Abstract
Wilson and King were among the first to recognize that the extent of phenotypic change between humans and great apes was dissonant with the rate of molecular change. Proteins are virtually identical1,2; cytogenetically there are few rearrangements that distinguish ape-human chromosomes3; rates of single-basepair change4-7 and retroposon activity8-10 have slowed particularly within hominid lineages when compared to rodents or monkeys. Here, we perform a systematic analysis of duplication content of four primate genomes (macaque, orangutan, chimpanzee and human) in an effort to understand the pattern and rates of genomic duplication during hominid evolution. We find that the ancestral branch leading to human and African great apes shows the most significant increase in duplication activity both in terms of basepairs and in terms of events. This duplication acceleration within the ancestral species is significant when compared to lineage-specific rate estimates even after accounting for copy-number polymorphism and homoplasy. We discover striking examples of recurrent and independent gene-containing duplications within the gorilla and chimpanzee that are absent in the human lineage. Our results suggest that the evolutionary properties of copy-number mutation differ significantly from other forms of genetic mutation and, in contrast to the hominid slowdown of single basepair mutations, there has been a genomic burst of duplication activity at this period during human evolution.
We began by developing a segmental duplication map for each of the four primate genomes (macaque, orangutan, chimpanzee and human) (Fig. S1). The approach is based on the alignment of whole-genome shotgun (WGS) sequence data against the human reference genome and predicts high-identity segmental duplications (SDs) based on excess depth of coverage and sequence divergence11 (Methods). Previous analyses have suggested excellent sensitivity and specificity for computational detection of duplications larger than 20 kbp in length11 (Table 1, Table S1 and Supplementary Note Table 2). By this criterion, we characterized 73 Mbp corresponding to the duplications identified in at least one of the four primate species, correcting for copy number in each primate (Methods). We furthermore characterized each duplication as “lineage-specific” or “shared”, depending on whether it was seen in only one or multiple genomes. This comparative map (Fig. S3, S4) is available as an interactive UCSC mirror browser, http://humanparalogy.gs.washington.edu, allowing researchers for the first time to interrogate the evolutionary history of any duplicated region of interest.
Table 1. Classes of primate segmental duplication.
| Copy-Number Corrected Duplicated Basepairs | |||||||
|---|---|---|---|---|---|---|---|
| Category | SD | SD >20 kbp | %Validation | HSA | PTR | PPY | MMU |
| HSA | 51,458,805 | 15,236,422 | 89-92% | 17,847,869 | |||
| PTR | 11,239,390 | 4,789,874 | 99% | 16,583,946 | |||
| PPY | 30,553,228 | 6,417,679 | 98% | 23,327,737 | |||
| MMU | 24,962,092 | 5,360,646 | 45% | 45,810,964 | |||
| MMU* | 35,493,466 | 7,715,410 | 85% | 18,266,656 | |||
| HSA / PTR | 32,392,480 | 21,061,194 | NA | 21,524,417 | 26,304,286 | ||
| HSA / PTR / PPY | 25,450,827 | 13,402,545 | NA | 11,259,061 | 14,012,351 | 11,541,148 | |
| HSA / PTR / PPY / MMU | 14,094,156 | 7,156,616 | NA | 8,092,997 | 12,820,607 | 6,176,876 | 12,542,691 |
| Total | 190,150,978 | 73,424,976 | 58,724,344 | 69,721,190 | 41,045,761 | 30,809,347 | |
Duplications were divided into eight categories based on the WSSD analysis of each primate genome (subsequent analysis were restricted to SDs >20 kbp in length). Lineage specific and shared duplication content are indicated. % validation indicates the fraction of species-specific duplications confirmed by cross-species array comparative genomic hybridization. Because the human genome was used, we corrected for copy number and examined sequence contigs not aligned to the human genome (see Methods). SDs assigned to the Y chromosome were not considered.
Macaque SDs detected in the macaque reference genome using WSSD and WGAC (<94% ID) approaches.
