Abstract
Telomeres are specialized heterochromatin at the ends of linear chromosomes. Telomeres are crucial for maintaining genome stability and play important roles in cellular senescence and tumor biology. Six core proteins—TRF1, TRF2, TIN2, POT1, TPP1 and Rap1 (termed the telosome or shelterin complex)—regulate telomere structure and function. One of these proteins, TIN2, regulates telomere length and structure indirectly by interacting with TRF1, TRF2 and TPP1, but no direct function has been attributed to TIN2. Here we present evidence for a TIN2 isoform (TIN2L) that differs from the originally described TIN2 isoform (TIN2S) in two ways: TIN2L contains an additional 97 amino acids, and TIN2L associates strongly with the nuclear matrix. Stringent salt and detergent conditions failed to extract TIN2L from the nuclear matrix, despite removing other telomere components, including TIN2S. In human mammary epithelial cells, each isoform showed a distinct nuclear distribution both as a function of cell cycle position and telomere length. Our results suggest a dual role for TIN2 in mediating the function of the shelterin complex and tethering telomeres to the nuclear matrix.
Keywords: alternative splicing, human fibroblasts, human mammary epithelial cells, telomerase, TRF1, TRF2, tankyrase
Introduction
Since McClintock’s seminal discovery of the breakage-fusion-bridge cycle,1 telomeres were recognized as crucial structures that cap and protect the ends of chromosomes and therefore prevent genomic instability. Human telomeres are composed of tandem TTAGGG repeats and several associated proteins, and range in size from 1–2 kb in some cancer cells to approximately 15 kb in the germ line. In most normal human somatic cells, the inability of DNA polymerases to completely replicate linear DNA segments causes telomeres to undergo progressive shortening with each round of DNA replication. When critically short telomeres fail to function, they trigger a DNA damage response and permanent cell cycle arrest termed cellular senescence.2 Because genomic instability put cells at risk for neoplastic transformation, and because many potentially oncogenic events induce a senescence response, cellular senescence is considered a tumor protective mechanism.3,4 In addition, because senescent cells can disrupt normal tissue microenvironments,5 the accumulation of senescent cells is thought to contribute to aging and age-related diseases such as atherosclerotic lesions,6 susceptibility to infection7 and ironically, late-life cancer.8 The senescence response is likely an example of antagonistic pleiotropy, the evolutionary theory that explains why some phenotypes that evolved to promote early life fitness (e.g., protection from cancer) can be detrimental later in life.9 Thus, telomeres play central roles in many important biological processes and diseases, ranging from cell cycle control and DNA damage responses to aging and cancer.
Mammalian telomeres form protective t-loop structures in which the single-stranded overhang invades the duplex,10 and are non-randomly distributed in the nucleus.11–13 The telomeric structure is regulated by a six-protein complex termed the telosome or shelterin. 14,15 Three of these proteins, TRF1, TRF2 and POT1, bind either single- or double-stranded telomeric DNA directly,16,17 whereas the other three, TIN2, TPP1/PTOP and RAP1 indirectly regulate telomeres through their interactions with TRF1, TRF2 and POT1.18–21 In budding yeast, telomeres are anchored within the nucleus; specifically, they are anchored to the nuclear envelope through Ku-and Sir-mediated pathways.22 Mammalian telomeres are also anchored within the nucleus,23,24 but how they are tethered to the nuclear matrix remains unclear.
We identified TIN2 as a TRF1-interacting protein.25 Subsequent studies showed that TIN2 also interacts with TRF2 and TPP1.15,19,25,26 TIN2 is an essential protein, as germ line inactivation in the mouse causes early embryonic lethality.27 It is likely a crucial regulator of a compact telomere structure, as it strongly facilitates interactions among telomeric DNA tracts,28 and the expression of TIN2 proteins that lack specific interaction domains increases telomere length25 or induces a robust DNA damage response.19 TIN2 also regulates tankyrase, a poly-ADP ribose polymerase that is important for proper telomere separation during mitosis.29,30 Here, we describe another potential function for TIN2: mediating interaction of the telomeric complex with the nuclear matrix. A portion of TRF1 was shown to co-fractionate with the nuclear matrix, but TRF1 does not appear to interact directly with nuclear lamina components.24,31 We identify here a new isoform of TIN2, which we term TIN2L because it contains an additional 97 amino acids. TIN2L retains its ability to bind TRF1, TRF2 and tankyrase. However, in contrast to the isoform we first identified, which we now term TIN2S, TIN2L strongly associates with the nuclear matrix. We hypothesize that TIN2L serves as a central anchoring protein for the organization and attachment of telomeres to the nuclear matrix.
Results
A comparison between the sequences encoding human (hTIN2) and mouse (mTIN2) proteins revealed a striking (85%) similarity between the 3' untranslated region (UTR) in the human gene (NM_012461.1) and C-terminal coding regions in mTIN2 (NM_145705.2). We previously reported that TIN2 protein sequences were highly (>95%) conserved between two murine species (M. musculus and M. spretus), but hTIN2 and mTIN2 sequences were, overall, only 67% conserved.32 Further, Swiss-Prot (Q9BSI4-1 and -2) identifies two human TIN2 proteins: one identical to the amino acid (aa) sequence reported by us,25 which now term hTIN2S, and a second that is predicted to contain additional aa amounting to an additional 10 kDa in mass (termed here hTIN2L).
