Abstract
Inherited BRCA1/2 mutations confer elevated ovarian cancer (OvCa) risk. Knowledge of factors that can improve OvCa risk assessment in BRCA1/2 mutation carriers is important because no effective early detection for OvCas exists. A cohort of 1,575 BRCA1 and 856 BRCA2 mutation carriers was used to evaluate SNPs and haplotypes at ATM, BARD1, BRIP1, CTIP, MRE11, NBS1, RAD50, RAD51, and TOPBP1 in OvCa risk. In BRCA1 carriers, no associations were observed with ATM, BARD1, CTIP, RAD50, RAD51, or TOPBP1. At BRIP1, an association was observed for one haplotype with a multiple testing corrected p-value (pcorr)=0.012, although no individual haplotype was significant. At MRE11, statistically significant associations were observed for one haplotype (pcorr=0.007). At NBS1, we observed a pcorr=0.024 for haplotypes. In BRCA2 carriers, no associations were observed with CTIP, NBS1, RAD50, or TOPBP1. Rare haplotypes at ATM (pcorr=0.044) and BARD1 (pcorr=0.012) were associated with OvCa risk. At BRIP1, two common haplotypes were significantly associated with OvCa risk (pcorr=0.011). At MRE11, we observed a significant haplotype association (pcorr=0.012), and at RAD51, one common haplotype was significantly associated with OvCa risk (pcorr=0.026). Variants in genes that interact biologically with BRCA1 and/or BRCA2 may be associated with modified OvCa risk in women who carry BRCA1/2 mutations.
INTRODUCTION
Mutations in BRCA1 and BRCA2 (BRCA1/2) are associated with an increased risk of developing breast and ovarian cancer (OvCa). However, there is substantial inter-individual variability in the age at diagnosis and site of cancer occurrence in BRCA1/2 mutation carriers. These observations imply that BRCA1/2 mutations may be necessary to explain the Mendelian pattern of cancer in some families, but are not sufficient to describe inter-individual variability in age- and site-specific cancer risk. Proper assessment of OvCa risk in BRCA1/2 mutation carriers is of clinical significance because no effective strategies for early detection of OvCa exist, and most ovarian tumors are diagnosed at a late stage with poor prognosis(1). Thus, women are counseled to strongly consider risk reducing salpingo-oophorectomy (RRSO). Although RRSO reduces ovarian and breast cancer risk and mortality (2, 3), the induction of surgical menopause is associated with menopausal symptoms which may affect quality of life, osteoporosis and cardiovascular disease. The goal of this research is to identify factors that modify OvCa risk in BRCA1/2 mutation carriers to improve risk assessment and disease prevention.
Modifying factors may influence cancer risk in BRCA1/2 mutation carriers. Begg et al. (4) reported that biases may exist in estimates of lifetime cancer risk if relevant covariates are ignored, and concluded that modifiers are likely to exist that affect BRCA1/2-associated cancer penetrance. Lee et al. (5) examined the lifetime risk of cancer in first-degree relatives of BRCA1/2 mutation carriers with breast or OvCas and concluded that there was more similarity in risks within families than would be expected by chance alone. A number of reports suggest that environmental exposures (e.g. oral contraceptives, smoking) affect OvCa penetrance in women who carry a germline BRCA1/2 mutation (6). Few genetic risk modifiers for BRCA1/2-associated OvCa have been studied (7, 8).
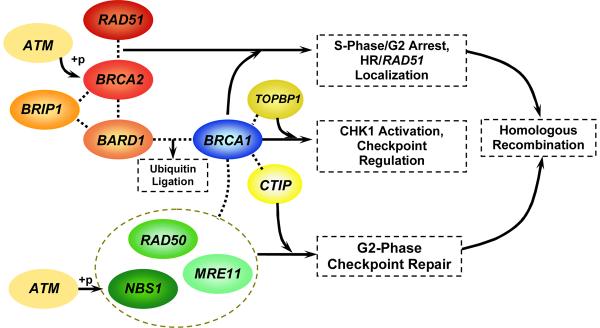
Genetic modifiers of OvCa risk in BRCA1/2 mutation carriers can be identified from our knowledge of BRCA1/2 function (9, 10). A number of proteins and protein complexes that interact with BRCA1/2 have been identified (11, 12) (Figure 1). BRCA1 has been found to interact with many DNA repair proteins including the RAD50-MRE11-NBS1 (MRN) protein complex (11, 13). The proteins associated with BRCA1 respond to aberrant DNA structures in a number of ways, including acting as DNA damage sensors, signal transducers, and repair effectors. BRCA1 has been hypothesized to work as a coordinator of the various functions of DNA damage, recognition, response and repair, and double strand break repair (11). While the functions of BRCA1 are not yet completely elucidated, we can hypothesize that the genes encoding the proteins that interact with BRCA1 could act as candidate modifiers of BRCA1-associated cancer penetrance.
Figure 1.
Biological Interactions of Candidate Proteins and BRCA1 or BRCA2
Fewer proteins are known to interact with BRCA2 (14). BRCA2 interacts with RAD51, which is involved in meiotic and mitotic recombination and in the repair of double-strand DNA breaks. The RAD51 protein interacts directly with the BRCA2 protein by binding to a series of repeats in BRCA2. As part of the cellular response to DNA damage, the BRCA2/RAD51 complex co-localizes to damage-induced foci, where double-strand break repair is thought to take place. It is hypothesized that BRCA2 plays a regulatory role with respect to RAD51 and prevents RAD51 from binding DNA and forming nucleoprotein filaments under normal circumstances (14). However, when DNA damage occurs, there is a change in the BRCA2/RAD51 complex (perhaps phosphorylation of either protein) resulting in the assembly of the recombination complex at the damage-induced foci for DNA repair, allowing Rad51 to bind to single strand DNA and participate in double strand break repair (15). BRCA2 is required for efficient Rad51 delivery to DNA damage sites for homologous recombination at single stranded-double stranded DNA junctions (15, 16). Mutations in either RAD51 or BRCA2 lead to severe defects in DNA repair and potentially to chromosomal rearrangements.
This evidence suggests that BRCA1 and BRCA2 are involved in super-complexes of proteins involved in networks responsible for tumor suppression (11). Therefore, we hypothesize that variation in the genes that encode BRCA1/2 interactors modulate BRCA1/2 penetrance as follows: RAD51 in BRCA1 mutation carriers via errors in HR/RAD51 localization; TOPBP1 or BRIP1 in BRCA1 or BRCA2 mutation carriers via errors in DNA replication associated DNA repair; MRN, CTIP in BRCA1 mutation carriers via errors in G2-phase checkpoint and CHK1 activation; or BARD1 in BRCA1 or BRCA2 mutation carriers by any of these mechanisms.
MATERIALS AND METHODS
Participants and Data Collection
Seventeen MAGIC centers contributed to this study: Baylor-Sammons Cancer Center; Beth Israel Deaconess Medical Center; City of Hope National Medical Center; Creighton University; Dana Farber Cancer Institute; NorthShore University HealthSystem; Fox Chase Cancer Center; Georgetown University; Jonsson Comprehensive Cancer Center at UCLA; Mayo Clinic College of Medicine; University of Chicago; University of California, Irvine; University of Pennsylvania; University of Texas, Southwestern; University of Vienna; Women's College Hospital; and the Kathleen Cunningham Consortium for Research into Familial Breast Cancer (kConFab)(17).
The protocol for this observational cohort was the same in each center. All participants were identified via high-risk programs for clinical and research purposes. Participants were referred by clinicians or self-referred because they were perceived to be at risk for hereditary breast and/or OvCa. Genetic counseling and testing was performed under clinical and/or research protocols specific to the IRB guidelines of each center. All centers identified women who had tested positive for BRCA1/2 mutations by commercial laboratory testing or, more rarely, research testing without clinical disclosure of test results.