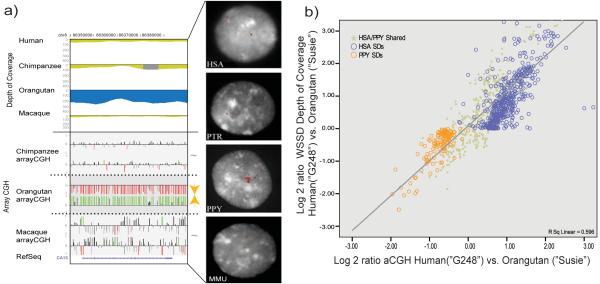
We validated our primate genomic duplication map using two different experimental approaches and, wherever possible, using DNA from the same individuals from which the computational predictions were generated. Using fluorescence in situ hybridization (FISH), we found that 86.5% of SDs were concordant with computational predictions when categorized as either lineage-specific (50/58) or shared duplications (40/46) (Figs. S1 and S2) (see below, Fig. 1 and Fig. S2 and Tables S2, S3 and S4). As a second approach, we designed a specialized oligonucleotide microarray (1 probe/585 bp) targeted to primate SDs (Table 1) and performed array comparative genomic hybridization (arrayCGH) between species (Table 1, Fig. 1 and S2). Among the great-ape genomes, we confirmed 89-99% of the lineage-specific duplications by interspecific arrayCGH (Table 1) with a very good correlation between computationally predicted and experimentally validated copy-number differences (Fig. 1 b). Since only 45% of macaque-specific duplications could be confirmed by interspecific arrayCGH, we performed an independent assessment of the macaque genome assembly and conservatively validated ~85% of macaque-specific duplications9,12 (unpublished results).
Fig. 1. Experimental validation of duplication map.
a) A computationally predicted orangutan-specific duplication (blue=excess depth of coverage of aligned WGS sequence) is confirmed by interspecific FISH and arrayCGH (oligonucleotide relative log2 ratios are depicted as red/green histograms and correspond to an increase and decrease in signal intensity when test/reference are reverse labeled) (see Supplementary Note for additional details). b) A comparison of duplication copy number as predicted by WSSD sequence analysis versus oligonucleotide arrayCGH between nonhuman and human primates showed a good correlation (r=0.77). Relative duplication copy number was computed as the log2 ratio of the number of aligned nonhuman primate reads against the human reference genome over the number of reads mapping to known single copy BACs.
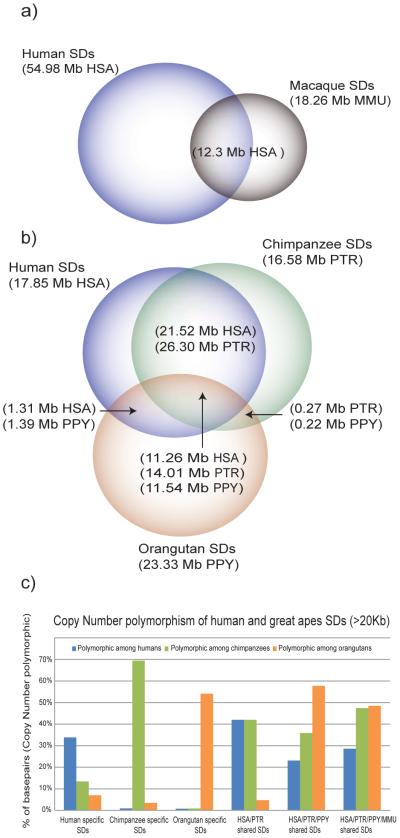
The comparative duplication map reveals several important features of primate SDs. As expected, most (80% or ~55 Mb) high-identity human segmental duplications arose after the divergence of the Old World and hominoid lineages (Fig. 2a). Humans and chimpanzees show significantly more duplications than either macaque or orangutan (Fig. 2b); with a large fraction being shared between chimpanzee and human. Based on our four-way primate genome analysis and leveraging arrayCGH data from gorilla and bonobo, we classify only ~10 Mb of duplication content as human-specific (210 duplications intervals with an average length of 53.1 Kb). The genomic distribution of great-ape segmental duplications is highly nonrandom (Fig. S5) with the presence of ancestral duplications being a strong predictor of “new”, lineage-specific events (P-value<0.001, randomization test, Supplementary Note, Table S5a,b). For example, 45% of human-chimp shared duplications map within 5 kbp of SDs shared among human-chimpanzee-orangutan, while 31% of human-chimpanzee-orangutan duplications map adjacent to human-chimpanzee-orangutan-macaque duplications. These observations emphasize that unique sequences flanking more ancient duplications have a much higher probability of segmental duplication11,13 and the duplication process itself is not random.
Fig. 2. Shared vs. lineage-specific duplications and great-ape polymorphism.