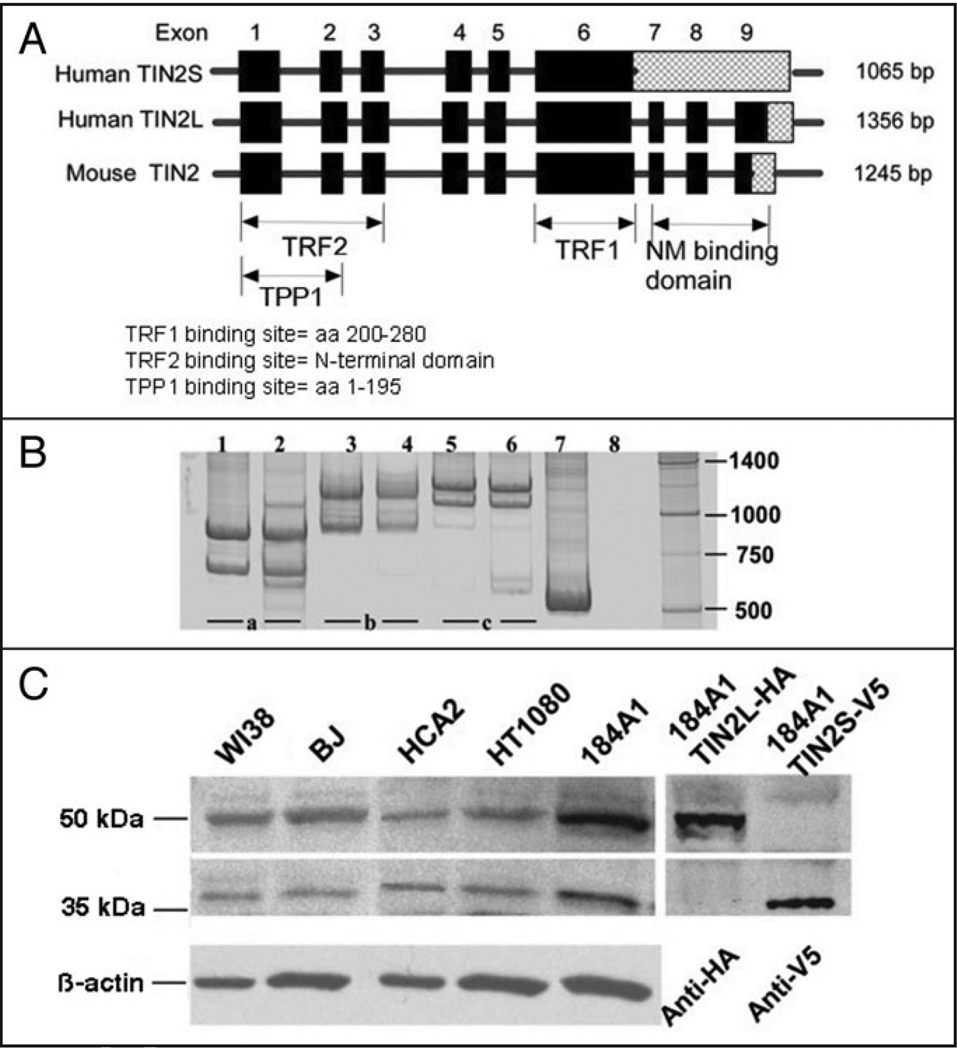
To better understand hTIN2, we aligned the exons predicted to encode the two hTIN2 and single mTIN2 proteins (Fig. 1A). Alignment between hTIN2S and both hTIN2L and mTIN2 identified three potential exons separated by three candidate introns in the 3' UTR of hTIN2S (see ENSEMBL transcript: ENST00000267415). Examination of the DNA sequence in this region showed that TIN2S likely results from retention of the intron that separates exons 6 and 7, which introduces a translational stop codon one nucleotide into the intron. Thus, hTIN2S and hTIN2L likely derive from alternative splicing, which generates two proteins that are identical over 354 aa residues and differ only by 97 additional residues present at the C-terminus of hTIN2L.
Figure 1.
Two isoforms of hTIN2 result from alternative splicing. (A) Alignment of exons encoding hTIN2 and mTIN2. Intron retention generates a translational stop sequence one nucleotide into the intron thereby encoding the smaller hTIN2S isoform. Hatched fill represents the 3' UTRs. Also shown are the coding regions known to be important for TIN2 interactions with TRF1, TRF2 and TPP1/PTOP, and the region deduced to be important for association with the nuclear matrix (NM). (B) RT-PCR products using primers spanning exons 6–9 (a), 5–9 (b) or 1–7 (c). We analyzed total RNA from human fibroblasts (lanes 1, 3 and 5) and epithelial cells (lanes 2, 4 and 6), described in the text. Sizes of the major PCR products were determined from the mobilities of markers (unlabeled right lane) and are consistent with transcripts that retain (upper bands) or splice (lower bands) introns separating exons 6–9. Lane 7 is a positive control using primers designed to identify β-actin mRNA and lane 8 is a negative control in which reverse transcription was omitted and PCR only was performed using primers spanning exons 5–9. (C) Western blot analysis confirming expression of two hTIN2 proteins in human cells. Total cell protein lysates were prepared from normal human fibroblasts (WI38, BJ, HCA2), human fibrosarcoma cells (HT1080), and immortal human mammary epithelial cells (184A1) using 2X Laemmli buffer (4% SDS). Blots were probed with an antibody raised against the N-terminal domain of hTIN2,25 then stripped and re-probed with an antibody against β-actin to control for protein loading. Identities of hTIN2L (~50 kDa) and hTIN2S (~40 kDa) were confirmed by expressing in 184A1 cells hTIN2L or hTIN2S cDNAs containing C-terminal epitope-tags (HA or V5, respectively) using the MSCV retroviral vector, as described in Materials and Methods, and analyzing lysates by western blotting using the indicated epitope-specific antibodies.
To confirm the alternative splicing, we designed oligonucleotide primers to span the potential exons and introns within the 3' UTR (spanning exons 6–9 or exons 5–9). We analyzed mRNA isolated from normal human fibroblasts (strain 82–6) and human mammary epithelial cells (HMECs, chemically immortalized line 184A1) by the reverse transcriptase-polymerase chain reaction (RT-PCR) (Fig. 1B). The sizes of the major PCR products produced by each primer pair were consistent with both cell types expressing two transcripts: one that retained the three introns that separate exons 6–9 (predicted to generate hTIN2S), and another that spliced these introns (predicted to generate hTIN2L). To eliminate the possibility that the mRNA preparations were contaminated with genomic DNA, which would generate PCR products indistinguishable from those generated by transcripts that retained the three 3' introns, we analyzed the mRNA using primers that span the invariably spliced introns located upstream of exon 6 (spanning exons 5–9 or exons 1–7) (Fig. 1B). The PCR product was consistent with amplification of mRNA, not DNA. Likewise, omitting the RT step indicated little or no DNA contamination. Finally, although some primer pairs produced minor PCR products, which could indicate non-specific PCR reactions or low levels of minor splice variants, all the primer pairs produced only two major products.
To determine whether human cells expressed both hTIN2S and hTIN2L proteins, we lysed several human cell strains and lines using relatively high (4%) concentrations of the denaturing detergent sodium dodecyl sulfate (SDS) and analyzed lysate proteins by western blotting using a polyclonal antibody raised against an N-terminal region of hTIN2S25 (Fig. 1C). The antibody detected two distinct proteins, which were present in approximately equal amounts: the expected 40 kDa protein corresponding to hTIN2S,25 and a 50 kDa protein consistent with TIN2L. To confirm the identities of these proteins as hTIN2S and hTIN2L, we constructed retroviral vectors to ectopically express either hTIN2S containing a C-terminal V5 epitope tag (hTIN2S-V5) or hTIN2L containing a C-terminal HA epitope tag (hTIN2L-HA). We expressed the tagged proteins in 184A1 cells, and analyzed high-detergent lysates by western blotting using epitope-specific antibodies (Fig. 1C). The epitope-tagged and endogenous hTIN2 proteins migrated to very similar positions. These gel conditions are not expected to detect small differences in molecular weight that are conferred by the epitope tags.
These data suggest that alternative splicing (intron retention) generates two major isoforms of hTIN2—a 40 kDa hTIN2S protein and a 50 kDa hTIN2L protein—and that both isoforms are expressed in several types of human cells.
