Abstract
We have obtained the 13Cα chemical shift tensors for each amino acid in the protein GB1. We then developed a CST force field and incorporated this into the Xplor-NIH structure determination program. GB1 structures obtained by using CST restraints had improved precision over those obtained in the absence of CST restraints, and were also more accurate. When combined with isotropic chemical shifts, distance and vector angle restraints, the root-mean squared error with respect to existing x-ray structures was better than ~1.0 Å. These results are of broad general interest since they show that chemical shift tensors can be used in protein structure refinement, improving both structural accuracy and precision, opening up the way to accurate de novo structure determination.
Introduction
Folding a protein into its native conformation generates a large range of chemical shifts.1–3 For 1H, the shift range is ~2–3 ppm, for 13C about 10 ppm, and for 15N, about 35 ppm. These shifts are often diagnostic of the type of secondary structure present; for example, Cα chemical shifts for residues in α-helices are downfield from those in β-sheet regions.4 There was, however, a gap of ~20 years between the first observation of folding-induced nonequivalence and the ability to actually predict 13C NMR shifts in proteins, using quantum chemistry.5–9 Now such calculations are more routine and open up the possibility of using quantum chemically derived chemical shift surfaces—how, e.g., the 13Cα shift varies with backbone torsion φ and ψ—in structure determination and refinement. In early work, it was shown that 13C isotropic chemical shifts could be used in protein structure refinement.10–12 Much more recently, Cavalli et al.13 have shown that complete three dimensional protein structures (having root mean squared deviation (RMSD) values of <2 Å from corresponding x-ray structures) can be determined by using solely isotropic 13C and 15N NMR chemical shifts (and empirical correlations with known structures) as experimentally determined properties. However, if there are N isotropic chemical shifts, there are 3N associated chemical shift tensor elements (as well as 3N orientations), so it should, at least in principle, be possible to obtain structures having improved accuracy and precision by using chemical shift tensor (CST) information.12,14–18
In this work, we report the structures of a solid protein, the β1-immunoglobulin binding domain of protein G (GB1), refined to high resolution through the use of Cα CST data. We first present complete measurements of the backbone Cα CST values in GB1 by combining our previous pulse sequence methods19,20 with isotopic labeling schemes that provide enhanced resolution.21–23 We then use Cα CST data to refine the structure of GB1 using theoretical Cα shielding surfaces incorporated into the Xplor-NIH program. The resulting structures have high precision (~0.2–0.3 Å backbone RMSD) and accuracy (~1.0 Å in comparison with the closest crystal structure, pdb entry 2QMT), opening up the way to using both isotropic13 as well as anisotropic chemical shift results in the de novo determination of high accuracy, solid-state protein structures.
Experimental and Computational Methods
Samples of GB1 were prepared using 1,3-13C-glycerol or 2-13C-glycerol as the primary carbon sources in the growth media.21–24 Hydrated nanocrystals (~18 mg) of each labeled sample were packed into the central 80% of limited speed, 36 μL, 3.2 mm Varian rotors (Varian, Inc., Palo Alto, California and Fort Collins, Colorado). NMR spectra were acquired using a 500 MHz Varian InfinityPlus spectrometer equipped with a 3.2 mm T3 Balun™ probe optimized for triple resonance 1H-13C-15N experiments. Pulse widths (π/2) for 1H, 13C, and 15N were 1.9 μs, 2.5 μs, and 5.0 μs, respectively. Spinning was maintained at 11.111 ± 0.002 kHz via a Varian MAS controller.
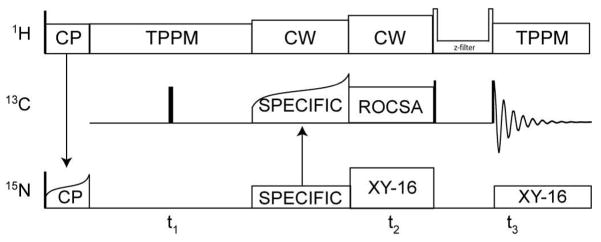
The ROCSA recoupling sequence25 was incorporated into a 3D experiment in a manner similar to that reported previously, but with a slightly modified set of isotropic (15N and 13C) and anisotropic (13C) dimensions. We refer to this pulse sequence as 15N-[13C CSA]-13C (Figure 1). Optimal resolution and sensitivity for the 15N-13C 2D planes were achieved as described,26 using the SPECIFIC CP27 condition near ωN = 5/2 ωr and ωC = 3/2 ωr. TPPM decoupling was utilized during acquisition,28 with a nutation frequency of ~75 kHz, a 14° total phase shift and 6.3 μs pulse width. SPECIFIC CP efficiency was approximately 55%, when comparing the 15N-filtered 13C spectrum to the 1D 13C spectrum based on 1H-13C CP.26 The direct 13C dimension was digitized as 3072 complex points with a 10 μs dwell time. The indirect 15N dimension (t1) was digitized as 160 real points, with an increment of 180 μs. The ROCSA dimension was digitized with 14 hypercomplex points using States-TPPI sampling29 and an increment of 180 μs (two rotor periods). The pulse delay was 3 s, resulting in a total measurement time of ~20 h per experiment. The experiment was repeated twice, to confirm that the measurements were stable to within the signal-to-noise ratio. The C441 element was used for ROCSA, sampling two complex points per ROCSA cycle.
Figure 1.
15N-[13C CSA]-13C 3D ROCSA pulse sequence. Transverse 15N coherence is created by adiabatic CP from 1H, followed by isotropic chemical shift evolution period (t1) and selective polarization transfer to 13Cα with SPECIFIC-CP.27 ROCSA25 recoupling is applied to the 13C spins, with a coherence filter (including States-TPPI hypercomplex sampling) prior to acquisition. During 13C ROCSA, 1H was decoupled with a CW field (~125 kHz nutation frequency) and 15N with rotor-synchronized π-pulses of 10 μs with XY-16 supercycling.
Spectra were processed using NMRPipe.30 The 15N interferogram was doubled by linear prediction (to 320 points) prior to Fourier transformation. Lorentzian-to-Gaussian apodization functions were applied in the two isotropic chemical shift dimensions, with a net line broadening matched to the typical linewidths observed (30 Hz for 13C and 15 Hz for 15N). Peak intensities in each 15N-13C (F1-F3) plane were determined and trajectories of the ROCSA time domain (t2) extracted using the autoFit.tcl package in NMRPipe.