Segmental duplications (>20 kbp) were classified as lineage-specific or shared based on a four-way comparison of human, chimpanzee, orangutan and macaque genomes. a) Human SDs were compared to Old World monkey SDs (based on a separate analysis of the macaque assembly9). b). Since nonhuman great-ape duplications were detected based on alignment of WGS sequence against the human genome, we corrected for copy number based on the depth of coverage of WGS sequence (in brackets with the name of the species for which the correction was performed, see Table 1). b) Copy-number polymorphic regions were estimated from the results of arrayCGH hybridizations between 8 samples each of human, chimpanzee and orangutan (using the same reference as the computational prediction). The proportion of duplicated bases that showed evidence of copy-number polymorphism (i.e. gain or loss for ≥ two individuals within each species) is shown for each class of SD.
Within the human-specific set of duplications, we identify 39 partial and 17 complete human genes (Table S7). As expected, we find that full-length hominid genes show greater evidence of positive selection when compared to similarly analyzed unique genes (Supplementary Note). Our analysis indicates that several genes associated with human adaptation (amylase (AMY1), aquaporin 7 and DUF1220) are shared with chimpanzee but humans show a general increase in copy number. Gene models associated with signal transduction, neuronal activities (e.g. neurotransmitter release, synaptic transmission), and muscle contraction are significantly enriched in human, chimpanzee and orangutan lineage-specific duplications (Table S7). Human and great-ape shared duplications or those shared with macaque are, in contrast, enriched for biological processes associated with amino acid metabolism (P-value=1.69e-2) (great-ape shared SDs) or oncogenesis (P-value=5.80e-13, 4.64e-6) (ape SDs shared with macaque). Although the number of such duplication events is few, these data suggest a shift in the types of genes that have been duplicated most recently during great-ape and human evolution.
There are two important caveats to the above analysis. First, we have analyzed a single individual in each case and it is unclear to what extent that single genome represents the duplication pattern of the species. Second, duplicated sequences shared by two or more species might have potentially been subjected to recurrent mutations (homoplasy) leading to an overestimate of the proportion of ancestral duplications. Both copy-number polymorphism and evolutionary homoplasy, in principle, will complicate classification of segmental duplications as “ancestral” or “lineage-specific”. We therefore performed a number of additional analyses to address the impact of polymorphism and recurrent events on our assignments.
First, we investigated the extent of copy-number variation for both shared and lineage-specific duplications. Using arrayCGH targeted to primate SDs, we assessed the extent of copy-number variation in a set of unrelated DNA samples (Fig. 2c) (Methods). As expected14,15, lineage-specific SDs are highly copy-number variant, with humans showing 1.5- to 2-fold less diversity in copy number when compared to chimps and orangutans (Fig. 2c; Supplementary Note Table S9). Surprisingly, we find that shared SDs are as copy-number variant as lineage-specific duplications and that humans show slightly greater copy-number variation for these (42% versus 34%) when compared to apes.
It is, however, important to distinguish between duplication copy-number variation versus duplication status. A segmental duplication may show a high level of copy-number variation while its status as duplicated remains relatively constant among different individuals within a species. To address this, we performed a series of 3-way arrayCGH comparisons (Supplementary Note Fig. 7; Methods) where we investigated how duplication status (human-specific, chimpanzee-specific status and orangutan-specific SDs) varied as function of copy-number polymorphism within a species. The results from these triangulations indicate that only 1-8% of the SDs change duplication status even though 18-32% of the duplications are copy-number polymorphic between two individuals within a species (Supplementary Note Fig. 8). As a second independent test, we compared the duplication maps of two human genomes (Venter or HuRef and Watson genomes)16,17 and found that 89% (595/666) of the regions are shared duplications between HuRef and the Watson genome. Although we predict copy-number differences between these shared duplications, the boundaries of the duplication intervals remain remarkably consistent (Fig. S7), suggesting again that duplication status is a relatively constant character state within a species.
To assess the potential impact of recurrent mutations leading to misclassification of ancestral events, we focused on shared duplications between human and chimpanzee that were not identified as duplicated in either orangutan or macaque. We examined 103 sets of chimpanzee-human shared duplications that mapped to two or more distinct locations in the human genome (Supplementary Note) and determined what fraction of these mapped to two or more orthologous positions between chimp and human. Using a paired end-sequence mapping approach18,19 (Supplementary Note, Figure 9), we find that 85% (88/103) of the chimpanzee-human shared duplications have two or more copies mapping to the same orthologous position in the two genomes. This implies that the majority of shared duplications were already duplicated in the human-chimp common ancestor (Supplementary Note Tables 6 and 7).
