Abstract
We explored two macromolecular scaffolds, bovine serum albumin (BSA) and polyvinyl alcohol (PVA), as chemically complementary platforms for immobilizing small molecule compounds on functionalized glass slides. We conjugated biotin molecules to BSA and amine-derivatized PVA and subsequently immobilized the conjugates on epoxy- functionalized glass slides through reaction of free amine residues on BSA and PVA with surface-bound epoxy groups. We studied binding reactions of such immobilized small molecule targets with solution-phase protein probes using an oblique-incidence reflectivity difference scanning optical microscope. The results showed that both BSA and amine-derivatized PVA were effective and efficient as carriers of small molecules with NHS residues and fluoric residues and for immobilization on epoxy-coated solid surfaces. A significant fraction of the conjugated small molecules retain their innate chemical activity.
1. Introduction
Microarrays are useful tools for discovery and functionality characterization in fundamental and applied research of genomics, proteomics, glycomics, and cytomics. They are high-throughput platforms that enable parallel studies of hundreds to tens of thousands of distinct biomolecular reactions [1, 2]. DNA microarrays have been successfully used for gene expression profiling [3, 4], sequencing, single nucleotide polymorphism (SNP) detection, and drug target validation [5]. Protein microarrays have been most useful in investigation and characterization of protein expression, post-transcription modification, and protein functionality, and discovery of protein ligands for specific purposes [6, 7]. Membrane proteins [8], antibodies [9], enzymes [10], and many other proteins can be directly immobilized on a suitably functionalized solid substrate [11, 12] in forms of microarrays for analysis. Since carbohydrates play key roles in cell adhesion and signaling processes and in controlling many disease pathways, glycan and saccharide microarrays have also been used for cell adhesion and functionality characterization [13–16]. Furthermore cells and tissues have been made directly into microarrays [17, 18] for high-throughput cellular and tissue-level analysis.
Our present work concerns small molecule microarrays that consist of synthetic and natural compounds. By performing binding reactions against purified proteins or protein complexes, cell lysates, human sera, viruses, or bacteria, [19, 20, 21, 22, 23], these microarrays offer a highly parallel assay platform for mapping out functionality of protein and for identifying ligands with particular sets of enabling and inhibiting functions from thousands or millions of molecular candidates.
One of the major challenges in small molecule microarray application is to efficiently immobilize a large variety of small molecule compounds with diverse structures on a singly or even multiply functionalized solid support while maintaining innate binding properties of the molecules. There exist a number of strategies for immobilization of small molecule compounds on functionalized glass slides or other solid supports [24–30]. We here explore two macromolecular scaffolds, bovine serum albumin (BSA) and amine-derivatized (or amine-modified) polyvinyl alcohol (PVA), for as carriers of small molecule compounds and for immobilization on epoxy-coated glass surfaces. BSA is one of the common carrier proteins of small molecules for producing antibodies in mouse, goat, or rabbit against the small molecule epitopes. As a result, conjugation of small molecules including peptides to BSA is relatively common, and many small molecules including drugs or drug candidates with suitable chemical residues have been successfully conjugated to BSA. BSA has been used as a scaffold [31] for immobilizing conjugated peptides or small molecules (less than 300 ~ 500 daltons) on glass slides to form microarrays. Since BSA is a relatively inert protein, it is also used often as a blocking agent in microarray-based application to prevent non-specific probe-surface binding. Yet there are pockets on the surface of a BSA molecule that render BSA reactive to some proteins. For example accessible hydrophobic pockets on BSA can accommodate non-specific binding of some solution-phase probes through hydrophobic interaction [32]. In this regard, amine-modified PVA, - a hydrophilic polymer, serves as an alternative to BSA. We will show that when PVA is derivatized with primary amine residues, it can be used as an equally effective macromolecular scaffold for small molecule conjugation and subsequent immobilization on epoxy functionalized glass surfaces.
Characterization of binding reactions between immobilized small molecule microarray and solution-phase proteins or other analytes routinely involves some form of fluorescence detection with the analytes labeled with fluorescent tags either extrinsically or intrinsically through genetic engineering (e.g., incorporation of green fluorescent protein or phytochrome proteins) [1, 7, 33]. However, labeling proteins inevitably changes innate properties of the molecules and in turn modify protein-ligand interactions in often uncharacterized ways [2, 34, 35]. We recently developed oblique-incidence reflectivity difference (OI-RD) scanning microscopes specifically for label-free, high-throughput detection of solution-phase analytes with small molecule microarrays on functionalized glass surfaces [35 – 39].
Using an OI-RD scanning optical microscope for microarray detection, we performed a set of experimental studies to evaluate BSA and PVA as chemically complementary scaffolds for small molecule microarray application. Specifically we address the following issues: (1) efficiency of small molecule conjugation to BSA and amine-modified PVA; (2) efficiency of immobilizing small-molecule-BSA conjugates and small-molecule-PVA conjugates on epoxy-functionalized glass surfaces; (3) binding affinity of immobilized small molecules to solution-phase protein probes compared to small molecules in solution; (4) efficacy of label-free characterization of small molecule microarrays without fluorescent labeling.
2. Materials and methods
Preparation of amine-modified polyvinyl alcohol (PVA)
PVA (average Mw = 13 kDa, 98% hydrolyzed) were purchased from Sigma Aldrich (St. Louis, MO). We first modified PVA by replacing 10% of hydroxyl groups on the polymer with amino groups. The percentage of the amino groups on an amine-modified PVA was confirmed with nitrogen elemental analysis.
Conjugation of biotin molecules to amine-modified PVA
Biotin molecules were purchased from Sigma Aldrich (St. Louis, MO) and used as received. We conjugated biotin molecules to amine-modified PVA by reacting biotin-LC-N-hydroxysuccinimidyl ester (NHS esters of biotin) with an aqueous amine-modified PVA solution in 0.1M NaHCO3.
Conjugation of biotin molecules to BSA
We conjugated biotin to BSA by reacting biotin-LC-N-hydroxysuccinimidyl ester (NHS ester of biotin) with an aqueous BSA solution in 0.1M NaHCO3. The loading of biotin was specified by the molar ratio of the NHS ester of biotin to BSA. It was chosen to be 5×, 10×, 20×, and 40× in our present study.