The ROCSA trajectories were fit (without apodization) to exact spin simulations in the time domain, using in-house FORTRAN-77 code that combines external MINUIT minimization libraries31 with the SPINEVOLUTION32 simulation package. For lineshape fitting, we used the notational convention described by Haeberlen,33 which is standard in simulation packages such as SPINEVOLUTION. In this convention, the three axes in the Principal Axis System (PAS) are defined as δxx, δyy, and δzz, and are ordered as:
| (1) |
Two other quantities are required in order to convert from Cartesian to polar coordinates: the reduced anisotropy (δ = δzz − δiso), and the asymmetry parameter (η = (δyy − δxx)/δ). Four parameters were fit iteratively using MINUIT minimization: δ, η, a phenomenological single exponential relaxation rate, Γ1, and an amplitude scaling factor. Standard errors were determined from the error matrix calculated by MINUIT during fitting. The measured values of δ and η were then converted into the Herzfeld-Berger convention,34,35 in which tensor elements are ordered from downfield (δ11) to upfield (δ33), with δ22 in the center. The tensor rhombicity is given by two parameters, the span (Ω = δ11 − δ33) and the skew (κ = 3(δ22 − δiso)/Ω).
The fitted tensor parameters were compared to ab initio chemical shielding surfaces12,15–18 which are available for all 20 common amino acids (http://feh.scs.uiuc.edu/amino_acid.php). As described previously,12,15–18 these surfaces were computed with Hartree-Fock (HF) theory and gauge-including atomic orbitals (GIAO) methodology as implemented in the Gaussian-98 program. Coupled Hartree-Fock shielding calculations used dense 6-311++G(2d,2p) basis sets on sites of interest with 6-311G basis sets used on neighboring atoms. Ab initio CST calculations provide shielding values referenced relative to the bare nucleus, which need to be re-referenced relative to DSS, for comparison with experimentally determined chemical shifts. This procedure has been performed in previous studies by regression analysis versus experimental data from samples of known geometry, such as tripeptides with highly resolved x-ray or neutron diffraction structures. 12,15–18 This fitting protocol produces two additional parameters: a scaling factor (least squares slope) and an offset (y-intercept). Ideally, the scaling factors should be −1.0, but in practice range from −0.7 to 1.1 for different amino acid residue types, due presumably to basis set deficiencies, and of course these scaling factors need to be taken into account by the structure calculation protocol, as described below. It is possible that discrepancies between x-ray and actual geometry make small contributions to these scaling factors; however, as illustrated below, scaling factors are in absolute terms uncertain, and therefore must be considered variable parameters in order to determine molecular geometry directly. Structure calculations were performed using a customized version of Xplor-NIH36 that includes a CST potential term.
Given a molecular structure, the three theoretical CST values δnn (n=1, 2, 3) can be determined for each Cα atom using chemical shielding surfaces, and the associated backbone torsion angle values ϕ and ψ, according to the linear equation:
| (2) |
where pscale and poffset are the scaling factor and offset, respectively, and are unique for each residue type; is a 2D cubic spline representation37 of the ab initio values implemented with derivatives continuous with respect to ϕ and ψ. To restrain calculated CST values to the experimental values ( ), the following pseudo-energy was used during simulated-annealing structure determination:
| (3) |
where the force constant kCST, is varied during the course of the simulated annealing, and the sum is over all tensor orientations of all Cα atoms of a given type. If pscale and poffset are known, they can be held constant during the structure calculation. However, because these parameters are generally not known with high precision, we determined them self-consistently within the structure calculation itself. To do this, a subroutine (to be included in a new Xplor-NIH release) was developed to compute pscale and poffset for each residue type. This routine uses singular value decomposition, which is computationally inexpensive and can thus be performed at each annealing temperature during the structure calculation.
The force constant kCST was chosen to be ~0.005 kcal/ppm2 at the beginning of structure calculation, so that the CST energy term made a negligible contribution. Then, during the course of annealing, kCST was increased geometrically, to a final value of ~0.5 kcal/ppm2. This value was calibrated by monitoring convergence of the structure calculations over a range of final kCST values, from 0.1 to 1.25 kcal/ppm2. The CST potential does not take experimental error into account, so even an error of 1 ppm between shift and shielding, (within the experimental error for most sites), can create a significant energy penalty. Because of this relatively stiff force field, and the variable reference between theory and experiment, setting the CST potential too high impedes convergence. This effect arises because each CST site produces three potential surfaces, which are relatively rough over the Ramachandran space; thus, if the CST energy is too high early in the annealing process, structures possibly will be trapped within local minima, with insufficient energy to traverse the barriers and arrive at the global minimum energy value. A subset of structures (or regions of the protein) may converge to high structure quality, but the overall convergence properties of the ensemble may be greatly compromised. These problems are especially pronounced in cases where pscale and poffset are allowed to vary. These issues are mitigated by the use of a small force constant value early in the annealing process, as detailed above.
For each set of parameters, ensembles of 200 structures were calculated by performing molecular dynamics at 3000 K for 40 ps, followed by slow cooling from 3000 K to 25 K in 25 K increments, with 2.5 ps of dynamics at each temperature, using a soft square NOE potential. Each structure was then refined by repeating the annealing protocol with only 15 ps of initial annealing using a hard square NOE potential with the kNOE force constant held constant at 30 kcal. Although no NOE restraints are used in SSNMR structure determinations, SSNMR distances (13C-13C, 15N-15N and 1H-1H) are included using the NOE potential function within Xplor-NIH. In calculations where pseudobond angles were used, the energy of bond and improper angles was reduced by an order of magnitude.
Results and Discussion
Chemical shift tensor determinations
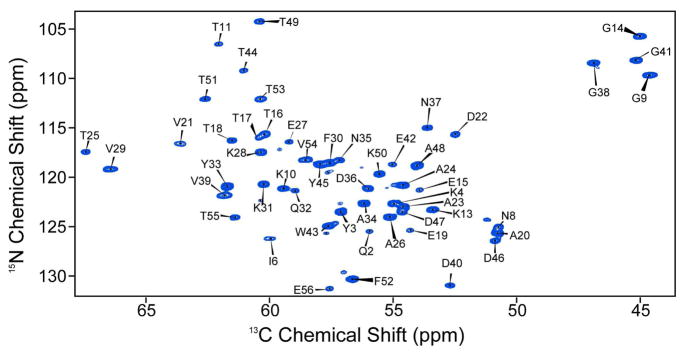
We first determined experimentally the 13Cα shift tensor elements δ11, δ22 and δ33, for all 56 residues in GB1, to provide a complete data set for CST backbone structure refinement. In earlier work20 we reported the Cα CSTs for 39 of these residues, but 17 were unresolved in the isotropic shift dimension, due to the relatively limited resolution in some regions of the 13C-13C correlation spectra. This is a consequence of using a uniformly 13C,15N-labelled GB1 sample in which homonuclear J-couplings and off rotational-resonance effects38–40 increased the effective line widths in the directly detected dimension. To remedy this, we used here a modified version of our 3D pulse sequence, together with sparse 13C labeling as derived from [2-13C]- or [1,3-13C]-glycerol. When using a [2-13C]-glycerol-[U-15N]-labeled GB1 sample, essentially all 15N-13Cα peaks can be resolved and assigned in the first 2D 15N-13C plane of the 3D ROCSA experiment (Figure 2). The exception is that Leu residues are not 13Cα-labeled in this sample, so we also acquired data on a sample prepared from [1,3-13C]-glycerol (and 15N ammonium chloride). The amino acids for which the Cα sites are fractionally labeled in both preparations (e.g., Thr, Asp, Lys, Ile) also provide an internal control in the data fitting, and we found agreement between the two data sets to be within experimental error (±1 ppm, see below).