The participating centers provided eligibility information to the University of Pennsylvania coordinating center, which in turn determined eligibility for all participants. Eligible participants included women over the age of 18, with documented disease-associated mutations in BRCA1 or BRCA2, who had never been diagnosed with cancer at any site prior to center ascertainment or were diagnosed with breast cancer only within five years or OvCa within three years of their clinic ascertainment in order to minimize the potential for survival bias. As only a small proportion of our cohort (<5%) includes minority groups, we included only participants who were white, including Hispanic, non-Hispanic, and Jewish. Selection was made without respect to RRSO or exposures, and no exclusions were applied based on any risk factors, surgeries, or cancer occurrences. BRCA1/2 mutation status of all subjects was confirmed by direct mutation testing and subjects provided full informed consent for this study under protocols approved by the human subjects review boards at each institution. Some participants were simultaneously consented for both research and clinical BRCA1/2 testing, while others were consented separately for clinical testing and for research participation. Women with BRCA1/2 variants of unknown clinical significance were excluded. Mutations were included in the analysis if they were pathogenic according to generally recognized criteria, including (i) mutations generating a premature termination codon (except truncating variants in exon 27 of BRCA2) as a result of a nonsense substitution, a frameshift due to small deletion or insertion, aberrant splicing or large genomic rearrangement; (ii) mutations resulting in loss of expression due to deletion of promoter and transcription start site; (iii) large in-frame deletions spanning one or more exons caused by aberrant splicing or large genomic rearrangement and (iv) missense mutations classified as pathogenic using the algorithms of Goldgar et al. (18) and Chenevix-Trench et al. (19).
Data were obtained on all eligible participants using medical records, telephone interviews, and/or self-administered questionnaires and included information on reproductive and exposure history, including hormone use and smoking. Vital status, cancer diagnoses, and prophylactic surgery data were verified by review of medical records, operative notes, and/or pathology reports. Follow-up from the time of ascertainment was conducted within each center on a periodic basis. This follow-up was active but did not occur at equivalent intervals for all individuals in this multicenter observational study. Follow-up was random with respect to RRSO use, cancer occurrence, or death. In addition, because this was not a randomized clinical trial of RRSO, both the case and the control groups underwent a variety of cancer surveillance programs that were not controlled for in this study. However, all occurrences of RRSO were verified by medical records if available, and these were carefully distinguished from ovarian surgeries that may have occurred in conjunction with an OvCa diagnosis. Any oophorectomy that was performed for therapeutic/symptomatic reasons and determined to be OvCa or was performed within one year of an OvCa diagnosis was not considered “prophylactic” but was determined to be related to OvCa diagnosis and treatment.
Genotype and Haplotype Data
We chose SNPs to tag haplotypes, as well as putative functional SNPs, in nine genes that interact with BRCA1 and/or BRCA2 (Figure 1; Supplementary Tables 1 and 2): ATM, BRIP1 (BRIP1/FANCJ), BARD1, CTIP, MRE11, NBS1, RAD50, RAD51, and TOPBP1. Haplotype tag SNPs (htSNPs) at each locus were selected using Tagging Wizard in SNPBrowser from publicly available HapMap data (release 16), if they had haplotype R2>95% and minor allele frequencies (MAF) of 5% or greater. We also excluded SNPs that had not been validated for the Taqman platform based on SNPBrowser data. We identified 127 htSNPs that met these criteria. In addition, we identified 51 putative functional non-synonymous SNPs (nsSNPs) that had been reported in HapMap (release 16) or in the literature. No MAF restrictions were placed on these nsSNPs. In order to generate pools for genotype analysis, 7 htSNPs were excluded because the sequence surrounding the SNP was incompatible with the primers or probes used for SNPlex, or would interfere with another SNP being queried. One nsSNP was excluded because one or more additional SNPs was found to be in close proximity to the SNP of interest, and therefore could not be interrogated by SNPlex. After completion of laboratory analysis, SNPs were also excluded from subsequent consideration if they had assay failure rates >20%, MAF <1% or if they showed statistically significant deviations from Hardy-Weinberg equilibrium in unrelated non-cancer Caucasian women were excluded with p<0.005 (Supplementary Table 1). At the conclusion of this process, 56 SNPs in BRCA1 carriers and 51 SNPs in BRCA2 carriers were included in the analyses presented here.
Genomic DNA samples were extracted from peripheral blood at each center and shipped to the Penn data coordinating center. Samples were genotyped using the SNPlex™ System genotyping kit (Applied Biosystems, Foster City, CA) using the standard protocol. Briefly, 40 ng of DNA extracted from peripheral blood was fragmented using heat. Samples then underwent the Oligo Ligation Assay (OLA) where allele specific oligos (ASOs), each containing a unique identifying code (ZIP code) were ligated to locus specific oligos (LSOs) to generate single stranded products. The products were cleaned using exonuclease to remove all unligated products. Cleaned OLA product then underwent PCR. PCR products were then bound to a plate coated with streptavadin and underwent several washes where reporters unique to each genotype (ZIP chutes) were hybridized to the products at the ZIP code. ZIP chutes were then eluted and run on a 3130xl Capillary Sequencer. Genotypes were read using GeneMapper 4.0.
Statistical Analysis
Analysis was undertaken using the weighted cohort approach of Antoniou et al. (20). The weighted approach was implemented to address the issue that study carriers may be ascertained from multiple-case families selected for genetic testing. In addition, since the presence of disease may influence the likelihood of testing, affected carriers may be over-represented in our cohort. The approach provides reasonably unbiased risk estimates (20). Five-year interval weights were applied based on published OvCa relative risks for BRCA1 and BRCA2 mutation carriers separately (21).
The primary event of interest was diagnosis of OvCa. Observations were censored at the earliest of the following events: RRSO, death, or having reached the end of follow-up without an OvCa or other censoring event. Time to event was computed from age at birth to age at first OvCa diagnosis or age at censoring. Analyses were adjusted for ethnicity (Jewish, Hispanic, or non-Hispanic non-Jewish white) and birth cohort (decade of birth). Breast cancer diagnosis was included in the Cox model as a censoring event. All analyses were undertaken in BRCA1 and BRCA2 mutation carriers separately. Finally, we present both the uncorrected p-values as well as p-values corrected for multiple hypothesis testing (denoted pcorr) to adjust the overall association between nine candidate genes in BRCA1 and BRCA2 mutation carriers separately, for a total of 18 hypothesis tests. The Benjamini-Hochberg method was used to generate the p-values corrected for multiple testing (22). All survival analyses were conducted in SAS version 9.1 (SAS Institute Inc., Cary, NC).
To investigate haplotype associations, the EM algorithm (23, 24) was used to estimate haplotype frequencies as implemented in R version 2.1.1 subroutine haplo.em (25). To assess the association between haplotypes and survival outcome, we created a user-defined model matrix to estimate haplotype associations. First, haplo.em was used to estimate haplotype frequencies under the null hypothesis of no association (in the pool of all data). This approach enumerated all possible haplotype pairs per subject along with the posterior probabilities of each haplotype pair, conditional on the genotype data. The posterior probabilities were then used to average the rows of the model matrix per subject and the resulting matrix was used in a Cox regression model. Global tests for association (to test the association between all haplotypes together with disease status) as well as haplotype-specific tests (to test the association between each haplotype and disease status) were conducted. Cox regression for haplotype analyses were conducted as implemented in SAS Genetics. Phase ambiguity was quantified by estimating the percentage of uncertainty in the imputed diplotypes. The majority of haplotypes had a maximum posterior probability of over 80%; hence we felt comfortable proceeding with the haplotype association method outlined above rather than assigning the most likely haplotype pair to each subject.