As part of our comparative analyses, we identified regions whose duplication patterns were inconsistent with the generally accepted human/great-ape phylogeny (Fig. S4, Table 2, S5 and S6). For example, we identified 43 intervals that are duplicated in human and gorilla but not chimpanzee (H+C-G+ duplications). Such a scenario may arise as a result of a deletion event in the chimpanzee lineage, incomplete lineage sorting or, less likely, recurrent duplication events in the human and gorilla lineages. Only the latter possibility would potentially lead to an overestimation of ancestral duplication events. We estimated the frequency of such events by mapping the location of the duplications in each species using paired end-sequence data19 (see Supplementary Note). If the duplicated sequence mapped to the same location in gorilla and human, we classified it as a chimpanzee-specific deletion event or incomplete lineage sorting. If mapping to different locations in the two genomes, we categorized it as a recurrent event. As expected, most of the informative H+C-G+ duplications (80% or 12/15) were the result of chimpanzee-specific deletions.
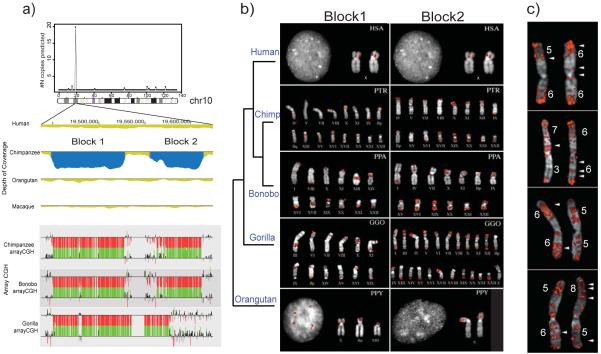
We investigated the most extreme example of recurrent African ape duplications in more detail (Fig. 3). We identified a region (~150 kbp in length) mapping to human chromosome 10 that had expanded in the chimpanzee genome but was largely single copy in human and orangutan. It consists of two distinct duplication blocks (~86 and 66 kbp in length). Both arrayCGH and FISH (Fig. 3a,b) confirm that the segments had been duplicated multiple times (~5-100 copies depending on the block and species) in the chimpanzee, bonobo and gorilla genomes but are single copy in all humans tested. Notably, the duplication boundaries (as delimited by arrayCGH) differ between the gorilla and chimpanzee lineages. With the exception of the chromosome 10 locus, we find that the map locations between gorilla and chimpanzee are non-orthologous (Supplementary Note and Methods) suggesting that this duplication expansion has occurred independently in both lineages.
Fig. 3. Convergent gene duplication expansion in African great apes but not humans.
a) Two regions on chromosome 10 have expanded in chimpanzee, gorilla and bonobo when compared to human based on computational and interspecific arrayCGH experiments (see Fig. 1 legend). b) FISH confirms 23-50 copies in chimpanzee and bonobo, and >100 copies in gorilla (Methods). End-sequence pair analysis using gorilla and chimpanzee WGS sequences reveals that all but the ancestral location are nonorthologous, indicating independent expansions in both lineages. c) Detailed analysis of 8 homologues of gorilla chromosome 1 reveal interstitial locations of the block 2 duplication that show variation both in copy number and in terms of location.
Based on the large number of interstitial sites on gorilla chromosomes, we compared chromosome 1 from four unrelated gorillas for variation in copy number and location of this segmental duplication. Remarkably, we find that both copy number (10-14 copies per homologous chromosome) as well as map location for this segmental duplication vary among these eight gorilla homologues with as many as 50% of the map locations being unoccupied by a duplication in another homologue (Fig. 3c and Supplementary Fig. 13). We conclude that this ancestral region of chromosome 10 has served as a preferred donor of chimpanzee/great-ape duplications and that the chimpanzee and gorilla genomes have been restructured by independent bursts of duplication activity. Interestingly, we detect and confirm by RT-PCR (reverse transcription PCR) at least one previously uncharacterized gene (14 exons, 141 Kb of genomic sequence, 1311 nt of CDSs and 437 a.a.) mapping to duplication block 1, which shows significant similarity to endosomal glycoprotein genes (Supplementary Note, Fig. 14-17). Thus, these duplications, in principle, may have led to African ape gene family expansions while remaining conspicuously a single copy in the human lineage. Although the mechanism by which such events have occurred is unclear, our data highlight the rapidity by which segmental duplications have restructured hominid genomes and emphasize their nonrandom nature both temporally and spatially.