Figure 2.
First 15N-13Cα plane from the 15N-[13C CSA]-13C 3D experiment. Spectra was acquired and processed with parameters discussed in the Experimental Section. Resonance assignments are based upon published values.24
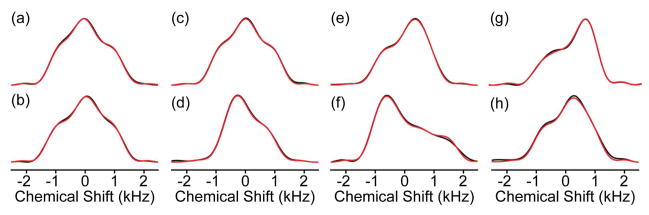
Spectra were then assigned,24 the 2D planes integrated, and recoupled CSA trajectories fit as discussed above. The experimental spectra have high sensitivity and the best fit simulations are in excellent agreement (Figure 3), with RMSD values for each fitted spectrum of 3%, typically <2%. The resulting CSA tensor parameters (Table 1) are presented in both the Haeberlen and Herzeld-Berger conventions. Within amino acid type, variations in tensor parameters are characteristic of secondary structures. For example, Cα tensors of α-helical residues are systematically narrower than those of β-sheet residues. This trend has a common origin to previously observed isotropic secondary chemical shifts,4 yet the overall dependence of the CST on molecular geometry captures additional subtleties within secondary structure type, as the tensor elements can be measured with sufficient precision to identify changes of ~5° in dihedral angles.
Figure 3.
ROCSA experimental and simulated lineshapes. Experimental ROCSA pseudostatic powder patterns (black) are overlaid with the best-fit simulations (red). Sites presented are: (a) A26, (b) A48, (c) D36, (d) D47, (e) K28, (f) K50, (g) T25, (h) T51. Fit values and experimental errors for each site are provided in Table 1.
Table 1.
Experimental chemical shift tensor (CST) values for protein GB1.
| Res | δ (ppm) | σδa (ppm) | η | σηa | δ11 (ppm) | σδ11a (ppm) | δ22 (ppm) | σδ22a (ppm) | δ33a (ppm) | σδ33 (ppm) |
|---|---|---|---|---|---|---|---|---|---|---|
| Q2 | 15.80 | 1.45 | 0.90 | 0.06 | 71.70 | 1.45 | 55.14 | 1.36 | 40.86 | 1.54 |
| Y3 | −21.34 | 1.20 | 0.78 | 0.02 | 76.03 | 1.20 | 59.31 | 1.17 | 35.66 | 1.22 |
| K4 | 18.20 | 0.90 | 0.74 | 0.01 | 73.10 | 0.90 | 52.51 | 0.89 | 39.09 | 0.90 |
| L5 | 19.55 | 1.48 | 0.93 | 0.07 | 72.55 | 1.48 | 52.30 | 1.38 | 34.15 | 1.59 |
| I6 | 15.23 | 1.14 | 0.99 | 0.14 | 75.23 | 1.14 | 59.96 | 0.98 | 44.81 | 1.30 |
| L7 | −20.32 | 1.48 | 0.52 | 0.07 | 70.14 | 1.48 | 59.59 | 1.38 | 34.38 | 1.59 |
| N8 | 21.35 | 1.25 | 0.76 | 0.10 | 72.05 | 1.25 | 48.12 | 1.12 | 31.93 | 1.38 |
| G9 | 26.44 | 1.82 | 0.94 | 0.09 | 71.04 | 1.82 | 43.85 | 1.65 | 18.91 | 1.98 |
| K10 | −17.24 | 0.49 | 0.69 | 0.01 | 73.83 | 0.49 | 62.01 | 0.49 | 42.06 | 0.49 |
| T11 | −21.02 | 2.31 | 0.48 | 0.06 | 77.47 | 2.31 | 67.35 | 2.17 | 40.88 | 2.44 |
| L12 | 17.44 | 1.03 | 0.82 | 0.03 | 71.84 | 1.03 | 52.79 | 1.00 | 38.57 | 1.06 |
| K13 | 20.02 | 3.04 | 0.84 | 0.13 | 73.32 | 3.04 | 51.70 | 2.63 | 34.88 | 3.45 |
| G14 | −19.26 | 1.55 | 0.73 | 0.01 | 61.58 | 1.55 | 47.47 | 1.54 | 25.64 | 1.56 |
| E15 | 17.81 | 1.82 | 0.73 | 0.08 | 71.71 | 1.82 | 51.52 | 1.68 | 38.48 | 1.96 |
| T16 | 16.89 | 0.54 | 0.30 | 0.07 | 76.99 | 0.54 | 54.19 | 0.50 | 49.12 | 0.57 |
| T17 | 17.83 | 1.73 | 0.65 | 0.13 | 78.13 | 1.73 | 57.18 | 1.51 | 45.59 | 1.95 |
| T18 | 16.40 | 1.42 | 0.22 | 0.19 | 77.70 | 1.42 | 54.89 | 1.15 | 51.31 | 1.69 |
| E19 | 16.95 | 1.64 | 0.87 | 0.07 | 71.25 | 1.64 | 53.23 | 1.52 | 38.41 | 1.75 |
| A20 | −22.54 | 0.51 | 0.86 | 0.09 | 71.69 | 0.51 | 52.25 | 0.47 | 28.16 | 0.56 |
| V21 | −13.42 | 1.01 | 0.58 | 0.04 | 74.08 | 1.01 | 66.34 | 0.97 | 50.08 | 1.05 |
| D22 | 21.39 | 1.00 | 0.81 | 0.04 | 73.89 | 1.00 | 50.51 | 0.96 | 33.09 | 1.04 |
| A23 | −20.75 | 1.19 | 0.91 | 0.20 | 74.37 | 1.19 | 55.57 | 0.95 | 33.85 | 1.42 |
| A24 | 18.59 | 1.72 | 0.85 | 0.16 | 73.09 | 1.72 | 53.07 | 1.45 | 37.34 | 1.99 |
| T25 | −18.68 | 0.87 | 0.37 | 0.04 | 80.02 | 0.87 | 73.06 | 0.84 | 48.52 | 0.91 |
| A26 | 19.45 | 0.61 | 0.90 | 0.03 | 74.45 | 0.61 | 54.00 | 0.59 | 36.55 | 0.63 |
