Abstract
Several commonly occurring polymorphisms in the IL-4Rα have been associated with atopy in humans; the Q576R and the S503P polymorphisms reside in the cytoplasmic domain, while the I50 to V (V50) polymorphism resides in the extracellular domain of the IL-4Rα. The effects of these polymorphisms on signaling remain controversial. To determine the effect of the polymorphisms on IL-4 signaling in human cells, we stably transfected the human monocytic cell line U937 with muIL-4Rα cDNA bearing the I or V at position 50 and the P503/R576 double mutant. Each form of the muIL-4Rα mediated tyrosine phosphorylation of STAT6 in response to murine IL-4 treatment similar to the induction of tyrosine phosphorylation by human IL-4 signaling through the endogenous human IL-4Rα. After IL-4 removal, tyrosine-phosphorylated STAT6 rapidly decayed in cells expressing I50 or P503R576 muIL-4Rα. In contrast, STAT6 remained significantly phosphorylated for several hours after muIL-4 withdrawal in cells expressing the V50 polymorphism. This persistence in pSTAT6 was associated with persistence in CIS mRNA expression. Blocking IL-4 signaling during the decay phase using the JAK inhibitor AG490 or the anti-IL-4Rα antibody M1 abrogated the persistence of pSTAT6 observed in the V50-IL-4Rα expressing cells. These results indicate that the V50 polymorphism promotes sustained STAT6 phosphorylation and that this process is mediated by continued engagement of the IL-4Rα suggesting enhanced responses of V50 IL-4 receptors when IL-4 is limiting.
Keywords: IL-4, STAT6, IL-4Rα, polymorphisms
Introduction
Interleukin 4 (IL-4) is a cytokine that plays a critical role in immune responses (1). IL-4 is produced by activated T lymphocytes, basophils, eosinophils, and mast cells and acts upon many cell types. For example, IL-4 acts on T cells to drive Th2 cell differentiation and it directs B lymphocytes to produce IgG4 and IgE in humans and IgG1 and IgE in mice (2). IL-4 induces expression of MHC class II on macrophages and induces the expression of genes associated with parasitic infections (2,3). IL-4 also impacts non-hematopoietic cells. IL-4 increases the expression of VCAM1 on endothelial cells, while suppressing E-selectin and can induce the expression of eotaxin by lung epithelial cells (4,5,6). IL-4 mediates many of these responses by binding to and signaling through either the Type I IL-4 receptor complex, which is composed of the IL-4Rα chain and the common gamma (γC) chain, and/or the Type II IL-4 receptor complex composed of the IL-4Rα chain and the IL-13Rα1 chain (7,8,9). Binding of IL-4 to the IL-4Rα chain leads to the subsequent activation of Janus kinases (JAKs) and downstream signaling pathways including the signal transducer and activator of transcription (STAT)-6 pathway and the insulin receptor substrate (IRS) pathway (3).
Single nucleotide polymorphisms leading to amino acid changes have been described for the mouse and human IL-4Rα (10,11,12). Several polymorphisms in the IL-4Rα chain have been associated with allergy and asthma in humans (13,14,15). Two of the polymorphisms located in the cytoplasmic domain, S503 to P and the Q576 to R, are frequently linked (15). This double mutation (S503P/Q576R) has been associated with lower total IgE concentrations, similar to the single S503P polymorphism, and an increase in the phosphorylation of IRS molecules (15). An I50 in the extracellular domain of the IL-4Rα chain was linked with enhanced signal transduction culminating in an increase in the production of IgE (14,16). On the other hand, several other studies reported no correlation of this polymorphism with enhanced IgE levels in patients (17,18). Furthermore, the V50 polymorphism has been linked with enhanced CD23 expression, an increase in atopic asthma, and an increase in allergic bronchopulmonary aspergillosis (19,20). Experiments designed to analyze the effects of these polymorphisms on receptor signaling have been contradictory (14,16,19,21,22–24).
Previous studies on the effects of the IL-4Rα chain polymorphisms have focused on linkage analyses in patients, functional differences in B cells or T cells isolated from patients, or the analysis of signaling by human IL-4Rα expressed in the background of murine B or T-cells (13,19,22,24–28). The analysis of responses in patient-derived cells can be impacted by the variable genetic backgrounds of the patients and therefore such studies may provide confounding results. Furthermore, the effects of the polymorphisms may not be apparent in murine lymphocytes. Therefore, in this study we investigated signaling through murine IL-4Rα engineered to express the polymorphisms described for the human IL-4Rα in human monocytic cells. We have found that the V50 polymorphism enhances the persistence of STAT6 phosphorylation.
MATERIALS AND METHODS
Reagents
Recombinant murine IL-4 (mIL-4) and recombinant human IL-4 (hIL-4) were obtained from R & D Systems (Minneapolis, MN). AG490 was obtained from Calbiochem (San Diego, CA) and was stored at −20°C in dimethyl sulfoxide (DMSO). Sodium orthovanadate (Na3VO4) was obtained from ICN (Aurora, OH). Cell lysis buffer (50 mM HEPES, pH 7.5; 0.5% NP-40; 5 mM EDTA; 50 mM NaCl; 10 mM sodium pyrophosphate; 50 mM sodium fluoride) was supplemented prior to each experiment with 1X protease inhibitor cocktail (10 mM benzamidine HCL, 20 mM Iodoacetamide, 10 µg/mL leupeptin, 1 µg/mL pepstatin A, 100 µg/mL tyrpsin inhibitor, and 2 mM 1,10 phenanthroline), 1mM Na3VO4, and 1 mM phenylmethylsulphonyl fluoride (PMSF). Geneticin (G418) was obtained from Gibco (Carlsbad, CA). Rat anti-mouse IL-4Rα (CD124) M1 blocking antibody was obtained from BD Pharmingen (Franklin Lakes, NJ).
Cell culture and transfection
Human monocytic U937 cells were derived from malignant cells from a patient with histiocytic lymphoma (29) and were maintained in RPMI-1640 (BioWhittaker, Inc.) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, 100 µg/mL streptomycin, and 2 mM glutamine (complete medium). The Murine Stem Cell Virus (MSCV) 2.2 plasmids containing polymorphisms in the murine IL-4Rα chains analogous to those described in human patients (I50-, V50-, Q576R-, S503P-, and P503R576-IL-4Rα) linked to EGFP by an IRES were previously described (22). For transfection, U937 cells were resuspended at 1.0 ×107 cells per 300 µl of PBS. U937 cells were mixed with MSCV 2.2 I50-, V50- (both are also S503 and Q576), or P503/R576-IL-4Rα and the pBH NeoR plasmid (encoding the neomycin gene for chemical selection with G418) at a 10:1 (10 µg:1 µg) ratio respectively and incubated on ice for 5 minutes. The U937 cells were electroporated at 950 µFa and 250 volts using the Gene Pulser® II (Bio-Rad Laboratories, Hercules, CA) and incubated on ice for 5 minutes. Transfected U937 cells were cultured in complete media for 24 hours after which time the transfected U937 cells were collected by centrifugation, resuspended in media containing G418 (800 µg/ml) and cultured in 24 well plates. Transfected U937 cells were grown through limiting dilution in selection media. Transfected U937 cells that grew in selection media were analyzed for the expression of GFP and the murine IL-4Rα chain by flow cytometry.
FACS analysis
U937 cells were prepared for flow cytometric analysis in the following manner. Cells were collected by centrifugation and washed three times in a 1X PBS/2% FBS solution (FACS buffer). The cells were resuspended in FACS buffer and pushed through a filter cap. Fc receptors were blocked at 4°C for 15 minutes using purified human IgG (Sigma-Aldrich, St. Louis, MO). Cells were then incubated for 30 minutes at 4°C in the presence of rat anti-murine IL-4Rα-linked to Phycoerythrin (PE) (M1, BD Pharmingen, Franklin Lakes, NJ). Cells were collected by centrifugation, washed three times and resuspended in FACS buffer. Flow cytometric analysis was performed on Becton Dickinson FACScan™ and Becton Dickinson FACSCalibur™ machines.
For the analysis of IL-4 binding to cell surface receptors, I50- and V50-IL-4Rα U937 cells were treated with 10 ng/mL of mIL-4 for 15 minutes. Cells were collected by centrifugation and either immediately resuspended in FACS buffer supplemented with 1% sodium azide (Azide FACS buffer) or they were incubated an additional 30 minutes in RPMI at 37°C before washing three times and resuspending in Azide FACS buffer. Fc receptors were blocked as previously described. Cells were incubated for 30 minutes at 4°C in the presence of biotin rat anti-murine IL-4 (BD Pharmingen, Franklin Lakes, NJ). Cells were incubated for 20 minutes at 4°C in the presence of PE streptavidin (BD Pharmingen, Franklin Lakes, NJ). Cells were collected by centrifugation, washed three times and resuspended in Azide FACS buffer. Flow cytometric analysis was performed as previously described.
Cell signaling
The U937 cell lines expressing various forms of murine IL-4Rα were treated in the absence or presence of various concentrations of mIL-4 for 15 minutes. The reactions were stopped by adding 25 mL ice cold 1X PBS. Cells were collected by centrifugation, resuspended in 500 µL cell lysis buffer, and incubated on ice for 20 minutes. The cells were centrifuged at 14,000 RPM for 10 minutes at 4°C. Cleared lysates were transferred to a new micro-centrifuge tube and stored at −70°C. For the wash-out experiments, the cells were stimulated in the absence or presence of 10 ng/mL mIL-4 for 15 minutes. The reactions were stopped by adding 25 mL ice cold 1X PBS. Cells were collected by centrifugation, washed three times in plain RPMI, and resuspended in RPMI and incubated at 37°C. Every 30 minutes over the course of 3 hours, the cells were mixed, and aliquots of the cells were collected by centrifugation and lysed. Where indicated, the cells were treated in the absence or presence of M1 anti-murine IL-4Rα blocking antibody or the JAK inhibitor AG490 before the addition of IL-4 or after IL-4 treatment during the wash-out phase. Every 30 minutes over the course of 3 hours an aliquot of cells from the indicated time points was removed, mixed with trypan blue and analyzed for viable cell number.
