Abstract
Background
The combined sequences encoding a partial and putative rpsI open reading frame (ORF), non-coding (NC) region, a putative ORF for the Campylobacter adhesin to fibronectin-like protein (cadF), a putative Cla_0387 ORF, NC region and a partial and putative Cla_0388 ORF, were identified in 16 Campylobacter lari isolates, using two novel degenerate primer pairs. Probable consensus sequence at the -35 and -10 regions were identified in all C. lari isolates, as a promoter.
Results
Thus, cadF (-like) gene is highly conserved among C. lari organisms. Transcription of the cadF (-like) gene in C. lari cells in vivo was also confirmed and the transcription initiation site was determined. A peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins was completely conserved amongst the putative cadF (-like) ORFs from the C. lari isolates.
Conclusion
The putative cadF (-like) ORFs from all C. lari isolates were nine amino acid larger than those from C. jejuni, and showed amino acid residues 137 -140 of FALG (50% identity), instead of the FRLS residues of the maximal fibronectin-binding activity site demonstrated within C. jejuni CadF. A neighbor joining tree constructed based on cadF (-like) gene sequence information formed a major cluster consisting of C. lari isolates, separating from the other three thermophilic campylobacters.
Background
Thermophilic Campylobacter species, primarily Campylobacter jejuni and C. coli, are curved, Gram-negative organisms, belonging to the ε-Proteobacteria, and are the most commonly recognized cause of acute bacterial diarrhea in the Western world [1-3].
Campylobacter lari is a relatively recently discovered thermophilic Campylobacter species that was first isolated from mammalian and avian species, particularly seagulls of the genus Larus [1,4]. C. lari has also been shown to be a cause of clinical infection [5-9].
In addition, an atypical group of isolates of urease-positive thermophilic Campylobacter (UPTC) have been isolated from the natural environment in England in 1985 [10]. Thereafter, these organisms were described as a biovar or variant of C. lari [11,12]. Subsequent reports described four human isolates in France [11,13]. Some additional isolates of UPTC have also been reported in Northern Ireland [14-16] in The Netherlands [17] and in Japan [18,19]. Thus, these two representative taxa, namely urease-negative (UN) C. lari and UPTC occur within the species of C. lari [20].
Bacterial pathogens have the ability to bind to fibronectin (Fn; a component of the extracellular matrix) [21-24]. Konkel et al. identified and cloned a gene encoding a fibronectin-binding protein (Campylobacter adhesin to Fn; CadF) from C. jejuni [22]. In C. jejuni and C. coli, the cadF virulence gene encodes a 37 kDa outer membrane protein that promotes the binding of these pathogens to intestinal epithelial cells [15].
In relation to cadF of thermophilic Campylobacter other than C. jejuni and C. coli described above, cadF and outer membrane protein gene F (OprF) have been identified in C. coli RM2228 (DDBJ/EMBL/GenBank accession number AAFL01000010 and ZP_00368187), C. lari RM2100 (AAFK01000002 and YP_002574995) and C. upsaliensis RM3195 (AAFJ01000008 and ZP_00371707), following whole genome shotgun sequence analysis [26]. However, no detailed descriptions of the cadF (oprF) gene have yet appeared for these thermophilic Campylobacter strains. In addition, no reports on the cadF (-like) gene in C. lari organisms have yet appeared.
Therefore, the aim of the present study was to clone, sequence and analyze the full-length gene encoding the Fn-binding (-like) protein (CadF) and its adjacent genetic loci from several C. lari organisms (UN C. lari and UPTC). We also aimed to confirm the expression of the gene in the C. lari cells.
Results
TA cloning, sequencing and sequence analyses of the full-length cadF gene and its adjacent genetic loci from the 16 isolates of C. lari
The two primer pairs (f-/r-cadF1 and f-/r-cadF2; Figure1) successfully amplified PCR products of approximately 1.4 and 1.2 [kilo base pairs (kbp)], respectively, with all 16 isolates of C. lari employed (data not shown). Following TA cloning and sequencing, the combined nucleotide and deduced amino acid sequence data from the 16 isolates of C. lari determined have been made accessible in the DDBJ/EMBL/GenBank, with the accession numbers indicated in Table 1.
Table 1.
C. lari isolates and other thermophilic Campylobacter reference strains analyzed in the present study and their accession numbers of the nucleotide sequence data accessible in DDBJ/EMBL/GenBank
| Isolate no. | Source | Country | Accession number |
|---|---|---|---|
| C. lari JCM2530T | Seagull | Japan | AB465344 |
| C. lari 298 | Human | Canada | AB465345 |
| C. lari 300 | Seagull | USA | AB465346 |
| C. lari 84C-1 | Human | N. Ireland | AB465347 |
| UPTC 99 | Sea water | N. Ireland | AB465348 |
| UPTC NCTC12892 | River water | England | AB295430 |
| UPTC NCTC12893 | River water | England | AB295431 |
| UPTC NCTC12894 | Sea water | England | AB295432 |
| UPTC NCTC12895 | Mussel | England | AB295433 |
| UPTC NCTC12896 | Mussel | England | AB295434 |
| UPTC CF89-12 | River water | Japan | AB295435 |
| UPTC A1 | Seagull | N. Ireland | AB295436 |
| UPTC A2 | Seagull | N. Ireland | AB295437 |
| UPTC A3 | Seagull | N. Ireland | AB295438 |
| UPTC 89049 | Human | France | AB295439 |
| UPTC 92251 | Human | France | AB295440 |
| C. lari RM2100 | Human | USA | AAFK01000002 |
| C. jejuni NCTC11168 | Human | USA | NC_002163 |
| C. jejuni RM1221 | Chicken | USA | NC_003912 |
| C. jejuni 81-176 | Human | USA | NC_008787 |
| C. jejuni 260.94 | Human | South Africa | AANK01000004 |
| C. jejuni CF93-6 | Human | Japan | AAFJ01000005 |
| C. jejuni HB93-13 | Human | China | AANQ01000001 |
| C. jejuni 84-25 | Human | Unknown | AANT02000001 |
| C. jejuni ss doylei 269.97 | Human | Unknown | AARB01000000 |
| C. coli RM2228 | Chicken | USA | AAFL01000008 |
| C. upsaliensis RM3195 | Human | USA | AAFJ01000005 |
The combined sequences of an approximately 2.3 kbp region encoding a partial and putative ribosomal protein SI rpsI open reading frame (ORF) (165 bp), a NC region downstream of the ORF (approximately 250 bp), a putative cadF (-like) ORF (984 bp), a Cla_0387 ORF (642 bp), a NC region (approximately 120 bp) and a partial and putative Cla_0388 ORF (126 or 128 bp) were identified with all 16 C. lari isolates examined.