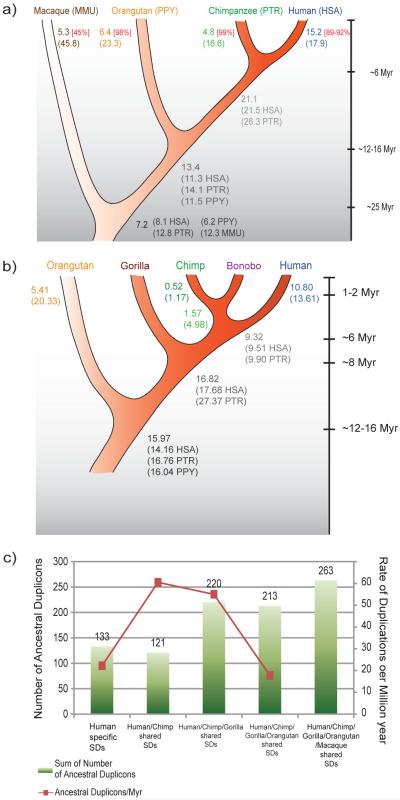
Based on our genome-wide assessment of segmental duplications in each of four primate species and our estimate of 20% homoplasy (see above), we calculated rates of segmental duplication both in events20 and basepairs along each lineage and ancestral node (Fig. 4, Supplementary Note Tables 13-16). We developed a maximum likelihood model to test if the rate of accumulation of segmental duplication has remained constant during the course of human/great-ape evolution. We compared the likelihood that the rate of segmental duplication has been uniform versus the likelihood of differential rates within specific lineages (Fig. 4). We find a significant increase (Likelihood Ratio Test (LRT), P-value<1e-10) in both the number of events and basepairs in the human/African great-ape lineage when compared to macaque/Old World monkey lineage. While terminal hominid lineages show an excess of duplications, the most significant burst of activity (4-10-fold, LRT P-value=1e-10) occurs in the common ancestor of human/chimpanzee and gorilla and after divergence of gorilla from the human-chimpanzee lineage (Supplementary Note Table 17). Our prediction is in strong agreement with the degree of sequence divergence among human intrachromosomal segmental duplications that shows a mode at 97-99% sequence identity. We note that this burst of duplication activity corresponds to a time when other mutational processes, such as point substitutions and retrotransposon activity, were slowing along the hominoid lineage. This apparent burst of activity may be the result of changes in the effective population size, generation time or imply a genomic destabilization at a period prior and perhaps during hominid speciation. In light of the importance of segmental duplications in contributing to copy-number changes associated with neurocognitive disease21-24 and disease susceptibility25-27, we predict that this apparent acceleration has had a profound impact on the reproductive success, adaptability and evolution of ancestral hominid populations.
Fig. 4. Rates of segmental duplication.
a) By basepair: We parsimoniously assigned the number of Mb to different branches, correcting for copy number in each species (shown in brackets). 89-99% of great-ape segmental duplications were validated by arrayCGH (square brackets). b) Further categorization of the segmental duplication data, based on arrayCGH from bonobo and gorilla, shows the greatest accumulation in the ancestor of humans and great apes. c) By event: We assigned 950 evolutionarily distinct human segmental duplication events20 to the human/great-ape phylogeny based on arrayCGH results. The red line estimates the duplication rate (per million years) and suggests an excess of large duplications in the common ancestor of human and chimpanzee but after the separation from gorilla.
METHODS
We estimated the duplication content of human, chimpanzee, orangutan and macaque by the whole-genome shotgun sequence detection (WSSD) method11,28. We mapped high-quality whole-genome shotgun (WGS) sequence reads for all species against the human reference assembly (NCBI build35) and identified regions of excess depth of coverage and divergence (see Supplementary Note). We also mapped macaque WGS reads to the macaque assembly (v 1.0). In this analysis, we considered SDs >20 Kb and >94% of identity (88% of identity for macaque reads against the human genome). We used read depth to estimate the number of copies for each duplication due to the excellent correlation (r2=0.953)11 between probes of known copy number and WGS depth-of-coverage.
We constructed an oligonucleotide microarray (n=385,000) targeted to regions of primate segmental duplication (~180 Mbp) and performed cross-species arrayCGH (with human as a reference) (GEO accession number: GSE13884). With the exception of human, we used DNA derived from the same genome that was sequenced as part of primate genome sequencing projects. The same microarray was used to assess copy-number polymorphism in DNA samples from 8 humans, 8 chimpanzees and 8 orangutans (GSE13885). We also used fluorescent in situ hybridizations (FISH) to further validate a subset of our duplications among the great apes.