RESULTS
Sample Set Description
We identified a cohort of 1575 BRCA1 and 856 BRCA2 female mutation carriers. Of BRCA1 mutation carriers, 179 (11%) had OvCa and 1,396 were censored. Among BRCA2 mutation carriers, 47 (5.5%) had OvCa and 809 were censored. The characteristics of the participants are described in Table 1. OvCa cases were significantly more likely to be members of an older birth cohort than censored controls for both BRCA1 and BRCA2 (p<0.0001). Data for oral contraceptive (OC) use was available for 1391 (88%) of participants with a BRCA1 mutation. Of those who were diagnosed with an OvCa, 98 (62%) had ever used OC and 1021 (83%) of non-OvCa cases ever used OC. Significantly more women without an OvCa diagnosis used OC (p<0.001). For BRCA2 carriers, data for OC use was available for 777 (91%). Of those who were diagnosed with an OvCa, 25 (57%) ever used OC and 592 (81%) of non-OvCa cases ever used OC. The difference in OC use between the two groups was significant (p<0.001) in BRCA2 carriers. Censored controls were more likely to have undergone risk-reducing salpingo-oophorectomy (RRSO), to have had a breast cancer diagnosis, and were more likely to be alive at the end of follow up.
Table 1.
Descriptive Characteristics of Study Participants
| BRCA1 | BRCA2 | |||||
|---|---|---|---|---|---|---|
| Variable | OvCa (N=179) | No OvCa (N=1,396) | P-value | OvCa (N=47) | No OvCa (N=809) | P-value |
| Mean Birth Year (range) | 1946 (1921–1967) | 1958 (1899–1986) | <0.001 | 1941 (1917–1960) | 1955 (1911–1985) | <0.001 |
| Jewish | 36 (20%) | 223 (16%) | 0.164 | 11 (23%) | 108 (13%) | 0.078 |
| Ascertainment Year (range) | 1998 (1978–2006) | 1998 (1973–2006) | 0.724 | 2000 (1978–2005) | 1999 (1978–2006) | 0.481 |
| Risk-Reducing Salpingo-Oophorectomy (%) | 8 (4%) | 655 (47%) | <0.001 | 1 (2%) | 318 (39%) | <0.001 |
| Alive at End of Follow Up | 112 (63%) | 1302 (94%) | <0.001 | 33 (70%) | 749 (93%) | <0.001 |
| Also have a BRCA2 deleterious mutation | 1 (1%) | 2 (0.1%) | - | NA | NA | NA |
| Also have a BRCA1 deleterious mutation | NA | NA | NA | 1 (2%) | 2 (0.2%) | - |
| Breast Cancer Diagnosis (%) | 51 (28%) | 648 (46%) | <0.001 | 18 (38%) | 418 (52%) | 0.098 |
| Mean Age at Breast Cancer Diagnosis (range) | 45.6 (23.6–73.2) | 40.0 (22.2–74.0) | <0.001 | 52.3 (29.5–70.2) | 44.1 (23.6–77.3) | <0.001 |
| Mean Age at Ovarian/Primary Peritoneal Cancer Diagnosis (range) | 50.5 (23.6–81.0) | NA | - | 57.6 (41.5–77.7) | NA | - |
| Age at Censoring (range) | 50.5 (23.6–81.0) | 42.4 (15–100) | 0.00 | 57.6 (41.5–77.7) | 47.2 (19.7–94.0) | 0.00 |
BRCA1
In BRCA1 mutation carriers, no statistically significant associations were observed between haplotypes (Table 2) at ATM, BARD1, CTIP, RAD51, or TOPBP1. At ATM, a significant relationship among rare haplotypes was observed but no overall significance across all haplotypes was observed.
Table 2.
Haplotype Analysis: BRCA1
| Locus | SNP | Frequency | Haplotype | HR | 95%CI | p-value (pcorr) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATM | 7 | 5 | 1 | 2 | 6 | 0.038 (0.058) | ||||||||
| T | T | G | A | C | 0.425 | reference | 1.00 | |||||||
| T | C | G | G | T | 0.003 | rare | 0.01 | 0.00 | 0.35 | |||||
| A | C | G | G | T | 0.411 | A | 0.93 | 0.70 | 1.24 | |||||
| A | T | G | G | C | 0.011 | B | 1.24 | 0.49 | 3.10 | |||||
| T | T | A | A | C | 0.149 | C | 0.90 | 0.62 | 1.31 | |||||
| BRIP1 | 4 | 21 | 33 | 6 | 15 | 2 | 26 | 7 | 0.003 (0.012) | |||||
| T | A | C | C | A | G | G | C | 0.206 | reference | 1.00 | ||||
| C | G | C | C | G | G | G | T | 0.008 | rare | 0.60 | 0.26 | 1.35 | ||
| T | A | C | T | A | A | G | T | 0.003 | rare | |||||
| T | G | C | T | G | G | A | T | 0.006 | rare | |||||
| C | G | C | T | G | G | G | T | 0.003 | rare | |||||
| C | G | C | T | A | A | A | T | 0.005 | rare | |||||
| T | G | C | T | G | G | G | C | 0.005 | rare | |||||
| C | G | C | C | A | A | G | T | 0.010 | A | 0.44 | 0.06 | 3.15 | ||
| C | G | C | C | A | G | A | T | 0.061 | B | 0.71 | 0.34 | 1.49 | ||
| C | G | C | C | A | G | G | C | 0.097 | C | 0.97 | 0.54 | 1.75 | ||
| C | G | C | C | A | G | G | T | 0.167 | D | 0.80 | 0.47 | 1.36 | ||
| C | G | C | T | A | A | G | C | 0.018 | E | 1.94 | 0.45 | 8.33 | ||
| C | G | C | T | A | A | G | T | 0.116 | F | 0.77 | 0.42 | 1.43 | ||
| C | G | C | T | A | G | G | C | 0.033 | G | 0.78 | 0.39 | 1.57 | ||
| C | G | C | T | A | G | G | T | 0.030 | H | 1.07 | 0.44 | 2.65 | ||
| C | G | G | T | A | A | G | T | 0.034 | I | 0.73 | 0.42 | 1.27 | ||
| T | A | C | C | A | G | A | T | 0.052 | J | 0.62 | 0.30 | 1.26 | ||
| T | A | C | C | A | G | G | T | 0.059 | K | 0.95 | 0.45 | 2.04 | ||
| T | G | C | T | G | G | G | T | 0.061 | L | 0.86 | 0.51 | 1.47 | ||
| BARD1 | 7 | 18 | 12 | 15 | 6 | 17 | 2 | 8 | 0.042 (0.058) | |||||
| T | C | T | G | C | A | T | T | 0.268 | reference | 1.00 | ||||
| T | T | A | G | C | G | T | T | 0.008 | rare | 0.88 | 0.50 | 1.55 | ||
| C | T | T | A | C | G | T | C | 0.009 | rare | |||||
| T | C | A | G | C | A | C | T | 0.009 | rare | |||||
| T | C | A | G | C | A | T | T | 0.006 | rare | |||||
| T | T | T | A | C | G | T | C | 0.008 | rare | |||||
| C | T | A | A | C | A | T | C | 0.038 | A | 0.95 | 0.48 | 1.88 | ||
| C | T | A | G | C | A | T | T | 0.035 | B | 1.01 | 0.50 | 2.06 | ||
| T | C | T | G | C | A | C | C | 0.019 | C | 1.27 | 0.50 | 3.19 | ||
| T | T | A | A | T | A | T | T | 0.048 | D | 0.83 | 0.38 | 1.82 | ||