The present sequence analyses identified the putative ORF for cadF (-like) gene to be 984 bp [nucleotide position (np) 414-1,397 bp for the C. lari JCM2530T] with all 16 C. lari isolates (n = 4 UN C. lari; n = 12 UPTC) and UN C. lari RM 2100. With regard to the cadF-like gene, the sequence commenced with an ATG start codon for all isolates and terminated with a TAA for 13 isolates and with a TGA for the other three isolates (NCTC12894, 12895 and 99). Regarding putative ORFs for cadF (-like) gene, apparent size differences occurred amongst the four thermophilic Campylobacter species examined, 984 bp (328 amino acid residues) for 16 C. lari isolates and C. lari RM2100 strain, 957 (319) for C. jejuni RM1221 and NCTC11168, 996 (332) for C. coli RM2228, and 948 (316) for C. upsaliensis RM3195, as shown in Table 2, although in this limited study a small number of reference strains of C. jejuni, C. coli and C. upsaliensis were examined. Probable ribosome-binding (RB) sites, AGGA (np 404-407 bp) [Shine-Dalgarno (SD) sequences] [27], that are complementary to a highly conserved sequence of CCUCCU, close to the 3' end of 16S rRNA, were also identified in all the C. lari isolates examined.
Table 2.
Putative ORFs of cadF (-like)and Cla_0387 genes from C. lariisolates and
| cadF (-like) | Cla_0387 | |||||
|---|---|---|---|---|---|---|
| Campylobacter | ORF | Number of amino acid | CMW(Da) | ORF | Number of amino acid | CMW(Da) |
| C. lari JCM2530T | 984 | 328 | 36,781 | 642 | 214 | 23,689 |
| C. lari 298 | 984 | 328 | 36,693 | 642 | 214 | 23,717 |
| C. lari 300 | 984 | 328 | 36,708 | 642 | 214 | 23,730 |
| C.lari 84C-1 | 984 | 328 | 36,578 | 642 | 214 | 23,689 |
| UPTC 99 | 984 | 328 | 36,707 | 642 | 214 | 23,717 |
| UPTC NCTC12892 | 984 | 328 | 36,674 | 642 | 214 | 23,695 |
| UPTC NCTC12893 | 984 | 328 | 36,672 | 642 | 214 | 23,695 |
| UPTC NCTC12894 | 984 | 328 | 36,695 | 642 | 214 | 23,695 |
| UPTC NCTC12895 | 984 | 328 | 36,718 | 642 | 214 | 23,695 |
| UPTC NCTC12896 | 984 | 328 | 36,836 | 642 | 214 | 23,845 |
| UPTC CF89-12 | 984 | 328 | 36,817 | 642 | 214 | 23,692 |
| UPTC A1 | 984 | 328 | 36,869 | 642 | 214 | 23,838 |
| UPTC A2 | 984 | 328 | 36,869 | 642 | 214 | 23,838 |
| UPTC A3 | 984 | 328 | 36,802 | 642 | 214 | 23,815 |
| UPTC 89049 | 984 | 328 | 36,803 | 642 | 214 | 23,845 |
| UPTC 92251 | 984 | 328 | 36,850 | 642 | 214 | 23,875 |
| C. lari RM2100 | 984 | 328 | 36,707 | 642 | 214 | 23,689 |
| C. jejuni NCTC11168 | 957 | 319 | 35,996 | 639 | 213 | 23,637 |
| C. jejuni RM1221 | 957 | 319 | 35,998 | 639 | 213 | 23,794 |
| C. coli RM2228 | 996 | 332 | 37,447 | 636 | 212 | 23,878 |
| C. upsaliensis RM3195 | 948 | 316 | 35,624 | 648 | 216 | 24,279 |
ORF, open reading frame; CMW, calculated molecular weights; Da, daltons.
In the region upstream of the cadF-like gene, a most probable promoter consensus sequence at the -10 region (TATAAT) (TAGAAT for UPTC isolates (271-276 for UPTC CF89-12)) was identified at the locus between np 272 and 277 bp, with all 16 C. lari isolates and the C. lari RM2100 strain. In addition, probable -35 regions (np 243-248) upstream of the -10 region were also identified, in all C. lari isolates examined.
A putative ORF for the Cla_0387 gene was also estimated to be 642 bp with all 16 C. lari isolates examined (np 1,404 - 2,045 bp). The Cla_0387 gene commenced with a TTG and terminated with a TAA with all 16 C. lari isolates and the C. lari RM2100 strain. Apparent small size differences of the putative ORFs for the Cla_0387 also occurred amongst the four thermophilic Campylobacter species examined (Table 2).
As shown in Table 3, the nucleotide sequences of the full-length cadF (-like) structural gene from the 17 C. lari isolates showed 89.4-100.0% similarities to each other (Table 3). The nucleotide sequences of the full-length Cla_0387 structural gene from the 17 C. lari isolates showed 85.1 - 100.0% similarities to each other (Table 4). Thus, the nucleotide sequence similarities of the cadF-like gene appear to be slightly higher than those of the Cla_0387 gene, amongst the 16 C. lari isolates and the C. lari RM2100 strain examined.
Table 3.
Nucleotide (upper right) and deduced amino acid (lower left) sequence similarities (%) of full-length cadF (-like) gene in C. lari isolates and other thermo- philic Campylobacter reference strains
| Campylobacter | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C. lari JCM2530T | 99.2 | 99.2 | 99.3 | 92.8 | 92.4 | 92.3 | 92.6 | 91.9 | 90.1 | 91.0 | 89.4 | 89.4 | 89.5 | 89.9 | 89.5 | 99.1 | 68.7 | 68.5 | 65.8 | 65.0 | |