We used end-sequence pair data from fosmid clones from a single human and a single chimpanzee as well as plasmid clones from a gorilla to map the location of segmental duplications within great-ape genomes (sequence data available from NIH trace repository). To estimate rates of segmental duplication along the hominoid phylogeny, we modeled the accumulation of segmental duplications in each branch as a pure birth process within a maximum likelihood framework. Nested models of segmental duplication were tested against each other by means of likelihood ratio tests (Supplementary Note).
Supplementary Material
Comparative primate segmental duplication analysis. The figure shows how segmental duplications were classified based on the WSSD computational analysis within the context of the UCSC genome browser. A ~500 kbp region is depicted corresponding to the fascioscapulohumeral muscular dystrophy on human chromosome 4. The depth of sequence read coverage (number of reads in five kbp windows is shown for (human (HSA), chimpanzee (PTR), orangutan (PPY) and macaque (MMU)) based on the alignment of these reads against the human genome. Regions of excess depth of coverage (blue, putative duplication) contrast with regions showing a depth-of-coverage within 3 s.d. of the mean of single copy regions (yellow). Three examples of SD classification are shown: from left to right, a HSA/PTR shared duplication, a HSA/PTR/PPY shared duplication (including partially the FRG1 gene) and a HSA/PTR/PPY/MMU SDs (including the full TUBB4Q gene). Any duplicated primate region can be viewed along with supporting experimental data using our customized map of primate segmental duplications displayed on a UCSC browser mirror (http://humanparalogy.gs.washington.edu).
FISH vs. cross-species arrayCGH data. This figure shows the specificity of our combined computational and experimental approach. a) An example of a human-chimpanzee shared duplication (predicted by WSSD analysis) that is single copy in gorilla and orangutan as determined by FISH and arrayCGH data (a replicate experiment is shown for each arrayCGH experiment as a dye-swap with the human reference DNA sample and test non-human primate DNA sample). b) An arrayCGH experiment showing a human-chimpanzee shared duplication that is duplicated in gorilla but single copy in orangutan based on experimental validation. c) A complex region mapping to one of the breakpoints of the Prader-Willi Syndrome that is duplicated in all four primate species showing patches of shared and species-specific duplications. ArrayCGH and FISH results confirm copy-number differences in regions of shared duplication (the region shown correspond to the extent of fosmid probe used in FISH experiment (WIBR2-0877G19)).
Construction of a primate segmental duplication map. We combined computational and experimental predictions to construct a primate segmental duplication map on the human reference genome. Three real examples are shown depicting a) a chimpanzee-specific duplication, b) an orangutan-specific duplication and c) a human-specific duplication. The top panel shows the “in silico” prediction (WSSD computational analysis) while the middle panel shows the results by replicate dye-swap arrayCGH for each non-human primate against human. These results were concatenated across the genome and summarized in the duplication map (Fig. 2) as follows: Regions of segmental duplication are shown in red while black denotes single copy sequence in each of the species. The next 5 rows summarize the results of cross-species arrayCGH hybridization experiments. Regions of increased signal intensity in human (blue) contrast with regions of increased signal intensity in each of the nonhuman primate species: green (chimp), purple (bonobo), dark red (gorilla), orange (orang) and pink (macaque). Grey regions show no significant difference in signal intensity. The extent of pericentromeric duplications (<5 Mb of centromere) and subtelomeric (<1000 kbp) are highlighted in purple and blue respectively based on human genome organization.
Comparative primate duplication map.
Computationally predicted regions of SDs (>20 kbp) (human, HSA; chimpanzee, PTR; orangutan, PPY and macaque, MMU) were concatenated and compared based on the human reference sequence (build35). SDs are shown in red while black denotes single copy sequence. The next 5 rows summarize the results of cross-species arrayCGH hybridization. Regions of increased signal intensity in human (blue) contrast with regions of increased signal intensity in chimp (green), bonobo (purple), gorilla (dark red), orang (orange) and pink (macaque). Grey regions show no significant difference in signal intensity (Fig. S3 for a schematic representation of the construction of the duplication map). Pericentromeric duplications (<5 Mb of centromere) and subtelomeric (<1000 kbp) are highlighted in purple and blue respectively based on human genome organization.