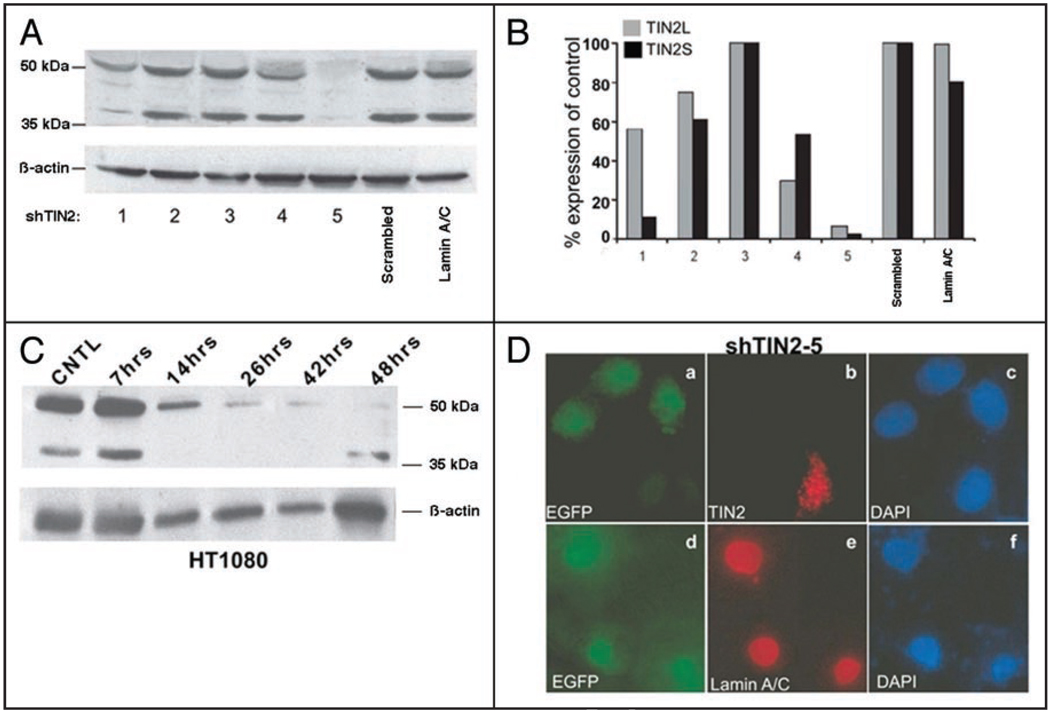
To further validate the expression of two hTIN2 isoforms, we used RNA interference (RNAi) to determine the relationship between hTIN2 transcripts and the 40 kDa and 50 kDa proteins. Because inactivation of mTIN2 in the mouse germline and RNAi depletion of hTIN2 in human cells showed that TIN2 is essential for cell viability,19,27 we transiently transfected HT1080 cells with expression vectors containing shRNA sequences that target hTIN2 and enhanced green fluorescent protein (EGFP). We failed to identify a shRNA that efficiently reduced the expression of one isoform but not the other (Fig. 2A and B) most likely because the hTIN2S and hTIN2L transcript sequences are so similar. However, we identified one shRNA (shTIN2-5) that reduced the expression of both isoforms by >90%. This shRNA markedly reduced the abundance of both the 40 kDa and 50 kDa proteins recognized by the hTIN2 antibody (Fig. 2A and B). A shRNA that targeted lamin A/C or contained a scrambled sequence had no effect on the abundance of these proteins. shTIN2-5 reduced the abundance of both proteins within 14 h after transfection (Fig. 2C), but, as expected,19,27 there was significant cell death within 48 h after transfection; shRNA vectors containing scrambled or lamin A/C sequences caused little cell death. Cells that expressed shTIN2-5, identifiable by EGFP expression (Fig. 2D), also lacked punctate nuclear hTIN2 immunostaining33 but normal lamin A/C immunostaining was detectable (Fig. 2D), supporting the specificity of the hTIN2 knockdown.
Figure 2.
RNAi reduces both TIN2S and TIN2L. (A) Western analyses of hTIN2 proteins in cells expressing different shRNA constructs. HT1080 cells were transiently transfected with vectors expressing different shRNAs designed to target hTIN2 (shTIN2-1 to 5). The cells were lysed in 2X Laemmli buffer 36 h later and analyzed for hTIN2 proteins and β-actin (protein loading control). Controls for non-specific shRNA effects include a scrambled shRNA sequence and a pre-validated lamin A/C shRNA. (B) Quantification of the western blot shown in (A). The signals were quantified by densitometry and the hTIN2 signals were normalized to the β-actin signals. The normalized signals are displayed as a percentage of signals generated by the scrambled shRNA control. (C) Time course of hTIN2 knockdown. HT1080 cells were transiently transfected with the shRNA 5 vector. At the indicated intervals, the cells were lysed and analyzed for hTIN2 proteins and β-actin by western blotting. (D) Immunofluorescence analysis of hTIN2 depletion by shRNA. HT1080 cells were transfected with the vector expressing shTIN2-5. Transfected cells were identified by EGFP expression (green; a and d); the nuclei of all cells were identified by DAPI staining (blue; c and f). The cells were immunostained for hTIN2 (red; b) or lamin A/C (red; e). Cells were visualized by fluorescence microscopy.
Taken together, the data strongly suggest that the gene encoding hTIN2 undergoes alternative splicing to generate two distinct protein isoforms.
In analyzing the expression of hTIN2 isoforms by western blotting, we noted that very little endogenous hTIN2L was detected when cells were lysed by non-denaturing detergent or low (<2%) concentrations of SDS. Because the TIN2 interacting protein TRF1 was reported to associate with the nuclear matrix, although not as an integral component of the relatively insoluble matrix structure,24 we tested the idea that hTIN2L might differ from hTIN2S in its ability to bind the nuclear matrix.
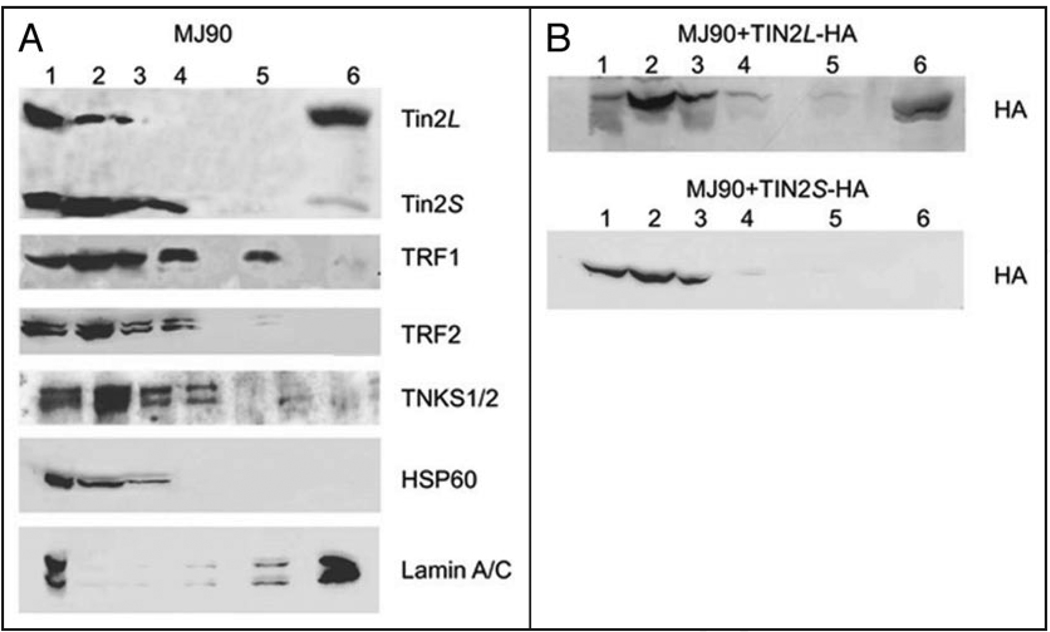
To analyze components of the nuclear matrix and other subcellular fractions, we subjected normal human fibroblasts to a fractionation protocol of increasing salt concentrations and nuclease digestion34 (Fig. 3A; Materials and Methods). We analyzed the fractions by western blotting using antibodies to detect endogenous TRF1, TRF2, tankyrases and hTIN2 isoforms. The fractionation showed that relatively large quantities of TRF1, TRF2, tankyrases and hTIN2S were recovered in the most soluble fraction (non-ionic detergent, low ionic strength; fraction 2), identified by the presence of HSP60 (heat shock protein 60). However, only a small quantity of hTIN2L was recovered in this fraction. After DNase digestion (fraction 3), DNA binding proteins solubilize.35 This step also solubilized significant quantities of TRF1, TRF2, tankyrases and hTIN2S, but only a small quantity of hTIN2L. Higher ionic strength (0.25 M NH4SO4; fraction 4), which extracts the majority of the histones, solubilized additional quantities of TRF1, TRF2, tankyrases and hTIN2S, but no detectable hTIN2L. Proteins that resist solubilization by 2 M NaCl (fraction 5) are considered nuclear matrix components, although not part of the relatively insoluble matrix structure.35 Fraction 5 contained a significant fraction of TRF1, as reported,24 but very little of the other proteins, including hTIN2L. The majority of hTIN2L was recovered in the relatively insoluble matrix fraction (solubilized by >2% SDS; fraction 6), identified by the presence of lamin A/C. This relative insolubility sharply distinguished hTIN2L from TRF1, TRF2, tankyrases and TIN2S, and suggests that TIN2L is tightly bound to the nuclear matrix structure.