| E27 | −15.26 | 1.34 | 0.95 | 0.13 | 73.97 | 1.34 | 59.49 | 1.17 | 43.84 | 1.51 |
| K28 | −17.79 | 1.78 | 0.63 | 0.03 | 74.72 | 1.78 | 63.47 | 1.73 | 42.41 | 1.83 |
| V29 | −15.87 | 0.95 | 0.29 | 0.04 | 76.53 | 0.95 | 71.94 | 0.92 | 50.43 | 0.99 |
| F30 | −19.09 | 0.38 | 0.93 | 0.04 | 75.88 | 0.38 | 58.22 | 0.36 | 38.41 | 0.39 |
| K31 | −19.54 | 0.97 | 0.54 | 0.07 | 75.12 | 0.97 | 64.61 | 0.90 | 40.56 | 1.04 |
| Q32 | 15.11 | 0.90 | 0.95 | 0.11 | 74.01 | 0.90 | 58.53 | 0.80 | 44.16 | 1.00 |
| Y33 | 13.94 | 3.14 | 0.80 | 0.06 | 75.54 | 3.14 | 60.21 | 2.93 | 49.05 | 3.34 |
| A34 | 19.07 | 0.85 | 0.96 | 0.05 | 75.17 | 0.85 | 55.74 | 0.81 | 37.39 | 0.89 |
| N35 | 17.70 | 1.01 | 0.77 | 0.07 | 74.70 | 1.01 | 54.96 | 0.94 | 41.34 | 1.08 |
| D36 | 18.65 | 0.27 | 0.97 | 0.05 | 74.55 | 0.27 | 55.64 | 0.25 | 37.51 | 0.28 |
| N37 | 22.99 | 1.48 | 0.74 | 0.11 | 76.49 | 1.48 | 50.49 | 1.33 | 33.52 | 1.64 |
| G38 | 26.18 | 0.85 | 0.68 | 0.00 | 72.98 | 0.85 | 42.65 | 0.85 | 24.77 | 0.86 |
| V39 | −15.08 | 2.10 | 0.82 | 0.11 | 75.41 | 2.10 | 63.06 | 1.87 | 46.62 | 2.33 |
| D40 | 16.18 | 1.46 | 0.69 | 0.07 | 68.98 | 1.46 | 50.26 | 1.36 | 39.16 | 1.56 |
| G41 | 22.20 | 0.44 | 1.00 | 0.07 | 67.30 | 0.44 | 45.10 | 0.41 | 22.90 | 0.47 |
| E42 | 18.52 | 0.35 | 1.00 | 0.08 | 73.62 | 0.35 | 55.10 | 0.32 | 36.58 | 0.38 |
| W43 | 19.51 | 2.34 | 1.00 | 0.09 | 77.01 | 2.34 | 57.50 | 2.13 | 37.99 | 2.55 |
| T44 | 19.54 | 2.57 | 0.79 | 0.02 | 80.44 | 2.57 | 58.83 | 2.51 | 43.44 | 2.63 |
| Y45 | 14.79 | 1.11 | 0.68 | 0.13 | 72.59 | 1.11 | 55.47 | 0.97 | 45.34 | 1.25 |
| D46 | 17.62 | 0.36 | 0.66 | 0.07 | 68.52 | 0.36 | 47.94 | 0.34 | 36.25 | 0.39 |
| D47 | 16.04 | 0.89 | 0.57 | 0.02 | 70.64 | 0.89 | 51.16 | 0.87 | 42.00 | 0.90 |
| A48 | −19.95 | 1.48 | 0.95 | 0.10 | 73.41 | 1.48 | 54.53 | 1.34 | 34.05 | 1.62 |
| T49 | −19.94 | 1.08 | 0.75 | 0.10 | 77.75 | 1.08 | 62.80 | 0.97 | 40.36 | 1.19 |
| K50 | 26.69 | 1.21 | 0.35 | 0.04 | 82.29 | 1.21 | 46.98 | 1.16 | 37.53 | 1.26 |
| T51 | −18.09 | 1.03 | 0.74 | 0.03 | 78.20 | 1.03 | 64.89 | 0.99 | 44.41 | 1.06 |
| F52 | 18.43 | 1.29 | 1.00 | 0.10 | 75.03 | 1.29 | 56.60 | 1.17 | 38.17 | 1.42 |
| T53 | 18.60 | 1.29 | 0.77 | 0.12 | 79.00 | 1.29 | 58.30 | 1.13 | 43.90 | 1.45 |
| V54 | 19.19 | 2.71 | 0.53 | 0.18 | 77.69 | 2.71 | 53.95 | 2.22 | 43.86 | 3.20 |
| T55 | −17.81 | 1.52 | 0.86 | 0.12 | 77.89 | 1.52 | 62.53 | 1.33 | 43.49 | 1.70 |
| E56 | 18.39 | 1.33 | 0.97 | 0.06 | 75.99 | 1.33 | 57.35 | 1.26 | 39.46 | 1.41 |
Estimated standard deviation for each subscripted quantity.
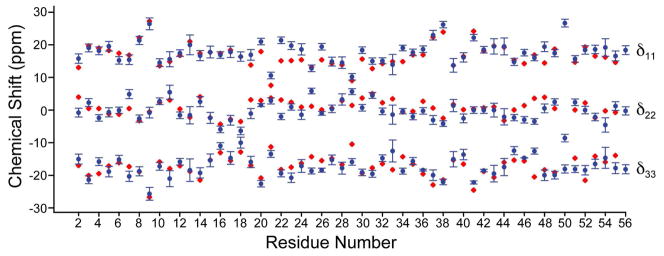
These results are of interest since they represent the first determination of the principal components of the 13Cα shift tensor for each residue in a protein. As can be seen in Figure 3 (and Figure S2 in the Supporting Information) there is a very broad range of lineshapes seen experimentally, reflecting the strong influence of φ, ψ (and χ1) on electronic structure and hence, the shift (or shielding) tensor. There is, however, good overall accord between the experimental and computed shift tensor elements. This is illustrated in Figure 4, where we show δ11, δ22 and δ33 for each residue in GB1 (as a function of residue number, in blue), together with the shift tensor elements predicted by using ab initio shielding surfaces (red, Figure 4). For shift predictions, we used backbone dihedral angles (φ, ψ) from the highest resolution (1.04 Å) crystal structure available for this mutant of GB1 (pdb entry 2QMT), a structure that is also the most closely related polymorph to the microcrystalline form used here.41 For convenience, the chemical shielding tensor elements were converted to shifts, and are shown in the traceless representation. Overall statistical agreement between theory and experimental δii is very good, with R2 = 0.99 over all residues and a RMSD of 2.27 ppm (see Figure 4).
Figure 4.