| T | T | A | G | C | A | C | T | 0.056 | E | 1.40 | 0.80 | 2.43 | ||
| T | T | A | G | C | A | T | T | 0.152 | F | 1.10 | 0.72 | 1.67 | ||
| T | T | A | G | C | G | C | C | 0.204 | G | 0.93 | 0.65 | 1.32 | ||
| T | T | T | A | T | A | T | T | 0.062 | H | 1.16 | 0.63 | 2.14 | ||
| T | T | T | G | C | A | T | T | 0.052 | I | 0.71 | 0.37 | 1.35 | ||
| CTIP | 7 | 8 | 5 | 3 | 6 | 4 | 2 | 0.046 (0.059) | ||||||
| A | A | G | A | G | C | A | 0.653 | reference | 1.00 | |||||
| A | A | G | G | A | C | A | 0.006 | rare | 0.92 | 0.32 | 2.64 | |||
| A | A | A | G | G | C | A | 0.004 | rare | ||||||
| A | A | A | G | A | C | A | 0.107 | A | 1.17 | 0.79 | 1.71 | |||
| G | C | A | A | A | C | A | 0.018 | B | 1.42 | 0.74 | 2.72 | |||
| G | C | A | G | A | C | G | 0.013 | C | 1.81 | 0.79 | 4.15 | |||
| G | C | A | G | A | T | G | 0.193 | D | 1.20 | 0.93 | 1.55 | |||
| MRE11 | 1 | 3 | 5 | 6 | 7 | 14 | 18 | <0.0001 (0.007) | ||||||
| C | A | A | G | C | A | A | 0.314 | reference | 1.00 | |||||
| T | A | A | G | C | A | A | 0.004 | rare | 0.47 | 0.10 | 2.10 | |||
| T | G | A | G | C | A | A | 0.003 | rare | ||||||
| C | A | A | G | C | A | G | 0.031 | A | 0.93 | 0.43 | 2.00 | |||
| C | A | G | A | T | G | G | 0.049 | B | 0.63 | 0.28 | 1.44 | |||
| T | A | A | G | T | A | G | 0.021 | C | 1.63 | 0.71 | 3.73 | |||
| T | A | G | A | T | G | G | 0.187 | D | 0.92 | 0.57 | 1.47 | |||
| T | A | G | G | T | G | G | 0.060 | E | 1.25 | 0.73 | 2.15 | |||
| T | G | A | G | T | A | A | 0.015 | F | 1.09 | 0.43 | 2.76 | |||
| T | G | A | G | T | A | G | 0.149 | G | 0.55 | 0.34 | 0.91 | |||
| T | G | G | A | T | G | G | 0.038 | H | 1.00 | 0.50 | 2.02 | |||
| T | G | G | G | T | A | G | 0.116 | I | 0.70 | 0.40 | 1.23 | |||
| NBS1 | 2 | 4 | 7 | 8 | 11 | 12 | 0.008 (0.024) | |||||||
| C | G | C | T | G | G | 0.492 | reference | 1.00 | ||||||
| C | G | C | C | G | G | 0.005 | rare | 1.50 | 0.50 | 4.47 | ||||
| C | A | C | T | G | G | 0.132 | A | 0.93 | 0.60 | 1.43 | ||||
| C | G | C | T | G | C | 0.018 | B | 2.17 | 0.98 | 4.81 | ||||
| C | G | T | T | A | G | 0.026 | C | 1.33 | 0.68 | 2.60 | ||||
| G | A | C | T | G | G | 0.024 | D | 1.20 | 0.52 | 2.76 | ||||
| G | G | C | C | G | C | 0.071 | E | 1.90 | 0.96 | 3.74 | ||||
| G | G | C | T | G | C | 0.193 | F | 1.20 | 0.82 | 1.75 | ||||
| G | G | C | T | G | G | 0.031 | G | 1.06 | 0.35 | 3.25 | ||||
| RAD50 | 2 | 3 | 4 | 5 | 0.022 (0.044) | |||||||||
| G | G | C | C | 0.391 | reference | 1.00 | ||||||||
| G | G | T | C | 0.008 | rare | 1.04 | 0.42 | 2.60 | ||||||
| G | T | C | C | 0.293 | A | 0.94 | 0.67 | 1.32 | ||||||
| G | T | T | C | 0.069 | B | 0.74 | 0.37 | 1.51 | ||||||
| G | T | T | T | 0.020 | C | 1.59 | 0.70 | 3.63 | ||||||
| T | T | C | C | 0.084 | D | 0.87 | 0.48 | 1.57 | ||||||
| T | T | T | C | 0.063 | E | 0.53 | 0.27 | 1.04 | ||||||
| T | T | T | T | 0.067 | F | 0.85 | 0.42 | 1.71 | ||||||
| RAD51 | 2 | 7 | 5 | 1 | 10 | 0.179 (0.205) | ||||||||
| A | C | T | A | T | 0.416 | reference | 1.00 | |||||||
| C | C | T | A | T | 0.006 | rare | 1.63 | 0.61 | 4.33 | |||||
| C | C | T | G | C | 0.004 | rare | ||||||||
| A | C | C | A | C | 0.030 | A | 1.10 | 0.48 | 2.47 | |||||
| A | C | C | A | T | 0.044 | B | 0.96 | 0.39 | 2.36 | |||||
| C | C | C | G | C | 0.321 | C | 1.08 | 0.79 | 1.48 | |||||
| C | T | C | G | C | 0.177 | D | 0.87 | 0.60 | 1.27 | |||||
| TOPBP1 | 13 | 10 | 3 | 5 | 1 | 6 | 0.035 (0.058) | |||||||
| T | C | G | C | C | G | 0.533 | reference | 1.00 | ||||||
| rare | 3.68 | 0.74 | 18.24 | |||||||||||
| C | A | G | C | A | A | 0.136 | A | 1.10 | 0.76 | 1.60 | ||||
| C | A | G | C | C | G | 0.029 | B | 1.06 | 0.42 | 2.68 | ||||
| C | C | G | C | A | G | 0.103 | C | 1.37 | 0.89 | 2.10 | ||||
| T | C | A | T | C | G | 0.193 | D | 0.96 | 0.68 | 1.35 | ||||
We observed a significant multiple testing corrected global p-value (pcorr) of 0.012 for haplotypes at BRIP1, but no individual haplotype was significantly associated with risk (Table 2). It is possible that the combination of multiple haplotypes may be associated with risk (since a number of haplotype associations are in the same direction), but we did not attempt to combine haplotypes based on observed HR estimates and did not have any a priori justification for combining haplotypes. Therefore, we cannot conclude with a high degree of confidence that there is a relationship between BRIP1 haplotypes and OvCa risk in BRCA1 mutation carriers.
At MRE11, we observed a significant association of haplotypes (pcorr=0.007). Haplotype G, with a frequency of 14.9% and containing the variant G allele at SNP3, was inversely and significantly associated with risk (HR=0.55, 95% CI: 0.34–0.91).
At NBS1, we observed a significant association of haplotypes (pcorr=0.024). While some haplotypes containing the individually significant SNPs had HR associations in the same direction as these individual SNP associations, no single haplotype was significantly associated with risk.
At RAD50, we observed a significant global association of haplotypes (pcorr=0.044), but no individual haplotype was significantly associated with risk. These results do not support the hypothesis that a relationship between RAD50 and OvCa risk in BRCA1 mutation carriers exists.
While the primary analysis undertaken here involved haplotype-based associations, associations involved single SNPs that comprised these haplotypes are shown in Supplementary Table 3.