Real-Time PCR
The U937 cell lines expressing various forms of murine IL-4Rα were treated in the absence or presence of mIL-4 (10 ng/ml) for 15 minutes. The reactions were stopped by adding 25 mL 1X PBS. Cells were collected by centrifugation, washed three times in plain RPMI, and resuspended in RPMI and incubated at 37°C. Every 30 minutes over the course of 3 hours, the cells were mixed, and aliquots of the cells were collected by centrifugation. Total RNA was isolated using the RNeasy kit (Qiagen [Valencia, CA]) according to the manufacturer’s protocol and cDNA was generated using the SuperScript™ III First Strand Synthesis System (Invitrogen). Real-Time PCR was performed in triplicate with specific primer sets for human CIS (forward: 5’-GCTGTGCATAGCCAAGACCTT-3’ reverse: 5’-CTGGCATCTTCTGCAGGTGTT-3’ synthesized by Invitrogen) on an Applied Biosystems Inc. 7900HT machine. Relative mRNA levels for specific genes are reported as fold induction over background levels detected in untreated samples, with hypoxanthine guanine phosphoribosyl transferase (HPRT) as the internal reference gene (2−ΔΔCt method).
Immunoprecipitation and western blotting
Frozen cell lysates were thawed on ice. To precipitate STAT6, polyclonal rabbit anti-human STAT6, S-20 (Santa Cruz Biotechnology, Santa Cruz, CA) was added to 500µl of whole cell lysate and incubated at 4°C for 2 hours. To precipitate the murine IL-4Rα, monoclonal anti-mL-4Rα was used. The precipitates were collected by adding 80 µl of a 50% slurry of GIBCO BRL® Protein G Agarose (Invitrogen Life Technologies) in cell lysis buffer. Samples were placed in a slow rotation device and incubated for 1 hour at 4°C. The precipitates were washed in lysis buffer and solubilized in SDS sample buffer. The samples were separated on 7.5% SDS-polyacrylamide gels before transfer to a polyvinylidene difluoride (PVDF) membrane. The membranes were then probed with a monoclonal anti-phosphotyrosine antibody, RC20 or polyclonal anti-murine IL-4Rα. The bound antibodies were detected using enhanced chemiluminescence (Amersham, Arlington, IL). The blots were stripped and re-probed as necessary. The film was scanned and band intensities were analyzed using the public domain software NIH Image. Band intensities were used to determine phosphorylated STAT6/total STAT6 ratios. The student's T-test was used to determine the significance of differences between groups. The relative mobilities (Rf) of the mature IL-4Rα was calculated as follows. The distance (cm) migrated by the Molecular Weight Markers (Amersham) was divided by the total distance migrated by the dye front. These values were graphed. The distance (cm) migrated by the major species of the IL-4Rα was divided by the total distance migrated by the dye front. These Rf values were plotted against the MW markers to calculate a predicted molecular weight. N-linked oligosaccharides were removed from the IL-4Rα by enzymatic digestion. The precipitated samples were incubated with 2000 units of Peptide: N-glycosidase F (PNGase F; New England Biolabs, Ipswich, MA) or 5000 units of Endoglycosidase H (Endo H; New England Biolabs, Ipswich, MA) for one hour at 37°C. The samples were separated on a 4–20% SDS-polyacrylamide gel before transfer to (PVDF) membrane. The membrane was probed with a polyclonal anti-murine IL-4Rα antibody. The bound antibodies were detected using enhanced chemiluminescence (Amersham, Arlington, IL) and the relative mobilities of the receptors were calculated as previously described.
RESULTS
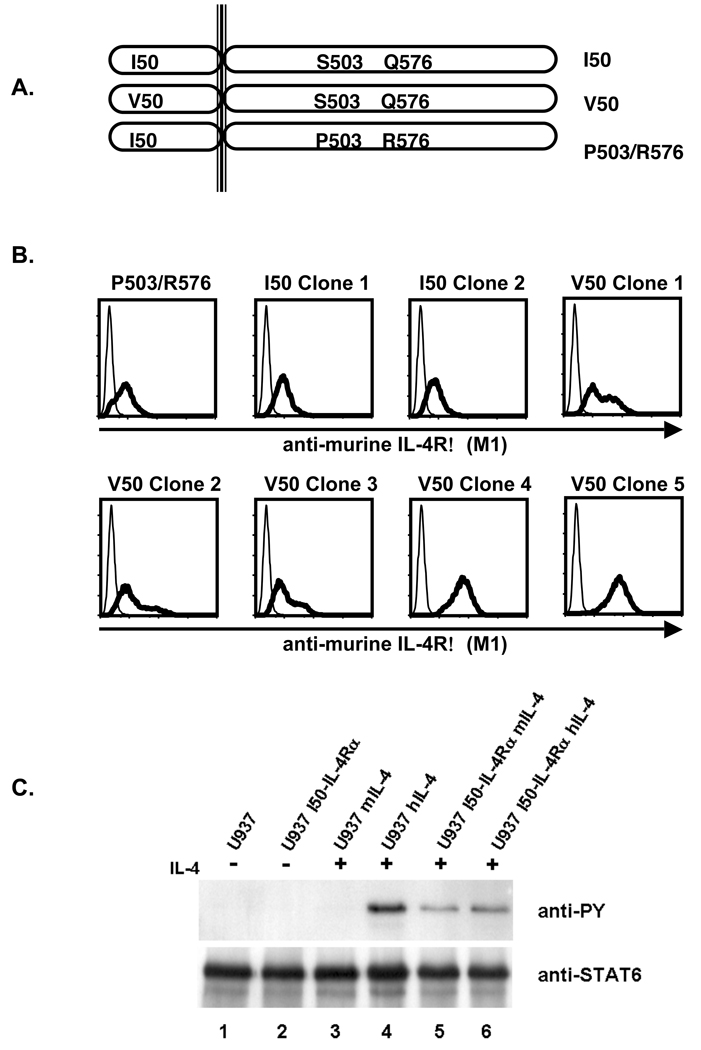
To analyze the effects of IL-4Rα polymorphisms on signal transduction in human cells, we stably transfected U937 cells with cDNA encoding the various IL-4Rα chain polymorphisms (Figure 1A). Post-transfection and selection, the U937 cells were analyzed for the expression of the murine IL-4Rα chain using flow cytometric analysis (Figure 1B). U937 clones transfected with MSCV cDNA encoding either the P503/R576-, I50-, or V50- polymorphic forms of the murine IL-4Rα (P503/R576-, I50-, or V50-IL-4Rα) demonstrated comparable staining with the anti-murine IL-4Rα monoclonal antibody M1. To determine if the introduced murine receptor was responsive to murine IL-4, U937 cells and U937 cells expressing the murine I50-IL-4Rα were stimulated with murine IL-4 or human IL-4. Tyrosine phosphorylation of STAT6 was analyzed by immunoprecipitation and immunoblotting (Figure 1C). Stimulation of parental U937 cells with human IL-4 resulted in the induction of STAT6 phosphorylation while stimulation with murine IL-4 failed to do so. Stimulation of U937 cells expressing the murine I50-IL-4Rα with either human or murine IL-4 resulted in the induction of STAT6 phosphorylation. These results indicate that murine IL-4 is incapable of stimulating U937 cells through the endogenous human IL-4Rα. The results also show that the introduced murine I50-IL-4Rα signals appropriately in response to murine IL-4, and at a level comparable to the response elicited by human IL-4. In contrast to IL-4, both human and murine IL-13 stimulated the tyrosine phosphorylation of STAT6 in parental and transfected U937 similarly (data not shown). Thus, this model was not suitable to analyze the effects of IL-4Rα polymorphisms on IL-13 signaling.
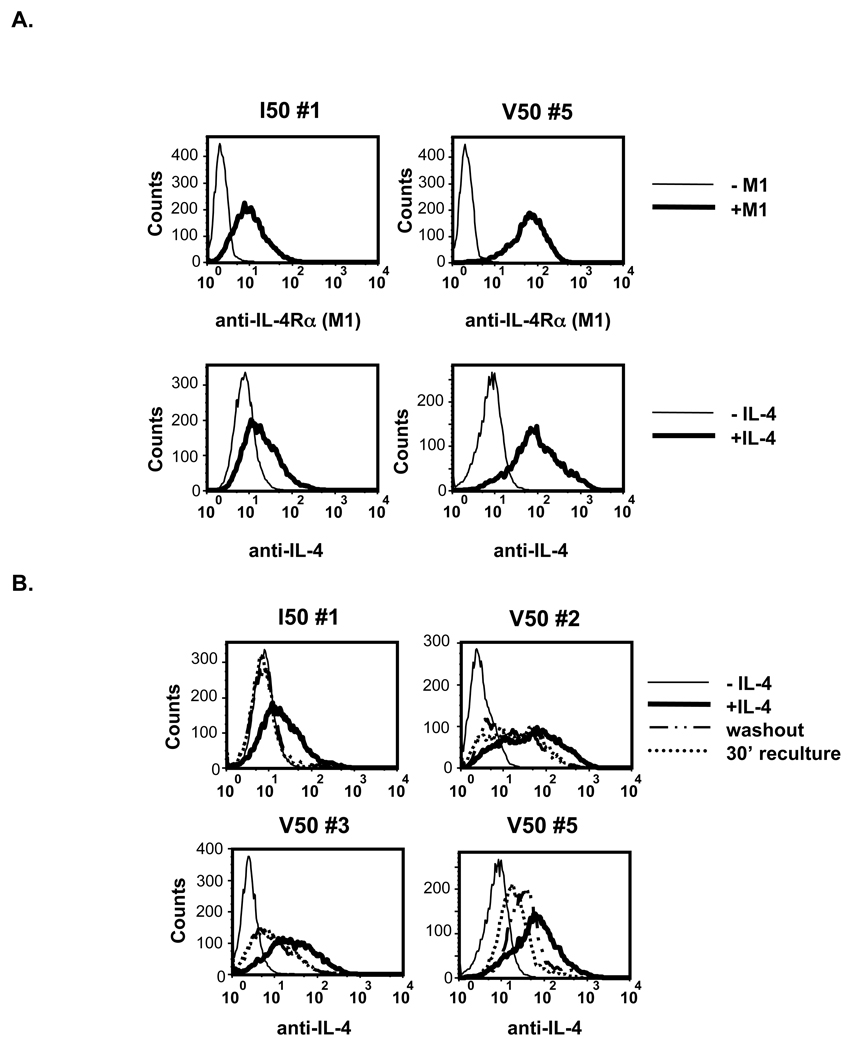
Figure 1. Murine IL-4Rα Expression on Transfected U937.
A. U937 cells were transfected with cDNA encoding various polymorphic forms of the murine IL-4Rα and neomycin resistance to allow selection of positive clones in G418. B. Following selection in G418, cells were screened by FACS for murine IL-4Rα expression using the M1 antibody. C. Parental U937 cells and U937 expressing the murine I50-IL-4Rα were stimulated in the absence or presence of 10 ng/mL murine and 10 ng/mL human IL-4 for 15 minutes. The cells were subsequently lysed, STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody to detect phosphotyrosine. The blot was stripped and reprobed with an anti-STAT6 antibody to detect STAT6.