Small molecule microarrays
Biotin-BSA and biotin-PVA conjugates were dissolved in 1×PBS (pH 7.4, 0.22 µm filtered) into printing solutions at decreasing concentrations from 18 µM to 0.14 µM for BSA conjugates, and from 77 µM to 0.6 µM for PVA conjugates. We printed microarrays of biotin-BSA and biotin-PAV conjugates on epoxy-functionalized glass slides (CEL Associates, Pearland, TX) using an OmniGrid 100 contact-printing robot (Genomic Solutions, Ann Arbor, MI). By contact printing once, approximately 1 nL of the solution was deposited over an area of 130 µm in diameter. The deposited conjugates bound covalently to the glass surface through exothermic amine-epoxy reaction.
We fabricated copies of a small molecule microarray consisting of differently loaded biotin-BSA conjugates (5×, 10×, 20×, 40×) and biotin-PVA conjugates (1%, 2%, and 4%) in decreasing printing concentrations. We also printed unmodified BSA in 1×PBS at a concentration of 18 µM and amine-modified PVA in 1×PBS at 77 µM as controls. The printed slide was assembled into a sample cartridge assembly. To block the remaining free epoxy groups on the glass surface from non-specific reaction with protein probes, we exposed the printed surface to a flowing BSA solution at 7.6 µM in 1×PBS for 10 min and then washed the surface wth a flow of 1×PBS for 5 min.
Conditions of solution-phase protein reaction with small molecule microarrays
For protein probes against biotin, we used unlabeled streptavidin tetramers and unlabeled monovalent Fab fragments of mouse IgG raised against biotin. They were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). For reaction, we replaced 1×PBS buffer in the sample cartridge assembly (0.4 mL active volume) with a solution of streptavidin or monovalent Fab fragments of anti-biotin mouse IgG in 1×PBS at a flow rate of 30 mL/min for a few seconds and then slowed down the flow rate to 0.05 mL/min during the remainder of the reaction. To observe the dissociation of the captured protein probes, we replaced the protein solution with 1×PBS at a flow rate of 30 mL/min for a few seconds and then slowed down the flow rate to 0.05 mL/min during observation.
Oblique-incidence reflectivity difference (OI-RD) scanning microscope for label-free detection of small molecule microarrays
The oblique-incidence reflectivity difference scanning microscope used in the present work was described in an earlier publication [35]. It employed a He-Ne laser at wavelength λ = 633 nm for illumination. We measure the complex differential reflectivity change (Δp - Δs) across a microarray-covered glass surface [40]. The physical properties of a surface-bound molecular layer on a glass surface are related to Δp - Δs by [37, 41, 42]
| (1) |
ϕinc is the incidence angle of illumination. ε0, εd, and εs are the respective optical constants of aqueous ambient, the molecular layer (e.g., printed targets and/or captured protein probes, blocking BSA), and the glass slide at λ = 633 nm. In our present study, ϕinc = 65°, εs = 2.307 for glass slide, ε0 = 1.788 for aqueous buffer, εd = 2.031 for packed protein in solution [41]. d is the thickness of the molecular layer, Θ is the coverage of the layer, defined as the ratio of the area covered by the layer to the total available area.
An image of a small molecule microarray was acquired with the pixel dimension of 20 µm × 20 µm. Due to the large incidence angle, the optical beam was 2.4 times long than wide so that the actual image of an otherwise “circular” microarray spot appeared elliptical in this study. To acquire binding curves, we selected one pixel from a printed spot (target pixel) and one pixel from the unprinted region adjacent to the printed target (reference pixel) and measure the optical signals from these pixels at a time interval shorter than the characteristic time of the reaction. We took the difference between the signal from a target pixel and the signal from the reference pixel as the binding curve. This significantly reduced the contribution of the drift in the optical system to the measurement.
Model for analysis of binding curves: - two-site Langmuir reaction model
The reaction of a solution-phase probe (e.g., proteins) with surface-immobilized small molecule targets is more reasonably described by treating each printed target as having more than one “configuration” that are stereochemically different. Multiple representations of an immobilized target come from (1) environmental variations at conjugation sites on a scaffold, both intrinsically and due to the orientation of BSA or the physical layout of PVA) in which the scaffold is immobilized on the glass surface; (2) multi-valence of a protein probe such as a whole IgG against an epitope [43 – 46]. We found it necessary and sufficient to assume two stereochemical configurations (Type-1 and Type-2) for each printed target such that the binding curve of a protein probe to the target are described by a two-site Langmuir reaction model. In our present study we designate the two stereochemical configurations for a given small molecule target as two binding sites for subsequent probes. Specifically, given a small-molecule conjugate, let N1 be numbers of the reacted targets and N1(0) the initially available targets of Type-1 per unit area, and accordingly N2 and N2(0) of Type-2. The total number of the captured protein probes per unit area during a binding reaction is given by [35]
| (2) |
t is the time lapse after a protein probe solution replaces the buffer. and are respective association rates at the two sites that are assumed to be linearly dependent on the probe concentration [c]. and are dissociation rates of the captured protein probes, assumed to be independent of [c]. The equilibrium dissociation constant is defined by KD = koff/kon. At t0 > 0, the protein probe solution is replaced by the buffer and the dissociation alone proceeds, and thus the number of the protein probes per unit area that remain bound to the targets is given by
| (3) |
The coverage of the captured protein probes Θ is proportional to N(t),
| (4) |
From Eq. (1), the OI-RD signal is expected to vary with time as
| (5) |
for 0 < t < t0; and for t > t0,
| (6) |
3. Results and Discussion
3.1 Efficiency of immobilization determined from end-point measurements
Streptavidin reactions with biotin-BSA and biotin-PVA conjugates
The experiment was performed on a 70-spot microarray of biotin-BSA, biotin-PVA, unmodified BSA and amine-modified PVA. Each column corresponds to a distinct conjugate with controls at the two ends; each row is printed at same target concentration for a scaffold. After BSA blocking step, the microarray was reacted with a solution of streptavidin in 1×PBS at [c] = 0.45 µM for 1 hour. Fig. 1 shows the differential OI-RD image of the microarray obtained by subtracting the image taken after BSA blocking (not shown) from the image taken after the reaction (with the microarray still in contact with the streptavidin solution.) Streptavidin reacted with the biotin-BSA and biotin-PVA conjugates while they did not with BSA and amine-modified PVA (negative controls). To quantify the captured streptavidin in terms of surface mass density Γ (in unit of g/cm2), we use a relationship Γ = Θdρprotein where Θ and d are the coverage and thickness of the streptavidin layer, and ρprotein is the volume mass density of streptavidin:. With Eq. (1) and the values of ϕinc = 65°, εs = 2.307 ε0 = 1.788, εd = 2.031 [41], and ρprotein = 1.33 g/cm2 for densely packed streptavidin in solution [49, 50], we arrive at
| (7) |
Using Eq. (7), we converted Im{Δp - Δs} in the differential OI-RD image into surface mass density Γ of the captured streptavidin.