Plot of chemical shift tensor principal elements compared with ab initio chemical shielding values. Values are presented in the traceless representation, with experimental values in blue (with error bars) and theoretical values in red. An ideal scaling between experimental chemical shift and theoretical shielding of −1.0 is assumed.
This good agreement might at first seem surprising, given that the amino-acid models used to construct the shielding surfaces were all simply N-formyl-amino acid amides, such as N-formyl valine amide in which the most populous side-chain confirmations (in the Protein Data Bank) were chosen. Also, there are likely to be small conformational differences between the 2QMT crystals and our nanocrystals, and protein x-ray structures do have their own associated φ,ψ errors. Moreover, the correlations we see here – especially when the shift tensor elements are compared for each individual amino-acid in GB1 (Figure S3, Supporting Information) are really no worse than those seen in tripeptides or even pure amino-acids12,14–18,42–44 – but of course the reason for the excellent results on GB1 is that there are (in general) no strong electrostatic interactions with nearly terminal CO2− or NH3+ groups, which greatly complicate shift calculations in the smaller molecules. There are, however, small differences in the slopes (pscale) and intercepts (poffset) for each amino-acid (Table 2), but as in previous work16 and as described in Equation 2 in the Experimental and Computational Methods section, these can be readily taken into account by a linear fitting procedure.
Table 2.
Statistical agreement of experimental CST data and ab initio chemical shielding surfaces.
| Residue Type | R2 | pscale | poffset (ppm) | RMSD (ppm) |
|---|---|---|---|---|
| Alanine | 0.99 | −1.18 | 222.8 | 1.64 |
| Asparagine | 0.99 | −0.97 | 196.8 | 1.59 |
| Aspartic Acid | 0.92 | −0.95 | 193.1 | 4.25 |
| Glutamine | 0.95 | −0.96 | 195.7 | 3.01 |
| Glutamic Acid | 0.93 | −1.00 | 202.3 | 3.64 |
| Glycine | 0.99 | −0.96 | 194.3 | 1.80 |
| Leucine | 0.98 | −1.04 | 207.6 | 1.89 |
| Lysine | 0.98 | −1.01 | 201.6 | 1.95 |
| Phenylalanine | 0.97 | −0.95 | 186.3 | 2.65 |
| Threonine | 0.97 | −1.06 | 212.4 | 2.39 |
| Tyrosine | 0.96 | −0.91 | 185.6 | 2.17 |
| Valine | 0.94 | −1.12 | 217.5 | 2.62 |
We now consider how shift tensor elements can be incorporated into solid-state protein structure refinement. To investigate to what extent it might be possible to refine solid-state NMR-derived protein structures, we first performed a series of structure determinations using previously reported distance restraints,45–47 together with the new CST restraints, as discussed in the Experimental Section. Important statistics for each structure are given in Table 3. Each structure is assigned a number and name defined by the information used during refinement. In this nomenclature “N” refers to distance information (from the NOE potential used), “T” refers to semi-empirical dihedral angles from TALOS, “C” refers to CST restraints, “V” refers to Vector Angles, and “semi-empirical” indicates that RAMA and HBDB potentials were included in the calculation. We first computed two “control” structures, incorporating either (a) distance or (b) both distance and semi-empirical torsion angles, deduced from the TALOS program,48 as reported previously.47 The family of ten lowest energy structures, derived with distances only (N, structure 1 in Table 3), had good precision (a backbone RMSD of 0.59 ± 0.08 Å) and accuracy (1.30 ± 0.08 Å), but the addition of TALOS dihedral angles (NT, structure 2 in Table 3) improved significantly in terms of both precision (0.29 ± 0.06 Å) and accuracy (1.15 ± 0.03 Å vs. 2QMT) (Supporting Figure 3).
Table 3.
Internal backbone RMSD values (ten lowest energy structures) and backbone RMSD values versus the x-ray crystal structure PDB entry 2QMT as a function of refinement method and statistical agreement with chemical shielding surfaces and isotropic chemical shifts predicted by SPARTA.
| Structurea | bbRMSDb (Å) | RMSD vs. c 2QMT (Å) | Most Favored d Regions | R2 e CST | σ CSTf (ppm) | σ Cα δisog (ppm) | |
|---|---|---|---|---|---|---|---|
| 1 | N | 0.59 ± 0.08 | 1.30 ± 0.08 | 56.0% | 0.92 | 4.38 | 1.28 |
| 2 | NT | 0.29 ± 0.06 | 1.15 ± 0.03 | 92.0% | 0.96 | 3.03 | 1.08 |
| 3 | NC (uncalibrated) | 0.38 ± 0.08 | 1.13 ± 0.04 | 82.0% | 0.99 | 1.56 | 1.12 |
| 4 | NCT | 0.23 ± 0.03 | 1.01 ± 0.04 | 93.8% | 0.99 | 1.66 | 1.08 |
| 5 | NCTV | 0.19 ± 0.04 | 1.01 ± 0.03 | 95.6% | 0.99 | 1.69 | 1.03 |
| 6 | NCTV (semi-empirical) | 0.19 ± 0.04 | 0.98 ± 0.04 | 96.0% | 0.99 | 1.81 | 1.08 |
| 7 | NTV (semi-empirical) | 0.27 ± 0.03 | 1.04 ± 0.03 | 98.4% | 0.97 | 2.52 | 1.18 |
| 8 | NC (calibrated) | 0.27 ± 0.06 | 1.09 ± 0.04 | 88.0% | 0.99 | 1.56 | 1.09 |
| 9 | 2QMT | 96.0% | 0.97 | 2.52 | 0.93 | ||
| 10 | 2JSV | 0.31 ± 0.06 | 1.43 ± 0.08 | 92.0% | 0.97 | 2.91 | 1.06 |
| Solid vs. Solution δiso | 0.59 | ||||||
Acronyms here indicate: N = distances, T = TALOS dihedral angles, C = Cα CST constraints, V = Vector Angles, and “semi-empirical” refers to calculations using RAMA and HBDB potentials.
RMSD of ten lowest energy structures.
RMSD between ten lowest energy structures and the 2QMT crystal structure.
The percentage of residues occupying most favored regions of Ramachandran space.
Correlation coefficient between experimental chemical shift tensor elements and theoretical chemical shielding.
RMSD between experimental anisotropic shifts and theoretical shieldings.
RMSD between experimental isotropic chemical shifts and chemical shifts predicted by SPARTA.
