of β-catenin was mostly found in the tumors (data not shown, and24). Furthermore, tumor formation and invasion into the smooth muscle layers was observed histologically in K19-Wnt1/C2mE mice at 50 weeks of age (Figure 5C).
Figure 5.
Tumor-associated expression of Hmga1 in K19-Wnt1/C2mE/Hmga1-EGFP mice. A: Expression of Hmga1 in mouse stomach tissues. RT-PCR analysis of Hmga1 was performed using stomach and tumors from age-matched wild-type and K19-Wnt1/C2mE mice. The relative mRNA levels in wild-type mice (33w) were normalized to 1. *P < 0.05, **P < 0.01 when compared with control cells. B: Expression of Hmga1 in the forestomach, glandular stomach, and tumor. Using quantitative RT-PCR, the mRNA levels of Hmga1 in the forestomach (Fs) were normalized to 1. Expression of Wnt1, cytokeratin 19 (K19), cyclin D1, and Ki-67 was also shown. **P < 0.01 when compared with control cells. C: H&E staining of stomach tumors in K19-Wnt1/C2mE mice at 50 weeks of age. The dysplastic tumors invaded into the smooth muscle layers. Scale bar = 100 μm. D: Induced expression of Hmga1-EGFP in tumor tissues. Fluorescent macroscopic analysis of the stomach tissues from Hmga1-EGFP mice at 30 weeks of age, and K19-Wnt1/C2mE/Hmga1-EGFP mice at 30 and 50 weeks of age was performed. Scale bar = 1 mm. The indicated region is magnified in the right panel. E: Expression of Hmga1-EGFP in the forestomach close to the glandular stomach and in most tumor cells. Frozen samples of the stomach from Hmga1-EGFP mice and K19-Wnt1/C2mE/Hmga1-EGFP mice were examined by fluorescence microscopy. Scale bar = 100 μm. F: Expression of Hmga1-EGFP in non-cancerous glandular stomach from K19-Wnt1/Hmga1-EGFP mice. H&E staining (left) and immunostaining of Hmga1-EGFP (right) of the glandular stomach from Hmga1-EGFP mice and K19-Wnt1/Hmga1-EGFP mice at 30 weeks of age. Scale bar = 100 μm.
To visualize Hmga1 expression in vivo, we mated K19-Wnt1/C2mE mice with Hmga1-EGFP mice and investigated whether the development of gastric tumors was related to Hmga1 expression in the K19-Wnt1/C2mE mice using fluorescent macroscopic analysis (Figure 5D). As in the Hmga1-EGFP mice, Hmga1-EGFP expression was found in the forestomach, rather than the glandular stomach, in the K19-Wnt1/C2mE/Hmga1-EGFP mice. In contrast, tumor tissues raised in the glandular stomach of the K19-Wnt1/C2mE/Hmga1-EGFP mice showed increased expression of Hmga1-EGFP at 30 and 50 weeks of age. These observations demonstrated the oncogenic induction of Hmga1 in vivo, and were consistent with the mRNA levels of Hmga1 found in the forestomach, glandular stomach and tumor tissues (Figure 5B). With fluorescence microscopy using sliced tissues, we further analyzed the expression of Hmga1-EGFP at the cellular level (Figure 5E). Hmga1-EGFP signals, as well as 4,6-diamidino-2-phenylindole-stained nuclei, were detected in the forestomach, especially along the upper border of the glandular stomach, in Hmga1-EGFP mice at 30 weeks of age, while most of tumor cells emerging in the upper glandular stomach showed Hmga1-EGFP expression in K19-Wnt1/C2mE/Hmga1-EGFP mice (See Discussion).
To confirm the induction of HMGA1 by the Wnt/β-catenin pathway in vivo, we finally examined the expression of Hmga1-EGFP in K19-Wnt1/Hmga1-EGFP mice with K19 promoter-induced expression of Wnt1 alone (Figure 5F). K19-Wnt1 mice had the expansion of undifferentiated progenitor cell population in the glandular stomach but did not show the dysplastic tumors in stomach.24 An immunohistochemical analysis using anti-GFP antibodies demonstrated enhanced expression of Hmga1-EGFP at the middle and lower fundic glands in the glandular stomach of these mice. Collectively, these results suggest that HMGA1 expression is induced by the Wnt/β-catenin pathway and maintains the proliferation of gastric tumor cells.