| 2 | C. lari 298 | 98.8 | 99.8 | 99.9 | 92.9 | 92.5 | 92.4 | 92.7 | 92.0 | 90.0 | 91.2 | 89.4 | 89.4 | 89.5 | 89.9 | 89.5 | 99.8 | 68.8 | 68.6 | 65.8 | 65.4 | |
| 3 | C. lari 300 | 99.1 | 99.7 | 99.9 | 92.9 | 92.5 | 92.4 | 92.7 | 92.0 | 90.0 | 91.2 | 89.4 | 89.4 | 89.5 | 89.9 | 89.5 | 100.0 | 68.8 | 68.6 | 65.8 | 65.6 | |
| 4 | C. lari 84C-1 | 99.1 | 99.7 | 100.0 | 93.0 | 92.6 | 92.5 | 92.8 | 92.1 | 90.1 | 91.3 | 89.5 | 89.5 | 89.6 | 90.0 | 89.6 | 99.9 | 68.9 | 68.7 | 65.9 | 65.5 | |
| 5 | UPTC 99 | 93.0 | 93.0 | 93.3 | 93.3 | 98.6 | 98.6 | 99.6 | 99.0 | 92.4 | 94.5 | 91.0 | 91.0 | 91.0 | 91.1 | 90.9 | 92.9 | 69.2 | 69.0 | 66.2 | 65.3 | |
| 6 | UPTC NCTC12892 | 93.0 | 93.0 | 93.3 | 93.3 | 99.1 | 99.4 | 98.1 | 97.5 | 92.1 | 94.0 | 90.9 | 90.9 | 90.9 | 91.0 | 90.8 | 92.5 | 68.9 | 68.7 | 65.7 | 65.4 | |
| 7 | UPTC NCTC12893 | 92.7 | 92.7 | 93.0 | 93.0 | 98.8 | 99.1 | 98.1 | 97.7 | 92.1 | 94.1 | 90.9 | 90.9 | 90.9 | 91.0 | 90.8 | 92.4 | 69.1 | 68.9 | 65.9 | 65.3 | |
| 8 | UPTC NCTC12894 | 92.4 | 92.4 | 92.7 | 92.7 | 99.4 | 98.5 | 98.2 | 98.6 | 92.1 | 94.4 | 90.7 | 90.7 | 90.7 | 90.8 | 90.6 | 92.6 | 69.1 | 68.8 | 66.1 | 65.3 | |
| 9 | UPTC NCTC12895 | 91.8 | 91.8 | 92.1 | 92.1 | 98.8 | 97.9 | 98.2 | 98.2 | 91.4 | 93.6 | 90.0 | 90.0 | 90.4 | 90.3 | 89.9 | 91.9 | 68.9 | 68.7 | 65.9 | 64.9 | |
| 10 | UPTC NCTC12896 | 90.9 | 90.9 | 91.2 | 91.2 | 95.4 | 94.8 | 94.5 | 95.4 | 94.2 | 91.9 | 98.0 | 98.0 | 98.4 | 98.3 | 98.5 | 90.1 | 68.3 | 68.2 | 66.3 | 65.0 | |
| 11 | UPTC CF89-12 | 91.8 | 91.8 | 92.1 | 92.1 | 95.4 | 94.8 | 94.5 | 95.4 | 94.5 | 93.3 | 91.3 | 91.3 | 91.2 | 91.4 | 91.2 | 91.2 | 69.2 | 69.1 | 66.3 | 65.6 | |
| 12 | UPTC A1 | 91.2 | 91.2 | 91.5 | 91.5 | 94.5 | 94.2 | 93.9 | 94.5 | 93.3 | 97.9 | 93.6 | 100.0 | 99.0 | 99.3 | 99.3 | 89.5 | 68.5 | 68.4 | 66.0 | 64.8 | |
| 13 | UPTC A2 | 91.2 | 91.2 | 91.5 | 91.5 | 94.5 | 94.2 | 93.9 | 94.5 | 93.3 | 97.9 | 93.6 | 100.0 | 99.0 | 99.3 | 99.3 | 89.5 | 68.5 | 68.4 | 65.8 | 64.8 | |
| 14 | UPTC A3 | 91.5 | 91.5 | 91.8 | 91.8 | 94.8 | 94.5 | 94.2 | 94.8 | 93.6 | 98.8 | 93.9 | 99.1 | 99.1 | 99.5 | 99.5 | 89.6 | 68.3 | 68.2 | 66.4 | 65.0 | |
| 15 | UPTC 89049 | 91.8 | 91.8 | 92.1 | 92.1 | 95.1 | 94.8 | 94.5 | 95.1 | 93.9 | 98.5 | 94.2 | 99.4 | 99.4 | 99.7 | 99.4 | 90.0 | 68.5 | 68.4 | 66.4 | 64.7 | |
| 16 | UPTC 92251 | 91.5 | 91.5 | 91.8 | 91.8 | 94.5 | 94.2 | 93.9 | 94.5 | 93.2 | 98.5 | 93.9 | 98.8 | 98.8 | 99.7 | 99.4 | 89.6 | 68.3 | 68.2 | 66.2 | 64.7 | |
| 17 | C. lari RM2100 | 99.1 | 99.7 | 100.0 | 100.0 | 93.0 | 93.3 | 93.0 | 92.7 | 92.1 | 91.5 | 91.8 | 91.2 | 91.2 | 91.5 | 91.8 | 91.5 | 68.9 | 68.7 | 65.9 | 65.6 | |
| 18 | C. jejuni NCTC11168 | 57.0 | 57.3 | 57.6 | 57.6 | 57.6 | 57.6 | 57.3 | 57.9 | 57.3 | 56.7 | 57.7 | 57.1 | 57.1 | 56.8 | 56.8 | 56.5 | 57.1 | 99.8 | 82.8 | 74.7 | |
| 19 | C. jejuni RM1221 | 56.4 | 56.7 | 57.0 | 57.0 | 57.3 | 57.0 | 56.7 | 57.6 | 57.0 | 56.7 | 57.4 | 57.1 | 57.1 | 56.8 | 56.8 | 56.5 | 56.5 | 99.4 | 82.6 | 74.5 | |
| 20 | C. coli RM2228 | 55.5 | 55.5 | 55.8 | 55.8 | 55.2 | 55.8 | 55.5 | 55.2 | 54.9 | 55.3 | 55.3 | 55.5 | 55.5 | 55.8 | 55.8 | 55.5 | 55.3 | 81.3 | 81.0 | 71.0 | |
| 21 | C. upsaliensis RM3195 | 53.6 | 54.0 | 54.2 | 54.2 | 54.0 | 53.6 | 53.6 | 54.2 | 53.6 | 53.6 | 54.9 | 53.5 | 53.5 | 53.8 | 53.8 | 53.8 | 54.1 | 74.1 | 73.8 | 68.6 |
Table 4.
Nucleotide (upper right) and deduced amino acid (lower left) sequence similarities (%) of full-length CLA0749 in C. lari isolates and other thermophilic Campylobacterreference strains employed
| Campylobacter | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C. lari JCM2530T | 99.9 | 100.0 | 99.7 | 89.4 | 90.0 | 90.0 | 89.4 | 89.4 | 85.5 | 90.0 | 85.5 | 85.5 | 85.4 | 85.5 | 85.5 | 100.0 | 61.7 | 61.6 | 61.8 | 62.5 | |
| 2 | C. lari 298 | 99.5 | 99.8 | 99.8 | 89.3 | 89.9 | 89.9 | 89.3 | 89.3 | 85.4 | 89.9 | 85.4 | 85.4 | 85.2 | 85.4 | 85.4 | 99.8 | 61.6 | 61.4 | 61.6 | 62.3 | |
| 3 | C. lari 300 | 100.0 | 99.5 | 99.7 | 89.4 | 90.0 | 90.0 | 89.4 | 89.4 | 85.5 | 90.0 | 85.5 | 85.5 | 85.4 | 85.5 | 85.5 | 100.0 | 61.7 | 61.6 | 61.8 | 62.5 | |
| 4 | C. lari 84C-1 | 99.5 | 100.0 | 99.5 | 89.1 | 89.7 | 89.7 | 89.1 | 89.4 | 85.2 | 89.7 | 85.2 | 85.2 | 85.1 | 85.2 | 85.2 | 99.7 | 62.2 | 62.1 | 61.6 | 62.3 | |
| 5 | UPTC 99 | 92.1 | 92.1 | 92.1 | 92.1 | 98.0 | 98.0 | 98.4 | 98.9 | 88.6 | 95.3 | 88.6 | 88.6 | 88.5 | 88.6 | 88.6 | 89.4 | 62.4 | 62.2 | 63.3 | 64.1 | |
| 6 | UPTC NCTC12892 | 93.0 | 93.0 | 93.0 | 93.0 | 99.1 | 100.0 | 97.7 | 97.8 | 89.4 | 95.1 | 89.1 | 89.1 | 89.2 | 89.4 | 89.4 | 90.0 | 61.8 | 61.6 | 63.1 | 64.1 | |