Landscape of great-ape and human SDs in the human genome. The map shows the actual distribution of all great-ape SDs (>20 kbps) placed in the context of the human genome (build35). For each human chromosomal ideogram, there are 8 rows grouped by grey blocks into 3 groups: a) The union of all SDs; b) Species-specific SDs, from 2nd to 5th row, human (HSA), chimpanzee (PTR), orangutan (PPY) and macaque (MMU) specific duplications respectively; and c) Shared SDs, from 6th to 8th row, HSA/PTR, HSA/PTR/PPY and HSA/PTR/PPY/MMU duplications. Duplications cluster within the pericentromeric and subtelomeric regions as well as other regions of the genome.
Copy-number distribution of primate segmental duplications. A non-redundant set of human segmental duplications20 were classified as lineage-specific or shared among the four primate species and the copy-number of each duplicon was estimated by the depth-of-coverage analysis (WSSD). The percentage of each category distributed across different copy-number bins is indicated. Lineage-specific duplications (colored histograms) show significantly fewer copies than shared duplications (in different intensities of grey). The copy number of every SD was calculated independently according to their species’ depth of coverage.
WSSD duplication analysis of two human genomes. We performed the depth-of-coverage analysis of two human genomes (Venter/HuRef Levy et al. 2007 and Watson Wheeler et al. 2008) and constructed two independent duplication maps for each to assess the extent of variation. We found that 95% of the duplication intervals (>20 kbp in length) were confirmed between these two genomes with the boundaries showing remarkable specificity. We depict 8 different intervals of the human assembly (build35) comparing the computationally predicted regions of duplication (blue) and unique sequence (yellow) with an assembly based analysis of human segmental duplications (WGAC analysis, top bar).
ACKNOWLEDGMENTS
We thank Heather Mefford, Andy Itsara, Greg Cooper, Tonia Brown and Graham McVicker for valuable comments in the preparation of this manuscript. The authors are also grateful to James Sikela and Laura Dumas for assistance with the comparison to cDNA microarray datasets. We are grateful to Lisa Faust, Jeffrey Rogers and Peter Parham for providing some of the primate material used in this study and to Mark Adams for providing the alignments for the positive selection analysis. We are also indebted to the large genome sequencing centers for early access to the whole genome sequence data for targeted analysis of segmental duplications. This work was supported, in part, by an NIH grant HG002385 to E.E.E. and NIH grant U54 HG003079 to R.K.W. and E.R.M. T.M.-B. is supported by a Marie Curie fellowship and by Departament d’Educació i Universitats de la Generalitat de Catalunya. E.E.E. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Goodman M. The role of immunochemical differences in the phyletic development of human behavior. Hum Biol. 1961;33:131–62. [PubMed] [Google Scholar]
- 2.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–16. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 3.Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982;215:1525–30. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- 4.Wu CI, Li WH. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 1985;82:1741–5. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li WH, Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;326:93–6. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- 6.Elango N, Thomas JW, Yi SV. Variable molecular clocks in hominoids. Proc Natl Acad Sci U S A. 2006;103:1370–5. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiper ME, Young NM, Sukarna TY. Genomic data support the hominoid slowdown and an Early Oligocene estimate for the hominoid-cercopithecoid divergence. Proc Natl Acad Sci U S A. 2004;101:17021–6. doi: 10.1073/pnas.0407270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 9.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 10.Consortium C.S.a.A. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Z, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Z, Hubley R, Smit A, Eichler EE. DupMasker: A tool for annotating primate segmental duplications. Genome Res. 2008;18:1362–8. doi: 10.1101/gr.078477.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Research. 2004;14:2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry GH, et al. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci U S A. 2006;103:8006–11. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee AS, et al. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 2008;17:1127–36. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 16.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 18.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 19.Newman TL, et al. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–56. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–8. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 21.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 22.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 23.Stone JL, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aitman TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–5. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 26.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 28.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparative primate segmental duplication analysis. The figure shows how segmental duplications were classified based on the WSSD computational analysis within the context of the UCSC genome browser. A ~500 kbp region is depicted corresponding to the fascioscapulohumeral muscular dystrophy on human chromosome 4. The depth of sequence read coverage (number of reads in five kbp windows is shown for (human (HSA), chimpanzee (PTR), orangutan (PPY) and macaque (MMU)) based on the alignment of these reads against the human genome. Regions of excess depth of coverage (blue, putative duplication) contrast with regions showing a depth-of-coverage within 3 s.d. of the mean of single copy regions (yellow). Three examples of SD classification are shown: from left to right, a HSA/PTR shared duplication, a HSA/PTR/PPY shared duplication (including partially the FRG1 gene) and a HSA/PTR/PPY/MMU SDs (including the full TUBB4Q gene). Any duplicated primate region can be viewed along with supporting experimental data using our customized map of primate segmental duplications displayed on a UCSC browser mirror (http://humanparalogy.gs.washington.edu).