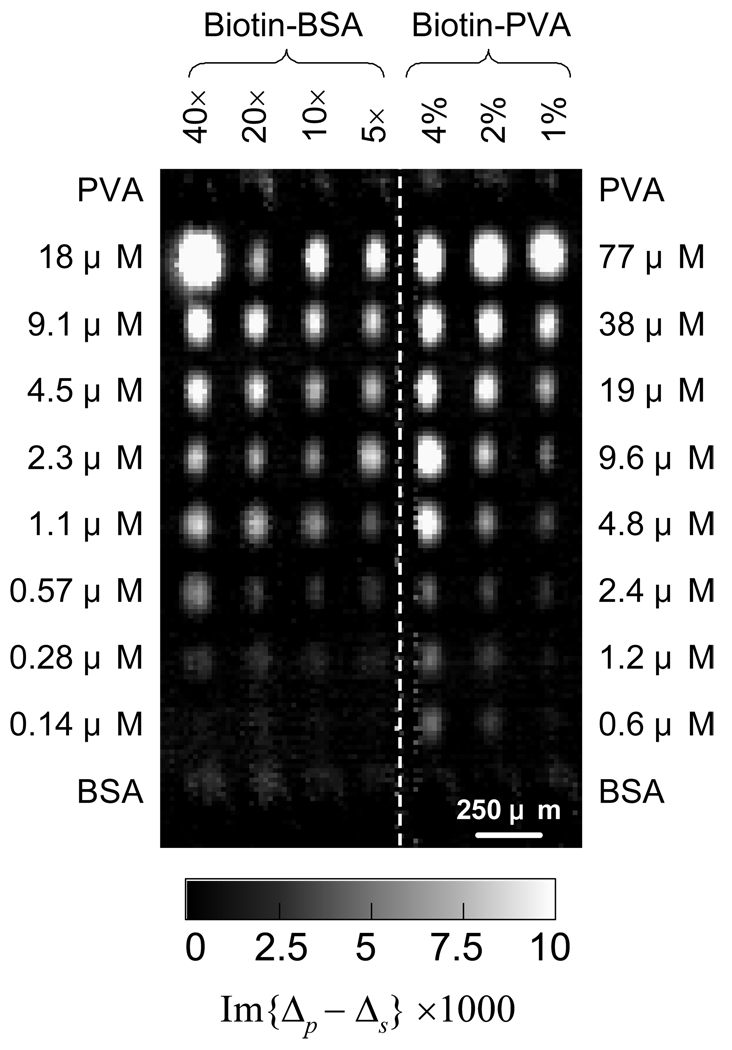
Fig. 1. Probe: Streptavidin.
Differential optical image in Im{Δp - Δs} of a 70-spot biotin-BSA and biotin-PVA conjugate microarray after reaction with a solution of streptavidin in 1×PBS at 0.45 µM for 1 hour. Printed BSA and amine-modified PVA were used negative controls.
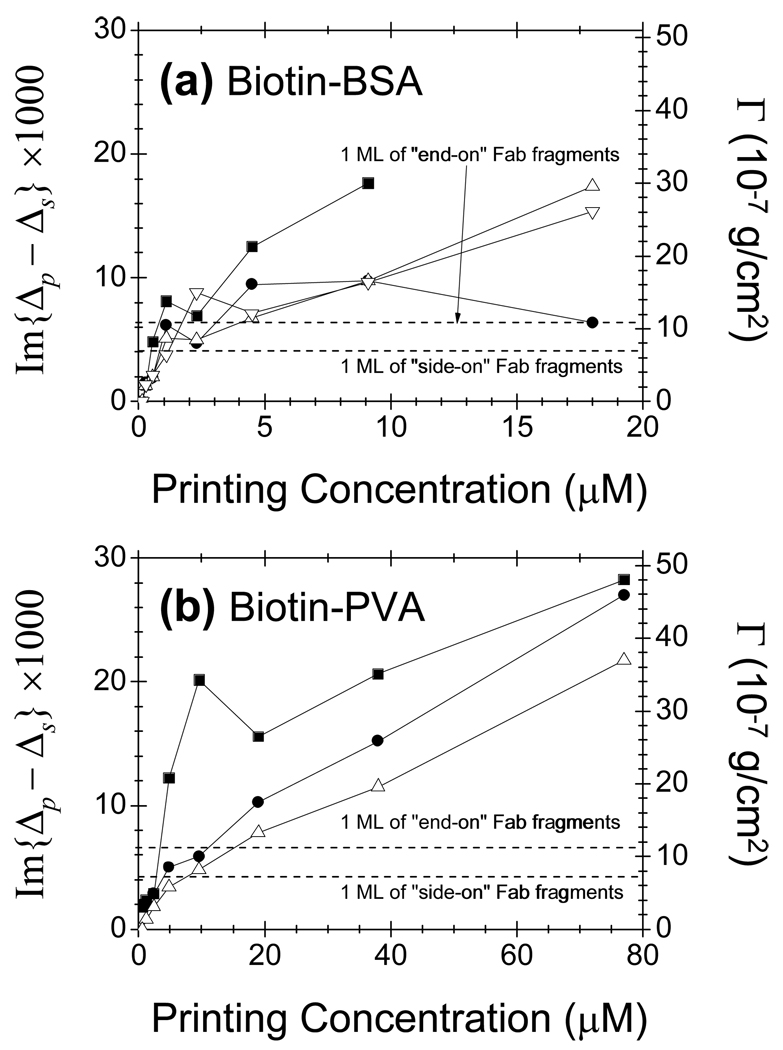
Fig. 2(a) and 2(b) show changes in Im{Δp - Δs} and Γ measured near the centers of printed targets vs. target printing concentration. For a tetrameric streptavidin that measures a×b×c = 5.4 nm × 5.8 nm × 4.8 nm [49], a fully packed monolayer of streptavidin has a surface mass density of Γ1ML Streptavidin = b × ρprotein = 7.7 × 10−7 g/cm2 with a thickness of b = 5.8 nm and a surface number density of 4×1012 /cm2. The dashed line corresponds to Γ1ML Streptavidin = 7.7 × 10−7 g/cm2 which is higher than the value of a dried monolayer of streptavidin [41]. The number of the captured streptavidin increases with conjugate printing concentration, indicating that the number of accessible conjugated biotin molecules increases. The surface mass density of captured streptavidin (Fig 2(a)) reaches a plateau around Γ1ML Streptavidin at close to 2.5 ~ 4 µM, regardless of biotin loading. This shows that there are sufficient immobilized biotin-BSA conjugates to capture one full monolayer of streptavidin with Γ1ML Streptavidin = 4×1012 /cm2. For 5× loaded biotin-BSA conjugates, if we assume that 5 biotin molecules were conjugated to one BSA and 3 to 4 of them were accessible to solution-phase streptavidin, at printing concentration of 2.5 ~ 4 µM, the immobilized biotin-BSA conjugates must have reached a surface density of 1×1012 /cm2 ~ 1.3×1012 /cm2. The latter corresponds to the density of a full side-on monolayer of BSA (the dimension of a BSA molecule is 8 nm × 8.7 nm × 6 nm [49].) Since the surface density of deposited biotin-BSA conjugates is roughly 6×1012 /cm2, the immobilization efficiency in the present study is over 20%. For 10× and 20× loaded biotin-BSA conjugates, the numbers of conjugated biotins are expectedly higher (though not necessarily proportional to the loading). The fact that the uptake of streptavidin probes vs. printing concentration are not much different from that for 5× loaded biotin-BSA conjugates shows that the accessible biotin epitopes to streptavidin on these higher loaded conjugates are more or less the same as on 5× loaded biotin-BSA. For 40× loaded biotin-BSA, there are clearly more accessible biotins. Beyond 4 µM, the surface mass density of the captured streptavidin continues to increase over Γ1ML Streptavidin but less vigorously, primarily due to non-specific binding.
Fig. 2. Probe: Streptavidin.
Change in Im{Δp - Δs} vs. conjugate printing concentration due to captured streptavidin by (a) biotin-BSA conjugates and (b) biotin-PVA conjugates . For biotin-BSA conjugates: (1) solid squares: 40× loading; (2) solid circles: 20× loading; (3) open up-pointing triangles: 10× loading; open down pointing triangles: 5× loading. For biotin-PVA conjugates: (i) solid squares: 4% loading; (ii) solid circles: 2% loading; (iii) open up-pointing triangles: 1% loading. Dashed line corresponds to a full monolayer of streptavidin. The optical signals are converted to surface mass density of streptavidin using Eq. (7).