| 7 | UPTC NCTC12893 | 92.6 | 92.6 | 92.6 | 92.6 | 98.6 | 99.6 | 97.7 | 97.8 | 89.4 | 95.1 | 89.1 | 89.1 | 89.2 | 89.4 | 89.4 | 90.0 | 61.8 | 61.6 | 63.1 | 64.1 | |
| 8 | UPTC NCTC12894 | 92.5 | 92.5 | 92.5 | 92.5 | 98.1 | 99.1 | 98.6 | 98.9 | 88.2 | 95.0 | 88.2 | 88.2 | 88.0 | 88.2 | 88.2 | 89.4 | 61.6 | 61.4 | 62.8 | 63.4 | |
| 9 | UPTC NCTC12895 | 93.0 | 93.0 | 93.0 | 93.0 | 99.1 | 100.0 | 99.6 | 99.1 | 88.3 | 95.5 | 88.3 | 88.3 | 88.2 | 88.3 | 88.3 | 89.4 | 62.1 | 61.9 | 63.0 | 63.5 | |
| 10 | UPTC NCTC12896 | 87.4 | 87.4 | 87.4 | 87.4 | 90.2 | 90.2 | 89.8 | 89.7 | 90.2 | 87.7 | 99.1 | 99.1 | 99.8 | 100.0 | 99.8 | 85.5 | 63.4 | 62.9 | 63.2 | 64.4 | |
| 11 | UPTC CF89-12 | 92.5 | 92.5 | 92.5 | 92.5 | 96.7 | 97.7 | 97.2 | 97.2 | 97.7 | 88.8 | 87.7 | 87.7 | 87.5 | 87.7 | 87.7 | 90.0 | 63.0 | 63.7 | 63.8 | 64.0 | |
| 12 | UPTC A1 | 87.9 | 87.9 | 87.9 | 87.9 | 90.7 | 90.7 | 90.2 | 90.2 | 90.7 | 98.6 | 89.3 | 100.0 | 98.9 | 99.1 | 98.9 | 85.5 | 63.5 | 63.1 | 63.2 | 64.6 | |
| 13 | UPTC A2 | 87.9 | 87.9 | 87.9 | 87.9 | 90.7 | 90.7 | 90.2 | 90.2 | 90.7 | 98.6 | 89.3 | 100.0 | 98.9 | 99.1 | 98.9 | 85.5 | 63.5 | 63.1 | 63.2 | 64.6 | |
| 14 | UPTC A3 | 86.9 | 86.9 | 86.9 | 86.9 | 89.7 | 89.7 | 89.3 | 89.2 | 89.7 | 99.5 | 88.3 | 98.1 | 98.1 | 99.8 | 99.7 | 85.4 | 63.2 | 62.8 | 63.0 | 64.3 | |
| 15 | UPTC 89049 | 87.4 | 87.4 | 87.4 | 87.4 | 90.2 | 90.2 | 89.8 | 89.7 | 90.2 | 100.0 | 88.8 | 98.6 | 98.6 | 99.5 | 99.8 | 85.5 | 63.4 | 62.9 | 63.2 | 64.4 | |
| 16 | UPTC 92251 | 87.4 | 87.4 | 87.4 | 87.4 | 90.2 | 90.2 | 89.8 | 89.7 | 90.2 | 99.5 | 88.8 | 98.1 | 98.1 | 99.1 | 99.5 | 85.5 | 63.2 | 62.8 | 63.4 | 64.3 | |
| 17 | C. lari RM2100 | 100.0 | 99.5 | 100.0 | 99.5 | 92.1 | 93.0 | 92.6 | 92.5 | 93.0 | 87.4 | 92.5 | 87.9 | 87.9 | 86.9 | 87.4 | 87.4 | 61.7 | 61.6 | 61.8 | 62.5 | |
| 18 | C. jejuni NCTC11168 | 51.2 | 51.2 | 51.2 | 51.2 | 52.1 | 52.1 | 51.9 | 51.6 | 52.1 | 52.1 | 52.1 | 52.1 | 52.1 | 51.6 | 52.1 | 52.1 | 51.2 | 99.1 | 81.6 | 63.3 | |
| 19 | C. jejuni RM1221 | 50.7 | 50.7 | 50.7 | 50.7 | 51.6 | 51.6 | 51.4 | 51.2 | 51.6 | 51.6 | 51.6 | 51.6 | 51.6 | 51.2 | 51.6 | 51.6 | 50.7 | 98.6 | 81.4 | 63.6 | |
| 20 | C. coli RM2228 | 52.6 | 52.6 | 52.6 | 52.6 | 54.0 | 54.0 | 53.7 | 53.5 | 54.0 | 52.6 | 54.0 | 52.1 | 52.1 | 52.1 | 52.6 | 52.6 | 52.6 | 81.2 | 80.8 | 63.8 | |
| 21 | C. upsaliensis RM3195 | 54.2 | 54.2 | 54.2 | 54.2 | 55.6 | 55.6 | 55.6 | 56.2 | 55.6 | 55.3 | 55.1 | 55.3 | 55.3 | 54.0 | 55.3 | 55.3 | 54.2 | 55.6 | 55.6 | 56.9 |
Moreover, the deduced amino acid sequence alignment analyses were also performed for putative ORFs of the full-length cadF (-like) gene of 16 C. lari isolates, as well as those of C. lari RM2100, C. jejuni, C. coli and C. upsaliensis strains. The putative ORFs from the 17 C. lari isolates showed 90.9 - 100.0% amino acid sequence similarities to each other, and 56.4 - 57.9% similarities, with those of two C. jejuni strains (Table 3). They also showed 53.5 - 55.8% similarities with those of other thermophilic Campylobacter organisms (two strains of C. coli and C. upsaliensis; Table 3).
Thus, the putative ORFs of the full-length cadF (-like) gene from the 17 C. lari isolates identified in the present study are identical size (984 bp and 328 amino acid residues) with sequence heterogeneity, at both nucleotide and amino acid levels.
As shown in Table 4, the deduced amino acid sequence similarities were also examined for the putative ORFs of the full-length Cla_0387 gene among the 17 C. lari isolates (86.9 - 100.0%) and other thermophilic Campylobacter organisms (50.7 - 56.2%), employed as references (data not shown).
Thus, cadF (-like) gene is highly conserved among C. lari organisms isolated from humans and natural environments in several countries of Asia, Europe and North America.
In relation to the NC regions, two NC regions of approximately 250 bp, including a promoter at the -10 region and 120 bp occurred upstream of the cadF (-like) gene and downstream of the Cla_0387 gene, respectively, when examined combined sequences from all 16 C. lari isolates. Nucleotide sequences of approximately 250 bp from the 16 C. lari isolates and C. lari RM2100 showed 85.0 - 100.0% sequences similarities to each other (Table 5). Nucleotide sequences of approximately 120 bp also showed 85.6 - 100.0% sequence similarities among the 17 C. lari isolates. Thus, a considerable genetic heterogeneity of nucleotide sequences in the 250 bp NC region, full-length cadF (-like) gene, full-length Cla_0387 gene and the 120 bp NC region identified in the present study also occurred among the 17 C. lari isolates including the C. lari RM2100 strain.
Table 5.