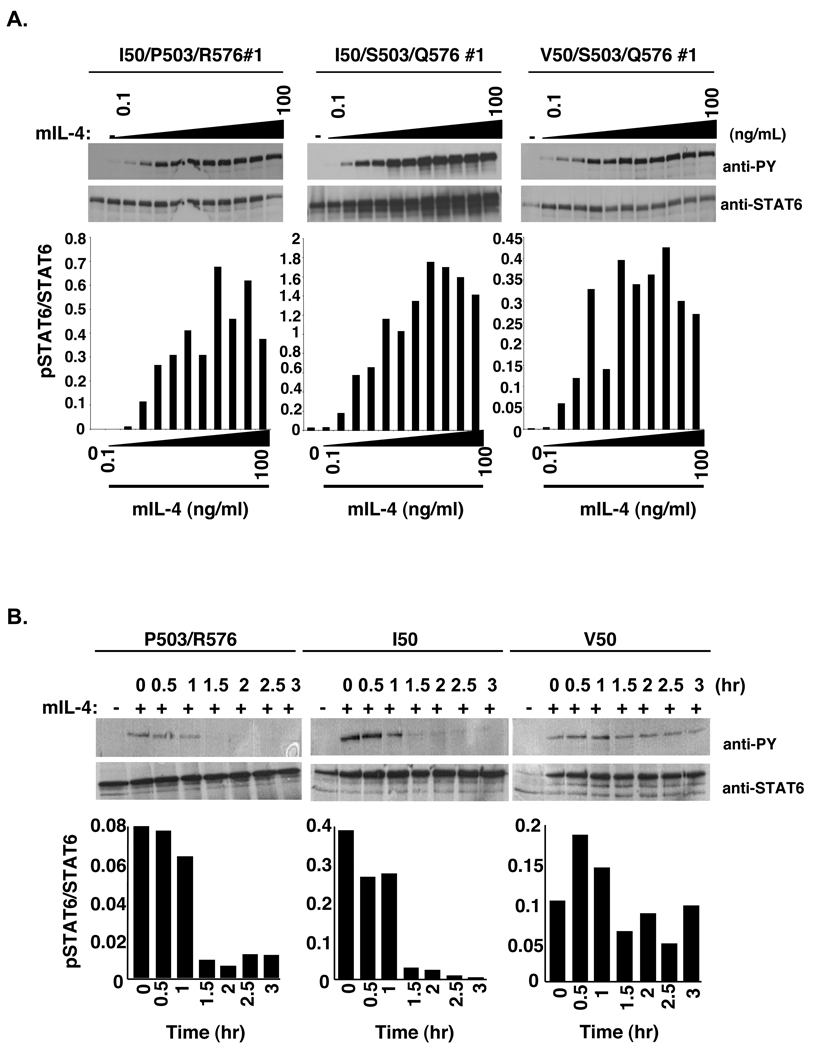
To determine the effects of the IL-4Rα polymorphisms on the activation of STAT6, a dose response was performed. P503R576-, I50-, and V50-IL-4Rα U937 clones were stimulated with increasing concentrations of murine IL-4 followed by analysis of STAT6 tyrosine phosphorylation (Figure 2A). Murine IL-4 stimulated the tyrosine phosphorylation of STAT6 with similar sensitivity in P503R576-, I50-, and V50-IL-4Rα U937 clones reaching half-maximal phosphorylation at 0.5–1 ng/ml. In contrast to studies in lymphocytes (22,24), these results indicate that P503/R576-, I50-, and V50-IL-4Rα do not exert their effects by substantially increasing sensitivity to IL-4. However, these results do not indicate whether the IL-4Rα polymorphisms regulate STAT6 activation levels through other means.
Figure 2. Analysis of STAT6 phosphorylation and dephosphorylation in the presence or absence of IL-4.
A. U937 cells expressing the murine P503/R576-, I50-, or V50-IL-4Rα were stimulated with various concentrations of murine IL-4 (0.1 ng/mL, 0.25 ng/mL, 0.5 ng/mL, 1.0 ng/mL, 2.5 ng/mL, 5.0 ng/mL, 10.0 ng/mL, 12.5 ng/mL, 25.0 ng/mL, 50.0 ng/mL, and 100.0 ng/mL) for 15 minutes. The cells were lysed, STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody to detect phosphotyrosine. The blots were stripped and reprobed with an anti-STAT6 antibody to detect STAT6. The film was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and graphed. B. U937 cells expressing the murine P503/R576-, I50-, or V50-IL-4Rα were stimulated in either the absence or presence of murine IL-4 for 15 minutes. Post stimulation, the IL-4 was washed out and the cells were re-cultured at 37°C in selection RPMI. The cells were lysed at the indicated time points and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blot was stripped and reprobed with an anti-STAT6 antibody to detect STAT6. The film was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and graphed.
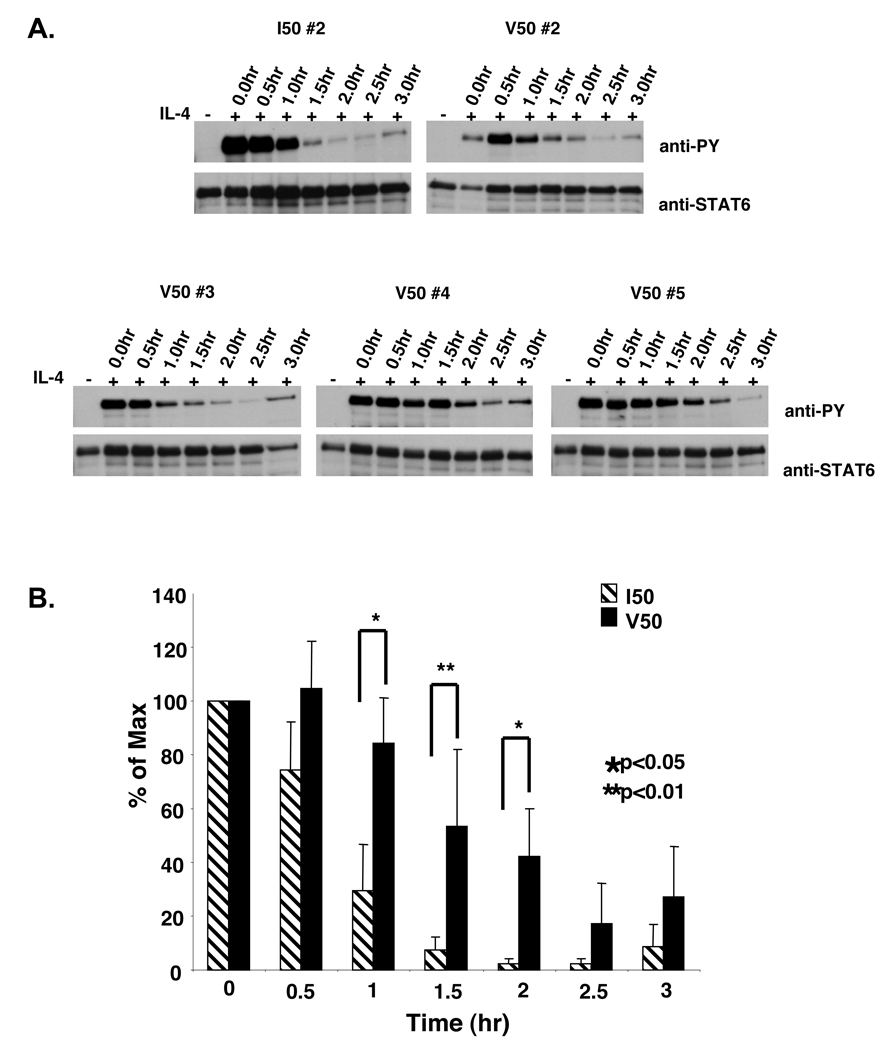
Studies analyzing the effects of a Y713F mutation in the IL-4Rα chain on IL-4 signaling revealed a decrease in the rate of dephosphorylation of STAT6 after excess free IL-4 was removed from the culture (30). These results were supported by studies comparing signaling in cells from viable motheaten mice and cells from WT mice, in which there was a decrease in the rate of dephosphorylation of STAT6 in cells from the viable motheaten mice after IL-4 removal (30). These findings suggested the possibility that the polymorphisms may impact the decay of STAT6 phosphorylation and not the efficiency of STAT6 activation per se. To determine whether the IL-4Rα polymorphisms exerted an effect on the decay of STAT6 phosphorylation, P503R576-, I50-, and V50-IL-4Rα U937 clones were stimulated with murine IL-4, the excess cytokine was washed away, and the cells were cultured in the absence of cytokine. Tyrosine phosphorylation of STAT6 was analyzed by immunoprecipitation and immunoblotting every thirty minutes over three hours (Figure 2B). After removal of IL-4, the levels of tyrosine phosphorylated STAT6 declined over time. Interestingly, the loss of tyrosine phosphorylated STAT6 occurred at approximately 1.5 hours after removal of IL-4 in both P503R576- and I50-IL-4Rα U937 clones. Similar results were obtained when parental U937 cells were first stimulated with human IL-4 (data not shown). However, the levels of tyrosine phosphorylated STAT6 remained elevated up to 3 hours after removal of murine IL-4 in V50-IL-4Rα U937 clones. Additional clones expressing the I50 or V50-IL-4Rα were also analyzed (Figure 3A,B). The enhanced phosphorylation of STAT6 observed in the V50-IL-4Rα expressing cells was highly significant at 1.0, 1.5, and 2.0 hours post-IL-4 removal.
Figure 3. Cytokine washout analysis of STAT6 phosphorylation in IL-4 stimulated I50- and V50-IL-4Rα U937 clones.
A. I50- and V50-IL-4Rα U937 clones were stimulated in either the absence or presence of murine IL-4. Post stimulation, the IL-4 was washed out and the cells were re-cultured at 37°C in selection RPMI for varying times. The cells were lysed at the indicated time points and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blot was stripped and reprobed with an anti-STAT6 antibody to detect STAT6. B. The film was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and the percent max was determined and graphed using Microsoft Excel. The average of 3 I50-IL-4Rα clones and 5 V50-IL-4Rα clones is shown +/− the SEM. The students T-test was used to calculate statistical significance.