Discussion
Although gastric cancer is one of the most common malignancies, the molecular basis of its oncogenesis remains to be elucidated. The present study revealed that activation of the Wnt/β-catenin pathway induces HMGA1 expression through c-myc, resulting in the maintenance of proliferation of gastric cancer cells. Our data indicate that: (i) HMGA1 promotes proliferation of gastric cancer cells, (ii) Wnt/β-catenin signaling induces expression of HMGA1, depending on c-myc, (iii) a correlation between HMGA1-positive cells and β-catenin-positive cells exists in human gastric cancer tissues, (iv) Hmga1-EGFP knock-in mice visualize endogenous Hmga1 expression in the forestomach, rather than the glandular stomach, and (v) K19-Wnt1/C2mE/Hmga1-EGFP mice show that the expression of Hmga1 coexists with high levels of Wnt1 in cancerous glandular stomach. These findings suggest that HMGA1 expression plays an important role in the maintenance of proliferative activities and tumor formation in gastric cancer.
Previous studies reported that HMGA1 was overexpressed in a broad range of human cancers, including gastric cancer,13,26,27 and that it correlated with the occurrence of metastasis and poor prognoses.11,12,14 We showed that HMGA1, but not HMGA2, was expressed in the gastric cancer cells studied, and that the depletion of HMGA1 significantly reduced the growth of these cells, suggesting that HMGA1 is involved in their proliferation. Despite the increasing evidence implicating HMGA1 in cancer development and progression, the molecular mechanisms of HMGA1 reactivation in malignant changes remain undetermined. Several factors responsible for the induction of HMGA1 expression have been identified, including serum, epidermal growth factor, fibroblast growth factor, hypoxia, and oncogenic Ras, in addition to transcription factors such as AP-1, c-myc and N-myc, which directly target the HMGA1 promoter.7,10,25,33,34 Our data first demonstrate that the Wnt/β-catenin pathway is linked to HMGA1 induction, leading to proliferation, in human gastric cancer. The Wnt/β-catenin pathway positively controls HMGA1 gene at least in part via c-myc. In addition, other factors may mediate the transcriptional up-regulation of HMGA1. As shown in Figure 2D, c-myc was involved in maintaining HMGA1 expression in the absence of Wnt3a. The high expression of HMGA1 in some patients completely lacked nuclear β-catenin expression in the gastric cancers (Figure 3C). For these reasons, we checked whether the COX2 signaling is involved in HMGA1 regulation. However, the use of COX2 inhibitors had little effect on HMGA1 expression in gastric cancer cells (data not shown). On the other hand, we recently reported that HMGA2, but not HMGA1, is involved in the maintenance of oncogenic RAS-induced epithelial–mesenchymal transition in human pancreatic cancer cells.15 This may be related with the fact that HMGA2 is primarily expressed in undifferentiated tissues of mesenchymal origin. Specific knockdown of HMGA2 inhibited proliferation of pancreatic cancer cells, notably leading to a transition to the epithelial state by up-regulation of E-cadherin. HMGA2 was induced by the oncogenic RAS pathway in pancreatic cancer, while this is not the case of gastric cancer. Thus HMGA proteins are likely to be induced by oncogenic pathways and contribute to malignant phenotypes in a cancer-specific manner.
The functional activation of Wnt/β-catenin may be responsible for gastrointestinal tumorigenesis,35,36,37 where c-myc and cyclin D are key downstream effectors of the canonical Wnt pathway. Mutations of the adenomatous polyposis coli, axin, and β-catenin genes are common in colorectal cancer, and alterations of the E-cadherin gene occur in familial gastric cancer.38 To demonstrate that HMGA1 is a downstream factor in the Wnt/β-catenin/c-myc pathway in gastric cancer cells, we showed that HMGA1 was induced by Wnt3a in proliferating cells, and that there was a close correlation between the expression of HMGA1 and β-catenin in gastric cancer tissues. In K19-Wnt1/Hmga1-EGFP mice, Hmga1 expression in the proliferative glandular stomach also increased during the overexpression of Wnt1 alone. Thus the expression of HMGA1 via the Wnt/β-catenin pathway may be one of the common mechanisms in oncogenesis. Since both Wnt/β-catenin and HMGA1 are actively expressed during organogenesis, these proteins would have an essential role in gut development, as well as tumorigenesis.
HMGA1 has previously been reported to be expressed during embryogenesis, whereas it has shown negligible expression in normal adult tissues.7 Our investigations using Hmga1-EGFP mice, however, detected Hmga1 in specific adult tissues, including the testis, brain, Payer’s patch, thymus, spleen, and forestomach, together with the very low expression in the kidney and liver. This is the first mouse model that can visualize the expression of endogenous Hmga1 in living tissues and organs. Although dominantly expressed during embryogenesis, this protein may retain a biological function during the postnatal and adult stages. Indeed, it was reported that HMGA1 is required for T cell differentiation through the regulation of interleukin-2 and interleukin-2 receptor α-chain expression,39,40,41 which may be related to its expression in lymphoid tissues in Hmga1-EGFP knock- in mice.