BRCA2
In BRCA2 mutation carriers, no haplotype associations were observed for CTIP, NBS1, RAD50, or TOPBP1 (Table 3). In ATM, we observed a significant association with haplotypes (pcorr =0.044; Table 3). The `rare' TTGGC haplotype at ATM (frequency=0.9%; Table 3), which represents a difference in SNPs 2 (rs664982) and 6 (rs664143) compared to the reference haplotype, was significantly associated with risk (HR=10.93, 95% CI 4.43–26.96).
Table 3.
Haplotype Analysis: BRCA2
| Locus | SNP | Frequency | Haplotype | HR | 95%CI | p-value (pcorr) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATM | 5 | 4 | 1 | 2 | 6 | 0.022 (0.044) | ||||||||
| T | T | G | A | C | 0.435 | reference | 1.00 | |||||||
| T | T | G | G | C | 0.009 | rare | 10.93 | 4.43 | 26.96 | |||||
| C | T | G | G | T | 0.415 | A | 1.09 | 0.59 | 2.01 | |||||
| T | C | A | A | C | 0.032 | B | 1.09 | 0.11 | 10.87 | |||||
| T | T | A | A | C | 0.107 | C | 1.28 | 0.53 | 3.11 | |||||
| BRIP1 | 21 | 6 | 25 | 2 | 18 | 29 | 14 | 27 | 0.001 (0.011) | |||||
| G | C | A | G | G | G | C | G | 0.305 | reference | 1.00 | ||||
| G | C | A | A | T | A | T | A | 0.009 | rare | 0.49 | 0.11 | 2.17 | ||
| A | C | A | G | G | G | C | A | 0.004 | rare | |||||
| G | T | G | A | T | A | C | G | 0.009 | rare | |||||
| G | C | A | G | G | A | T | A | 0.005 | rare | |||||
| A | T | G | A | T | A | T | A | 0.005 | rare | |||||
| G | T | A | G | T | G | C | G | 0.004 | rare | |||||
| A | C | A | G | T | A | T | A | 0.009 | rare | |||||
| A | C | A | G | G | G | C | G | 0.273 | A | 1.17 | 0.52 | 2.63 | ||
| G | C | A | G | G | G | C | A | 0.020 | B | 6.59 | 1.10 | 39.65 | ||
| G | C | A | G | T | A | T | A | 0.014 | C | 0.49 | 0.03 | 8.98 | ||
| G | T | A | G | G | G | C | G | 0.023 | D | 6.83 | 0.43 | 107.31 | ||
| G | T | A | G | T | A | T | A | 0.015 | E | 2.22 | 0.24 | 20.50 | ||
| G | T | A | G | T | G | C | A | 0.037 | F | 2.78 | 0.81 | 9.52 | ||
| G | T | G | A | G | G | C | G | 0.024 | G | 7.28 | 1.67 | 31.82 | ||
| G | T | G | A | T | A | T | A | 0.159 | H | 1.16 | 0.46 | 2.96 | ||
| G | T | G | G | G | G | C | G | 0.058 | I | 0.54 | 0.08 | 3.59 | ||
| BARD1 | 7 | 18 | 13 | 6 | 10 | 17 | 2 | 8 | 0.003 (0.012) | |||||
| T | C | T | C | G | A | T | T | 0.271 | reference | 1.00 | ||||
| T | T | C | C | G | A | T | C | 0.004 | rare | 4.62 | 1.31 | 16.31 | ||
| C | T | C | C | G | A | T | T | 0.008 | rare | |||||
| T | T | T | C | G | G | C | T | 0.004 | rare | |||||
| T | T | T | C | G | G | T | T | 0.008 | rare | |||||
| C | T | C | C | G | A | T | C | 0.043 | A | 2.16 | 0.75 | 6.23 | ||
| C | T | T | C | A | A | T | T | 0.019 | B | 0.78 | 0.11 | 5.52 | ||
| C | T | T | C | G | G | T | C | 0.011 | C | 0.68 | 0.06 | 7.28 | ||
| T | C | C | C | G | A | C | T | 0.020 | D | 0.68 | 0.09 | 4.96 | ||
| T | C | T | C | G | A | C | C | 0.019 | E | 2.92 | 0.48 | 17.83 | ||
| T | T | C | C | G | A | C | T | 0.049 | F | 2.33 | 0.68 | 8.02 | ||
| T | T | C | C | G | A | T | T | 0.017 | G | 0.55 | 0.08 | 4.01 | ||
| T | T | T | C | G | A | T | T | 0.170 | H | 1.88 | 0.75 | 4.72 | ||
| T | T | T | C | G | G | C | C | 0.192 | I | 1.41 | 0.49 | 4.10 | ||
| T | T | T | C | G | G | T | C | 0.018 | J | 0.30 | 0.02 | 5.59 | ||
| T | T | T | T | A | A | T | T | 0.062 | K | 0.59 | 0.22 | 1.57 | ||
| T | T | T | T | G | A | T | T | 0.064 | L | 0.74 | 0.16 | 3.42 | ||
| CTIP | 7 | 8 | 5 | 3 | 6 | 4 | 2 | 0.183 (0.205) | ||||||
| A | A | G | A | G | C | A | 0.647 | reference | 1.00 | |||||
| A | A | G | G | A | C | A | 0.007 | rare | 2.82 | 1.06 | 7.48 | |||
| A | C | A | G | A | T | G | 0.004 | rare | ||||||
| G | C | A | G | A | C | G | 0.009 | rare | ||||||
| A | A | A | G | A | C | A | 0.120 | A | 1.15 | 0.45 | 2.92 | |||
| G | C | A | A | A | C | A | 0.015 | B | 2.18 | 0.30 | 15.98 | |||
| G | C | A | G | A | T | G | 0.191 | C | 1.42 | 0.77 | 2.60 | |||
| MRE11 | 1 | 3 | 6 | 8 | 13 | 14 | 19 | 0.003 (0.012) | ||||||
| C | A | G | T | C | A | G | 0.318 | reference | 1.00 | |||||
| T | G | G | C | T | A | G | 0.008 | rare | 5.13 | 1.24 | 21.24 | |||
| T | A | G | T | C | A | A | 0.004 | rare | ||||||
| C | A | A | C | T | G | A | 0.047 | A | 2.09 | 0.40 | 10.76 | |||
| C | A | G | T | C | A | A | 0.020 | B | 0.00 | 0.00 | 0.30 | |||
| T | A | A | C | T | G | A | 0.184 | C | 2.33 | 1.39 | 3.91 | |||
| T | A | A | C | T | G | G | 0.010 | D | 0.00 | 0.00 | 23.37 | |||
| T | A | G | C | T | G | A | 0.047 | E | 1.09 | 0.23 | 5.23 | |||
| T | A | G | C | T | G | G | 0.012 | F | 1.28 | 0.18 | 9.06 | |||
| T | A | G | T | C | A | G | 0.029 | G | 1.49 | 0.29 | 7.58 | |||
| T | G | A | C | T | G | A | 0.023 | H | 2.19 | 0.18 | 26.88 | |||
| T | G | G | C | C | A | G | 0.012 | I | 1.47 | 0.14 | 15.63 | |||
| T | G | G | C | T | A | A | 0.077 | J | 0.67 | 0.16 | 2.92 | |||
| T | G | G | T | C | A | A | 0.015 | K | 0.00 | 0.00 | 0.0 | |||
| T | G | G | T | C | A | G | 0.171 | L | 1.12 | 0.27 | 4.69 | |||
| NBS1 | 1 | 3 | 4 | 0.040 (0.058) | ||||||||||
| A | A | G | 0.480 | reference | 1.00 | |||||||||
| G | A | A | 0.008 | rare | 3.15 | 0.66 | 15.04 | |||||||
| A | C | G | 0.005 | rare | ||||||||||
| A | A | A | 0.118 | A | 0.73 | 0.18 | 3.05 | |||||||
| G | A | G | 0.011 | B | 0.00 | 0.00 | ||||||||
| G | C | A | 0.07 | C | 1.51 | 0.10 | 22.46 | |||||||
| G | C | G | 0.3613 | D | 0.93 | 0.49 | 1.75 | |||||||
| RAD50 | 2 | 3 | 4 | 5 | 0.342 (0.342) | |||||||||
| G | G | C | C | 0.401 | reference | 1.00 | ||||||||
| G | G | T | C | 0.005 | rare | 0.43 | 0.03 | 5.98 | ||||||
| G | T | C | C | 0.300 | A | 1.19 | 0.54 | 2.59 | ||||||
| G | T | T | C | 0.068 | B | 1.38 | 0.31 | 6.06 | ||||||
| G | T | T | T | 0.0180 | C | 0.83 | 0.16 | 4.42 | ||||||
| T | T | C | C | 0.080 | D | 0.87 | 0.23 | 3.35 | ||||||
| T | T | T | C | 0.048 | E | 0.88 | 0.10 | 7.76 | ||||||
| T | T | T | T | 0.075 | F | 0.69 | 0.19 | 2.48 | ||||||
| RAD51 | 8 | 2 | 1 | 10 | 0.010 (0.026) | |||||||||
| C | A | A | T | 0.445 | reference | 1.00 | ||||||||
| T | C | A | T | 0.006 | rare | 4.49 | 0.54 | 37.69 | ||||||
| C | A | A | C | 0.012 | A | 1.16 | 0.11 | 12.69 | ||||||