Figure 3.
Nuclear compartmentalization of hTIN2 isoforms. (A) Association of endogenous hTIN2 isoforms with nuclear fractions. Normal human fibroblasts (strain MJ90) were subjected to increasingly stringent extractions consisting of: 0.5% triton (lane 2), DNAse I digestion (lane 3), 0.25 M ammonium sulfate (lane 4) and 2 M NaCl (lane 5) extraction. The remaining pellet was solubilized in 2X Laemmli buffer (lane 6). Half the cell equivalents were solubilized directly in 2X Laemmli buffer (whole cell lysate) (lane 1) and used to determine the input. Equal cell equivalents were analyzed for each of the other fractions. Proteins were analyzed by western blotting using antibodies to detect the N-terminus of hTIN2, TRF1, TRF2, tankyrases (TNKS1/2), heat shock protein 60 (HSP60, a mitochondrial chaperone and cytoplasmic marker) and lamin A/C (insoluble nuclear matrix marker). (B) Solubilities of epitope-tagged hTIN2 isoforms. C-terminal epitope (HA)-tagged hTIN2 isoforms (hTIN2L-HA, hTIN2S-HA) were expressed in MJ90 cells, as described in the text and Materials and Methods. The cells were subjected to fractionation and analysis as described in A except that anti-HA antibodies were used for detection.
To confirm this tight binding, we expressed HA epitope-tagged hTIN2 isoforms (hTIN2S-HA, hTIN2L-HA) in MJ90 fibroblasts. We subjected the cells to the fractionation protocol described above, and analyzed the fractions by western blotting using anti-HA antibodies (Fig. 3B). Similar to the behavior of the endogenous hTIN2 isoforms, hTIN2S-HA was recovered primarily in the relatively soluble fractions 2–4, whereas the majority of hTIN2L-HA was recovered in the relatively insoluble fraction 6.
These findings support the conclusion that two TIN2 isoforms are expressed by human cells, and suggest that the isoforms differ markedly in their ability to associate with the nuclear matrix.
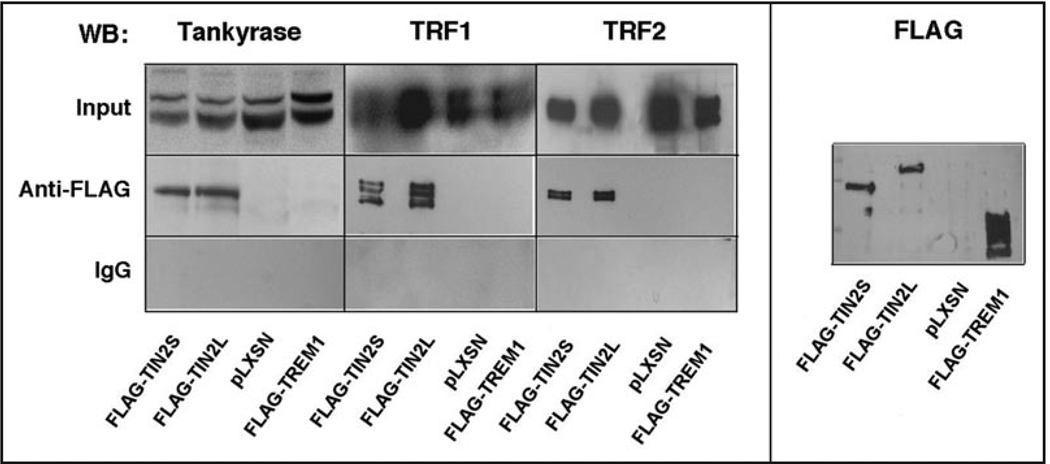
The addition of 97 aa to the C-terminus of hTIN2 could potentially alter its ability to interact with other telomere-associated proteins, such as tankyrases, TRF1 and TRF2.15,19,25,26 To determine whether this was the case, we used retroviral vectors to stably express FLAG epitope-tagged forms of TIN2S or TIN2L in HT1080 cells, and isolated FLAG-specific immune complexes from cell lysates. We analyzed the complexes for the presence of TRF1, TRF2 and tankyrase-1 by western blotting (Fig. 4). Regardless of whether the FLAG epitope was attached to TIN2S or TIN2L, the anti-FLAG antibody precipitated similar amounts of tankyrase-1, TRF1 and TRF2. These proteins were not precipitated by non-specific IgG, nor were they precipitated by anti-FLAG when we used lysates from cells expressing an insertless vector (pLXSN) or an irrelevant FLAG-tagged protein (FLAG-TREM1). Addition of ethidium bromide to the immune complexes did not alter the results, indicating the interactions were not mediated by DNA (data not shown).
Figure 4.
hTIN2 isoforms interact similarly with known telomere-associated partners. HT1080 cells were infected with retroviruses expressing either FLAG-TIN2S, FLAG-TIN2L, no insert (pLXSN) or FLAG-TREM1 (negative control), as indicated in the bottom panel. After selection, the cells were lysed and immunoprecipitated using either anti-FLAG or non-specific IgG resin. Proteins were eluted from resin as described in Materials and Methods, and analyzed by western blotting using antibodies (WB) against tankyrase, TRF1, TRF2 or FLAG. 2.5% of the lysate (input) were also analyzed by western blotting.