In this study, Hmga1-EGFP was densely detected in the forestomach, especially along the upper boundary of the glandular stomach, in Hmga1-EGFP mice. Previous and our studies showed that K19-Wnt1/C2mE mice usually develop dysplastic tumors in the proximal glandular stomach, close to the boundary with the forestomach.24 There is the possibility that a cancer-initiated microenvironment is present in the proximal glandular stomach near the forestomach, where epithelial cells and the surrounding interstitial cells substantially transit and are exposed to the gastric acid. Since Hmga1 inhibits the retinoblastoma protein, leading to the activation of E2F-target genes,42 Hmga1 may be likely implicated in the tumor development. Epithelial cells in the glandular stomach were also reported to exhibit intermediate characteristics between those of the forestomach and the duodenum in response to growth factors.43 Hmga1-EGFP knock-in mice will prove useful for further investigations into the tissue-specific function of Hmga1 and the role of this protein in cancer and stem cell biology.
Supplementary Material
Acknowledgments
We thank Dr. Takaya Ichimura (Graduate School of Medical Sciences, Kumamoto University) for immunohistochemistry, Dr. Shinji Takada (National Institutes of Natural Sciences, Japan) for the L9 cells stably expressing Wnt3a, Dr. Kristel Peeters (University of Leuven, Belgium) for the human HMGA1 promoter-luciferase construct, and members of our laboratory for helpful discussions.
Footnotes
Address reprint requests to Mitsuyoshi Nakao, M.D., Ph.D., Department of Medical Cell Biology, Institute of Molecular Embryology and Genetics, Kumamoto University, 2-2-1 Honjo, Kumamoto 860-0811, Japan. E-mail: mnakao@gpo.kumamoto-u.ac.jp.
Supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (M.N. and S.W.), and by a Grant-in-Aid for Global Center of Excellence, “Cell Fate Regulation Research and Education Unit,” Kumamoto University.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121–125. doi: 10.1016/s1535-6108(04)00033-9. [DOI] [PubMed] [Google Scholar]
- Kapadia CR. Gastric atrophy, metaplasia, and dysplasia: a clinical perspective. J Clin Gastroenterol. 2003;36:S29–S36. doi: 10.1097/00004836-200305001-00006. [DOI] [PubMed] [Google Scholar]
- Clouston AD. Timely topic: premalignant lesions associated with adenocarcinoma of the upper gastrointestinal tract. Pathology. 2001;33:271–277. doi: 10.1080/00313020120070830. [DOI] [PubMed] [Google Scholar]
- Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A, Manfioletti G, Giancotti V. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 2004;574:1–8. doi: 10.1016/j.febslet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti V, Santoro M, Simeone A, Fusco A. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–2446. [PubMed] [Google Scholar]
- Wood LJ, Mukherjee M, Dolde CE, Xu Y, Maher JF, Bunton TE, Williams JB, Resar LM. HMG-I/Y, a new c-myc target gene and potential oncogene. Mol Cell Biol. 2000;20:5490–5502. doi: 10.1128/mcb.20.15.5490-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N, Watanabe T, Masaki T, Mori T, Sugiyama M, Uchimura H, Fujioka Y, Chiappetta G, Fusco A, Atomi Y. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000;60:3117–3122. [PubMed] [Google Scholar]
- Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21:575–594. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam ES, Kim DH, Cho SJ, Chae SW, Kim HY, Kim SM, Han JJ, Shin HS, Park YE. Expression of HMGI(Y) associated with malignant phenotype of human gastric tissue. Histopathology. 2003;42:466–471. doi: 10.1046/j.1365-2559.2003.01618.x. [DOI] [PubMed] [Google Scholar]
- Evans A, Lennard TW, Davies BR. High-mobility group protein 1(Y): metastasis-associated or metastasis-inducing? J Surg Oncol. 2004;88:86–99. doi: 10.1002/jso.20136. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol. 2009;174:854–868. doi: 10.2353/ajpath.2009.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, Sasaki Y, Imaoka S, Murata M, Shimano T, Yamaoka Y, Nakamura Y. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- Liu W, Dong X, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, Halling KC, Cunningham JM, Boardman LA, Qian C, Christensen E, Schmidt SS, Roche PC, Smith DI, Thibodeau SN. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating β-catenin/TCF signalling. Nat Genet. 2000;26:146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- Clements WM, Wang J, Sarnaik A, Kim OJ, MacDonald J, Fenoglio-Preiser C, Groden J, Lowy AM. β-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–3506. [PubMed] [Google Scholar]