Nucleotide sequence similarities (%) of the NC regions upstream of cadF (-like) gene(250 bp; upper right) and downstream of Cla_0387 (120 bp; lower left) among C. lari isolates
| Campylobacter lari | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C.lari JCM2530T | 98.8 | 98.8 | 98.4 | 87.3 | 89.7 | 89.7 | 88.1 | 88.6 | 89.1 | 86.5 | 87.5 | 87.5 | 87.9 | 87.8 | 87.9 | 98.8 | |
| 2 | C.lari 298 | 100.0 | 100.0 | 99.6 | 88.1 | 89.7 | 89.7 | 88.2 | 88.6 | 88.8 | 86.9 | 87.2 | 87.2 | 87.5 | 87.5 | 87.5 | 100.0 | |
| 3 | C.lari 300 | 100.0 | 100.0 | 99.6 | 88.1 | 89.7 | 89.7 | 88.2 | 88.6 | 88.8 | 86.9 | 87.2 | 87.2 | 87.5 | 87.5 | 87.5 | 100.0 | |
| 4 | C.lari 84C-1 | 100.0 | 100.0 | 100.0 | 87.8 | 89.3 | 89.3 | 87.8 | 88.2 | 88.4 | 86.5 | 86.8 | 86.8 | 87.1 | 87.0 | 87.1 | 99.6 | |
| 5 | UPTC 99 | 93.2 | 93.2 | 93.2 | 93.2 | 95.6 | 95.6 | 96.0 | 96.0 | 90.0 | 89.0 | 85.0 | 85.0 | 85.9 | 85.4 | 85.3 | 88.1 | |
| 6 | UPTC NCTC12892 | 93.2 | 93.2 | 93.2 | 93.2 | 98.3 | 100.0 | 96.8 | 97.6 | 91.3 | 89.7 | 86.6 | 86.6 | 87.0 | 87.0 | 87.3 | 89.7 | |
| 7 | UPTC NCTC12893 | 93.2 | 93.2 | 93.2 | 93.2 | 98.3 | 100.0 | 96.8 | 97.6 | 91.3 | 89.7 | 86.6 | 86.6 | 87.0 | 87.0 | 87.3 | 89.7 | |
| 8 | UPTC NCTC12894 | 93.2 | 93.2 | 93.2 | 93.2 | 100.0 | 98.3 | 98.3 | 98.4 | 93.2 | 89.0 | 86.3 | 86.3 | 86.7 | 86.6 | 87.0 | 88.2 | |
| 9 | UPTC NCTC12895 | 93.2 | 93.2 | 93.2 | 93.2 | 99.2 | 97.4 | 97.4 | 99.2 | 92.5 | 89.4 | 85.6 | 85.6 | 85.9 | 85.9 | 86.2 | 88.6 | |
| 10 | UPTC NCTC12896 | 88.1 | 88.1 | 88.1 | 88.1 | 92.4 | 90.7 | 90.7 | 92.4 | 91.5 | 86.5 | 92.3 | 92.3 | 92.7 | 92.7 | 93.1 | 88.8 | |
| 11 | UPTC CF89-12 | 89.7 | 89.7 | 89.7 | 89.7 | 91.5 | 91.5 | 91.5 | 91.5 | 90.6 | 85.6 | 85.5 | 85.5 | 85.5 | 85.4 | 85.7 | 86.9 | |
| 12 | UPTC A1 | 88.1 | 88.1 | 88.1 | 88.1 | 92.4 | 90.7 | 90.7 | 92.4 | 91.5 | 100.0 | 85.6 | 100.0 | 99.2 | 98.8 | 99.2 | 87.2 | |
| 13 | UPTC A2 | 88.1 | 88.1 | 88.1 | 88.1 | 92.4 | 90.7 | 90.7 | 92.4 | 91.5 | 100.0 | 85.6 | 100.0 | 99.2 | 98.8 | 99.2 | 87.2 | |
| 14 | UPTC A3 | 88.1 | 88.1 | 88.1 | 88.1 | 92.4 | 90.7 | 90.7 | 92.4 | 91.5 | 100.0 | 85.6 | 100.0 | 100.0 | 99.2 | 99.2 | 87.5 | |
| 15 | UPTC 89049 | 88.1 | 88.1 | 88.1 | 88.1 | 92.4 | 90.7 | 90.7 | 92.4 | 91.5 | 100.0 | 85.6 | 100.0 | 100.0 | 100.0 | 98.8 | 87.5 | |
| 16 | UPTC 92251 | 88.1 | 88.1 | 88.1 | 88.1 | 92.4 | 90.7 | 90.7 | 92.4 | 91.5 | 100.0 | 85.6 | 100.0 | 100.0 | 100.0 | 100.0 | 87.5 | |
| 17 | C. lari RM2100 | 100.0 | 100.0 | 100.0 | 100.0 | 93.2 | 93.2 | 93.2 | 93.2 | 93.2 | 88.1 | 89.7 | 88.1 | 88.1 | 88.1 | 88.1 | 88.1 |
NC, non-coding.
Northern blot hybridization, reverse transcription-PCR and primer extension analysis
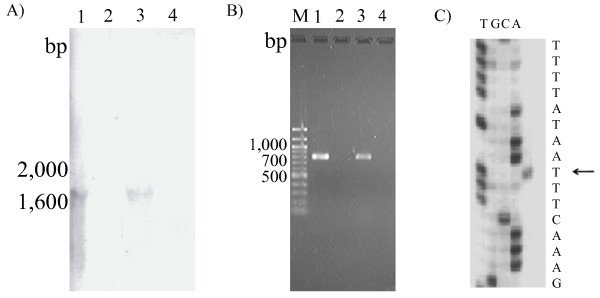
Northern blot hybridization analysis detected the cadF (-like) gene transcription in the two C. lari isolates cells, UN C. lari JCM2530T and UPTC CF89-12 (Figure 2A). Since the positive signals of the hybridization were shown at around 1,600 bp (Figure 2A), the cadF (-like) gene may possibly be transcribed together with the Cla_0387 gene. Thus, cadF (-like) gene transcription was confirmed in the C. lari organisms. When RT-PCR analysis was carried out for the RNA components extracted from the UN C. lari JCM2530T and UPTC isolates CF89-12 cells with the primer pair of f-cadF2 in the cadF (-like) gene and r-cadF3 in the Cla_0387 gene, as shown in Figure 1, a positive RT-PCR signal was detected at around 800 bp region with both isolates, respectively (Figure 2B).
Figure 2.
Northern blot hybridization (A) and RT-PCR (B) analyses of the cadF (-like) and Cla_0387 structural gene transcripts expressed in the C. lari isolates. Lane M, 100 bp DNA ladder; Lane 1, C. lari JCM2530T with the reverse transcriptase (RTase); lane 2, C. lari JCM2530T without the RTase.; lane 3, UPTC CF89-12 with the RTase; lane 4, UPTC CF89-12 without the RTase. Primer extension analysis (C) of the cadF (-like) and Cla_0387 mRNA transcript in the C. lari JCM2530T isolate cells. The arrow indicates the transcription initiation site.
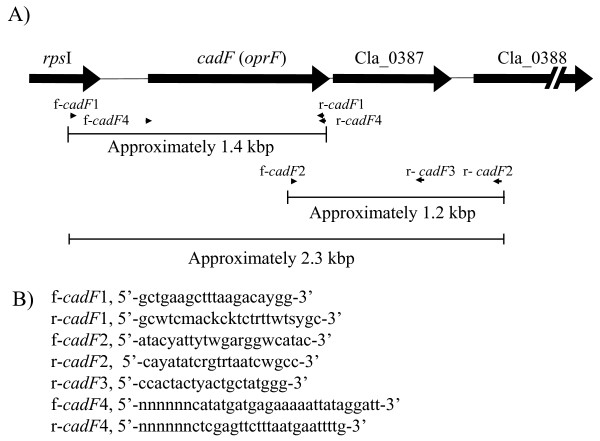
Figure 1.