We next investigated the effects of incorporating CST restraints in structure refinement, using the new potential for Xplor-NIH described above in the Experimental and Computational Methods sections. In a first set of calculations (NC (uncalibrated), structure 3 in Table 3), we allowed poffset and pscale to vary during the annealing and cooling process. The inclusion of this CST constraint again improved both the precision (0.38 ± 0.08 Å) and accuracy (1.13 ± 0.04 Å vs. 2QMT) of the structure ensemble compared to that obtained by using distances restraints alone, with the precision being slightly worse than that obtained with the TALOS dihedral angle restraints, while the accuracy (versus 2QMT) was statistically equivalent. We then solved the structure of GB1 by combining CST constraints with TALOS dihedral angles (NCT, structure 4 in Table 3), while allowing pscale and poffset to vary. Combining Cα CST restraints with TALOS resulted in a substantial improvement in both the precision of the ensemble (RMSD = 0.23 ± 0.03 Å) as well as accuracy (a 1.01 Å ± 0.04 Å RMSD versus 2QMT). This ensemble exhibited comparable accuracy, but slightly worse precision compared to the ensemble including Cα CST, TALOS, and vector angle restraints (NCTV, structure 5 in Table 3; 0.19 ± 0.04 and 1.01 ± 0.03 Å). Based upon these results, we decided to calibrate pscale and poffset to a structural ensemble combining Cα CSTs, TALOS, vector angles, and empirical database potentials which improve Ramachandran quality (RAMA)49,50 and hydrogen bonding networks (HBDB).51 When these potentials were included in the calculation the precision (NCTV (semi-empirical), structure 6 in Table 3) remained at 0.19 ± 0.04 Å, while the RMSD relative to 2QMT improved by one standard deviation, to 0.98 ± 0.04. As a control, CST restraints were then removed (NTV (semi-empirical), structure 7 in Table 3): precision decreased to 0.27 ± 0.03, and the RMSD relative to 2QMT increased nearly two standard deviations, to 1.04 ± 0.03 Å. In the final structure solved, pscale and poffset were set to the values provided in Table 4, which correspond to the structure with the lowest total CST energy in the structure 6 ensemble. In this final structure (NC (calibrated), structure 8 in Table 3) the precision and accuracy are comparable to the structure solved using TALOS dihedral angles (bbRMSD = 0.27 ± 0.6 Å, 1.09 ± 0.04 Å vs. 2QMT).
Table 4.
pscale and poffset determined from structure determinations using distance, CST, TALOS, and VEAN data sets.
| Residue Type | pscale | poffset (ppm) |
|---|---|---|
| Alanine | −1.21 | 226.5 |
| Asparagine | −0.98 | 198.0 |
| Aspartic Acid | −1.09 | 212.9 |
| Glutamine | −0.89 | 186.0 |
| Glutamic Acid | −1.00 | 201.3 |
| Glycine | −1.00 | 200.2 |
| Isoleucine | −0.81 | 173.4 |
| Leucine | −1.05 | 208.7 |
| Lysine | −1.09 | 211.9 |
| Phenylalanine | −1.04 | 198.8 |
| Threonine | −1.08 | 215.2 |
| Tryptophan | −0.98 | 199.4 |
| Tyrosine | −0.87 | 181.2 |
| Valine | −1.12 | 217.7 |
When each of the eight structures shown in Table 3 are used to back-calculate experimental isotropic (using the SPARTA program)52 and anisotropic shifts (from the shielding surfaces), we find that agreement between experimental and computational isotropic as well as anisotropic shifts improves with the addition of Cα CST information, as shown in Table 3. The worst results are all found with the distance-only structure (structure 1), where, the RMSD between experimental isotropic Cα (1.28 ppm) and anisotropic shifts (4.38 ppm) are significantly worse than the results obtained by using all of the other structures. Likewise, the structure solved with distances and TALOS (structure 2) had only a fair agreement with anisotropic constraints (R2 = 0.96, RMSD = 3.03 ppm). Overall, structures which included Cα CST information exhibited RMSDs between experimental and predicted anisotropic chemical shifts of 1.56–1.81 ppm and R2 values of 0.99, with the best agreement being in structures solved with only distance and CST constraints, as expected. The agreement between CST values predicted from 2QMT were slightly worse vs. experiment (R2 = 0.97, RMSD = 2.52 ppm), and equivalent to values predicted from structure 7, where CST restraints were removed. Likewise, the isotropic shifts exhibited clear trends depending upon constraints used. The agreement between predicted Cα isotropic chemical shifts and experiment for structures solved using CST constraints range from 1.03 to 1.12 ppm, slightly worse than shifts predicted using the x-ray structure where the RMSD between predicted and experimental shifts is 0.93 ppm. It should be noted that all reported deviations for the isotropic Cα shifts are within the reported reliability of the SPARTA program and the agreement between shifts measured in solid vs. solution for comparable samples of GB1 (Table 3, last row).
These results are of course not wholly unexpected since the structures were based at least in part of isotropic and anisotropic shift information. However, it is not unreasonable to believe that a significant part of the ~1Å RMSD found between the NMR and x-ray structures arises from uncertainties in the x-ray coordinates themselves. How big are these errors? As noted by Blundell,53 “The accuracy of atomic positions in x-ray crystal structures remains an open and contentious question, with quoted errors ranging form 0.1–0.3 Å to around 0.5 Å to 0.6–1.0 Å.” The errors would have major effects on shift and shift tensor calculations, since bond length and bond angle shielding derivatives are large, as are φ,ψ effects on shielding. The origins of the ~1.0 Å RMSD between the x-ray and NMR determined structures is, therefore, an open question. However, as more shift and shift tensors elements are determined, for both backbone and side-chain atoms, it may soon be possible to deduce which structures are indeed the most accurate, in much the same way that the combination of solid-state 13C and 17N NMR, 57Fe NMR, Mössbauer and infra-red spectroscopies, combined with quantum chemistry, enabled the first accurate prediction of the CO ligand geometry in carbonmonoxy-myoglobin,54 in advance of its revised crystallographic structures.53,55–57
At present, the major outliers we find in our chemical shift predictions are those for aspartic acid residues. We attribute this observation to the fact these residues may have ionized sidechains (in GB1), creating a complex dependence of the shielding tensors on local electrostatics. The shifts are hard to compute precisely, since the dielectric constant and solvation state are not precisely known, and so, interpretation of shielding tensors for readily ionized residues should be utilized with caution, and such uncertainties are therefore included in the structure calculations. We anticipate the same effects for Lys and Arg, but in GB1 (which has no Arg residues) the Lys residues fit quite well with theory, due most likely to the fact that the location of the sidechain charge is more distal to Cα than it is in Asp. As noted above in Table 2, Asp residues exhibit the greatest deviation from the theoretical prediction, due to the high likelihood of the ionized carboxylate and its proximity to the backbone. Nevertheless, the overall agreement throughout GB1 is better than 3 ppm in comparison to the theoretically computed tensor parameters, in the case of all the available high-resolution structures. Indeed, as shown in Figure 6, tensor parameters calculated from the high-resolution structures (even those lacking CST information in refinement) demonstrated superior agreement with the experimentally demonstrated CST values, illustrating the self-consistency of the CST method and a rigorous cross-validation with other structure refinement techniques.