For 4% loaded biotin-PVA conjugates (Fig. 2(b)), the plateau in surface mass density of the captured streptavidin near Γ1ML Streptavidin occurred at 4 µM, close to those for biotin-BSA conjugates. For 1% and 2% loaded biotin-PVA conjugates, the plateau was reached at higher printing concentrations, close to 20 µM. The amount of the captured streptavidin continued to increase significantly beyond Γ1ML Streptavidin when the printing concentration increased beyond ~ 20 µM. Though a small fraction of the additionally captured streptavidin was due to non-specific binding, the majority was due to specific reaction with accessible biotins that clearly grew in number with increasing printing concentration. This has to do with the flexible linear structure of 13-kDa amine-modified PVA (compared to BSA) that enabled immobilized biotin-PVA conjugates to extend into the aqueous ambient and form an effectively thicker and still accessible layer with more biotin targets available to streptavidin.
Reactions of monovalent Fab fragments of anti-biotin mouse IgG with immobilized biotin-BSA and biotin-PVA conjugates
The experiment was performed on another 70-spot microarray of biotin-BSA and biotin-PVA conjugates with the same layout as illustrated in Fig. 1. After BSA blocking step, the microarray was reacted with a solution of monovalent Fab fragments of the mouse IgG in 1×PBS at 87 nM for 4 hours. Fig. 3 shows the differential OI-RD image obtained by subtracting the image after BSA blocking from the image taken after the reaction (with the microarray still in contact with the probe solution.) The strong reaction between biotin and anti-biotin mouse IgG was unambiguous while the reaction with BSA and amine-modified PVA was negligible. We converted the optical signal Im{Δp - Δs} to surface mass density Γ of the captured Fab fragments using Eq. (7). We assume that the volume mass density of packed monovalent fragments in 1×PBS is still ρprotein = 1.33 g/cm2 (i.e., same as packed streptavidin in 1×PBS) and the optical constant of the packed Fab fragments in 1×PBS is also εd = 2.031 at 633 nm [41]. The surface mass density of the captured Fab fragments vs. target printing concentration is shown in Fig. 4. For reference, an Fab fragment of an IgG molecule measures 8.5 nm (h’) × 6 nm (w’) × 5 nm (t’) [47, 50]. If a monolayer of Fab fragments consists of packed molecules that lay flat on the surface, the surface mass density of such a “side-on” monolayer is . If instead a monolayer of Fab fragments consists of packed molecules that stand upright, the surface mass density of such an “end-on” monolayer should be . In Fig. 4 we mark with dash lines the surface mass densities of a monolayer of Fab fragments in these two configurations.
Fig. 3. Probe: Anti-Biotin.
Differential optical image in Im{Δp - Δs} of a 70-spot biotin-BSA and biotin-PVA conjugate microarray immobilized with a solution of monovalent Fab fragments of anti-biotin mouse IgG in 1×PBS at 87 nM for 4 hours.
Fig. 4. Probe: Anti-Biotin.
Change in Im{Δp - Δs} vs. conjugated printing concentration due to captured Fab fragments of anti-biotin mouse IgG by (a) biotin-BSA and (b) biotin-PVA . For biotin-BSA conjugates: (1) solid squares: 40× loading; (2) solid circles: 20× loading; (3) open up-pointing triangles: 10× loading; open down pointing triangles: 5× loading. For biotin-PVA conjugates: (i) solid squares: 4% loading; (ii) solid circles: 2% loading; (iii) open up-pointing triangles: 1% loading. The optical signal was converted to surface mass density of the Fab fragments. Dashed lines correspond to a full monolayer of “side-on” and “end-on” Fab fragments.