A schematic representation of the cadF gene and its adjacent genetic loci for C. lari RM2100, including locations of the novel primers designed in silico (A). Nucleotide sequences of the primers are also shown (B).
The transcription initiation site for the cadF (-like) gene was determined by the primer extension analysis (Figure 2C). The +1 transcription initiation site for the cadF (-like) gene is underlined in the following sequence; 5'-TTTTATAATTTCAAAG-3', as shown in Figure 2C.
Deduced amino acid sequence alignment analysis and phylogenetic analyses of the cadF (-like) ORF
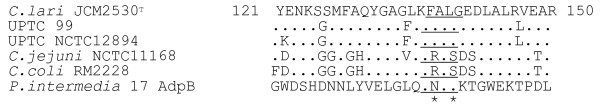
We carried out deduced amino acid sequence alignment analysis to elucidate the differences in CadF (-like) protein amongst the thermophilic Campylobacter. As shown in Figure 3, the C. coli RM2228 strain carried a strech of 12 amino acid (VVTPAPAPVVSQ) from amino acid positions 190 to 201, as well as a Q at amino acid position 180, and regarding the nine larger amino acid for C. lari isolates than C. jejuni strains, four amino acid sequences (THTD) from amino acid positions 80 to 83 and five [A(T for UPTC 99) KQID] from 193 to 197 were identified to occur.
Figure 3.
Amino acid sequence alignment analysis of parts (around larger CadF sequences for C. coli and C. lari) of the putative cadF (-like) ORF from the thermophilic Campylobacter isolates examined in the present study. Since nine amino acid larger sequences of the other 14 C. lari isolates were identical to either those from the C. lari JCM2530T or UPTC isolates, alignment analysis data were omitted from the Figure.
When, in retation to a single Fn-binding domain localized at four amino acid (FRLS; CadF amino acid positions 134-137 for C. jejuni) [28], amino acid sequence alignment analysis was carried out, the putative cadF (-like) ORFs from all 17 C. lari isolates examined showed amino acid residues of FALG (50% identity) within the amino acid positions 137-140 instead of the FRLS residues, as shown in Figure 4.
Figure 4.
Amino acid sequence alignment analysis of part (around a single-Fn binding domain within C. jejuni CadF) of the putative ORF for cadF (-like) gene from the 17 C. lari isolates. Amino acid sequences of those from the C. jejuni and C. coli reference strains were aligned for comparison. FALG residues of C. lari and FRLS residues of C. jejuni and C. coli strains were underlined, respectively. In this Figure, amino acid sequence of AdpB (aa 201-230) from Prevotella intermedia 17 [32] was also aligned for comparison. FNLG residues of P. intermedia 17 were also underlined. The alignment analysis data from the UN C. lari isolates RM2100, 298, 300 and 84C-1, from the UPTC isolates NCTC12892, 12893, 12895, 12896, CF89-12, A1, A2, A3, 89049 and 92251, and from C. jejuni strains RM1221, 81-176, 260.94, CF93-6, HB93-13, 8425 and ss doylei 269.97 were omitted from the Figure, because of the occurrence of the identical sequences.
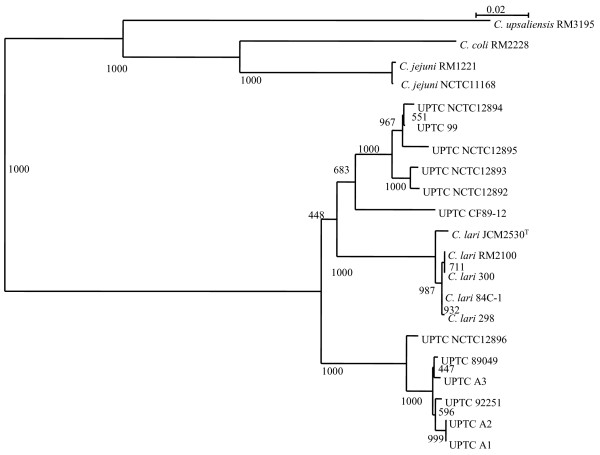
A dendrogram showing phylogenetic relationships constructed by the NJ method [29] based on nucleotide sequence information of full-length cadF (-like) gene from 16 C. lari isolates and C. lari RM2100 and other thermophilic Campylobacter reference strains, the 17 C. lari isolates forming a major cluster separating from the other three thermophilic Campylobacter spp. (Figure 5). In addition, UN C. lari and UPTC organisms were not different and similar based on the nucleotide sequence data of the cadF (-like) gene, as shown in Figure 5.
Figure 5.
A phylogenetic tree constructed based on nucleotide sequence information of full-length cadF (-like) gene from 17 C. lari isolates and other thermophilic campylobacters. The tree was constructed by the NJ method [29]. values, 0.02, in the figure represent evolutionary distances. Boot-strap values of 1,000 are shown at the branch point. Out-group is C. upsaliensis RM3195.
Discussion
This is the first demonstration of the structural analysis of the full-length gene encoding a CadF (-like) protein and its adjacent genetic loci within C. lari.
Regarding the NC region upstream of the cadF (-like) gene, this region is approximately 250 bp in length with all 16 C. lari isolates and C. lari RM2100 strain. However, the NC regions from the eight C. jejuni and a C. coli reference strains shown in Table 1 examined, are shorter than those and approximately 150 bp in length with unknown reason(s).
In 1995, Koebnik described a peptidoglycan-associating alpha-helical consensus motif in the C-terminal regions of 16 bacterial cell-surface proteins (NX2 LSX2 RAX2 VX3L) [30]. When we compared the corresponding amino acid sequences of the putative cadF (-like) ORF from the 17 C. lari and some C. jejuni isolates with this consensus motif, the motif was completely conserved amongst the cadF (-like) ORFs from the isolates (data not shown).
As shown in Table 2, the CMW of the putative cadF (-like) ORF was estimated to be 36,578 to 36,869 Da for the 16 C. lari isolates and C. lari RM2100 reference strain (data not shown). In addition, the value was also estimated to be approximately 36 kDa for the two C. jejuni reference strains (Table 2). These estimated CMW values are in agreement with the previous description of the immunodetection of the CadF protein from five C. jejuni and C. coli isolates [25].
When the nucleotide and deduced amino acid sequence alignment analyses were carried out for the putative cadF (-like) ORF, apparent size differences occurred amongst the four thermophilic Campylobacter species, as described above. Regarding the putative ORFs for cadF (-like) gene between C. lari and C. jejuni organisms, nine amino acid residues are shorter in C. jejuni strains than in C. lari isolates.
Recently, Krause-Gruszczynska et al. (2007) described that the CadF protein from C. coli strains was 13 amino acid larger than those from C. jejuni strains, based on the deduced amino acid sequence alignment analysis [31]. This is consistent with our present results (Table 2). They also indicated that C. coli strains carried a stretch of 13 amino acid in the middle region of the protein [31]. In addition, in the present study, the deduced CadF (-like) protein was shown to be 328 amino acid from all 17 C. lari isolates and were nine amino acid larger than CadF from two C. jejuni strains (319 amino acid) (Table 2). Then, we carried out deduced amino acid sequence alignment analysis to elucidate the differences in CadF (-like) protein between C. lari and C. jejuni organisms. As shown in Figure 3, the C. coli RM2228 strain carried a stretch of 12 amino acid (VVTPAPAPVVSQ) from amino acid positions 190 to 201 as well as a Q at amino acid position 180 (Figure 3). In relation to the nine larger amino acid for C. lari isolates than C. jejuni strains, interestingly, four amino acid sequences (THTD) from amino acid positions 80 to 83 and five [A(T for UPTC99) KQID] from 193 to 197 were identified, as shown in Figure 3.