These findings suggest that hTIN2L binds the same major protein partners that are bound by hTIN2S. This result is also expected given that both isoforms have identical sequences over 354 aa, which includes the tankyrase, TRF1 and TRF2 interaction domains. However, given that distinct hTIN2S-containing subcomplexes have been identified in cells,36 we cannot rule out the possibility that TIN2S and TIN2L exist in subcomplexes that contain different components of the telosome/shelterin complex.
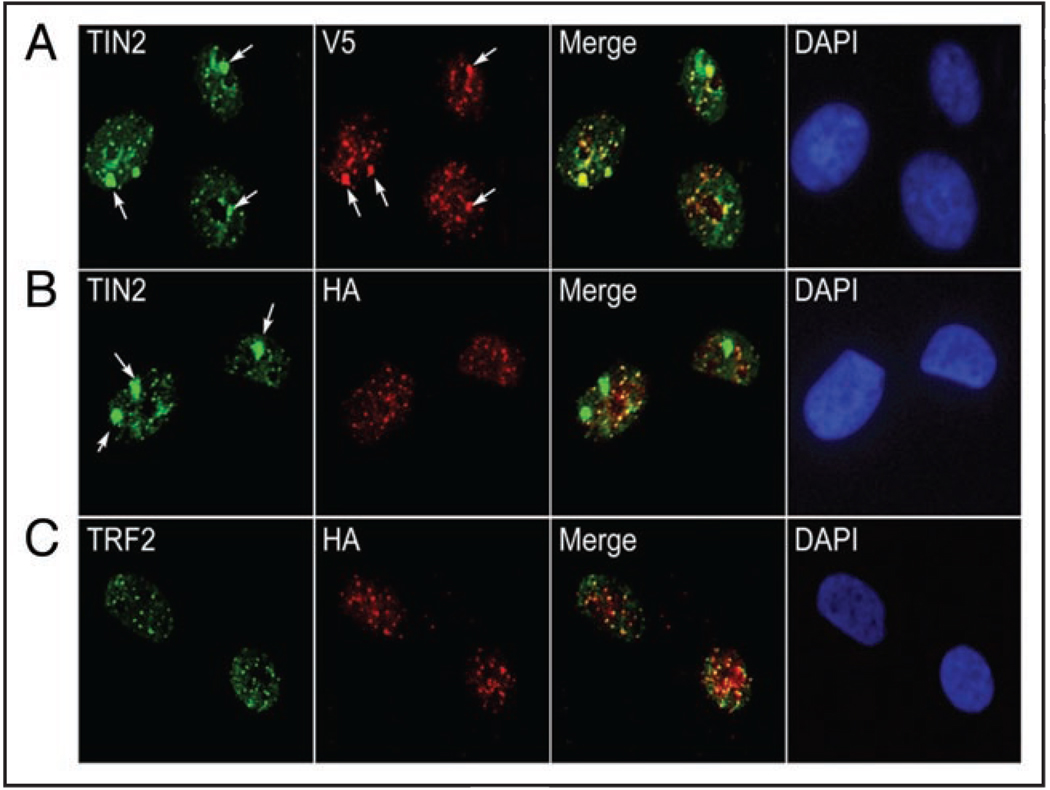
We previously found that hTIN2 localized not only to telomeres, but also to non-telomeric nuclear domains in growth-arrested HMECs.33 To determine whether these domains contained hTIN2S, hTIN2L or both, we expressed the hTIN2S-V5 and hTIN2L-HA proteins (Fig. 1C) in 184A1 HMECs, induced growth arrest by preventing epidermal growth factor (EGF) signaling,37 and immunostained the cells using antibodies to detect either both hTIN2 isoforms (N-terminal TIN2 antibody) or only one isoform (anti-V5 or anti-HA). Immunostaining for V5 and hTIN2 showed that TIN2S localized to both small foci, which we previously identified as telomeres, as well as large foci, which we previously identified as telomere-independent domains33 (Fig. 5A). By contrast, immunostaining for HA and hTIN2 showed that hTIN2L localized only to small foci (Fig. 5B). These foci were identified as telomere, as determined by coincident staining for TRF2 (Fig. 5C). To rule out the possibility that the HA epitope prevented localization to non-telomeric domains, we tagged hTIN2S with HA, but saw no difference in distribution compared to V5-tagged hTIN2S (data not shown).
Figure 5.
hTIN2L localizes to telomeres. (A) Immunolocalization of TIN2S-V5 expressed in 184A1 HMECs using antibodies against the hTIN2 N-terminal domain (TIN2; green) and the V5 epitope (V5; red). Arrows identify the telomere-independent nuclear domains,33 which co-stain (Merge; yellow). DAPI staining (blue) identifies nuclei. (B) Immunolocalization of TIN2L-HA expressed in 184A1 HMECs using antibodies against the hTIN2 N-terminal domain (green) and the HA epitope (red). Arrows identify telomere-independent nuclear domains,33 and DAPI staining identifies nuclei (blue). (C) Co-localization (Merge; yellow) of TIN2L-HA (HA; red) with TRF2 (TRF2; green) in nuclei (DAPI; blue).
This result shows that hTIN2L, like hTIN2S, localizes to telomeres. Further, in contrast to hTIN2S, which localizes to both telomeric and non-telomeric sites, hTIN2L localization is restricted to telomeres.