For biotin-BSA conjugates (Fig. 4(a)), the captured Fab fragments reach a plateau around at around ~ 2 µM. In end-on geometry, a full monolayer of Fab fragments has a surface number density of 1/t’w’ ~ 3.3×1012 /cm2, close to that of accessible biotin targets (~ 4×1012 /cm2) from a monolayer of side-on biotin-BSA conjugates formed at ~ 2 µM. The excess captured Fab fragments are mostly due to non-specific binding.
For 4% loaded biotin-PVA conjugates (Fig. 4(b)), the surface mass density of the captured monovalent Fab fragments reaches at around 4 µM, close to that for biotin-BSA conjugates. For 1% and 2% loaded biotin-PVA conjugates, the surface mass density of the captured Fab fragments reaches at around 20 µM. Similar to streptavidin, the captured Fab fragments of anti-biotin mouse IgG continue to increase beyond with printing concentrations, indicating that the immobilized biotin-PVA conjugates on epoxy-coated glass surface became more extended into the aqueous ambient and thus made more biotin epitopes available per unit area than a full end-on monolayer of Fab fragments, a distinct feature of a polymer chain as a scaffold.
From these end-point measurements, we can conclude that both BSA and amine-modified PVA are effective scaffolds for conjugation of small molecule targets and subsequent immobilization on epoxy-functionalized glass surfaces. We next examine how well these two scaffolds fare when protein-small molecule reaction kinetics is concerned.
3.2 Binding kinetics of protein-ligand reactions
Kinetics of streptavidin reaction with immobilized biotin-BSA and biotin-PVA conjugates
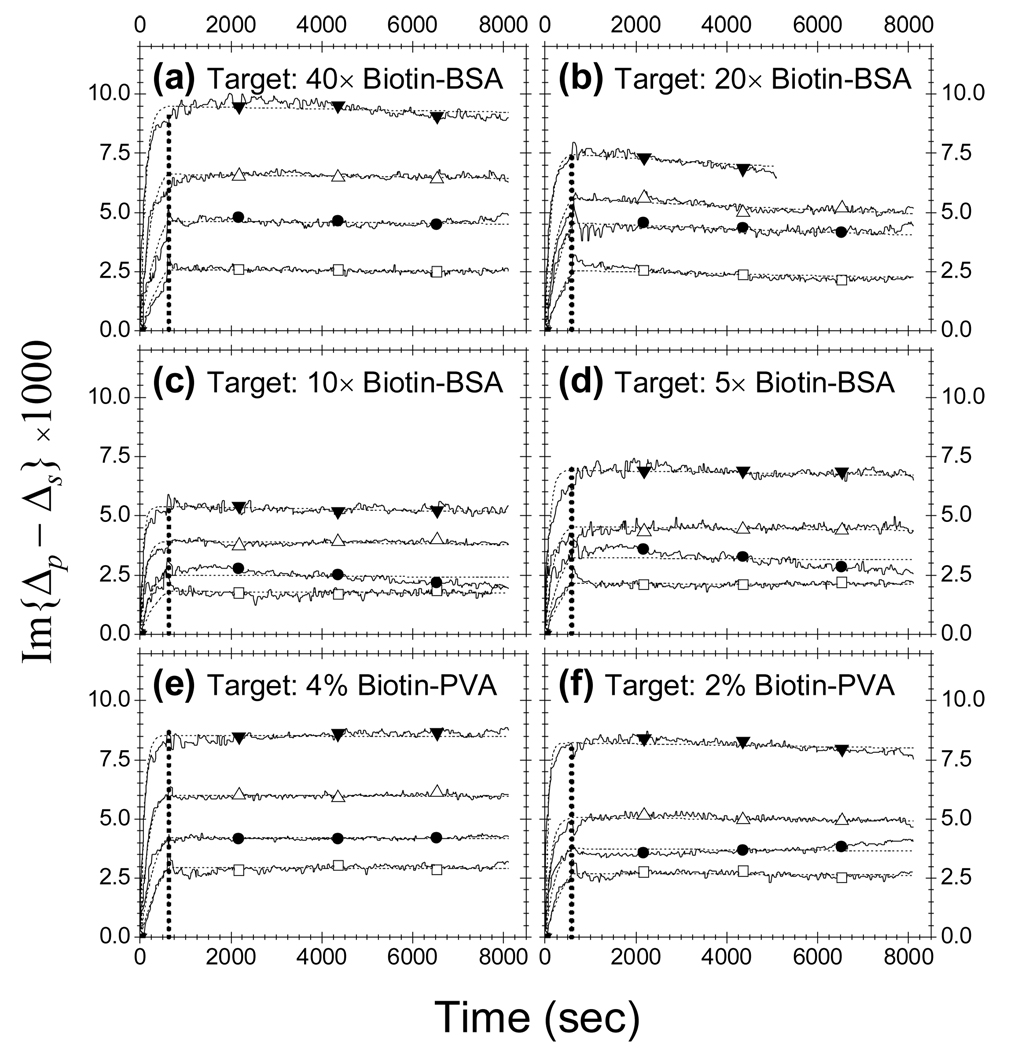
We repeated the protein-ligand reactions by following in real time the amount of the protein probes captured by surface-bound targets. In Fig. 5, we show 6 sets of binding curves for streptavidin reaction with (a) 40× loaded biotin-BSA; (b) 20× loaded biotin-BSA; (c) 10× loaded biotin-BSA; (d) 5× loaded biotin-BSA; (e) 4% loaded biotin-PVA; (f) 2% loaded biotin-PVA. The binding curves for each target were measured at 4 probe concentrations: [c] = 400, 200, 100, and 50 nM. For procedure of a real time experiment (see section 2), we exposed the protein solution to the microarray for 600 seconds starting from t = 0, then replaced the protein solution with 1×PBS and observed for another 7400 seconds. Each set of binding curves were fitted to the two-site Langmuir reaction model (Eq. (5) and Eq. (6)) with , and as the fitting parameters. For biotin-PVA conjugates, the binding curves were better fit if we allow θ1 variable for each curve in a set. This is reasonable since amine-modified PVA is a linear flexible polymer. When printed on different glass slides as necessary for performing binding reactions at different probe concentrations, the relative distribution of two target configurations (Type-1 and Type-2 sites) for conjugated biotins is prone to change. The fits are shown in dotted lines in Fig. 5 with the fitting parameters listed in Table 1. It is noteworthy that for high affinity reactions the smallest dissociation rate koff extractable from a binding curve is limited by the observation time tm (8000 seconds in our present study) and the uncertainty in the coverage measurement as follows,
| (8) |
Here Nm = tm/Δt is the total number of samplings (400 in our present study) and σ is the standard deviation in determined from the standard deviation of Im{Δp - Δs}. If the dissociation rate koff for a binding reaction is less than koff,minimum from the fit, we list in Table 2 koff,minimum as the upper bound of koff and KD = koff,minimum/kon as the upper bound of the equilibrium dissociation constant.