| C | C | G | C | 0.418 | B | 1.45 | 0.66 | 3.17 | ||||||
| T | C | G | C | 0.119 | C | 3.53 | 1.77 | 7.05 | ||||||
| TOPBP1 | 10 | 8 | 4 | 3 | 5 | 6 | 0.241 (0.255) | |||||||
| C | G | G | G | C | G | 0.334 | reference | 1.00 | ||||||
| rare | 0.03 | 0.00 | 1.44 | |||||||||||
| A | A | A | G | C | A | 0.122 | A | 0.60 | 0.24 | 1.48 | ||||
| A | G | A | G | C | G | 0.021 | B | 2.77 | 0.59 | 12.96 | ||||
| C | G | A | A | T | G | 0.213 | C | 0.99 | 0.50 | 1.98 | ||||
| C | G | A | G | C | G | 0.308 | D | 0.83 | 0.39 | 1.73 | ||||
At BRIP1, we observed a significant association of haplotypes (pcorr =0.011; Table 3). The B and G haplotypes (frequencies of 2.0% and 2.4%, respectively; Table 3) were significantly associated with risk (HR=6.59, 95% CI 1.10–39.65 and HR=7.28, 95%CI: 1.67–31.82, respectively). Haplotype B differs from the reference haplotype in SNP 27 (rs4988340) only. Haplotype G differs from the reference haplotype in SNPs 2 (rs12453935), 6 (rs169456280), and 25 (rs10515211).
At BARD1, we observed a significant association of haplotypes (pcorr=0.012; Table 3). The `rare' haplotypes at BARD1 were significantly associated with risk (HR=4.62, 95% CI: 1.31–16.31; ptrend=0.012; Table 3). One of the rare haplotypes (TTTCGGCT) differs from the reference haplotype by only SNP2 (rs6712055). These results support the hypothesis that a relationship of rare BARD1 haplotypes and OvCa risk in BRCA2 mutation carriers exists.
At MRE11, we observed a significant association of haplotypes (pcorr=0.012; Table 3). The `rare' haplotypes at MRE11 were significantly associated with risk (HR=5.13, 95% CI: 1.24–21.24). In addition, the C haplotype in MRE11 with a frequency of 18.4% was significantly associated with risk (HR=2.33, 95% CI 1.39–3.91). Haplotype C differed for all SNPs except SNP 3 (rs6483327) compared to the reference haplotype.
Finally, at RAD51, we observed a significant association of haplotypes (pcorr=0.026). Haplotype C at RAD51 (frequency=11.9%; Table 3) was significantly associated with risk (HR=3.53, 95% CI: 1.77–7.05). Haplotype C differs from the reference haplotype at all individual SNPs. Consistent with what has been observed in BRCA2-associated breast cancer (26–28), a relationship of rare RAD51 haplotypes and OvCa risk in BRCA2 mutation carriers may exist. Given that rs1801320 (135G>C) in RAD51 has been previously reported as a modifier of breast cancer risk in BRCA2 mutation carriers, we also evaluated this SNP as a candidate modifier of OvCa risk. This SNP was not included as an htSNP for haplotype analysis. The HR associated with carriage of the C allele, previously associated with breast cancer risk modification, was 0.40 (95%CI: 0.05–3.40). However, the variant was relatively rare, occurring in only 5% of 192 BRCA2 mutation carriers, so that the power to detect associations with this sample size was small. This SNP was in strong linkage disequilibrium (i.e., D' > 0.95) with other SNPs at this locus for which haplotype associations were observed, including rs11070291, rs2619680, rs957603, and rs2619681. Therefore, it is likely that we have detected the same association in OvCa as has been previously reported at this locus for breast cancer.
While the primary analysis undertaken here involved haplotype-based associations, associations involved single SNPs that comprised these haplotypes are shown in Supplementary Table 4.
nsSNP Analysis
In addition to haplotype-based analyses, we evaluated 28 candidate nsSNPs in the nine loci studied here. No polymorphic variation was detected in a number of SNPs reported in HapMap data and genotyped here, including K312N, R658C, N470S, Q564H, N295S in BARD1; Q540L, C832Y, L195P, F531V, P47A, or V193I in BRIP1; D488Y in CTIP; V31A or M670V in MRE11; T497A or P672L in NBS1; T191I in RAD50; L109V or K313Q (K216Q) in RAD51; or H1140P in TOPBP1. Among SNPs that showed polymorphic variation, we found no association with OvCa risk for D1853N (D505N) or F858L in ATM; C557S or R378S in BARD1; K370Q, S730L, or N955S in TOPBP1. The only SNP for which a significant trend was observed was Q185E (rs1805794) in NBS1 for BRCA1 mutation carriers (HR=1.40, 95%CI: 0.92 –2.14 for the QE genotype and HR=1.91, 95%CI: 1.02–3.56 for the EE genotype relative to the QQ genotype; p-value for linear trend=0.026).
DISCUSSION
We identified a number of biologically plausible associations in genes that are involved in DNA damage response, interact with BRCA1 and/or BRCA2 (Figure 1), and may act in concert with a mutated BRCA1/2 to modify cancer risk. Rare haplotypes at ATM were associated with OvCa risk in BRCA2 mutation carriers. In response to the formation of double-strand breaks, ATM kinase phosphorylates the BRCA2 protein, leading to activation of an S-phase checkpoint (29, 30). Thus, the regulation of BRCA2 by ATM suggests a plausible mechanism by which rare ATM haplotypes may influence BRCA2-associated OvCa risk.
BARD1 and BRIP1 associate with BRCA1 as cells progress through the S phase of the cell cycle (31, 32). Although BRIP1 probably does not associate with BRCA2, BARD1 is a stoichiometric partner of BRCA1 and remains associated with BRCA1 throughout the cell cycle. BARD1 also interacts with BRCA2 in a substoichimetric manner. This observation provides a plausible explanation for the observation in our data that BRIP1 is associated with OvCa in BRCA1 and BRCA2 mutation carriers, and that BARD1 is associated with OvCa in BRCA2 mutation carriers. BRIP1 is a DNA helicase that interacts with the C-terminal BRCT repeat of BRCA1. BRIP1 is associated with GM1/2 checkpoint and CHK1 activation as well as regulation of entry into the S phase of the cell cycle and maintenance of genomic stability (32). Thus, if the complex involving BRCA1, BRCA2, BARD1, and BRIP1 is involved in tumor suppression, mutations in the genes that encode these proteins should be associated with altered cancer risk. Our data indicating that variants in BRIP1 and BARD1 modify BRCA1/2-associated OvCa support this hypothesis.