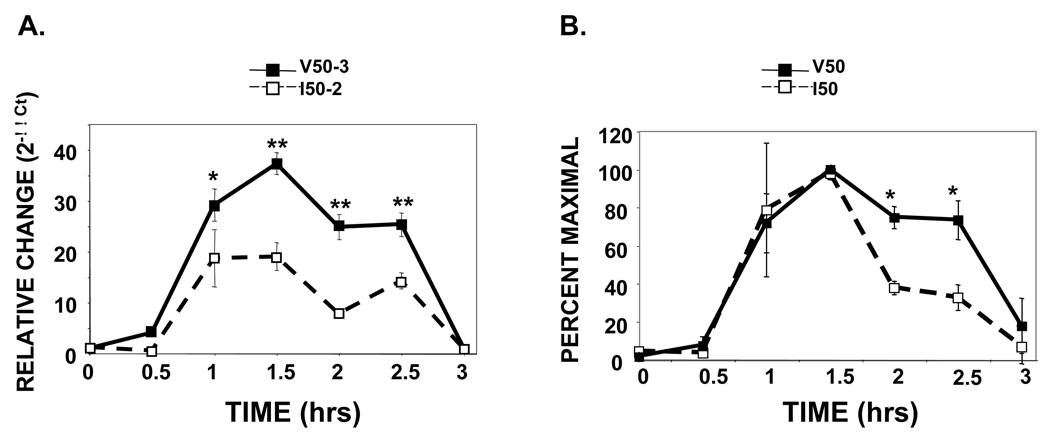
To determine whether this enhanced persistence of STAT6 phosphorylation observed in the V50-IL-4Rα U937 clones had biological implications, we analyzed the expression of a STAT6-dependent gene that would be regulated during this time frame. In previous studies, we found that the SOCS family member CIS was rapidly induced by IL-4 in U937 cells (30). To determine whether the IL-4Rα polymorphisms exerted an effect on the induction and decay of CIS mRNA, I50- and V50-IL-4Rα U937 clones were stimulated with murine IL-4 for 15 minutes, the excess cytokine was washed away, and the cells were cultured in the absence of cytokine. The relative abundance of CIS mRNA was analyzed by q-PCR (Figure 4). In both the I50- and V50-IL-4Rα U937 clones, we observed the induction of CIS mRNA after 1 hour of further culture, with maximal induction occurring after 1.5 hours. As expected, these kinetics represented a lag between the phosphorylation of STAT6 (maximal after 15 minutes of IL-4 treatment) and the induction of CIS mRNA (Figure 4A). In absolute terms, the fold enhancement in CIS mRNA was typically greater in the V50-IL-4Rα-expressing cells. The relative abundance of CIS mRNA remained elevated for 2.5 hours in the V50-IL-4Rα expressing cells while it declined in the I50-IL-4Rα expressing cells. The induction of CIS mRNA in several clones expressing the I50- or V50-IL-4Rα was analyzed in independent experiments and expressed as percent maximal response (Figure 4B). Using this representation, which controls for the differential levels of CIS mRNA induction, the persistence of CIS mRNA observed in the V50-IL-4Rα expressing cells was significantly greater at 2.0 and 2.5 hours post-IL-4 removal.
Figure 4. U937 cells expressing the V50-IL-4Rα show prolonged elevation of CIS mRNA after a short IL-4 treatment followed by cytokine washout.
I50- and V50-IL-4Rα U937 clones were stimulated in either the absence or presence of 10 ng/mL murine IL-4 for 15 minutes. Post stimulation, the IL-4 was washed out and the cells were re-cultured for various times. Total RNA was harvested and complimentary DNA was generated. Real-time PCR analysis using specific primers for human CIS was performed in triplicate. A. The results are expressed as average fold increase of CIS mRNA +/− the standard deviation compared to that of unstimulated cells. HPRT was used as the internal reference. A representative experiment analyzing the I50-2 and the V50-3 clones is shown. The students T-test was used to calculate statistical significance (*, p< 0.05; **, p< 0.001). B. Similar experiments were performed using 2 different clones of each variant multiple times. The percentage of maximal CIS induction was calculated for each clone in individual experiments. The average percent maximal induction (n=4) +/− the standard deviation is shown. The students T-test was used to calculate statistical significance (*, p< 0.05).
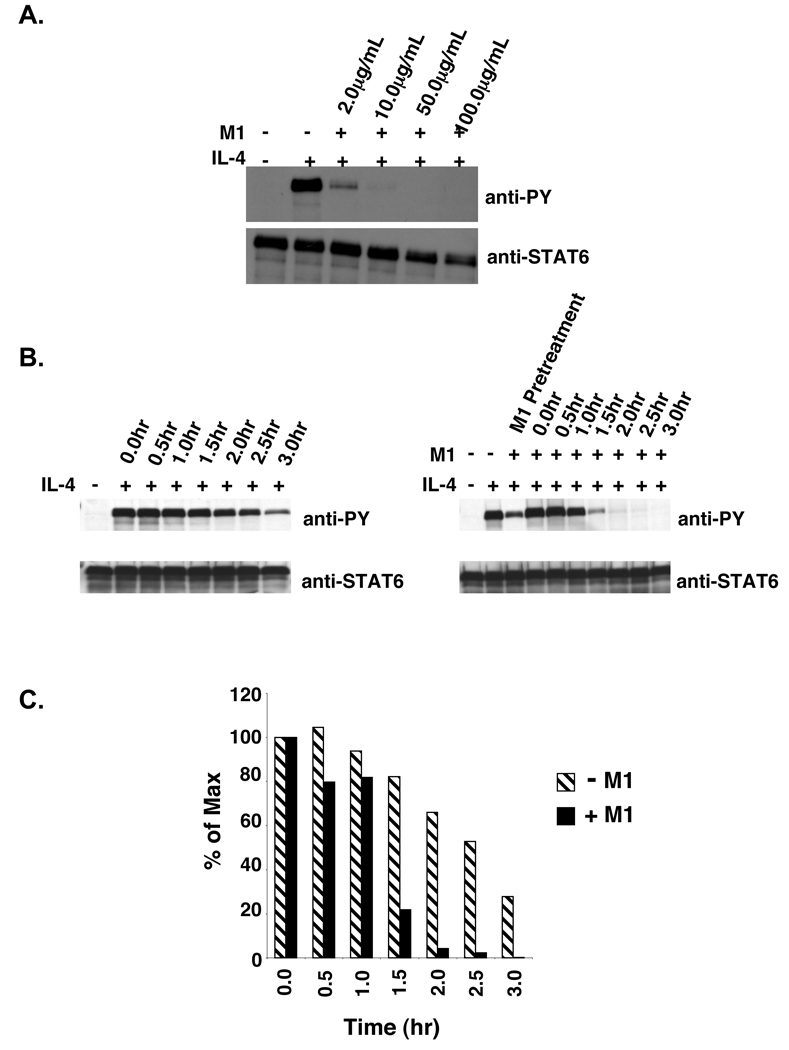
The persistence of STAT6 phosphorylation observed in V50-IL-4Rα U937 clones compared to I50-IL-4Rα U937 clones and the location of these polymorphisms in the extracellular domain of the IL-4Rα chain suggested that the differences in STAT6 phosphorylation could be due to repeated binding of IL-4 to the polymorphic IL-4Rα chain and continual signaling. The binding of murine IL-4 to the murine IL-4Rα can be blocked using the M1 anti-IL-4Rα blocking antibody and vice-versa (31). To determine the optimal concentration of the M1 blocking antibody that results in complete inhibition of IL-4 induced STAT6 phosphorylation, V50-IL-4Rα U937 clones were pretreated for 60 minutes with various concentrations of the M1 anti-IL-4Rα blocking antibody followed by treatment with murine IL-4 (Figure 5A). The optimal concentration of the M1 anti-IL-4Rα blocking antibody that induced complete inhibition of STAT6 phosphorylation in V50-IL-4Rα U937 clones was 10 µg/mL. To determine if renewed IL-4 binding to the V50-IL-4Rα was necessary for the prolonged phosphorylation of STAT6, V50-IL-4Rα U937 clone #5 was stimulated in the absence or presence of 10 ng/mL murine IL-4. The cytokine was washed away and the cells were cultured in the absence or presence of M1 anti-IL-4Rα blocking antibody. Tyrosine phosphorylation of STAT6 was analyzed by immunoprecipitation and immunoblotting over time (Figure 5B,C). IL-4 stimulated V50-IL-4Rα U937 cells treated with the M1 blocking antibody showed an increase in the rate of dephosphorylation of STAT6 in comparison to untreated V50-IL-4Rα U937 cells. We further compared the rate of STAT6 dephosphorylation in I50- and V50-IL-4Rα U937 clones treated with the M1 blocking antibody (Figure 6A,B). The rate of dephosphorylation of STAT6 in IL-4 stimulated V50-IL-4Rα U937 cells treated with the M1 blocking antibody was similar to the rate of STAT6 dephosphorylation observed in I50-IL-4Rα U937 clones. These observations indicate that the effect of the V50 polymorphism on the persistence of STAT6 phosphorylation is dependent on renewal of IL-4 binding to unoccupied receptors.
Figure 5. M1 blocking of murine IL-4 stimulated V50-IL-4Rα U937 clones.
A. V50-IL-4Rα U937 cells were treated with various concentrations of the M1 anti-mIL-4Rα blocking antibody (2.0 µg/mL, 10.0 µg/mL, 50.0 µg/mL, and 100.0 µg/mL) for 60 minutes. The cells were then stimulated with 10 ng/mL murine IL-4 for 15 minutes. The cells were lysed and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blot was stripped and reprobed with an anti-STAT6 antibody to detect STAT6. B. V50-IL-4Rα U937 clone #5 was stimulated in either the absence or presence of 10 ng/mL murine IL-4 for 15 minutes. Post stimulation, the IL-4 was washed out and the cells were re-cultured in the presence or absence of 10 µg/mL of the anti-murine IL-4Rα blocking antibody M1. The cells were lysed at the indicated time points and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blots were stripped and reprobed with an anti-STAT6 antibody to detect STAT6. C. The film shown in Panel B was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and the percent max was determined and graphed.