Fig. 5. Probe: Streptavidin.
Simultaneously measured binding curves vs. probe concentration for streptavidin reaction with (a) 40× loaded biotin-BSA; (b) 20× loaded biotin-BSA; (c) 10× loaded biotin-BSA; (d) 5× loaded biotin-BSA; (e) 4% loaded biotin-PVA; (f) 2% loaded biotin-PVA. For probe concentration, (1) open squares: 50 nM; (2) solid circles: 100 nM; (3) open up-pointing triangles: 200 nM; (3) solid down-pointing triangles: 400 nM. The curves in each panel were fitted globally using a two-site Langmuir reaction model (indicated in dashed lines). The fitting parameters are listed in Table 1.
Table 1.
Global fitting parameters to experimentally measured binding curves (association-dissociation) for solution-phase streptavidin reaction with surface-immobilized biotin-BSA and biotin-PVA conjugates. The listed equilibrium dissociation constants KD = koff/kon are computed from the fitting parameters.
| θ(1) | kon(1) (Ms)−1 | koff(1) (s)−1 | KD(1) (nM) | kon(2) (Ms)−1 | koff(2) (s)−1 | KD(2) (nM) | |
|---|---|---|---|---|---|---|---|
| 40× BSA | 0.44 | 1.65×103 | 2.99×10−5 | 18.09 | 2.92×104 | < 1.2×10−6 | < 0.041 |
| 20× BSA | 0.52 | 4.01×102 | 1.86×10−4 | 464.1 | 2.40×104 | < 1.3×10−6 | < 0.054 |
| 10× BSA | 0.44 | 1.65×105 | 1.68×10−5 | 0.102 | 1.26×104 | < 1.5×10−6 | < 0.119 |
| 5× BSA | 0.47 | 5.44×103 | < 2.2×10−6 | < 0.404 | 9.44×104 | 7.78×10−6 | 0.083 |
| 4% PVA | 0.31 a | 1.68×103 | < 3.8×10−6 | < 2.26 | 4.01×104 | < 1.2×10−6 | < 0.030 |
| 0.28 b | |||||||
| 0.43 c | |||||||
| 0.40 d | |||||||
| 2% PVA | 0.34 a | 1.37×103 | 4.85×10−5 | 35.30 | 5.73×104 | < 9×10−7 | < 0.016 |
| 0.39 b | |||||||
| 0.43 c | |||||||
| 0.40 d | |||||||
| 1% PVA | 0.53 a | 1.57×103 | 5.42×10−5 | 34.59 | 7.88×104 | < 1.4×10−6 | < 0.018 |
| 0.29 b | |||||||
| 0.44 c | |||||||
| 0.46 d | |||||||
[c] = 50 nM.
[c] = 100 nM.
[c] = 200 nm.
[c] = 400 nM.
Table 2.
Fitting parameters to binding curves for reaction of monovalent Fab fragments of anti-biotin mouse IgG with immobilized biotin-BSA and biotin-PVA conjugates. The equilibrium dissociation constants KD = koff/kon are computed from the fitting parameters. Coverage of high-affinity targets are shown in bold face.
| θ(1) | kon(1) (Ms)−1 | koff(1) (s)−1 | KD(1) (nM) | θ(2) | kon(2) (Ms)−1 | koff(2) (s)−1 | KD(2) (nM) | |
|---|---|---|---|---|---|---|---|---|
| 40×BSA | 0.30 | 4.87×104 | 3.19×10−4 | 6.55 | 0.70 | 9.80×103 | < 9×10−7 | < 0.092 |
| 20×BSA | 0.35 | 2.10×104 | 3.69×10−4 | 17.57 | 0.65 | 2.12×104 | < 2×10−6 | < 0.094 |
| 10×BSA | 0.41 | 1.53×104 | 4.91×10−4 | 32.09 | 0.59 | 6.96×104 | < 3.6×10−6 | < 0.052 |
| 5×BSA | 0.34 | 4.12×104 | 5.25×10−4 | 12.74 | 0.66 | 3.04×104 | < 4.9×10−6 | < 0.161 |
| 4%PVA | 0.16 a | 2.25×104 | 8.03×10−4 | 35.69 | 0.84 | 2.04×104 | < 1.9×10−6 | < 0.093 |
| 0.53 b | 0.47 | |||||||
| 0.61 c | 0.39 | |||||||
| 0.55 d | 0.45 | |||||||
| 2%PVA | 0.27 a | 3.67×104 | 8.14×10−4 | 22.18 | 0.73 | 3.06×104 | < 1×10−6 | < 0.033 |
| 0.61 b | 0.39 | |||||||
| 0.58 c | 0.42 | |||||||
| 0.39 d | 0.41 | |||||||
| 1%PVA | 0.61 a | 2.46×104 | 1.04×10−3 | 40.81 | 0.39 | 6.31×104 | < 7×10−7 | < 0.011 |
| 0.60 b | 0.40 | |||||||
| 0.35 c | 0.65 | |||||||
| 0.30 d | 0.70 | |||||||
[c] = 27 nM.
[c] = 80 nM.
[c] = 240 nm.
[c] = 480 nM.
From Table 1, we see that immobilized biotin-BSA and biotin-PVA conjugates both present two types of binding sites with nearly equal coverage: one site has a high affinity to streptavidin with KD having upper-bounds of 20 pM to 100 pM and thus more or less preserves the integrity of biotin as a streptavidin ligand; the other site has a significantly lower affinity with KD in the range of nM to tens of nM. At probe concentrations much less than 1 nM, only high-affinity sites are chemically significant. To examine the effectiveness of BSA and PVA as macromolecular scaffolds for protein-ligand binding kinetics assays, it is only sensible to compare binding kinetics measured at the high affinity sites. Table 1 shows that the fraction (coverage) of conjugated biotins with high affinity to streptavidin is comparable on BSA and PVA; and KD of streptavidin to these conjugated biotin are less than tens of pM on both scaffolds. Given the flexibility of a PVA molecule, the fact that the equilibrium dissociation constants for streptavidin to conjugated biotins on amine-modified PVA has a smaller upper bound (limited by the observation time and the uncertainty of the optical measurement) validates PVA as an equally effective scaffold as BSA.