FISH vs. cross-species arrayCGH data. This figure shows the specificity of our combined computational and experimental approach. a) An example of a human-chimpanzee shared duplication (predicted by WSSD analysis) that is single copy in gorilla and orangutan as determined by FISH and arrayCGH data (a replicate experiment is shown for each arrayCGH experiment as a dye-swap with the human reference DNA sample and test non-human primate DNA sample). b) An arrayCGH experiment showing a human-chimpanzee shared duplication that is duplicated in gorilla but single copy in orangutan based on experimental validation. c) A complex region mapping to one of the breakpoints of the Prader-Willi Syndrome that is duplicated in all four primate species showing patches of shared and species-specific duplications. ArrayCGH and FISH results confirm copy-number differences in regions of shared duplication (the region shown correspond to the extent of fosmid probe used in FISH experiment (WIBR2-0877G19)).
Construction of a primate segmental duplication map. We combined computational and experimental predictions to construct a primate segmental duplication map on the human reference genome. Three real examples are shown depicting a) a chimpanzee-specific duplication, b) an orangutan-specific duplication and c) a human-specific duplication. The top panel shows the “in silico” prediction (WSSD computational analysis) while the middle panel shows the results by replicate dye-swap arrayCGH for each non-human primate against human. These results were concatenated across the genome and summarized in the duplication map (Fig. 2) as follows: Regions of segmental duplication are shown in red while black denotes single copy sequence in each of the species. The next 5 rows summarize the results of cross-species arrayCGH hybridization experiments. Regions of increased signal intensity in human (blue) contrast with regions of increased signal intensity in each of the nonhuman primate species: green (chimp), purple (bonobo), dark red (gorilla), orange (orang) and pink (macaque). Grey regions show no significant difference in signal intensity. The extent of pericentromeric duplications (<5 Mb of centromere) and subtelomeric (<1000 kbp) are highlighted in purple and blue respectively based on human genome organization.
Comparative primate duplication map.
Computationally predicted regions of SDs (>20 kbp) (human, HSA; chimpanzee, PTR; orangutan, PPY and macaque, MMU) were concatenated and compared based on the human reference sequence (build35). SDs are shown in red while black denotes single copy sequence. The next 5 rows summarize the results of cross-species arrayCGH hybridization. Regions of increased signal intensity in human (blue) contrast with regions of increased signal intensity in chimp (green), bonobo (purple), gorilla (dark red), orang (orange) and pink (macaque). Grey regions show no significant difference in signal intensity (Fig. S3 for a schematic representation of the construction of the duplication map). Pericentromeric duplications (<5 Mb of centromere) and subtelomeric (<1000 kbp) are highlighted in purple and blue respectively based on human genome organization.
Landscape of great-ape and human SDs in the human genome. The map shows the actual distribution of all great-ape SDs (>20 kbps) placed in the context of the human genome (build35). For each human chromosomal ideogram, there are 8 rows grouped by grey blocks into 3 groups: a) The union of all SDs; b) Species-specific SDs, from 2nd to 5th row, human (HSA), chimpanzee (PTR), orangutan (PPY) and macaque (MMU) specific duplications respectively; and c) Shared SDs, from 6th to 8th row, HSA/PTR, HSA/PTR/PPY and HSA/PTR/PPY/MMU duplications. Duplications cluster within the pericentromeric and subtelomeric regions as well as other regions of the genome.
Copy-number distribution of primate segmental duplications. A non-redundant set of human segmental duplications20 were classified as lineage-specific or shared among the four primate species and the copy-number of each duplicon was estimated by the depth-of-coverage analysis (WSSD). The percentage of each category distributed across different copy-number bins is indicated. Lineage-specific duplications (colored histograms) show significantly fewer copies than shared duplications (in different intensities of grey). The copy number of every SD was calculated independently according to their species’ depth of coverage.
WSSD duplication analysis of two human genomes. We performed the depth-of-coverage analysis of two human genomes (Venter/HuRef Levy et al. 2007 and Watson Wheeler et al. 2008) and constructed two independent duplication maps for each to assess the extent of variation. We found that 95% of the duplication intervals (>20 kbp in length) were confirmed between these two genomes with the boundaries showing remarkable specificity. We depict 8 different intervals of the human assembly (build35) comparing the computationally predicted regions of duplication (blue) and unique sequence (yellow) with an assembly based analysis of human segmental duplications (WGAC analysis, top bar).