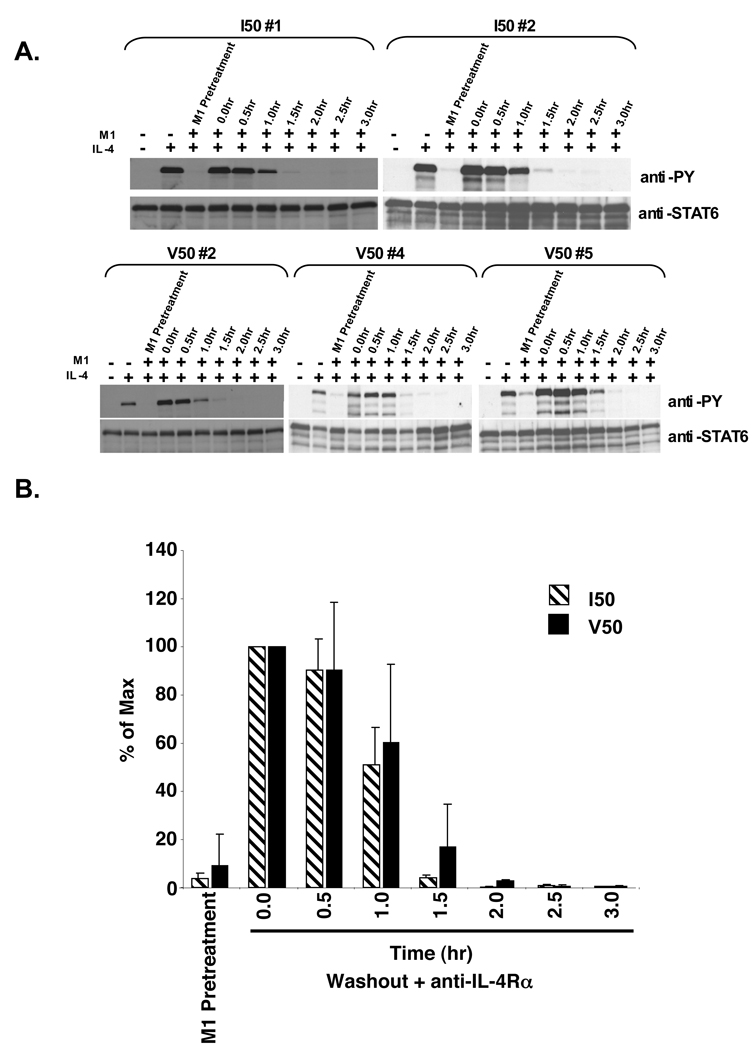
Figure 6. Cytokine washout analysis of STAT6 phosphorylation levels in IL-4 stimulated and M1 treated I50- and V50-IL-4Rα U937 clones.
A. I50- and V50-IL-4Rα U937 clones were stimulated in either the absence or presence of 10 ng/mL murine IL-4 and M1 (10 µg/ml) as indicated for 15 minutes. Post stimulation, the IL-4 was washed out and the cells were re-cultured in the presence of 10 µg/mL of the anti-murine IL-4Rα blocking antibody M1. The cells were lysed at the indicated time points and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blots were stripped and reprobed with an anti-STAT6 antibody to detect STAT6. B. The film was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and the percent max was determined and graphed using Microsoft Excel. The average of 3 I50-IL-4Rα clones and 3 V50-IL-4Rα clones is shown +/− the SEM.
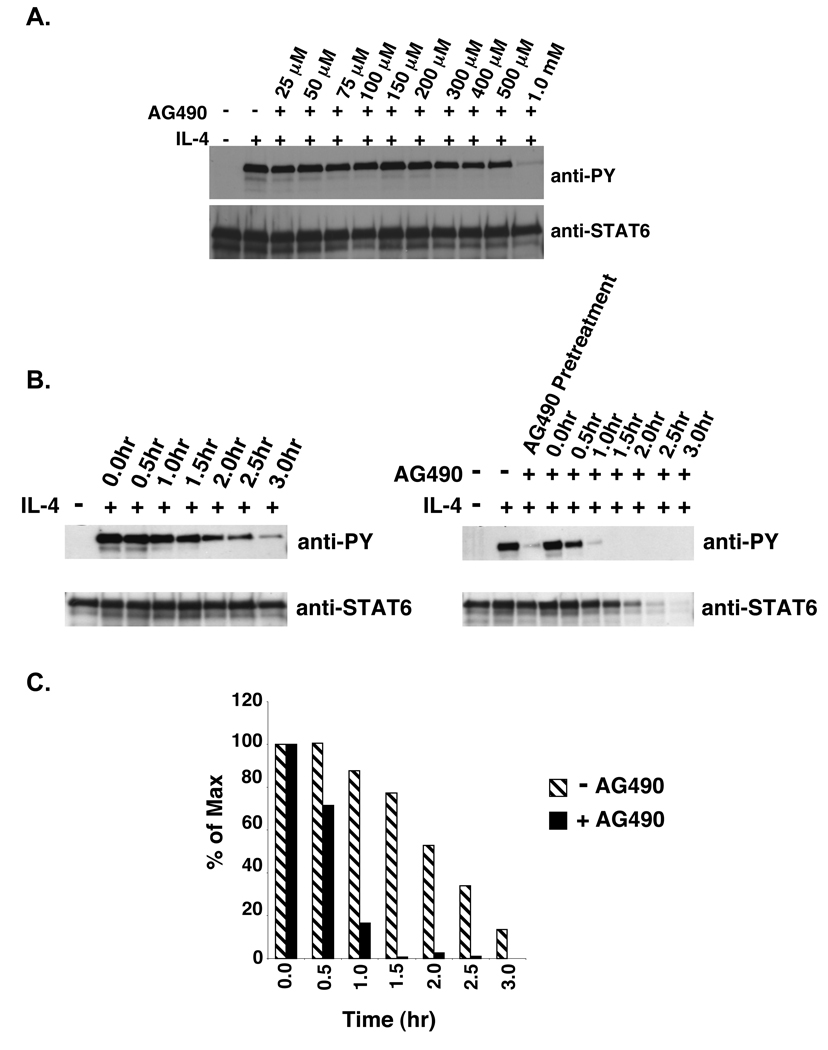
To determine if the continual activation of JAK is necessary to support the prolongation of STAT6 phosphorylation in V50-IL-4Rα U937 clones, the JAK inhibitor AG490 was added after the removal of IL-4 to abrogate JAK activity during the washout stage. To determine the optimal concentration of AG490 that results in complete inhibition of IL-4 induced STAT6 phosphorylation, U937 cells were pretreated for 60 minutes with various concentrations of AG490 followed by stimulation with human IL-4 (Figure 7A). The optimal concentration of AG490 that induced complete inhibition of STAT6 phosphorylation in U937 cells was 1.0 mM. This concentration is considerably higher than that needed to block STAT6 phosphorylation in the murine B lymphoma cell line M12.4.1 (50 µM), and may be a property of these leukemic U937 cells (30). To assess STAT6 tyrosine phosphorylation during the cytokine washout stage while in the presence of AG490, V50-IL-4Rα U937 clone #5 was stimulated with murine IL-4. The cytokine was washed away, and the cells were cultured in the absence or presence of 1.0 mM AG490. Tyrosine phosphorylation of STAT6 was analyzed by immunoprecipitation and immunoblotting over time (Figure 7B,C). Treatment of V50-IL-4Rα U937 cells with AG490 resulted in an increase in the rate of dephosphorylation of STAT6 in comparison to untreated V50-IL-4Rα U937 cells. Again, we compared the rate of STAT6 dephosphorylation in I50- and V50-IL-4Rα U937 clones treated with AG490 (Figure 8A,B). There was no loss of viable cell number under these conditions for up to 2.5 hours (data not shown). In the presence of AG490, there was no significant difference in the levels of STAT6 phosphorylation found in V50- or I50-IL-4Rα expressing cells after IL-4 removal (Figure 8A,B). These results indicate that the prolongation of STAT6 phosphorylation observed in V50-IL-4Rα U937 clones is a result of new binding of IL-4 to the V50-IL-4Rα and new JAK-dependent signaling.
Figure 7. Cytokine washout analysis of STAT6 phosphorylation in V50-IL-4RαU937 clones stimulated with IL-4 in the absence or presence of AG490.
A. U937 cells were treated with various concentrations of the JAK inhibitor AG490 for 60 minutes. The cells were then stimulated with 10 ng/mL IL-4 for 15 minutes. The cells were lysed and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blot was stripped and reprobed with an anti-STAT6 antibody to detect STAT6. B. V50-IL-4Rα U937 clones were stimulated in either the absence or presence of 10 ng/mL murine IL-4 for 15 minutes. Post stimulation, the IL-4 was washed out and the cells were re-cultured at 37°C in the presence or absence of 1.0 mM of AG490 for various times. STAT6 was immunoprecipitated from cell lysates and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blots were stripped and reprobed with an anti-STAT6 antibody to detect STAT6. C. The film in Panel B was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and the percent max was determined and graphed.
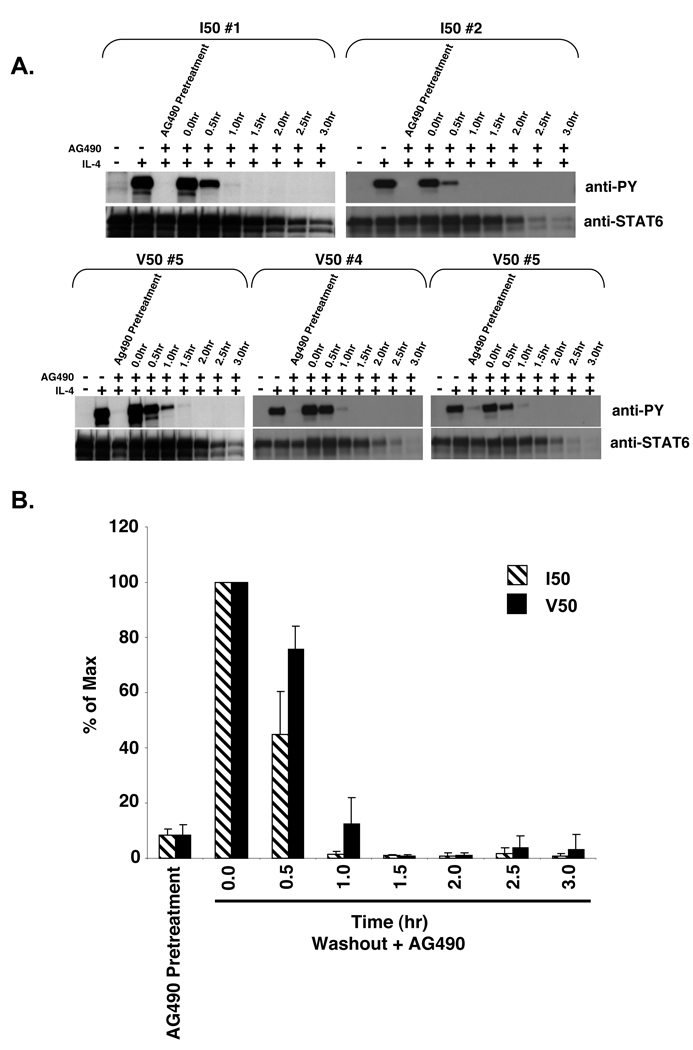
Figure 8. Cytokine washout analysis of STAT6 phosphorylation in IL-4 stimulated and AG490 treated I50- and V50-IL-4Rα U937 clones.
