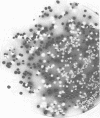
Abstract
Cryptococcus neoformans produced pigments when p-diphenols were substrates in a glucose-amino acid-salts medium. The best substrates were 2.5-dihydroxybenzoic acid and 2,5-dihydroxybenzenesulfonic acid. In contrast to the cellular pigment production from o-diphenols (hydroxyl groups in the 2,3- or 3,4-position of phenyl ring), the p-diphenols (1,4- or 2,5-positions for the hydroxyl groups) produced large amounts of soluble pigments that diffused into the medium. When an optimal source of nitrogen (glutamine, glycine, and asparagine) was used, 89% of the C. neoformans strains produced pigments from p-diphenols. In contrast, 0 to 67% of the strains produced pigments when a suboptimal nitrogen source (proline, ammonium sulfate, ornithine, and methionine) was used. When glutamine-glycine-asparagine was the nitrogen source, 100% of the C. neoformans strains produced pigments from o0diphenols, whereas 77 to 100% of the strains produced pigment when proline-ammonium sulfate-ornithine-methionine was the nitrogen source. Cryptococcus species other than C. neoformans and all tested Candida species failed to produce pigments from any of the substrates except when hydroquinone was used. A combination of glutamine-glycine-asparagine and 3,4-dihydroxyphenylalanine allowed differentiation of colonies of C. neoformans from C. albicans in 3 to 6 days. These data showed that pigment production from o- and p-diphenols served as an excellent biochemical test for the identification of C. neoformans.