Reaction kinetics of monovalent Fab fragments of anti-biotin mouse IgG with immobilized biotin-BSA and biotin-PVA conjugates
In Fig. 6, we show 6 sets of the binding curves for reaction of monovalent fragments of anti-biotin mouse IgG with (a) 40× loaded biotin-BSA; (b) 20× loaded biotin-BSA; (c) 10× loaded biotin-BSA; (d) 5× loaded biotin-BSA; (e) 4% loaded biotin-PVA; (f) 1% loaded biotin-PVA. Again each set corresponds to binding reactions at 4 protein probe concentrations: [c] = 480, 240, 80 and 27 nM. The measurement procedure was the same as for streptavidin-biotin reactions. Each set of the binding curves was fitted using Eq. (5) and Eq. (6) with , and as the fitting parameters. Again for biotin-PVA conjugates, we let θ(1) be variable for each binding curve of a set. Table 2 lists the fitting parameters.
Fig. 6. Probe: Anti-Biotin.
Simultaneously measured binding curves vs. probe concentration for reaction of monovalent Fab fragments of anti-biotin mouse IgG with (a) 40× loaded biotin-BSA; (b) 20× loaded biotin-BSA; (c) 10× loaded biotin-BSA; (d) 5× loaded biotin-BSA; (e) 4% loaded biotin-PVA; (f) 2% loaded biotin-PVA. For probe concentration, (1) open squares: 27 nM; (2) solid circles: 80 nM; (3) open up-pointing triangles: 240 nM; (4) solid down-pointing triangles: 480 nM. The curves in each panel were fitted globally using a two-site Langmuir reaction model (shown in dashed lines). The fitting parameters are listed in Table 2.
Similar to streptavidin-biotin reactions, immobilized biotin-BSA and biotin-PVA conjugates present two types of biotin targets with comparable coverage: one type has a high affinity to monovalent fragments of anti-biotin mouse IgG with KD less than 100 pM; the other has a significantly lower affinity with KD in the range of tens of nM. If we only concern ourselves with the high-affinity biotin targets, the fraction (coverage) of these conjugated biotins on BSA and amine-modified PVA are comparable, and their KD to Fab fragments of anti-biotin mouse IgG are also comparable. This again validates BSA and amine-modified PVA as equally effective scaffolds for characterization of protein-small molecule ligand binding kinetics.
4. Summary
Bovine serum albumin (BSA) and amine-modified polyvinyl alcohol (PVA) are effective anchors to epoxy-functionalized glass surface with the immobilization efficiency in the vicinity of 20%. As a result, to immobilize a monolayer of BSA conjugates or amine-modified PVA on an epoxy-coated glass surface only requires single printing and solutions of conjugates at as low as a few µM. The single printing is necessary in high-throughput microarray fabrication. By conjugating small molecules to BSA or amine-modified PVA, the conjugates are effective for microarray-based characterization of protein-small molecule binding reactions. As long as the total number of the accessible small molecule targets is no more than needed to capture a full monolayer of solution-phase protein probes, the small-molecule targets conjugated on BSA and amine-modified PVA have a significant fraction whose affinity to solution-phase protein probes essentially retain the liquid-phase value. Binding kinetics of a protein probe to the high-affinity fraction of immobilized targets can be separated from those to the low affinity fraction by using a two-site Langmuir reaction model. As for the high affinity fraction of conjugated biotins, the equilibrium dissociation constants for streptavidin and monovalent Fab fragments of mouse IgG raised against biotin are in the range of tens of pM or less.
Not surprising and yet noteworthy is that amine-modified PVA, being a linear polymeric scaffold, can accommodate far more accessible small molecule targets to protein probes by extending into the aqueous ambient from a solid surface. Such an extended scaffold quickly reduces the efficiency of mass transport of solution-phase protein probes, rendering extraction of binding kinetics from real-time measurements unreliable or even irrelevant. However if one is only concerned with the steady-state of a binding reaction and in pursuit of better sensitivity of an optical measurement, amine-modified PVA is superior over BSA. Eq. (5) shows that given the optical measurement methodology (fixed γ) and protein probe concentration [c], the absolute optical response increases with and . The latter can be increased by extending the effective thickness of the target layer to a quasi-3D porous matrix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health under R01-HG3827.