A. I50- and V50-IL-4Rα U937 clones were stimulated in either the absence or presence of 10 ng/mL murine IL-4 or 1 mM AG490 as indicated for 15 minutes. Post stimulation, the IL-4 was washed out and the cells were re-cultured in the presence of 1.0 mM of AG490. The cells were lysed at the indicated time points and STAT6 was immunoprecipitated and subjected to western blot analysis using an anti-phosphotyrosine antibody. The blots were stripped and reprobed with an anti-STAT6 antibody to detect STAT6. B. The film was scanned and NIH-Image 1.63 was used to determine the densities of the bands developed on the western blots. The ratio of phosphorylated STAT6 to total STAT6 was calculated and the percent max was determined and graphed. The average of 3 I50-IL-4Rα clones and 3 V50-IL-4Rα clones is shown +/− the SEM.
Since the excess free IL-4 was washed away after the initial stimulation phase, any new IL-4 binding to unoccupied receptors must come from IL-4 remaining at the cell surface during the secondary culture. To determine if murine IL-4 was detectable on the surface of the cells, I50- and V50-IL-4Rα U937 clones were stimulated in the absence or presence of murine IL-4 for 15 minutes. The cytokine was washed away and the cells were cultured for an additional 30 minutes at 37°C. The presence of murine IL-4 on the surface of the cells was analyzed by FACS using an anti-IL-4 antibody that is not influenced by binding to the IL-4Rα (Figure 9). The combination of murine IL-4 followed by anti-IL-4 detected cell surface IL-4 on I50- and V50-IL-4Rα expressing cells (Figure 9A); the staining intensity was similar to the intensity observed using the M1 antibody to directly detect IL-4Rα on the surface of these cells. Interestingly, murine IL-4 was not detected on I50-IL-4Rα U937 cells after the cytokine washout or secondary culture. However, murine IL-4 was detected on the surface of V50-IL-4Rα U937 cells immediately after washing and after an additional 30 minutes of culture at 37°C (Figure 9B).
Figure 9. Prolonged binding of IL-4 to V50-IL-4Rα.
A. U937 clones expressing the I50- or V50-IL-4Rα were analyzed for expression of IL-4Rα by M1 staining or by IL-4 binding. Cells were treated with 10.0 ng/mL of murine IL-4 for 15 minutes at 4°C. The cells were isolated and stained for murine IL-4 bound to the receptors using the biotin rat anti-mouse IL-4 monoclonal antibody. B. U937 clones expressing the I50- or V50-IL-4Rα were treated with 10.0 ng/mL of murine IL-4 for 15 minutes. The cells were isolated and separated into three groups: (1) IL-4 treatment, (2) IL-4 removal, (3) 30 minutes post-IL-4 removal. The cells were stained for murine IL-4 bound to the polymorphic receptors using the biotin rat anti-mouse IL-4 monoclonal antibody.
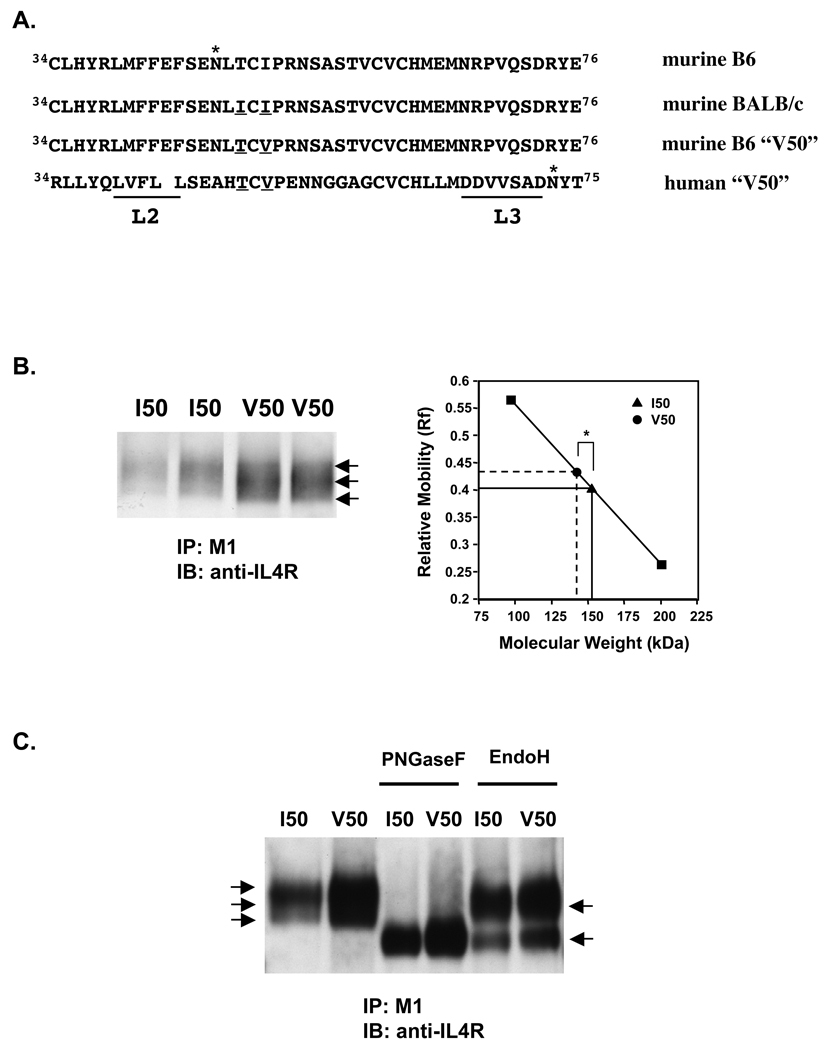
The prolongation of STAT6 phosphorylation observed in V50-IL-4Rα U937 cells appears to require new binding of murine IL-4 to the IL-4Rα and JAK activation. These results suggest differences in interactions with the I50- and V50-IL-4Rα. It has been suggested that binding of IL-4 to the IL-4Rα may be affected by the glycosylation of either IL-4 or IL-4Rα (10,32). Expression of Ile49 in murine IL-4Rα either as a natural polymorphism found in BALB/c mice or as an induced mutation leads to the abrogation of an N-glycosylation site at Asn47, a marked enhancement of mobility on SDS-gels, and a 7–16 fold increase in the dissociation rate when compared to murine IL-4Rα expressing Thr49 (10). Based on the crystal structure of human IL-4Rα, this region of the receptor is not predicted to make direct contact with IL-4 but it is near the binding loop 2 (33) (Figure 10A). While the conservative change from isoleucine to valine at position 50 in the extracellular domain of the murine IL-4Rα would not directly affect the consensus glycosylation site Asn47XThr, the position of the residue is near this glycosylation site and could influence oligosaccharide processing (Figure 10A). Therefore, to determine if there are glycosylation differences between I50- and V50-IL-4Rα, the murine IL-4Rα was immunoprecipitated from I50- and V50-IL-4Rα U937 clones using M1 and analyzed by immunoblotting. Both forms of murine IL-4Rα migrated as broad diffuse bands as is expected for a cell surface glycoprotein with complex and heterogenous oligosaccharides. However, the major species of V50-IL-4Rα precipitated from U937 cells migrated significantly faster when compared to the migration of I50-IL-4Rα (Figure 10B). Similar results were obtained using a rabbit anti-carboxyterminal specific antibody or wheat germ agarose for the immunoprecipitation (data not shown). This difference correlates with a smaller predicted molecular weight for V50-IL-4Rα in comparison with the predicted molecular weight of ~150 kDa for I50-IL-4Rα (Figure 10B). The enhanced mobility of the V50-IL-4Rα on SDS-gels suggests an alteration in glycosylation patterns.
Figure 10. Alteration of mobility of V50-IL-4Rα on SDS-PAGE.
A. The amino acid residues of the extracellular domain of the murine and human IL-4Rα surrounding position 50 are shown. The threonine and isoleucine polymorphic residues located at position 49 are underlined. The IL-4-binding regions of the human IL-4Rα, loop 2 (L2) and loop 3 (L3), are underlined. The asterisk above Asn47 in the C57BL/6 IL-4Rα sequence and above Asn73 in the human IL-4Rα sequence denotes them as confirmed sites of glycosylation. The Asn47 site is not glycosylated in the BALB/c-IL-4Rα. B. Lysates were prepared from several clones of U937 expressing I50- or V50-IL-4Rα. The IL-4Rα was immunoprecipitated with M1 anti-IL-4Rα monoclonal antibody. The samples were separated by SDS-PAGE and western blotted with rabbit anti-IL-4Rα. Arrows indicate 3 different migrating species of the IL-4Rα. The relative mobility (Rf) for the major band in each lane was calculated as described in Materials and Methods. The averages (n=3 for I50, n=5 for V50) of the Rf values from several clones run in 2 independent experiments +/− SEM is shown (*p<0.05). The SEM is too low to be seen in the graph. C. Lysates were prepared from several clones of U937 expressing I50- or V50-IL-4Rα. IL-4Rα was immunoprecipitated with M1 anti-IL-4Rα monoclonal antibody and subjected to enzymatic digest with either PNGase F or Endo H. The samples were separated by SDS-PAGE and western blotted with rabbit anti-IL-4Rα.
To determine if the smaller predicted molecular weight of the V50-Il-4Rα is due to an alteration in glycosylation, the murine IL-4Rα was immunoprecipitated from I50- and V50-IL-4Rα U937 clones and subjected to enzymatic digest of the glycoproteins using PNGase F or Endo H (Figure 10C). Digestion of murine IL-4Rα from I50- or V50-IL-4Rα U937 cells with PNGase F, an enzyme that cleaves all N-linked oligosaccharides, resulted in a single band that migrated similarly on SDS-Gels. The predicted molecular weight of the digested receptor was ~130 kDa, indicating that ~20 kDa of the IL-4Rα is comprised of N-linked sugars. Digestion of both receptor types with Endo H, an enzyme that only cleaves high mannose N-linked oligosaccharides, resulted in 2 bands indicating the IL-4Rα does contain some high mannose oligosaccharide chains. The similar migration patterns of PNGase F- or Endo H- digested mIL-4Rα from I50- and V50-IL-4Rα U937 suggest that the enhanced mobility of V50-IL-4Rα is due to glycosylation differences. Taken together, these results suggest that subtle glycosylation differences may modulate IL-4 binding parameters leading to alterations in the persistence of STAT6 tyrosine phosphorylation.