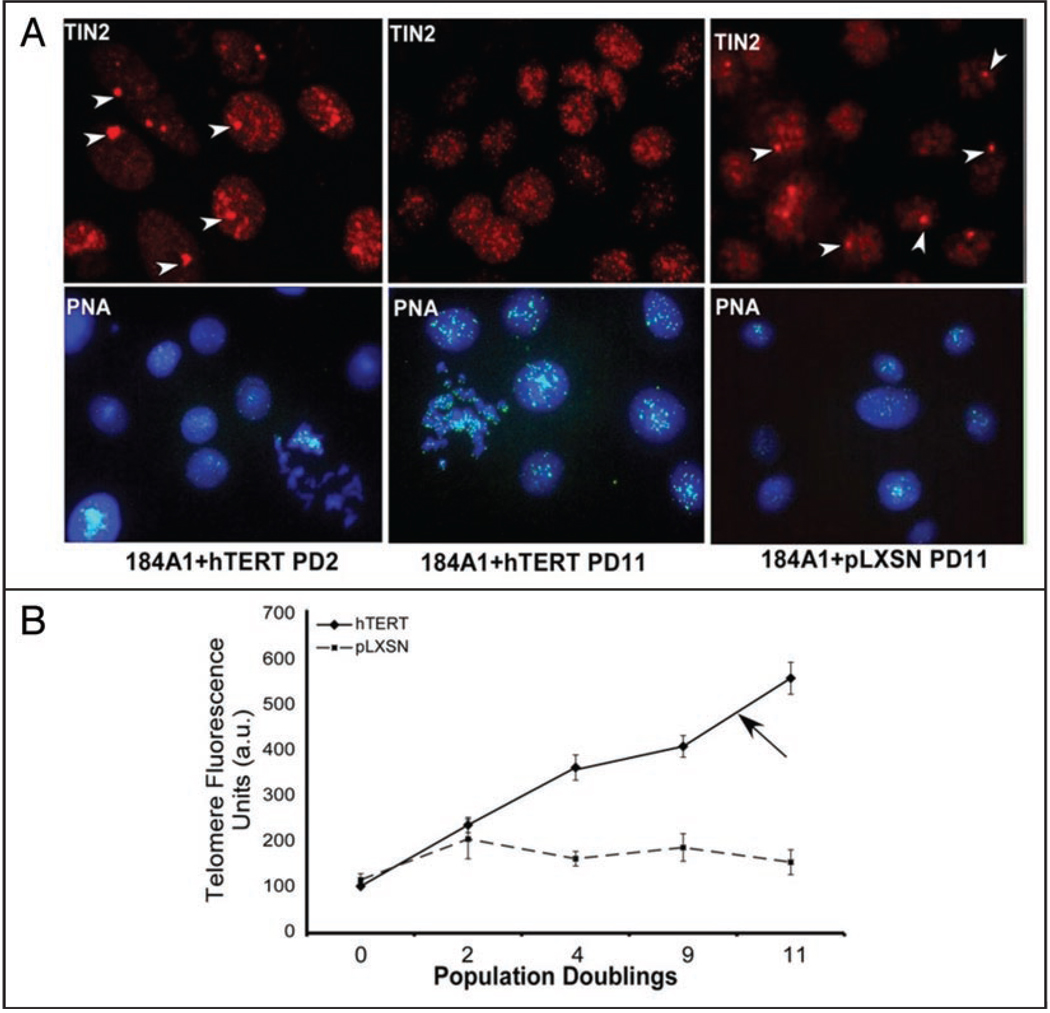
Telomere length is regulated by TRF1, TRF2, Rap1 and TIN218,25,38,39 and was recently shown to affect the epigenetic status of telomeres.40 Because hTIN2S localized to both telomeric and non-telomeric heterochromatic regions,33 we asked whether telomere length altered the distribution of hTIN2L. 184A1 HMECs have a mean terminal restriction length of ~5 kb, which increase rapidly upon overexpression of the catalytic subunit of telomerase (hTERT).41 We therefore overexpressed hTERT in 184A1 HMECs. We then measured the distribution of hTIN2, using the N-terminal antibody that recognizes both isoforms, and telomere length, using quantitative fluorescence in situ hybridization (qFISH), after increasing population doublings (Fig. 6A), during which time telomere lengths progressively increased (Fig. 6B). After 10 population doublings and an approximately 4-fold increase in telomere length, the large, non-telomere33 TIN2 domains disappeared, leaving only punctate (telomeric) staining (Fig. 6B). These findings suggest that telomere length influences the distribution of hTIN2S from non-telomeric heterochromatin to telomeres, but does not influence the telomeric distribution of hTIN2L.
Figure 6.
hTIN2S, but not hTIN2L, localization depends on telomere length. (A) 184A1 HMECs were infected with a control retrovirus (pLXSN) or retrovirus expressing the catalytic subunit of telomerase (hTERT). The cells were immunostained for TIN2 (red) 2 and 11 population doublings (PD) after infection. Arrows indicate non-telomeric TIN2 domains. Parallel cultures were analyzed for telomere length using qFISH (green), as described in Materials and Methods. Nuclei were counterstained with DAPI (blue). (B) Quantification of average fluorescence from qFISH analyses. Fluorescence intensities from >300 cells were measured. The results are expressed in arbitrary units (a.u.) relative the fluorescence intensities of uninfected cells (PD 0). Arrow indicates the telomere length at which non-telomeric TIN2 domains are no longer visible.
Discussion
TIN2 is an important component of the telosome/shelterin complex, which is crucial for maintaining the structure, and hence function, of telomeres.14,15 The interaction between TIN2 and TRF1, and between TRF1 and tankyrase, is important for the regulation of telomere length.25,30 Further, the interaction between TIN2 and TRF2 is responsible for maintaining the end-capping function of telomeres,19,26 and the TIN2-TPP1 interaction is critical for the higher order assembly of the entire telomeric complex.42 Recently, sister telomeres—as opposed sister chromatid arms and centromeres—were shown to require a distinct set of proteins for cohesion and separation during mitosis.29 TIN2 and TRF1 were each shown to bind the SA1 ortholog of the cohesin Scc3 subunit and to be important for regulating the dynamics of sister telomere associations during mitosis.43 It will now be of interest to determine whether one or both hTIN2 isoforms are important for these varied functions.
Our data provides the first evidence that two hTIN2 proteins are generated through alternative splicing in human cells. The only distinction between the mRNAs encoding each of the hTIN2 isoforms is the retention of a small intron between exons 6 and 7, which generates a stop codon that produces the smaller, previously identified25 40 kDa TIN2S isoform. This similarity in transcript sequence hampered our ability to selectively silence the expression of one isoform but not the other by RNAi. Nonetheless, one shRNA strongly reduced the expression of both isoforms, arguing against the possibility that the 50 kDa hTIN2L isoform was a non-specific cross-reactant to the hTIN2 antibody. Two other shRNAs reduced the expression of one isoform more than the other, but the differences in level of depletion were modest—too small to allow us to determine the effects of a sustained decrease in one isoform but not the other.
The most striking difference we observed between hTIN2S and hTIN2L was in their ability to associate with the insoluble fraction of the nuclear matrix. The additional 97 aa present in hTIN2L clearly provides the ability to tightly bind the nuclear matrix, since the two isoforms are identical in sequence except for additional aa in hTIN2L. These 97 aa also make it difficult to solubilize hTIN2L, and therefore also difficult to immunoprecipitate. We were able to immunoprecipitate epitope-tagged hTIN2L only after overexpression, which produces excess soluble protein (Fig. 3B). The relatively insoluble properties of hTIN2L may explain why this isoform was not discovered earlier by us and others. The matrix-binding properties of hTIN2L suggest that its biological importance lies in tethering telomeres to the nuclear matrix.
Both hTIN2 isoforms bind TRF1 and TRF2. Within the telo-some/shelterin complex, TRF1 and TRF2 bind telomeric DNA as homodimers.16 The differential recruitment of each hTIN2 isoform to DNA-bound TRF1 or TRF2 dimers may determine the number of telomere-nuclear matrix tethering sites. Dominant negative mutations that disrupt either the TRF1 or TRF2 binding domains of hTIN2 produce a variety of phenotypes, including telomere elongation, cell death and cellular senescence. It remains to be determined now whether one or more of these phenotypes results from loss of telomere tethering, or disruption of the stoichiometric relationship between each isoform and other members of the telosome/shelterin complex. Immunofluorescence staining of interphase nuclei showed that hTIN2 localizes exclusively to telomeres in human fibroblasts,25 although it also localizes to heterochromatin rich domains in human epithelial cells.33 Therefore, at least in fibroblasts, the association of hTIN2L with the nuclear matrix appears to be coincident with its association with telomeres. Recently, the nuclear localization of TPP1 and POT1 was shown to depend on TIN2,44 suggesting that TIN2—possibly hTIN2L in particular—acts as a central anchoring protein in the nucleus.