Regarding the CadF in Campylobacter, the cadF virulence gene, encoding 37 kDa outer membrane protein that promotes the binding of the pathogens to intestinal epithelial cells, was identified and cloned [22,25]. In relation to identification of the binding domain within C. jejuni CadF, Konkel et al. (2005) recently described that a single Fn-binding domain was localized at four amino acids (CadF amino acid positions 134 -137), consisting of the residues, phenylalanine-arginine-leucine-serine (FRLS) [28]. However, when amino acid sequence alignment analysis was carried out, the putative cadF (-like) ORFs from all 17 C. lari isolates examined in the present study showed amino acid residues of FALG (50% identity) within the amino acid positions 137 - 140, instead of the FRLS residues (Figure 4). No FRLS residues were also detected within any other regions of the cadF (-like) ORF from all 17 C. lari isolates examined. Interestingly, FNLG residues within AdpB (Ad-adhesin in p-Prevotella, B-second identified adhesin) in Prevotella intermedia (a black-pigmented gram-negative anaerobe) [32] was 75% identical to the FALG from C. lari (Figure 4). Therefore, it may be important to clarify if the CadF (-like) protein from C. lari isolates can bind to fibronectin or not. An experiment is now in progress to resolve this.
In the present study, for the first time, we have described the cloning, sequencing and characterization of full-length Cla_0387 from the 16 C. lari isolates. The CMW values were estimated to be 23,689 - 23,875 Da for the 16 C. lari isolates and C. lari RM2100 strain and these values were also equivalent to those from two C. jejuni and a C. coli reference strains (Table 2). In addition, the cadF (-like) gene and the Cla_0387 gene may possibly be functional within C. lari isolates, based on the present northern blot hybridization and RT-PCR observations, as shown in Figure 2A and 2B.
Thus, the cadF (-like) gene and the Cla_0387 gene could be co-transcribed within C. lari organisms, consisting of an operon. Since the Cla_0387 showed a high deduced amino acid sequence similarity to the Escherichia coli haloacid dehalogenase-like phosphatase [33], these two may have an important biological relationship within the C. lari cells.
In the present study, the authors designed two novel primer pairs (f-/r-cadF1 and f-/r-cadF2) in silico for amplification of an approximate 2.3 kbp region, including the full-length cadF (-like) gene and its adjacent genetic loci, based on sequence information of C. lari RM2100, C. jejuni RM1221 and C. coli RM2228 strains, resulting in successful amplification, TA-cloning and sequencing of those from the 16 C. lari isolates isolated from differencet sources and in several countries. Therefore, the present novel PCR primer pairs would be likely of value for, C. jejuni and C. coli organisms, as well as for other C. lari isolates.
A dendrogram showing phylogenetic relationships was constructed by the NJ method [29], based on nucleotide sequence information of full-length cadF (-like) gene from 16 C. lari isolates and C. lari RM2100 and other thermophilic Campylobacter reference strains. As shown in Figure 5, the 17 C. lari isolates form a major cluster separating from the other three thermophilic Campylobacter spp. In addition, the 17 C. lari isolates form some minor clusters, respectively, based on nucleotide sequence information from cadF (-like) gene (Figure 5). Thus, nucleotide sequence information of full-length cadF (-like) gene can be regarded as reliable in the molecular discrimination of C. lari organisms from the other three thermophilic campylobacters. In addition, Figure 5 also indicated that NJ dendrogram of UN C. lari and UPTC organisms were not different and similar based on the nucleotide sequence data of the cadF-like gene.
Conclusion
The combined sequences encoding a partial and putative rpsI open reading frame (ORF), non-coding (NC) region, a putative ORF for the Campylobacter adhesin to fibronectin-like gene, a putative Cla_0387 ORF, NC region and a partial and putative Cla_0388 ORF, were identified in 16 Campylobacter lari isolates, using two novel degenerate primer pairs. Transcription of the cadF-like gene in C. lari cells in vivo was also confirmed and the transcription initiation site was determined. The putative cadF (-like) ORFs from all C. lari isolates were nine amino acid larger than those from C. jejuni, and showed amino acid residues 137 -140 of FALG (50% identity), instead of the FRLS residues of the maximal fibronectin-binding activity site demonstrated within C. jejuni CadF.
Methods
Campylobacter isolates and culture conditions
C. lari isolates (n = 4 UN C. lari; n = 12 UPTC), which were isolated from different sources and in several countries of Asia, Europe and North America and used in the present study, are shown in Table 1.
These isolates were cultured on Mueller-Hinton broth medium at 37°C for 48 h in an aerobic jar on Blood Agar Base No. 2 (Oxoid, Hampshire, UK) containing 7% (v/v) defibrinated horse blood (Nippon Bio-Test, Tokyo, Japan) and Campylobacter selective medium (Virion, Zurich, Switzerland). An atmosphere of 5% (v/v) O2 and 10% (v/v) CO2 was produced by BBL Campypak Microaerophilic System Envelopes (Bacton Dickinson, NJ, USA).
Genomic DNA preparation, primer design and PCR amplification
Genomic DNA was prepared using sodium dodecyl sulfate and proteinase K treatment, phenol-chloroform extraction and ethanol precipitation [34].
Two novel degenerate primer pairs (f-/r-cadF1 and f-/r-cadF2) were first designed in silico to generate two PCR products of approximately 1.4 and 1.2 kbp respectively, corresponding to the full-length cadF-like gene and its adjacent genetic loci, including full-length Cla_0387 (approximately 2.3 kbp) for the C. lari isolates, based on the sequence information of C. lari RM2100, C. jejuni RM1221 and C. coli RM2228 strains. A schematic representation of the cadF gene and its adjacent genetic loci for C. lari RM2100 (AAFK01000002) [26], including the locations of the two primer pairs and nucleotide sequences of the primers designed in silico in the present study, are shown in Figure 1.
PCR mixtures contained 50 ng of template DNA, 10 mM Tris-HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 400 μM of each dNTP, 0.6 μM of each primer, and a total of 1 unit of rTaq DNA polymerase (Takara Bio Inc., Shiga, Japan). PCR was performed in 50 μl reaction volumes, for 30 cycles of 94°C for 1.0 min, 50°C for the f-/r-cadF1 and 45°C for the f-/r-cadF2 for 0.5 min, and 72°C for 1.5 min, followed by a final extension at 72°C for 5.0 min.
TA cloning, nucleotide sequencing and sequence analyses
Amplified PCR products were separated by 1.0% (w/v) agarose gel electrophoresis in 0.5× TBE at 100 V and detected by staining with ethidium bromide. PCR products amplified by the newly constructed two primer pairs were purified using a QIAquick PCR Purification Kit (QIAGEN, Tokyo, Japan) and inserted into the pGEM-T vector using the pGEM-T Easy Vector System (Promega Corp. Tokyo, Japan). Sequencing of the cloned cadF (-like) gene fragments was performed with a Hitachi DNA autosequencer (SQ5500EL; Hitachi Electronics Engineering Co. Tokyo, Japan), after dideoxy nucleotide sequencing using a Thermo Sequenase Pre-Mixed Cycle Sequencing Kit (Amersham Pharmacia Biotech, Tokyo, Japan). Sequence analysis of the PCR amplicons was carried out using the computer software GENETYX-MAC version 9 (GENETYX Co., Tokyo, Japan).