DISCUSSION
Since the first report in 1997 (13) there have been numerous studies linking IL-4Rα polymorphisms to allergic phenotypes in patient populations. However, a direct causal link between a specific polymorphism and an effect on IL-4-induced signaling has proved difficult to demonstrate and the literature is rife with controversial reports (13–16,18–20,22,24–28,34,35). One of the strongest associations is that of a hydrophobic residue in the ectodomain of the IL-4Rα; this association is particularly challenging to understand in conceptual terms due to the uncertainty of how such a change could alter allergy-related signaling. Here, we have shown a direct effect of the V50 polymorphism in the IL-4Rα on the persistence of STAT6 activation in response to IL-4 and in the persistence of a STAT6-dependent gene. Previous studies focused on the effects of polymorphisms on STAT6 phosphorylation in the continuous presence of saturating concentrations of IL-4. The induction of STAT6 phosphorylation under these conditions analyzes differences in STAT6 phosphorylation at equilibrium and does not account for the regulation of STAT6 dephosphorylation. STAT6 is rapidly phosphorylated and this activity remains elevated in cells cultured in the continuous presence of IL-4 for 24 hours (36). However, Hanson et al. showed that STAT6 is dephosphorylated in a time dependent manner after the removal of excess free IL-4 (30). Under such conditions, we observed enhanced longevity of phosphoSTAT6 in V50-IL-4Rα U937 cells as compared to I50-IL-4Rα or endogenous human IL-4Rα in U937 cells. This enhanced longevity of STAT6 phosphorylation over time indicates that the cycle of STAT6 activation/deactivation is altered in the V50-IL-4Rα U937 cells.
This alteration was associated with enhanced induction and persistence of CIS mRNA in U937 cells suggesting the V50 polymorphism may indeed influence biological responses.
STAT6 dephosphorylation is regulated through several different mechanisms including the action of tyrosine phosphatases, induction of SOCS family members, and the action of the proteosome. Since the V50 polymorphism is located in the extracellular domain of the IL-4Rα chain, we suspected that the prolongation of STAT6 phosphorylation in V50-IL-4Rα U937 cells was likely not due to modulation of these largely intracellular mechanisms. We found that the prolongation of STAT6 phosphorylation in V50-IL-4Rα U937 cells was dependent upon continued JAK activity. This result is in contrast to the effect of JAK inhibition using AG490 on the prolonged STAT6 phosphorylation seen in cells expressing a Y713F-IL-4Rα (30). This mutation abrogates a cytoplasmic SHP-1 docking site in the IL-4Rα, preventing the dephosphorylation of STAT6 by SHP-1 thus enhancing the persistence of STAT6 phosphorylation. AG490 had no effect on this phenotype indicating that the effect of the Y713F mutation was independent of continued JAK activation.
The prolongation of STAT6 phosphorylation in V50-IL-4Rα U937 cells and its abrogation by a JAK inhibitor suggested that the prolongation of STAT6 phosphorylation could be a result of renewed IL-4 binding to cell surface receptors. While most of the free IL-4 was removed, IL-4 bound to a receptor could dissociate and bind to a nearby unoccupied receptor starting a new signaling cascade. Indeed, IL-4 binding to the IL-4Rα is characterized by a fast on and a fast off rate. Several investigators previously proposed that this fast off rate of IL-4 could result in new binding and new signaling (10,37). We found that culturing V50-IL-4Rα U937 cells in the presence of the M1 anti-IL-4Rα blocking antibody led to the abrogation of the persistence of STAT6 phosphorylation and a similar rate of decrease to that observed in I50-IL-4Rα U937 cells. This occurred in all clones regardless of the expression levels of receptor. Since M1 cannot bind to an occupied receptor, these results suggest that the persistence of STAT6 phosphorylation in V50-IL-4Rα U937 cells requires new IL-4 binding to an unoccupied IL-4 receptor inducing new JAK and STAT6 activation. This effect became apparent when excess free IL-4 was washed away. In support of this hypothesis, cell surface bound IL-4 was detected on cells expressing V50-IL-4Rα 30 minutes after free IL-4 was washed away, while bound IL-4 was undetectable on I50-IL-4Rα- expressing cells at the same time point.
In addition to activating signal transduction, IL-4 binding to its receptor induces receptor-mediated internalization of IL-4 followed by its degradation (37). The receptors are then recycled to the plasma membrane. Galizzi et al. reported that the rate of IL-4 internalization in the human B-cell line JiJoye was twice as fast as the rate of IL-4 dissociation from the cell surface (t1/2 13 minutes vs t1/2 25 minutes, respectively) (37). Furthermore, mutation of IL-4 in the region that makes contact with γc resulted in less IL-4 internalization due to an increased dissociation rate (38). Several investigators have proposed that alterations in dissociation rates could lead to a reduction in internalization and degradation of IL-4, allowing IL-4 to remain available for new receptor binding on the surface of the cells (10,37). While this model remains controversial, our results in this transfected U937 cell system support the possibility. However, in vivo studies in allergic IL-13−/− mice do not support such a model. The T49I polymorphism in the extracellular domain of the IL-4Rα derived from BALB/c mice eliminates the N-linked glycosylation site at N47, a site present in the IL-4Rα derived from C57Bl/6 mice; this change in glycosylation was associated with an increase in the dissociation rate of IL-4 (10). Webb et al reported that the ability of IL-4 to promote allergic lung inflammation in the absence of IL-13 was substantially greater in mice expressing the C57Bl/6 polymorphism (T49) with the slower dissociation rate (39).
The question remains as to the mechanism by which the conservative substitution of an amino acid that is not in the IL-4 binding domain (I to V at position 50) might influence ligand binding parameters and signal transduction (33,40). Such a change in the murine IL-4Rα or the human IL-4Rα would not be expected to directly impact the addition of the core oligosaccharide to the closest N-linked site (N47 or N73 respectively). However, the processing of an individual N-linked sugar is determined by the configuration of the protein and its accessibility to the processing enzymes as the nascent protein travels through the endoplasmic reticulum and the golgi in a particular cell type. We identified differences in the mobility of I50-IL-4Rα and V50-IL-4Rα derived from U937 cells on SDS-gels; these mobility differences were abrogated by removal of N-linked oligosaccharides suggesting that the polymorphisms influence the processing of glycosylation.
However, the nature of the differences is unclear; it does not appear to be limited to differences in terminal sialic acid residues as neuraminidase treatment did not alter the persistence of phosphorylated STAT6 in I50- or V50-IL-4Rα-expressing cells (data not shown). Characterization of the processing of the carbohydrate side-chains will require extensive analyses of the carbohydrate biochemistry of the I50-IL-4Rα and V50-IL-4Rα.
While glycosylation of IL-4 or IL-4Rα is not required for binding, it has been shown that the glycosylation state of either the IL-4Rα chain or IL-4 can influence ligand binding. Unglycosylated murine IL-4 was more active than its glycosylated counterpart in stimulating the D10 cell line (32). As discussed above, the abrogation of an N-glycosylation site in the murine IL-4Rα near loop 2 lead to an increase in the dissociation rate of IL-4 (10). Therefore it is possible the differences in signaling between the I50- and V50-IL-4Rα are ultimately a result of glycosylation differences between the receptors. Early equilibrium binding studies did not detect any differences in overall binding affinities of human IL-4 for the I50- or V50-humanIL-4Rα (14,16,19). A detailed evaluation of glycosylation patterns has not been performed on the polymorphic human IL-4Rα. It is interesting to note that the human IL-4Rα has an N-linked site at position 73, a site confirmed to be glycosylated (41). This site is adjacent to loop 3 which contains amino acids important for interacting with IL-4. Our comparison of the I50- vs V50-polymorphisms was performed in a defined system using the murine IL-4Rα expressed in a human cell line U937. This system certainly does not recapitulate the more complex genetic background found in the human population. Nevertheless, we speculate that the V50 polymorphism may also impact glycosylation of the human IL-4Rα and that the relative impact may be cell type specific. This glycosylation could influence IL-4 binding parameters leading to alterations in persistence of IL-4 signaling and gene expression in human patients. We are currently designing model systems to evaluate these possibilities and to test the contribution of the V50 polymorphism on IL-13 induced signaling.
ACKNOWLEDGEMENTS
We would like to acknowledge Ms. Elena Semenova and Xiulan Qi for excellent technical support. We also thank Dr. Alexander Mackerell for discussions of the potential effects of the polymorphisms on IL-4Rα structure.
Footnotes
This work was supported by PHS grant AI038985 (ADK) and T32HL007698 (AQF and NMH).
This work was submitted in partial fulfillment of the requirements for the Doctor of Philosophy degree, Immunology Program, Institute for Biomedical Sciences of the Columbian School of Arts and Sciences, The George Washington University.