We found that hTIN2 can localize to telomere-independent chromatin domains. These domains are DNase but not RNase sensitive, frequently found in the heterochromatin-dense perinucleolar regions, and contain an abundance of heterochromatin protein-1γ.33 We found that hTIN2S, but not hTIN2L, localized to these domains. This finding suggests that hTIN2S, but not hTIN2L, might function in heterochromatin formation or organization. Further, increasing telomere length by overexpression of hTERT caused hTIN2S to redistribute from dual localization at both non-telomeric chromatin domains and telomeres to an exclusively telomeric localization. Recently, telomere length and the epigenetic status of telomeres and subtelomeric DNA were shown to reciprocally influence each other,40,45 suggesting that hTIN2S but not hTIN2L participates in linking telomere length to chromatin organization. However, because hTIN2L binds tightly to the nuclear matrix, which is also an important chromatin organizing structure,46 it is possible that this hTIN2 isoform also plays a role in chromatin organization or function.
Materials and Methods
Reagents and antibodies
Antibodies obtained commercially were as follows: monoclonal anti-HA (Covance Research Products), rabbit polyclonal HA (Santa Cruz Biotechnology), monoclonal anti-TRF2 (Upstate Biotechnologies), monoclonal anti-V5 (Invitrogen) and anti-β-Actin (Abcam). We generated TIN2, TRF1, and tankyrase antibodies as described.19,47 Fluorescent-conjugated secondary antibodies were obtained from Invitrogen.
Cell culture
Normal human fibroblasts and HT1080 fibro-sarcoma cells were obtained from the American Type Culture Collection (ATCC) or previously described sources, and cultured described.25,47,48 184A1 HMECs are immortalized but non-malignant epithelial cells,49 provided by Martha Stampfer (Lawrence Berkeley National Laboratory). For the growth arrest experiments, 184A1 cells were cultured in MEGM medium (Lonza) supplemented with transferrin (5 µg/ml) and isoproterenol (10−5 M) (Sigma) and seeded at 50% confluence. After 24 h, the medium was replaced with MEGM lacking EGF and supplemented with 8 µg/ml EGF blocking antibody (MAb 225, ATCC hybridoma clone HB-8508); the cells arrested growth with a G1 DNA content 48 h later.37
TIN2L sequence
We isolated RNA from A549 lung carcinoma and HT1080 fibrosarcoma cell lines and amplified cDNAs encoding both isoforms by PCR. cDNAs encoding the individual isoforms were isolated by gel purification and inserted into the pCR4 vector by TA cloning. We verified the sequence of the cDNAs using multiple overlapping internal and flanking primers. We deposited the sequence into GenBank (accession number EU851975).
Expression vectors
We generated epitope-tagged TIN2L cDNAs by PCR amplification of MGC clone 12628 (ATCC) using N-terminal FLAG or C-terminal HA linked primers (Operon Technologies). We generated epitope-tagged TIN2S cDNAs as described.19,33 MSCV retroviral vectors were purchased from Clontech. MSCV and LXSN retroviruses were produced and used as described25 with the following exception: we harvested the hTERT-expressing retrovirus from packaging cells after overnight incubation in MEGM medium supplemented with 0.1% BSA and 1% EX-CYTE (Celliance). We verified expression by western analyses for the appropriate proteins, or by measuring telomerase activity using the TRAP-EZ (Millipore).
RT-PCR
We isolated total RNA from one 100 mm plate of normal human fibroblasts (82–6) and 184A1 HMECs using RNAWiz (Ambion) according to manufacturer’s protocol. We chose three forward oligo primers that covered either the full-length coding region or common exons upstream of the alternative splicing site and two reverse primers within exon 7 and 9 to distinguish between the splice variants. The forward primers were (5'→3'): CAC TTC AAT CTG GCC CCT CTA GG, GCC TAC ACC GCA GGC ACA GC and ACC ATG GCT ACG CCC CTG GTG G. The reverse primers were (3'→5'): GCA GTG TAG TTA GGC AAT CCA AGC C and GCT GCA CAG AGA CGG AGG ACA CA. We amplified hTIN2 mRNA segments from 1 µg of total RNA using the Titanium one-step RT-PCR reaction (BD Biosciences) according to the manufacturer’s instructions. PCR products were separated on 12% native polyacrylamide gels (Invitrogen) and analyzed using a Typhoon densitometer (GE Healthcare).
Western blot analyses
We lysed cells directly in Laemmli buffer (4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.125 M Tris HCl, pH 6.8) and separated the proteins on 8% or 10% Tris-glycine polyacrylamide gels (Invitrogen). Separated proteins were transferred to membranes and probed with antibodies as described.25
Short hairpin (shRNAs) experiments
We generated five shRNAs that target different regions of the hTIN2 mRNA and contained flanking BamHI and HindIII sites. The sequences are as follows (5'→3'): shTIN2-1, GGT CAT ATC TAA TCC TGA G; shTIN-2, GAA TTG CTT GGA TTG CTA CAT; shTIN-3, GGT AGT CTG AGT CAG GAT TGG; shTIN-4, AGC CTG TGT TCA TCT AGA A; shTIN2–5, GTG CAG CTC CGT CAT TAC CAT; scrambled shRNA, GTC CGT TCC GGT TCA AAC ACT; lamin A/C shRNA, ATA CCA AGA AGG AGG GTG ACC TG (Invitrogen). Oligomers were annealed and inserted into the pENTR-H1 vector (Invitrogen) and transferred into a destination vector containing a modified pBlock-iT6 backbone (Invitrogen) and a CMV driven EGFP cassette, excised from EGFP-C1 (BD Biosciences). We transfected HT1080 cells using Fugene 6 (Roche Applied Sciences) according to the manufacturer’s protocol and used the EGFP signal to determine transfection efficiency. We harvested cells in 2X Laemmli buffer.
FLAG immunoprecipitation
We stably expressed FLAG-TIN2S, FLAG-TIN2L or FLAG-TREM1 (triggering receptor expressed on myeloid cells) in HT1080 cells using pLXSN retroviral vectors, and analyzed complexes containing FLAG-tagged proteins as described.19 Briefly, 107 were lysed in 1 ml NP-40 buffer (50 mM Tris pH7, 200 mM NaCl, 1 mM EDTA, 1% NP40, 10% glycerol, and protease inhibitor cocktail) and pre-cleared for 1 h using 2 µg of Protein A/G agarose (Santa Cruz Biotechnologies). We incubated cleared lysates with 40 µl of either anti-FLAG-M2 or non-specific IgG gel (Sigma) at 4°C overnight. Immune complexes were collected by centrifugation, washed four times in NP-40 buffer, and eluted with either 3X FLAG peptides (150 ng/µl in NP-40 buffer) or glycine (pH 3.5). The eluted proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by western blotting.
Immunolocalization
Indirect immunofluorescence staining was performed as described.33 Briefly, four-well chamber slides (Nalge Nunc International, Naperville, IL) were seeded with 1 × 104 cells. After 24 h, the cells were treated with cytoskeletal buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES pH 6.8, 5 mM MgCl2, 1 mM pefabloc, 10 µg/ml aprotinin, 250 µM NaF) containing 0.5% Trition X-100 (Sigma) for 5 min prior to fixation with 4% paraformaldehyde (EM Sciences). Cells were blocked with 5% goat serum (Rockland Immunochemicals) in phosphate buffered saline for 1 h, followed by incubation with the indicated antibodies in 2.5% goat serum and anti-goat secondary antibodies conjugated to Alexa Fluor dyes (Invitrogen). DAPI (4'-6diamidino-2-phenylindole; Sigma) staining was used to identify nuclei.