- Oshima H, Matsunaga A, Fujimura T, Tsukamoto T, Taketo MM, Oshima M. Carcinogenesis in mouse stomach by simultaneous activation of the Wnt signaling and prostaglandin E2 pathway. Gastroenterology. 2006;131:1086–1095. doi: 10.1053/j.gastro.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Cleynen I, Huysmans C, Sasazuki T, Shirasawa S, Van de Ven W, Peeters K. Transcriptional control of the human high mobility group A1 gene: basal and oncogenic Ras-regulated expression. Cancer Res. 2007;67:4620–4629. doi: 10.1158/0008-5472.CAN-06-4325. [DOI] [PubMed] [Google Scholar]
- Xiang YY, Wang DY, Tanaka M, Suzuki M, Kiyokawa E, Igarashi H, Naito Y, Shen Q, Sugimura H. Expression of high-mobility group-1 mRNA in human gastrointestinal adenocarcinoma and corresponding non-cancerous mucosa. Int J Cancer. 1997;74:1–6. doi: 10.1002/(sici)1097-0215(19970220)74:1<1::aid-ijc1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Lee S, Baek M, Yang H, Bang YJ, Kim WH, Ha JH, Kim DK, Jeoung DI. Identification of genes differentially expressed between gastric cancers and normal gastric mucosa with cDNA microarrays. Cancer Lett. 2002;184:197–206. doi: 10.1016/s0304-3835(02)00197-0. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Ijichi H, Kato N, Kanai F, Masaki T, Rengifo W, Okamoto M, Matsumura M, Kawabe T, Shiratori Y, Omata M. Analysis of the β-catenin/T cell factor signaling pathway in 36 gastrointestinal and liver cancer cells. Jpn J Cancer Res. 2002;93:1213–1220. doi: 10.1111/j.1349-7006.2002.tb01226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S, Michiue T, Asashima M, Kikuchi A. Casein kinase Iε enhances the binding of Dvl-1 to Frat-1 and is essential for Wnt-3a-induced accumulation of β-catenin. J Biol Chem. 2003;278:14066–14073. doi: 10.1074/jbc.M213265200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Mazor M, Kawano Y, Walker MM, Leung HY, Armstrong K, Waxman J, Kypta RM. Analysis of Wnt gene expression in prostate cancer: mutual inhibition by WNT11 and the androgen receptor. Cancer Res. 2004;64:7918–7926. doi: 10.1158/0008-5472.CAN-04-2704. [DOI] [PubMed] [Google Scholar]
- Pedulla ML, Treff NR, Resar LM, Reeves R. Sequence and analysis of the murine Hmgiy (Hmga1) gene locus. Gene. 2001;271:51–58. doi: 10.1016/s0378-1119(01)00500-5. [DOI] [PubMed] [Google Scholar]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- Liu J, Schiltz JF, Shah PC, Benson KF, Chada KK. Genomic structure and expression of the murine Hmgi (y) gene. Gene. 2000;246:197–207. doi: 10.1016/s0378-1119(00)00073-1. [DOI] [PubMed] [Google Scholar]
- Giannini G, Cerignoli F, Mellone M, Massimi I, Ambrosi C, Rinaldi C, Dominici C, Frati L, Screpanti I, Gulino A. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res. 2005;65:8308–8316. doi: 10.1158/0008-5472.CAN-05-0607. [DOI] [PubMed] [Google Scholar]
- Taketo MM. Wnt signaling and gastrointestinal tumorigenesis in mouse models. Oncogene. 2006;25:7522–7530. doi: 10.1038/sj.onc.1210058. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- Mishra L, Shetty K, Tang Y, Stuart A, Byers SW. The role of TGF-β and Wnt signaling in gastrointestinal stem cells and cancer. Oncogene. 2005;24:5775–5789. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- Himes SR, Reeves R, Attema J, Nissen M, Li Y, Shannon MF. The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J Immunol. 2000;164:3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- John S, Reeves RB, Lin JX, Child R, Leiden JM, Thompson CB, Leonard WJ. Regulation of cell-type-specific interleukin-2 receptor α-chain gene expression: potential role of physical interactions between Elf-1. HMG-I(Y), and NF-κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista S, Pentimalli F, Baldassarre G, Fedele M, Fidanza V, Croce CM, Fusco A. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J. 2003;17:1496–1498. doi: 10.1096/fj.02-0977fje. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Watanabe S, Tei S, Saitoh N, Kuratsu J, Nakao M. The high mobility group protein HMGA1 inhibits retinoblastoma protein-mediated cellular G0 arrest. Cancer Sci. 2007;98:1893–1901. doi: 10.1111/j.1349-7006.2007.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamachi H, Ichinose M, Tsukada S, Kurokawa K, Shiokawa K, Miki K, Takeuchi S. Growth of fetal rat gastro-intestinal epithelial cells is region-specifically controlled by growth factors and substrata in primary culture. Develop Growth Differ. 1995;37:11–19. doi: 10.1046/j.1440-169X.1995.00002.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.