Figure 6.
Comparison between experimental and theoretical CST elements, obtained by using different backbone geometries. In all cases, theoretical chemical shielding tensor principal components were converted to chemical shift tensor elements using regression analysis. Backbone geometries represented, and R2 and RMSD between theory and experiment are: (a) Structure calculated with distance only (structure 1, Table 3), R2 = 0.92, RMSD = 4.4 ppm. (b) 2QMT crystal structure, R2 = 0.97, RMSD = 2.5. (c) Previously published SSNMR structure of GB1 (structure 10, Table 3. pdb cod 2JSV), R2 = 0.97, RMSD = 2.9 ppm. (d) Structure solved with calibrated CST data (structure 8) R2 = 0.99, RMSD = 1.6 ppm.
Conclusions
The results we have presented above are of interest for several reasons. First, they represent the first experimental determination of the 13Cα shift tensor elements for all amino acids in a protein, GB1. Second, we find that the experimentally determined shielding tensor elements are in good accord with those obtained by using quantum chemistry. Third, we have incorporated these shielding tensor restraints into the Xplor-NIH program and used them, together with additional restraints (distances, TALOS-derived torsion angles and vector angles) to produce refined solid-state NMR structures of the GB1 protein. Both isotropic chemical shifts as well as the 13Cα shift tensor elements improve the agreement between the NMR and x-ray deduced structures, and vector angles improve the agreement even further. Fourth, we find that both back-calculated isotropic, as well as anisotropic, shift predictions improve considerably using the refined structures, which themselves have no major distance or other violations. Fifth, the data acquired in this study required less than 48 hours of experiment time, suggesting comparable results might be acquired on larger systems using 4D spectroscopy in less than a week. In the future, it seems likely that CST restraints will prove most informative in unusual geometric situations, since these are by definition, under-represented in isotropic shift data bases. Properly constraining these residues, which often occur in turns and loops, may be of particular use in larger systems, such as membrane proteins, where turn residues between α-helices are difficult to constrain. In such cases, knowledge of backbone 15N and 13C′ tensors or sidechain resonances where already, γ-gauche effects are known to dominate isotropic chemical shifts,44,58 would complement Cα information. It has been shown previously that Cα tensor orientation is also a powerful indicator of molecular geometry, and it should also be possible to use shift tensor element orientations in structure refinement14 of high-resolution SSNMR structures.
The importance of CST constraints extends beyond MAS SSNMR. Spectra of samples in aligned bilayers or other aligned media are strongly dependent upon both tensor magnitude and orientation relative to the magnetic field.59–65 SSNMR techniques for aligned membrane protein samples exploit knowledge of the orientation and magnitudes of 1H-15N dipole, 15N CST,59,60 13C′ CST64,65 and other tensors. Therefore improved knowledge of tensor magnitude and orientations in peptides and proteins has the potential to enhance structure quality derived from such oriented sample experiments, as well as in the MAS techniques shown here.
Supplementary Material
Figure 5.
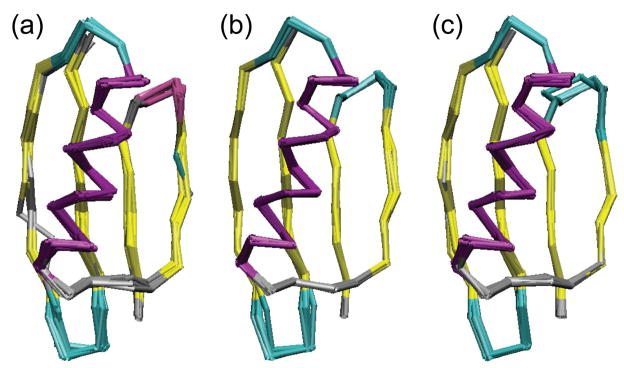
Ensembles of ten lowest energy structures of GB1 computed with CST restraints: (a) using distances and CST data and variable poffset and pscale; (b) using distances, CST, TALOS, and VEAN restraints and variable poffset and pscale; (c) using distances and CST with pscale and poffset fixed to values from the lowest energy structure in (b). The coloring is according to secondary structure (helix in purple, strands in yellow, turns in cyan, and coils in grey).
Acknowledgments
This research was supported by the National Science Foundation (CAREER Award, MCB 0347824 to C.M.R.), the CIT Intramural Research Program of the NIH (C.D.S.), and in part by the National Institutes of Health (GM73216 to E.O. and GM73770 to C.M.R.). We thank the NMR Facility at the School of Chemical Sciences, University of Illinois at Urbana-Champaign, for technical assistance.
Footnotes
Supporting Information Available: All experimental and best-fit simulation lineshapes, regression plots of all residue types represented more than twice in GB1, experimental restraint lists, and control structure figures are available in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Oldfield E, Norton RS, Allerhand A. J Biol Chem. 1975;250:6381–6402. [PubMed] [Google Scholar]
- 2.Oldfield E, Norton RS, Allerhand A. J Biol Chem. 1975;250:6368–6380. [PubMed] [Google Scholar]
- 3.Oldfield E, Allerhand A. J Biol Chem. 1975;250:6403–6407. [PubMed] [Google Scholar]
- 4.Spera S, Bax A. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 5.Oldfield EJ. Biomol NMR. 1995;5:217–225. doi: 10.1007/BF00211749. [DOI] [PubMed] [Google Scholar]
- 6.de Dios AC, Oldfield EJ. Am Chem Soc. 1994;116:5307–5314. [Google Scholar]
- 7.de Dios AC, Pearson JG, Oldfield E. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- 8.Oldfield E. Ann Rev Phys Chem. 2002;53:349–378. doi: 10.1146/annurev.physchem.53.082201.124235. [DOI] [PubMed] [Google Scholar]
- 9.Oldfield E. Phil Trans R Soc B Biol Sci. 2005;360:1347–1361. doi: 10.1098/rstb.2003.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le HB, Pearson JG, De Dios AC, Oldfield E. J Am Chem Soc. 1995;117:3800–3807. [Google Scholar]
- 11.Pearson JG, Wang JF, Markley JL, Le HB, Oldfield E. J Am Chem Soc. 1995;117:8823–8829. [Google Scholar]
- 12.Pearson JG, Le HB, Sanders LK, Godbout N, Havlin RH, Oldfield E. J Am Chem Soc. 1997;119:11941–11950. [Google Scholar]