REFERENCES
- 1.Seder RA, Paul WE. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 2.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 3.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300(5625):1527–1528. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 4.Thornhill MH, Wellicome SM, Mahiouz DL, Lanchbury JS, Kyan-Aung U, Haskard DO. Tumor necrosis factor combines with IL-4 or IFN-gamma to selectively enhance endothelial cell adhesiveness for T cells. The contribution of vascular cell adhesion molecule-1-dependent and -independent binding mechanisms. J Immunol. 1991;146(2):592–598. [PubMed] [Google Scholar]
- 5.Bennett BL, Cruz R, Lacson RG, Manning AM. Interleukin-4 suppression of tumor necrosis factor alpha-stimulated E-selectin gene transcription is mediated by STAT6 antagonism of NF-kappaB. J Biol Chem. 1997;272(15):10212–10219. doi: 10.1074/jbc.272.15.10212. [DOI] [PubMed] [Google Scholar]
- 6.Matsukura S, Stellato C, Plitt JR, Bickel C, Miura K, Georas SN, Casolaro V, Schleimer RP. Activation of eotaxin gene transcription by NF-kappa B and STAT6 in human airway epithelial cells. J. Immunology. 1999;163(12):6876–6883. [PubMed] [Google Scholar]
- 7.Russell SM, Keegan AD, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann MC, Miyajima A, Puri RK, Paul WE, Leonard WJ. Interleukin-2 receptor gamma chain: a functional component of the interleukin-4 receptor. Science. 1993;262(5141):1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 8.Leonard WJ, Noguchi M, Russell SM. Sharing of a common gamma chain, gamma c, by the IL-2, IL-4, and IL-7 receptors: implications for X-linked severe combined immunodeficiency (XSCID) Adv Exp Med Biol. 1994;365:225–232. doi: 10.1007/978-1-4899-0987-9_23. [DOI] [PubMed] [Google Scholar]
- 9.Callard RE, Matthews DJ, Hibbert L. IL-4 and IL-13 receptors: are they one and the same? Immunol Today. 1996;17(3):108–110. doi: 10.1016/0167-5699(96)80600-1. [DOI] [PubMed] [Google Scholar]
- 10.Schulte T, Kurrle R, Röllinghof M, Gessner A. Molecular characterization and functional analysis of murine interleukin 4 receptor allotypes. J Exp Med. 1997;186(9):1419–1429. doi: 10.1084/jem.186.9.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ober C, Leavitt SA, Tsalenko A, Howard TD, Hoki DM, Daniel R, Newman DL, Wu X, Parry R, Lester LA, Solway J, Blumenthal M, King RA, Xu J, Meyers DA, Bleecker ER, Cox NJ. Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet. 2000;66(2):517–526. doi: 10.1086/302781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirakawa I, Deichmann KA, Izuhara I, Mao I, Adra CN, Hopkin JM. Atopy and asthma: genetic variants of IL-4 and IL-13 signalling. Immunol Today. 2000;21(2):60–64. doi: 10.1016/s0167-5699(99)01492-9. [DOI] [PubMed] [Google Scholar]
- 13.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337(24):1720–1725. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuyasu H, Izuhara K, Mao XQ, Gao PS, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N, Shirakawa T, Hopkin JM. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet. 1998;19(2):119–120. doi: 10.1038/472. [DOI] [PubMed] [Google Scholar]
- 15.Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, Deichmann KA. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology. 1999;96(3):365–371. doi: 10.1046/j.1365-2567.1999.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsuyasu H, Yanagihara Y, Mao XQ, Gao PS, Arinobu Y, Ihara K, Takabayshi A, Hara T, Enomoto T, Sasaki S, Kawai M, Hamasaki N, Shirakawa T, Hopkin JM, Izuhara K. Cutting edge: dominant effect of Ile50Val variant of the human IL-4 receptor alpha-chain in IgE synthesis. J Immunol. 1999;162(3):1227–1231. [PubMed] [Google Scholar]
- 17.Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kobayashi K, Imoto N, Nakahara S, Matsui A, Hamaguchi H. No association between atopy/asthma and the ILe50Val polymorphism of IL-4 receptor. Am J Respir Crit Care Med. 1999;160(1):342–345. doi: 10.1164/ajrccm.160.1.9807130. [DOI] [PubMed] [Google Scholar]
- 18.Khoo SK, Zhang G, Backer V, Porsbjerg C, Nepper-Christensen S, Creegan R, Baynam G, de Klerk N, Rossi GA, Hagel I, Di Prisco MD, Lynch N, Britton J, Hall I, Musk AW, Goldblatt J, Le Souëf PN Greenlandic Population Study Group. Associations of a novel IL4RA polymorphism, Ala57Thr, in Greenlander Inuit. J Allergy Clin Immunol. 2006;118(3):627–634. doi: 10.1016/j.jaci.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Risma KA, Wang N, Andrews RP, Cunningham CM, Ericksen MB, Bernstein JA, Chakraborty R, Hershey GK. V75R576 IL-4 receptor alpha is associated with allergic asthma and enhanced IL-4 receptor function. J Immunol. 2002;169(3):1604–1610. doi: 10.4049/jimmunol.169.3.1604. [DOI] [PubMed] [Google Scholar]
- 20.Knutsen AP, Kariuki B, Consolino JD, Warrier MR. IL-4 alpha chain receptor (IL-4Ralpha) polymorphisms in allergic bronchopulmonary aspergillosis. Clin Mol Allergy. 2006;4:3. doi: 10.1186/1476-7961-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruse S, Braun S, Deichmann KA. Distinct signal transduction processes by IL-4 and IL-13 and influences from the Q551R variant of the human IL-4 receptor alpha chain. Respir Res. 2002;3:24. doi: 10.1186/rr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephenson L, Johns MH, Woodward E, Mora AL, Boothby M. An IL-4R alpha allelic variant, I50, acts as a gain-of-function variant relative to V50 for Stat6, but not Th2 differentiation. J Immunol. 2004;173(7):4523–4528. doi: 10.4049/jimmunol.173.7.4523. [DOI] [PubMed] [Google Scholar]
- 23.Franjkovic I, Gessner A, König I, Kissel K, Bohnert A, Hartung A, Ohly A, Ziegler A, Hackstein H, Bein G. Effects of common atopy-associated amino acid substitutions in the IL-4 receptor alpha chain on IL-4 induced phenotypes. Immunogenetics. 2005;56(11):808–817. doi: 10.1007/s00251-004-0763-1. [DOI] [PubMed] [Google Scholar]
- 24.Prots I, Skapenko A, Wendler J, Mattyasovszky S, Yoné CL, Spriewald B, Burkhardt H, Rau R, Kalden JR, Lipsky PE, Schulze-Koops H. Association of the IL4R single-nucleotide polymorphism I50V with rapidly erosive rheumatoid arthritis. Arthritis Rheum. 2006;54(5):1491–1500. doi: 10.1002/art.21832. [DOI] [PubMed] [Google Scholar]
- 25.Wang HY, Shelburne CP, Zamorano J, Kelly AE, Ryan JJ, Keegan AD. Cutting edge: effects of an allergy-associated mutation in the human IL-4R alpha (Q576R) on human IL-4-induced signal transduction. J Immunol. 1999;162(8):4385–4389. [PubMed] [Google Scholar]
- 26.Patuzzo C, Trabetti E, Malerba G, Martinati LC, Boner AL, Pescollderungg L, Zanoni G, Pignatti PF. No linkage or association of the IL-4Ralpha gene Q576R mutation with atopic asthma in Italian families. J Med Genet. 2000;37(5):382–384. doi: 10.1136/jmg.37.5.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donati M, Berglundh T, Hytönen AM, Hahn-Zoric M, Hanson LA, Padyukov L. Association of the −159 CD14 gene polymorphism and lack of association of the −308 TNFA and Q551R IL-4RA polymorphisms with severe chronic periodontitis in Swedish Caucasians. J Clin Periodontol. 2005;32(5):474–479. doi: 10.1111/j.1600-051X.2005.00697.x. [DOI] [PubMed] [Google Scholar]
- 28.Suppiah V, Goris A, Alloza I, Heggarty S, Dubois B, Carton H, Antigüedad A, Mendibe M, McDonnell G, Droogan A, Hawkins S, Graham C, Vandenbroeck K. Polymorphisms in the interleukin-4 and IL-4 receptor genes and multiple sclerosis: a study in Spanish-Basque, Northern Irish and Belgian populations. Int J Immunogenet. 2005;32(6):383–388. doi: 10.1111/j.1744-313X.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- 29.Sundström C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 30.Hanson EM, Dickensheets H, Qu CK, Donnelly RP, Keegan AD. Regulation of the dephosphorylation of Stat6. Participation of Tyr-713 in the interleukin-4 receptor alpha, the tyrosine phosphatase SHP-1, and the proteasome. J Biol Chem. 2003;278(6):3903–3911. doi: 10.1074/jbc.M211747200. [DOI] [PubMed] [Google Scholar]
- 31.Beckmann MP, Schooley KA, Gallis B, Vanden Bos T, Friend D, Alpert AR, Raunio R, Prickett KS, Baker PE, Park LS. Monoclonal antibodies block murine IL-4 receptor function. J Immunol. 1990;144:50–56. [PubMed] [Google Scholar]
- 32.Thor G, Brian AA. Glycosylation variants of murine interleukin-4: evidence for different functional properties. Immunology. 1992;75(1):143–149. [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller TD, Zhang JL, Sebald W, Duschl A. Structure, binding, and antagonists in the IL-4/IL-13 receptor system. Biochim Biophys Acta. 2002;1592(3):237–250. doi: 10.1016/s0167-4889(02)00318-x. [DOI] [PubMed] [Google Scholar]
- 34.Daley T, Metcalfe DD, Akin C. Association of the Q576R polymorphism in the interleukin-4 receptor alpha chain with indolent mastocytosis limited to the skin. Blood. 2001;98(3):880–882. doi: 10.1182/blood.v98.3.880. [DOI] [PubMed] [Google Scholar]
- 35.Soriano A, Lozano F, Oliva H, García F, Nomdedéu M, De Lazzari E, Rodríguez C, Barrasa A, Lorenzo JI, Del Romero J, Plana M, Miró JM, Gatell JM, Vives J, Gallart T. Polymorphisms in the interleukin-4 receptor alpha chain gene influence susceptibility to HIV-1 infection and its progression to AIDS. Immunogenetics. 2005;57(9):644–654. doi: 10.1007/s00251-005-0041-x. [DOI] [PubMed] [Google Scholar]
- 36.Schindler C, Kashleva H, Pernis A, Pine R, Rothman P. STF-IL-4: a novel IL-4 induced signal transducing factor. Embo J. 1994;13(6):1350–1356. doi: 10.1002/j.1460-2075.1994.tb06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galizzi JP, Zuber CE, Cabrillat H, Djossou O, Banchereau J. Internalization of human interleukin 4 and transient down-regulation of its receptor in the CD23-inducible Jijoye cells. J Biol Chem. 1989;264(12):6984–6989. [PubMed] [Google Scholar]
- 38.Friedrich K, Kammer W, Erhardt I, Brändlein S, Arnold S, Sebald W. The two subunits of the interleukin-4 receptor mediate independent and distinct patterns of ligand endocytosis. Eur J Biochem. 1999;265(1):457–465. doi: 10.1046/j.1432-1327.1999.00773.x. [DOI] [PubMed] [Google Scholar]
- 39.Webb DC, Matthaei KI, Cai Y, McKenzie AN, Foster PS. Polymorphisms in IL-4R alpha correlate with airways hyperreactivity, eosinophilia, and Ym protein expression in allergic IL-13−/− mice. J. Immunol. 2004;172(2):1092–1098. doi: 10.4049/jimmunol.172.2.1092. [DOI] [PubMed] [Google Scholar]
- 40.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132(2):259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajan N, Tsarbopoulos A, Kumarasamy R, O’Donnell R, Taremi SS, Baldwin SW, Seelig GF, Fan X, Pramanik B, Le HV. Characterization of recombinant human interleukin 4 receptor from CHO cells: role of N-linked oligosaccharides. Biochem Biophys Res Commun. 1995;206(2):694–702. doi: 10.1006/bbrc.1995.1098. [DOI] [PubMed] [Google Scholar]