Contributor Information
Y. S. Sun, Department of Physics, University of California, Davis, California 95616
J. P. Landry, Department of Physics, University of California, Davis, California 95616
Y. Y. Fei, Department of Physics, University of California, Davis, California 95616
X. D. Zhu, Department of Physics, University of California, Davis, California 95616.
J. T. Luo, School of Medicine, University of California at Davis, Sacramento, California 95817
X. B. Wang, School of Medicine, University of California at Davis, Sacramento, California 95817
K. S. Lam, School of Medicine, University of California at Davis, Sacramento, California 95817
Reference
- 1.Schena M. Microarray Analysis. Hoboken: John Wiley and Sons; 2003. [Google Scholar]
- 2.MacBeath G. Nat. Genet. 2002;32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 3.Lockhart DJ, Winzeler EA. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 4.Ramsay G. Nat. Biotechnol. 1998;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- 5.Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, Stoughton R, Burchard J, Slade D, Dai H, Bassett DE, Hartwell LH, Brown PO, Friend SH. Nat. Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 6.MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Moufek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 8.Fang Y, Frutos AG, Lahiri J. J. Am. Chem. Soc. 2002;11:2394–2395. doi: 10.1021/ja017346+. [DOI] [PubMed] [Google Scholar]
- 9.Angenendt P, Glokler J, Murphy D, Lehrach H, Cahill DJ. Anal. Biochem. 2002;309:253–260. doi: 10.1016/s0003-2697(02)00257-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen GYJ, Uttamchandani M, Zhu Q, Wang G, Yao SQ. ChemBioChem. 2003;4:336–339. doi: 10.1002/cbic.200390054. [DOI] [PubMed] [Google Scholar]
- 11.Xu Q, Lam KS. J. Biomed. Biotech. 2003;5:257–266. doi: 10.1155/S1110724303209220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uttamchandani M, Wang J, Yao SQ. Mol. BioSyst. 2006;2:58–68. doi: 10.1039/b513935j. [DOI] [PubMed] [Google Scholar]
- 13.Wang D. Proteomics. 2003;3:2167–2175. doi: 10.1002/pmic.200300601. [DOI] [PubMed] [Google Scholar]
- 14.Shin I, Park S, Lee MR. Chem. Eur. J. 2005;11:3194–3206. [Google Scholar]
- 15.Liang P-L, Wang S-K, Wong C-H. J. Am. Chem. Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
- 16.Liang P-L, Wu C-Y, Greenberg WA, Wong C-H. Curr. Opin. Chem. Biol. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziauddin J, Sabatini DM. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 18.Moch H, Kononen T, Kallioniemi OP, Sauter G. Adv. Anat. Pathol. 2001;8:14–20. doi: 10.1097/00125480-200101000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 20.Stockwell BR. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koehler AN, Shamji AF, Schreiber SL. J. Am. Chem. Soc. 2003;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 22.Urbina HD, Debaene F, Jost B, Bole-Feysot C, Mason DE, Kuzmic P, Harris JL, Winssinger N. ChemBiochem. 2006;7:1790–1797. doi: 10.1002/cbic.200600242. [DOI] [PubMed] [Google Scholar]
- 23.Tolliday N, Clemons PA, Ferraiolo P, Koehler AN, Lewis TA, Li X, Schreiber SL, Gerhard DS, Eliasof S. Cancer Res. 2006;66:8935–8942. doi: 10.1158/0008-5472.CAN-06-2552. [DOI] [PubMed] [Google Scholar]
- 24.Xu Q, Miyamoto S, Lam KS. Mol. Diversity. 2004;8:301–310. doi: 10.1023/b:modi.0000036238.42200.3d. [DOI] [PubMed] [Google Scholar]
- 25.Lee M, Shin I. Angew. Chem. Int. Ed. Engl. 2005;44:2881–2884. doi: 10.1002/anie.200462720. [DOI] [PubMed] [Google Scholar]
- 26.Kusnezow W, Pulli T, Witt O, Hoheisel JD. In: Protein Microarrays. Schena M, editor. Jones and Bartlett; 2005. pp. 247–283. [Google Scholar]
- 27.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung LS, Wagner P, Nock S. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 28.Bradner JE, McPherson OM, Koehler AN. Nat. Protoc. 2006;1:2344–2352. doi: 10.1038/nprot.2006.282. [DOI] [PubMed] [Google Scholar]
- 29.Bradner JE, McPherson OM, Mazitschek R, Barnes-Seeman D, Shen JP, Dhaliwal J, Stevenson KE, Duffner JL, Park SB, Neuberg DS, Nghiem P, Schreiber SL, Koehler AN. Chem. Biol. 2006;13:493–504. doi: 10.1016/j.chembiol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Kanoh N, Kyo M, Inamori K, Ando A, Asami A, Nakao A, Osada H. Anal. Chem. 2006;78:2226–2230. doi: 10.1021/ac051777j. [DOI] [PubMed] [Google Scholar]
- 31.Gildersleeve JC, Oyelaran O, Simpson JT, Allred B. Bioconjugate Chem. 2008;19:1485–1490. doi: 10.1021/bc800153t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avdulov NA, Chochina SV, Daragan VA, Schroeder F, Mayo KH, Wood WG. Biochemistry. 1996;35:340–347. doi: 10.1021/bi9513416. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell M. Nat. Biotechnol. 2002;20:225–229. doi: 10.1038/nbt0302-225. [DOI] [PubMed] [Google Scholar]
- 34.Kodadek T. Chem. Biol. 2001;8:105–115. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 35.Sun YS, Landry JP, Fei YY, Zhu XD, Luo JT, Wang XB, Lam KS. Langmuir. 2008;24:13399–13405. doi: 10.1021/la802097z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landry JP, Zhu XD, Gregg JP. Opt. Lett. 2004;29:581–583. doi: 10.1364/ol.29.000581. [DOI] [PubMed] [Google Scholar]
- 37.Zhu XD, Landry JP, Sun YS, Gregg JP, Guo XW. Appl. Opt. 2007;46:1890–1895. doi: 10.1364/ao.46.001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fei YY, Landry JP, Sun YS, Zhu XD, Luo JT, Lam KS. Rev. Sci. Instrum. 2008;79:1–7. doi: 10.1063/1.2830286. 013708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu XD. Opt. Comm. 2006;259:751–753. [Google Scholar]
- 40.Thomas P, Nabighian E, Bartelt MC, Fong CY, Zhu XD. Appl. Phys. A. 2004;79:131–137. [Google Scholar]
- 41.Landry JP, Sun YS, Guo XW, Zhu XD. Appl. Opt. 2008;47:3275–3288. doi: 10.1364/ao.47.003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu XD. Phys. Rev. B. 2004;69:1–5. 115407. [Google Scholar]
- 43.Morton TA, Myszka DG, Chaiken IM. Anal. Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 44.Heding A, Gill R, Ogawa Y, Meyts PD, Shymko RM. J. Biol. Chem. 1996;271:13948–13952. doi: 10.1074/jbc.271.24.13948. [DOI] [PubMed] [Google Scholar]
- 45.House-Pompeo K, Boles JO, Höök M. Methods. 1994;6:134–142. [Google Scholar]
- 46.Campagnolo C, Meyers KJ, Ryan T, Atkinson RC, Chen YT, Scanlan MJ, Ritter G, Old LJ, Batt CA. J. Biochem. Biophys. Methods. 2004;61 doi: 10.1016/j.jbbm.2004.05.006. 283-198. [DOI] [PubMed] [Google Scholar]
- 47.Höök F, Vörös J, Rodahl M, Kurrat R, Böni P, Ramsden JJ, Textor M, Spencer ND, Tengvall P, Gold J, Kasemo B, Colloid Surf B. Biointerfaces. 2002;24:155–170. [Google Scholar]
- 48.Vörös J. Biophys. J. 2004;87:553–561. doi: 10.1529/biophysj.103.030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendrickson WA, Pähler A, Smith JS, Satow Y, Merritt EA, Phizackerley RP. Proc. Natl. Acad. Sci. USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris LJ, Larson SB, Hasel KW, McPherson A. Biochemistry. 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.