- 13.Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Proc Natl Acad Sci U S A. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wi S, Sun HH, Oldfield E, Hong M. J Am Chem Soc. 2005;127:6451–6458. doi: 10.1021/ja042935b. [DOI] [PubMed] [Google Scholar]
- 15.Sun HH, Sanders LK, Oldfield E. J Am Chem Soc. 2002;124:5486–5495. doi: 10.1021/ja011863a. [DOI] [PubMed] [Google Scholar]
- 16.Havlin RH, Laws DD, Bitter HML, Sanders LK, Sun HH, Grimley JS, Wemmer DE, Pines A, Oldfield E. J Am Chem Soc. 2001;123:10362–10369. doi: 10.1021/ja0115060. [DOI] [PubMed] [Google Scholar]
- 17.Heller J, Laws DD, Tomaselli M, King DS, Wemmer DE, Pines A, Havlin RH, Oldfield E. J Am Chem Soc. 1997;119:7827–7831. [Google Scholar]
- 18.Havlin RH, Le HB, Laws DD, deDios AC, Oldfield E. J Am Chem Soc. 1997;119:11951–11958. [Google Scholar]
- 19.Wylie BJ, Franks WT, Rienstra CM. J Phys Chem B. 2006;110:10926–10936. doi: 10.1021/jp060507h. [DOI] [PubMed] [Google Scholar]
- 20.Wylie BJ, Franks WT, Graesser DT, Rienstra CM. J Am Chem Soc. 2005;127:11946–11947. doi: 10.1021/ja053862e. [DOI] [PubMed] [Google Scholar]
- 21.LeMaster DM. J Am Chem Soc. 1996;118:9255–9264. [Google Scholar]
- 22.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 23.Wylie BJ, Sperling LJ, Frericks HL, Shah GJ, Franks WT, Rienstra CM. J Am Chem Soc. 2007;129:5318–5319. doi: 10.1021/ja0701199. [DOI] [PubMed] [Google Scholar]
- 24.Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. J Am Chem Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 25.Chan JCC, Tycko R. J Chem Phys. 2003;118:8378–8389. [Google Scholar]
- 26.Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. J Biomol NMR. 2007;39:107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- 27.Baldus M, Petkova AT, Herzfeld JH, Griffin RG. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- 28.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 29.Marion D, Ikura M, Tschudin R, Bax A. J Magn Reson. 1989;85:393–399. [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.James F, Roos M. Comp Phys Comm. 1975;10:343–367. [Google Scholar]
- 32.Veshtort M, Griffin RG. J Magn Reson. 2006;178:248–282. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Haeberlen U. High Resolution NMR in Solids: Selective Averaging. Academic Press; New York: 1976. [Google Scholar]
- 34.Herzfeld J, Berger AEJ. Chem Phys. 1980;73:6021. [Google Scholar]
- 35.Mason J. Solid State Nucl Magn Reson. 1993;2:285–288. doi: 10.1016/0926-2040(93)90010-k. [DOI] [PubMed] [Google Scholar]
- 36.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 37.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes. 2. Cambridge University Press; Cambridge: 1992. [Google Scholar]
- 38.Raleigh DP, Levitt MH, Griffin RG. Chem Phys Lett. 1988;146:71–76. [Google Scholar]
- 39.Levitt MH, Raleigh DP, Creuzet F, Griffin RGJ. Chem Phys. 1990;92:6347–64. [Google Scholar]
- 40.Wylie BJ, Sperling LJ, Rienstra CM. Phys Chem Chem Phys. 2008;10:405–413. doi: 10.1039/b710736f. [DOI] [PubMed] [Google Scholar]
- 41.Frericks Schmidt HL, Sperling LJ, Gao YG, Wylie BJ, Boettcher JM, Wilson SR, Rienstra CMJ. Phys Chem B. 2007;111:14362–14369. doi: 10.1021/jp075531p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukkamala D, Zhang Y, Oldfield E. J Am Chem Soc. 2007;129:7385–7392. doi: 10.1021/ja071227y. [DOI] [PubMed] [Google Scholar]
- 43.Cheng F, Sun HH, Zhang Y, Mukkamala D, Oldfield E. J Am Chem Soc. 2005;127:12544–12554. doi: 10.1021/ja051528c. [DOI] [PubMed] [Google Scholar]
- 44.Sun HH, Oldfield E. J Am Chem Soc. 2004;126:4726–4734. doi: 10.1021/ja030612u. [DOI] [PubMed] [Google Scholar]
- 45.Zhou DH, Shea JJ, Nieuwkoop AJ, Franks WT, Wylie BJ, Mullen C, Sandoz D, Rienstra CM. Angew Chem Int Ed. 2007;46:8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou DH, Shah G, Cormos M, Mullen C, Sandoz D, Rienstra CM. J Am Chem Soc. 2007;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- 47.Franks WT, Wylie BJ, Frericks HL, Nieuwkoop AJ, Mayrhofer RM, Shah GJ, Graesser DT, Rienstra CM. Proc Natl Acad Sci U S A. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 49.Kuszewski J, Gronenborn AM, Clore GM. Protein Sci. 1996;5:1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clore GM, Kuszewski J. J Am Chem Soc. 2002;124:2866–2867. doi: 10.1021/ja017712p. [DOI] [PubMed] [Google Scholar]
- 51.Grishaev A, Bax A. J Am Chem Soc. 2004;126:7281–7292. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 52.Shen Y, Bax A. J Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 53.DePristo MA, de Bakker PIW, Blundell TL. Structure. 2004;12:831–838. doi: 10.1016/j.str.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 54.McMahon MT, deDios AC, Godbout N, Salzmann R, Laws DD, Le HB, Havlin RH, Oldfield EJ. Am Chem Soc. 1998;120:4784–4797. [Google Scholar]
- 55.Kachalova GS, Popov AN, Bartunik HD. Science. 1999;284:473–476. doi: 10.1126/science.284.5413.473. [DOI] [PubMed] [Google Scholar]
- 56.Vojtechovsky J, Chu K, Berendzen J, Sweet RM, Schlichting I. Biophys J. 1999;77:2153–2174. doi: 10.1016/S0006-3495(99)77056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stec B, Phillips GN. Acta Crystallogr D. 2001;57:751–754. doi: 10.1107/s0907444901001731. [DOI] [PubMed] [Google Scholar]
- 58.Paul EG, Grant DMJ. Am Chem Soc. 1963;85:1701–1702. [Google Scholar]
- 59.Wu CH, Ramamoorthy A, Gierasch LM, Opella SJ. J Am Chem Soc. 1995;117:6148–6149. [Google Scholar]
- 60.Ramamoorthy A, Opella SJ. Solid State Nucl Magn Reson. 1995;4:387–392. doi: 10.1016/0926-2040(95)00054-t. [DOI] [PubMed] [Google Scholar]
- 61.Hong M. Structure. 2006;14:1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi S, Hong M. J Magn Reson. 2002;155:244–250. doi: 10.1006/jmre.2002.2517. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi S, Hong T, Waring AJ, Lehrer RI, Hong M. Biophys J. 2002;82:538A–538A. [Google Scholar]
- 65.Doherty T, Waring AJ, Hong M. Biochemistry. 2006;45:13323–13330. doi: 10.1021/bi061424u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.