Full text
PDF

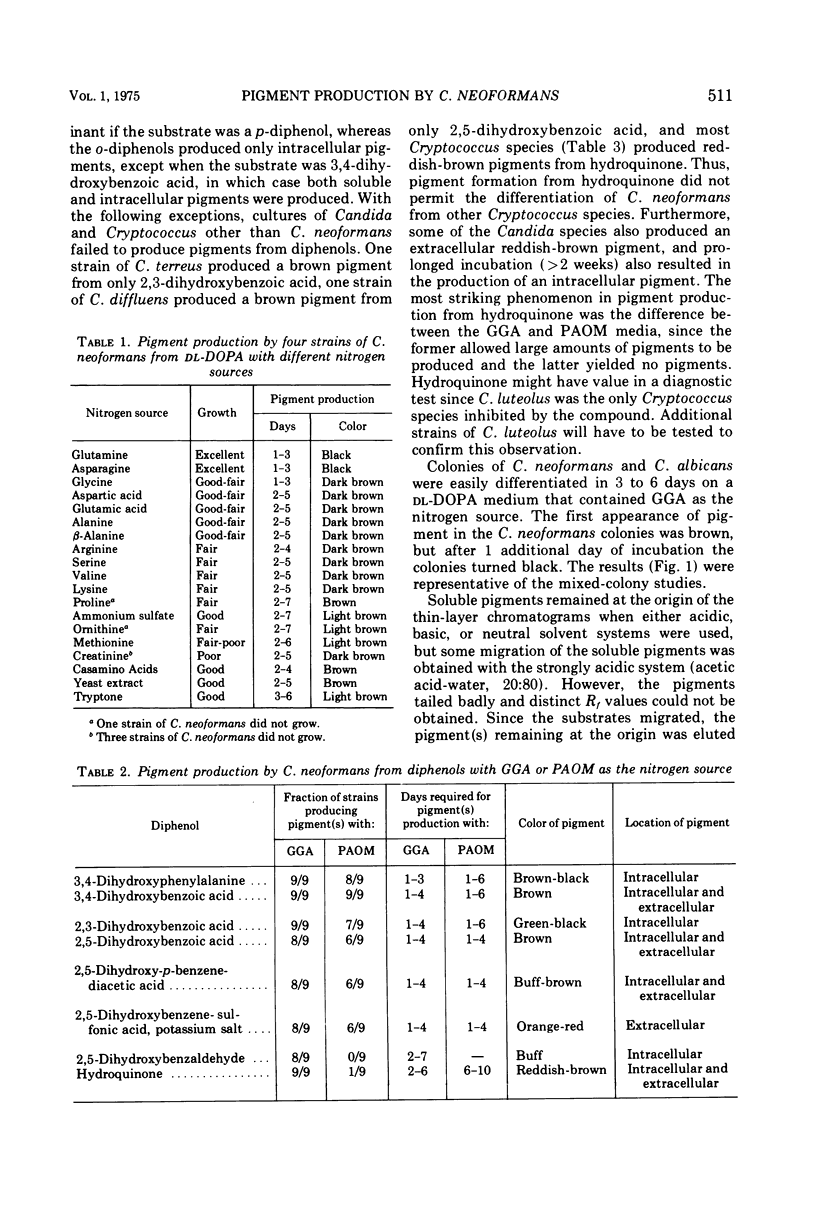
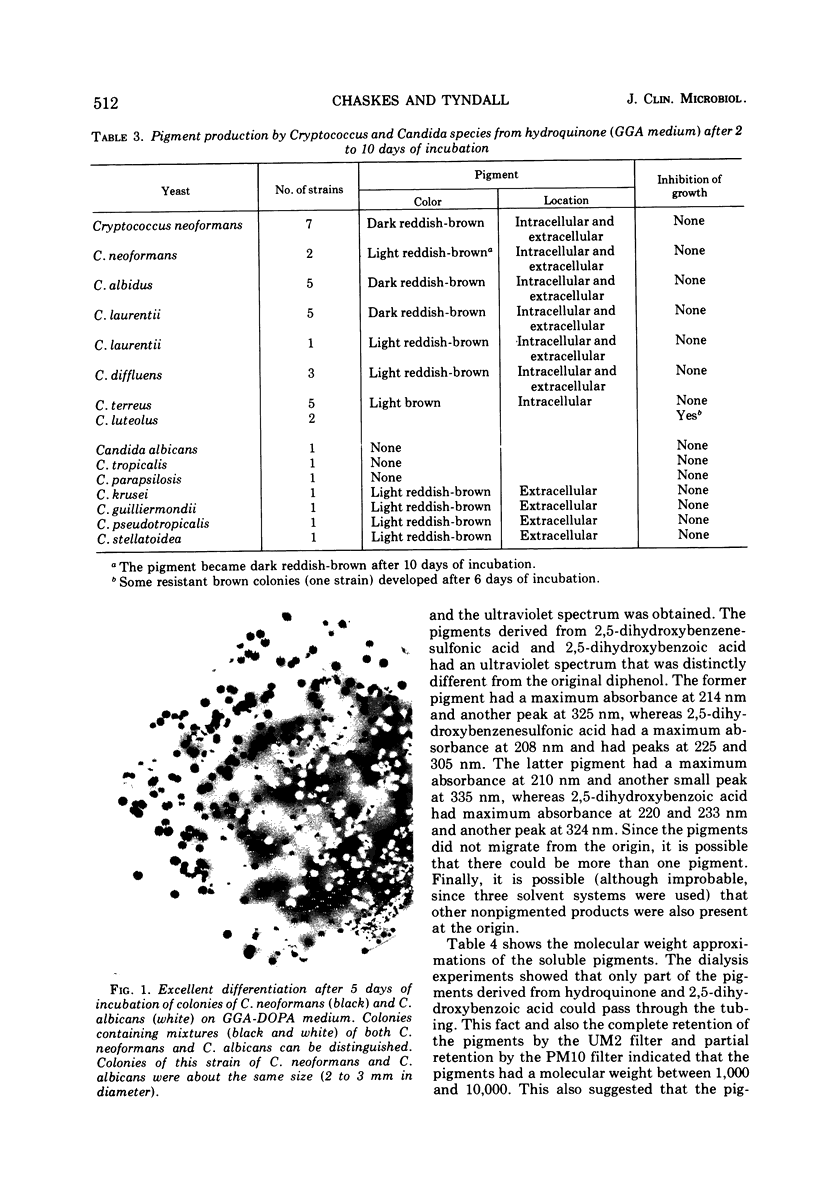


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chaskes S., Phillips A. W. Pigmentation and autofluorescence of Candida species after growth on tryptophan media. Can J Microbiol. 1974 Apr;20(4):595–603. doi: 10.1139/m74-091. [DOI] [PubMed] [Google Scholar]
- Korth H., Pulverer G. Pigment formation for differentiating Cryptococcus neoformans from Candida albicans. Appl Microbiol. 1971 Mar;21(3):541–542. doi: 10.1128/am.21.3.541-542.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C. E., Kapica L. Production of diagnostic pigment by phenoloxidase activity of cryptococcus neoformans. Appl Microbiol. 1972 Nov;24(5):824–830. doi: 10.1128/am.24.5.824-830.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields A. B., Ajello L. Medium for selective isolation of Cryptococcus neoformans. Science. 1966 Jan 14;151(3707):208–209. doi: 10.1126/science.151.3707.208. [DOI] [PubMed] [Google Scholar]
- Staib F., Randhawa H. S., Grosse G., Blisse A. Cryptococcose. Zur Identifizierung von Cryptococcus neoformans aus klinischem Untersuchungsmaterial. Zentralbl Bakteriol Orig A. 1973 Dec;225(2):211–222. [PubMed] [Google Scholar]
- Strachan A. A., Yu R. J., Blank F. Pigment production of Cryptococcus neoformans grown with extracts of Guizotia abyssinica. Appl Microbiol. 1971 Sep;22(3):478–479. doi: 10.1128/am.22.3.478-479.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]