We also identified associations of MRE11, RAD50 and NBS1 (MRN) in BRCA1 mutation carriers and MRE11 in BRCA2 mutations carriers. The MRN proteins interact directly with one another and interact with BRCA1 in a DNA damage inducible manner (11). NBS1 protein and the activated form of BRCA2 co-localize in subnuclear foci in response to mitomycin C-induced DNA damage and interact in the cellular response to DNA crosslink formation (33). The MRN complex therefore interacts with BRCA2 and the Fanconi Anemia pathway in S-phase checkpoint response (34). These observations are consistent with our findings that MRE11 and NBS1 are associated with OvCa risk in BRCA1 mutation carriers, and MRE11 was associated with OvCa in BRCA2 mutation carriers. Polymorphisms in NBS1 have been studied for an association with early onset breast cancer with varying results (35–42), but studies of MRE11 and RAD50 as OvCa susceptibility in BRCA1/2 mutation carriers have not been published.
We also report that RAD51 is associated with altered OvCa risk in BRCA2 mutation carriers. This result is consistent with previous validated studies of RAD51 as a modifier of BRCA2-associated breast cancer risk (26, 43, 44). RAD51 interacts with BRCA2 (Fig 1) (45). Levy-Lahad et al. (44) and Wang et al. (26) reported that a 135G→C substitution in the 5' untranslated region of the RAD51 gene was associated with increased breast cancer risk in BRCA2 mutation carriers. A large consortium study has validated the relationship of RAD51 genotypes with breast cancer risk (43). While studies did not detect an association of this variant in BRCA1 mutation carriers, one study has reported an inverse association of this SNP with breast cancer risk (46). In addition, the study of Antoniou et al. (28) reported a potential functional link between the 135G→C variant and RAD51 protein expression. Our findings, along with the previous validation of RAD51 in BRCA2-associated breast cancer, represent biologically plausible associations that have been validated in multiple studies as a BRCA2-associated cancer risk modifier.
To date, few OvCa risk modifiers have been identified. Use of oral contraceptives may reduce OvCa risk in BRCA1/2 mutation carriers (47). Candidate modifier genes include the PROGINS progesterone receptor allele and oral contraceptive use (8) and rare HRAS1 alleles (7). However, neither of these associations has been validated. Our study expands the possible OvCa risk modifier genes by reporting that a number of genes that encode BRCA1/2 interacting proteins explain interindividual variability in OvCa risk in BRCA1/2 mutation carriers.
Strengths of this study include a large cohort of BRCA1/2 mutation carriers and a focus on biologically plausible associations involving genes that encode proteins that interact with BRCA1/2. Despite the relatively large sample size used here, power was still low to detect some small associations involving rare haplotypes. Several significant results may be driven by “rare” haplotypes. This is of concern since the SNP selection strategy was not designed to fully capture rare haplotypes and combined with the low power of the study to evaluate rare events, these associations may represent false positive findings. Finally, we did not have the power to study interactions or higher-order associations among genes or with exposures. Therefore, additional large-scale studies should be undertaken to confirm the results reported here.
Despite the biological plausibility of our results, we cannot make strong inferences about the mechanism of these associations. The SNPs selected here are, for the most part, not functionally relevant, and we do not have information about the causative alleles that may be in LD with the haplotype or SNP associations identified here. Therefore, the inferences made here allow us to test the hypothesis that genomic variation in our candidate genes represents potential modifiers of OvCa risk in BRCA1/2 mutation carriers. Additional studies are required to evaluate the biological mechanism of these associations.
Approximately 10% of OvCa can be explained by BRCA1/2 mutations, and the majority of breast and OvCa families are attributable to BRCA1/2. Over 100,000 patients are currently tested for BRCA1/2 mutations each year. Thus, a substantial proportion of women at risk for OvCa because of a BRCA1/2 mutation could benefit from improved knowledge of factors that influence risk. Since there are no effective screening strategies and OvCa prevention revolves around the use of RRSO, reliable models of individualized OvCa risk assessment must be developed. Our limited understanding of factors that modify these risks in BRCA1/2 mutation carriers hampers our clinical decision-making ability, including decisions about the appropriate type and timing of preventive interventions. The long-term goal of this research is to inform OvCa risk prediction estimates that can be used to focus the timing and method of OvCa risk reduction. This type of information is currently unavailable to the field of hereditary OvCa. While our current results are insufficient to guide clinical practice, they may represent a first step in helping to improve our understanding of OvCa risk and prevention in BRCA1/2 mutation carriers. In addition, this research could motivate additional studies that can elucidate mechanisms of BRCA1/2-associated ovarian carcinogenesis.
Supplementary Material
Acknowledgements
The MAGIC Consortium includes the following centers and individuals: Baylor-Charles A. Sammons Cancer Center (Joanne L. Blum, M.D. PhD, Becky Althaus, R.N., C.G.C., Gaby Ethington), Baylor College of Medicine (Claire Noll, Sharon Plon, M.D., Ph.D.), Beth Israel Deaconess Medical Center (Nadine Tung, M.D.), City of Hope National Medical Center (Sharon Sand, Jeffrey N. Weitzel, M.D.), Creighton University (Carrie Snyder, B.A., Henry T. Lynch, M.D., Patrice Watson, Ph.D.), Dana Farber Cancer Institute (Kathryn Stoeckert, Judy E. Garber, M.D., M.P.H.,), Duke University (Sydnee Crankshaw, Joellen Schildkraut, Ph.D.), Evanston Northwestern Healthcare Center for Medical Genetics (Suzanne M. O'Neill, Ph.D., Christina Selkirk, Wendy S. Rubinstein, M.D., Ph.D.), Fox Chase Cancer Center (Mary B. Daly, M.D., Ph.D., Andrew Godwin, PhD), Queensland Institute of Medical Research (Georgia Chenevix-Trench), Georgetown University (Claudine Isaacs, M.D.), Jonsson Comprehensive Cancer Center at the University of California, Los Angeles (Joyce Seldon, Patricia A. Ganz , M.D.), Mayo Clinic College of Medicine (Linda Wadum, Fergus Couch, Ph.D.), University of Chicago (Shelly Cummings, Olufunmilayo Olopade, M.D.), University of California, Irvine (Susan L. Neuhausen, Ph.D., Linda Steele), University of Pennsylvania Health System: (Susan Domchek, MD, Katherine Nathanson M.D., Tara Friebel, M.P.H., Timothy Rebbeck, Ph.D.), University of Texas, Southwestern (Gail Tomlinson, M.D.), University of Vienna (Christian Singer, M.D.), Women's College Hospital (Steven A. Narod, MD). This publication was supported in part by revenue from Nebraska cigarette taxes awarded to Creighton University by the Nebraska Department of Health and Human Services. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Nebraska or the Nebraska Department of Health and Human Services. Support was also received from NIH grants 5UO1 CA86389 (to HTL) and R01-CA083855, R01-CA74415 (to SLN), P30-CA51008-19 (to CI) and R01-CA102776 and R01-CA083855 (to TRR) and a General Clinical Research Center grant from NIH (M01 RR00043) awarded to the City of Hope National Medical Center (JNW). We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (funded by NHMRC grants 145684, 288704 and 454508) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia.