Nuclear matrix fractionation
We isolated subcellular fractions of increasing insolubility using an established protocol.34 Briefly, we incubated 107 cells in cytoskeletal buffer (see above) containing 0.5% Triton X-100 for 5 min on ice. We recovered the supernatant by centrifugation (fraction 2). To the pellet, we added DNase (40 U/ml) in digestion buffer (CSK buffer containing 50 mM NaCl) and incubated for 50 min at 32°C. We recovered the supernatant by centrifugation (fraction 3). We resuspended the pellet in digestion buffer, slowly added ammonium sulfate (1 M) to a final concentration of 0.25 M, mixed gently for 5 min at room temperature, and again recovered the supernatant (fraction 4). We again resuspended the pellet slowly added NaCl to a final concentration of 2 M, and incubated for 5 min at room temperature. After recovering the supernatant (fraction 5), we washed the final pellet twice with digestion buffer and dissolved in 2X Laemmli buffer (fraction 6). Cells (107) that were solubilized directly in 2X Laemmli buffer served as the input reference (fraction 1). Equal cell equivalents from each fraction were analyzed by western blotting.
qFISH
In situ hybridization was performed using an established protocol (Current Protocols in Cell Biology, John Wiley & Sons, Inc.,) and 5 nmol FITC-conjugated (C3TA2)peptide nucleic acid probe (PE Biosystems). Relative fluorescence intensities were measured using TFL Telo software graciously provided by Peter Lansdorp (University of British Columbia).
Acknowledgements
We thank Dr. Martha Stampfer (Lawrence Berkeley National Laboratory, Berkeley, CA) for providing 184A1 human mammary epithelial cells. This work was supported by grants AG09909 and AG017242 from the National Institutes of Health.
Abbreviations
- aa
amino acid
- DAPI
4'-6 diamidino-2-phenylindole
- EGF
epidermal growth factor
- EGFP
enhanced green fluorescent protein
- RNAi
RNA interference
- RT-PCR
reverse transcriptase-polymerase chain reaction
- SDS
sodium dodecyl sulfate
- UTR
untranslated region
References
- 1.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reaper PM, di Fagagna F, Jackson SP. Activation of the DNA damage response by telomere attrition: a passage to cellular senescence. Cell Cycle. 2004;3:543–546. [PubMed] [Google Scholar]
- 3.Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:27–31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 4.Hornsby PJ. Senescence as an anticancer mechanism. J Clin Oncol. 2007;25:1852–1857. doi: 10.1200/JCO.2006.10.3101. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. Senescent cells, tumor suppression and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Yildiz O. Vascular smooth muscle and endothelial functions in aging. Ann N Y Acad Sci. 2007;1100:353–360. doi: 10.1196/annals.1395.038. [DOI] [PubMed] [Google Scholar]
- 7.Effros RB. T cell replicative senescence: pleiotropic effects on human aging. Ann N Y Acad Sci. 2004;1019:123–126. doi: 10.1196/annals.1297.022. [DOI] [PubMed] [Google Scholar]
- 8.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin GM. Modalities of gene action predicted by the classical evolutionary biological theory of aging. Ann NY Acad Sci. 2007;1100:14–20. doi: 10.1196/annals.1395.002. [DOI] [PubMed] [Google Scholar]
- 10.Griffith JD, Comea L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 11.Molenaar C, Wiesmeijer K, Verwoerd NP, Khazen S, Eils R, Tanke HJ, et al. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J. 2003;22:6631–6641. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, et al. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 2003;11:485–502. doi: 10.1023/a:1025016828544. [DOI] [PubMed] [Google Scholar]
- 13.Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, et al. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
- 14.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, O’Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 16.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 17.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem. 2004;279:43799–43804. doi: 10.1074/jbc.M408650200. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 21.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, et al. POT1- interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 23.de Lange T. Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T. Structure, subnuclear distribution and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 27.Chiang YJ, Kim SH, Tessarollo L, Campisi J, Hodes RJ. Telomere-associated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway. Mol Cell Biol. 2004;24:6631–6634. doi: 10.1128/MCB.24.15.6631-6634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SH, Han S, You YH, Chen DJ, Campisi J. The human telomere-associated protein TIN2 stimulates interactions between telomeric DNA tracts in vitro. EMBO Rep. 2003;4:685–691. doi: 10.1038/sj.embor.embor872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 30.Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet. 2004;36:618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- 31.Okabe J, Eguchi A, Wadhwa R, Rakwal R, Tsukinoki R, Hayakawa T, et al. Limited capacity of the nuclear matrix to bind telomere repeat binding factor TRF1 may restrict the proliferation of mortal human fibroblasts. Hum Mol Genet. 2004;13:285–293. doi: 10.1093/hmg/ddh032. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Parrinello S, Kim J, Campisi J. Mus musculus and Mus spretus homologues of the human telomere-associated protein TIN2. Genomics. 2003;81:422–432. doi: 10.1016/s0888-7543(02)00033-2. [DOI] [PubMed] [Google Scholar]
- 33.Kaminker P, Plachot C, Kim SH, Chung P, Crippen D, Petersen OW, et al. Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J Cell Sci. 2005;118:1321–1330. doi: 10.1242/jcs.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan KM, Nickerson JA, Krockmalnic G, Penman S. The nuclear matrix prepared by amine modification. Proc Natl Acad Sci USA. 1999;96:933–938. doi: 10.1073/pnas.96.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SH, Davalos AR, Heo SJ, Rodier F, Zou Y, Beausejour C, et al. Telomere dysfunction and cell survival: roles for distinct TIN2-containing complexes. J Cell Biol. 2008;181:447–460. doi: 10.1083/jcb.200710028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stampfer MR, Pan CH, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P, et al. Blockage of EGF receptor signal transduction causes reversible arrest of normal and immortal human mammary epithelial cells with synchronous reentry into the cell cycle. Exp Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- 38.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 39.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, et al. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 41.Stampfer MR, Garbe J, Levine G, Lichtsteiner S, Vasserot AP, Yaswen P. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor beta growth inhibition in p16INK4A(−) human mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci USA. 2006;103:11874–11879. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen LY, Liu D, Songyang Z. Telomere maintenance through spatial control of telomeric proteins. Mol Cell Biol. 2007;27:5898–5909. doi: 10.1128/MCB.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39 h1 and Suv39 h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 46.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 47.Kaminker PG, Kim SH, Taylor RD, Zebarjadian Y, Funk WD, Morin GB, et al. TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J Biol Chem. 2001;276:35891–35899. doi: 10.1074/jbc.M105968200. [DOI] [PubMed] [Google Scholar]
- 48.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stampfer MR, Yaswen P. Human epithelial cell immortalization as a step in carcinogenesis. Cancer Lett. 2003;194:199–208. doi: 10.1016/s0304-3835(02)00707-3. [DOI] [PubMed] [Google Scholar]