References
- 1.American Cancer Society . Cancer Facts and Figures. American Cancer Society; Atlanta, GA: 2008. [Google Scholar]
- 2.Rebbeck TR, Lynch HT, Neuhausen SL, et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N Engl J Med. 2002;346(21):1616–22. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- 3.Domchek SM, Friebel TM, Neuhausen SL, et al. Mortality after bilateral salpingo-oophorectomy in BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Lancet Oncol. 2006;7(3):223–9. doi: 10.1016/S1470-2045(06)70585-X. [DOI] [PubMed] [Google Scholar]
- 4.Begg CB. On the use of familial aggregation in population-based case probands for calculating penetrance. Journal of the National Cancer Institute. 2002;94(16):1221–6. doi: 10.1093/jnci/94.16.1221. see comment. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, John EM, McGuire V, et al. Breast and OvCa in relatives of cancer patients, with and without BRCA mutations. Cancer Epidemiol Biomarkers Prev. 2006;15(2):359–63. doi: 10.1158/1055-9965.EPI-05-0687. [DOI] [PubMed] [Google Scholar]
- 6.Narod SA. Modifiers of risk of hereditary breast and OvCa. Nature Reviews Cancer. 2002;2(2):113–23. doi: 10.1038/nrc726. [DOI] [PubMed] [Google Scholar]
- 7.Phelan CM, Rebbeck TR, Weber BL, et al. OvCa risk in BRCA1 carriers is modified by the HRAS1 variable number of tandem repeat (VNTR) locus. Nature Genetics. 1996;12(3):309–11. doi: 10.1038/ng0396-309. [DOI] [PubMed] [Google Scholar]
- 8.Runnebaum IB, Wang-Gohrke S, Vesprini D, et al. Progesterone receptor variant increases OvCa risk in BRCA1 and BRCA2 mutation carriers who were never exposed to oral contraceptives. Pharmacogenetics. 2001;11(7):635–8. doi: 10.1097/00008571-200110000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108(2):171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 10.Scully R, Xie A, Nagaraju G. Molecular functions of BRCA1 in the DNA damage response. Cancer Biol Ther. 2004;3(6):521–7. doi: 10.4161/cbt.3.6.842. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20(1):34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117(4):305–17. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes & Development. 2000;14(8):927–39. [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, West SC. Distinct functions of BRCA1 and BRCA2 in double-strand break repair. Breast Cancer Research. 2002;4(1):9–13. doi: 10.1186/bcr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orelli BJ, Bishop DK. BRCA2 and homologous recombination. Breast Cancer Research. 2001;3(5):294–8. doi: 10.1186/bcr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433(7026):653–7. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 17.Mann GJ, Thorne H, Balleine RL, et al. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 2006;8(1):R12. doi: 10.1186/bcr1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75(4):535–44. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenevix-Trench G, Healey S, Lakhani S, et al. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66(4):2019–27. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- 20.Antoniou AC, Goldgar DE, Andrieu N, et al. A weighted cohort approach for analysing factors modifying disease risks in carriers of high-risk susceptibility genes. Genet Epidemiol. 2005;29(1):1–11. doi: 10.1002/gepi.20074. [DOI] [PubMed] [Google Scholar]
- 21.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and OvCa associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. American Journal of Human Genetics. 2003;72(5):1117–30. doi: 10.1086/375033. erratum appears in Am J Hum Genet. 2003 Sep;73(3):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 23.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12(5):921–7. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 24.Fallin D, Cohen A, Essioux L, et al. Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer's disease. Genome Res. 2001;11(1):143–51. doi: 10.1101/gr.148401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinnwell JP, Schaid DJ. haplo.stats: Statistical Analysis of Haplotypes with Traits and Covariates when Linkage Phase is Ambiguous. In: Clinic M, editor. R package version 1.2.2. Rochester, MN: 2005. [Google Scholar]
- 26.Wang WW, Spurdle AB, Kolachana P, et al. A single nucleotide polymorphism in the 5' untranslated region of RAD51 and risk of cancer among BRCA1/2 mutation carriers. Cancer Epidemiology, Biomarkers & Prevention. 2001;10(9):955–60. [PubMed] [Google Scholar]
- 27.Kadouri L, Kote-Jarai Z, Hubert A, et al. A single-nucleotide polymorphism in the RAD51 gene modifies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. British Journal of Cancer. 2004;90(10):2002–5. doi: 10.1038/sj.bjc.6601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoniou A, Sinilnikova O, Simard J, et al. RAD51 135G>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. American Journal of Human Genetics. 2007 doi: 10.1086/522611. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi T, Garcia-Higuera I, Xu B, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109(4):459–72. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 31.Dong Y, Hakimi MA, Chen X, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Molecular Cell. 2003;12(5):1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 32.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27(19):6733–41. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi K, Taniguchi T, Ranganathan V, et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4(12):913–20. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- 34.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nature Reviews Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 35.Bogdanova N, Schurmann P, Waltes R, et al. NBS1 variant I171V and breast cancer risk. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9820-4. [DOI] [PubMed] [Google Scholar]
- 36.Buslov KG, Iyevleva AG, Chekmariova EV, et al. NBS1 657del5 mutation may contribute only to a limited fraction of breast cancer cases in Russia. Int J Cancer. 2005;114(4):585–9. doi: 10.1002/ijc.20765. [DOI] [PubMed] [Google Scholar]
- 37.Gorski B, Debniak T, Masojc B, et al. Germline 657del5 mutation in the NBS1 gene in breast cancer patients. Int J Cancer. 2003;106(3):379–81. doi: 10.1002/ijc.11231. [DOI] [PubMed] [Google Scholar]
- 38.Hsu HM, Wang HC, Chen ST, Hsu GC, Shen CY, Yu JC. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Biomarkers Prev. 2007;16(10):2024–32. doi: 10.1158/1055-9965.EPI-07-0116. [DOI] [PubMed] [Google Scholar]
- 39.Kanka C, Brozek I, Skalska B, Siemiatkowska A, Limon J. Germline NBS1 mutations in families with aggregation of Breast and/or OvCa from north-east Poland. Anticancer Res. 2007;27(4C):3015–8. [PubMed] [Google Scholar]
- 40.Roznowski K, Januszkiewicz-Lewandowska D, Mosor M, Pernak M, Litwiniuk M, Nowak J. I171V germline mutation in the NBS1 gene significantly increases risk of breast cancer. Breast Cancer Res Treat. 2008;110(2):343–8. doi: 10.1007/s10549-007-9734-1. [DOI] [PubMed] [Google Scholar]
- 41.Steffen J, Nowakowska D, Niwinska A, et al. Germline mutations 657del5 of the NBS1 gene contribute significantly to the incidence of breast cancer in Central Poland. Int J Cancer. 2006;119(2):472–5. doi: 10.1002/ijc.21853. [DOI] [PubMed] [Google Scholar]
- 42.Kuschel B, Auranen A, McBride S, et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11(12):1399–407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- 43.Antoniou AC, Sinilnikova OM, Simard J, et al. RAD51 135G-->C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81(6):1186–200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy-Lahad E, Lahad A, Eisenberg S, et al. A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3232–6. doi: 10.1073/pnas.051624098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scully R, Chen J, Plug A, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88(2):265–75. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 46.Jakubowska A, Gronwald J, Menkiszak J, et al. The RAD51 135 G>C polymorphism modifies breast cancer and OvCa risk in Polish BRCA1 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2007;16(2):270–5. doi: 10.1158/1055-9965.EPI-06-0562. [DOI] [PubMed] [Google Scholar]
- 47.Narod SA, Risch H, Moslehi R, et al. Oral contraceptives and the risk of hereditary OvCa. Hereditary OvCa Clinical Study Group. N Engl J Med. 1998;339(7):424–8. doi: 10.1056/NEJM199808133390702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.