Total cellular RNA purification, reverse transcription-PCR, northern blot hybridization and primer extension analysis
Total cellular RNA was extracted and purified from C. lari cells by using RNA protect Bacteria Reagent and RNeasy Mini Kit (QIAGEN). Reverse-transcription (RT)-PCR was carried out with a primer pair of f-cadF2 and r-cadF3 (Figure 1), by using the QIAGEN OneStep RT-PCR Kit (QIAGEN). This primer pair is expected to generate a RT-PCR product of the cadF (-like) structural gene segment of approximately 780 bp including the Cla_0387 region. Northern blot hybridization analysis was carried out according to the procedure described by Sambrook and Russell (2001) [34], using a PCR amplified cadF (-like) fragment as a probe. The fragment was amplified using a primer pair of f-/r-cadF4 (Figure 1). Random primer extension was performed in order to prepare the fragment probe using a DIG-High Prime (Roche Applied Science, Penzberg, Germany).
The transcription initiation site for the cadF (-like) gene was determined by the primer extension analysis with the purified total cellular RNA of C. lari JCM2530T cells. The primer that was selected for this assay was 5'-CTAAATTTCCTTCTGGMGTTGT-3', which corresponds to the reverse complementary sequence of np 504 through 525. The transcription initiation site was determined by primer extension with the sizes of DNA fragments generated by sequencing reactions. In the present study, the np which the authors used, are for those of C. lari JCM2530T.
Phylogenetic analysis
Nucleotide sequences of approximately 980 bp of the full-length cadF (-like) gene, from the isolates of C. lari and the C. lari RM2100 strain, were compared to each other and with the accessible sequence data from some other thermophilic campylobacters using CLUSTAL W software, respectively [35], which was incorporated in the DDBJ. Following this, a phylogenetic tree was constructed by the neighbor-joining (NJ) method [29].
List of abbreviations used
CadF: Campylobacter adhesin to fibronectin; C. lari: Campylobacter lari; kbp: kilo base pairs; np: nucleotide position; ORF: open reading frame; RT: reverse transcription; UPTC: urease-positive thermophilic Campylobacter; UN: urease-negative.
Authors' contributions
JH, TS, AT and IT were involved with cloning, sequencing and analysis of the rRNA gene sequences from Campylobacter strains. JEM and BCM participated in its design and coordination, and review of the manuscript. MM participated in design of the study, collected strains, drafted the manuscript and reviewed the manuscript. All authors read and approved the final version.
Contributor Information
Junichi Hirayama, Email: h05308@azabu-u.ac.jp.
Tsuyoshi Sekizuka, Email: sekizuka@nih.go.jp.
Akihiro Tazumi, Email: de0702@azabu-u.ac.jp.
Ikue Taneike, Email: ikuet1120@yahoo.co.jp.
John E Moore, Email: j.moore1@ulster.ac.uk.
B Cherie Millar, Email: bcmillar@niphl.dnet.co.uk.
Motoo Matsuda, Email: matsuda@azabu-u.ac.jp.
Acknowledgements
This research was partially supported by the Promotion and Mutal Aid Corporation For Private Schools of Japan, Grant-in-Aid for Matching Fund Subsidy for Private Universities and by a Grant-in-Aid for Scientific Research (C) (no. 20580346) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to MM). This study was also partially supported by a project grant (Start-Up Support for the Matching Fund Subsidy for Private Universities, 2007-2008) awarded by the Azabu University Research Services Division. MM and JEM were supported by a Butterfield Award from the Great Britain Sasakawa Foundation to jointly examine the role of campylobacter in food-poisoning in the UK and Japan.
References
- Benjamin J, Leaper S, Owen RJ, Skirrow MB. Description of Campylobacter laridis, a new species comprising the nalidixic acid resistant thermophilic Campylobacter (NARTC) group. Curr Microbiol. 1983;8:231–238. doi: 10.1007/BF01579552. [DOI] [Google Scholar]
- Blaser MJ, Taylor DN, Feldman RA. Epidemiology of Campylobacter jejuni infections. Epidemiol Rev. 1983;5:157–176. doi: 10.1093/oxfordjournals.epirev.a036256. [DOI] [PubMed] [Google Scholar]
- Stirling J, Griffith M, Blair I, Cormican M, Dooley JSG, Goldsmith CE, Glover SG, Loughrey A, Lowery CJ, Matsuda M, McClurg R, McCorry K, McDowell D, McMahon A, Millar BC, Nagano Y, Rao JR, Rooney PJ, Smyth M, Snelling WJ, Xu J, Moore JE. Prevalence of gastrointestinal bacterial pathogens in a population of zoo animals. Zoo Public Health. 2008;55:166–172. doi: 10.1111/j.1863-2378.2007.01099.x. [DOI] [PubMed] [Google Scholar]
- Skirrow MB, Benjamin J. '1001' campylobacters: cultural characteristics of intestinal campylobacters from man and animals. J Hyg (Camb) 1980;85:427–442. doi: 10.1017/s0022172400063506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinot M, Jaulhac B, Moog R, Martino SD, Kehrli P, Monteil H, Piemont Y. Campylobacter lari bacteremia. Clin Microbiol Infect. 2001;7:96–97. doi: 10.1046/j.1469-0691.2001.00212.x. [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Stowell C, Skalina D, Jones AM, Hoop RM, Smibert RM. Campylobacter laridis causing bacteremia in an immunosuppressed patient. Ann Int Med. 1984;101:55–57. doi: 10.7326/0003-4819-101-1-55. [DOI] [PubMed] [Google Scholar]
- Simor AE, Wilcox L. Enteritis associated with Campylobacter laridis. J Clin Microbiol. 1987;25:10–12. doi: 10.1128/jcm.25.1.10-12.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauxe RV, Patton CM, Edmonds P, Brenner DJ, Blake PA. Illness associated with Campylobacter laridis, a newly recognized Campylobacter species. J Clin Microbiol. 1985;21:222–225. doi: 10.1128/jcm.21.2.222-225.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werno AM, Klena JD, Shaw GM, Murdoch DR. Fatal case of Campylobacter lari prosthetic joint infection and bacteremia in an immunocompetent patient. J Clin Microbiol. 2002;40:1053–1055. doi: 10.1128/JCM.40.3.1053-1055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton FJ, Holt A, Hutchinson DN. Urease-positive thermophilic campylobacters. Lancet. 1985;I:1217–1218. doi: 10.1016/S0140-6736(85)92898-3. [DOI] [PubMed] [Google Scholar]
- Mégraud F, Chevrie D, Desplaces N, Sedallian A, Guesdon JL. Urease-positive thermophilic Campylobacter (Campylobacter laridis variant) isolated from an appendix and from human feces. J Clin Microbiol. 1988;26:1050–1051. doi: 10.1128/jcm.26.5.1050-1051.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RJ, Costas M, Sloss L, Bolton FJ. Numerical analysis of electrophoretic protein patterns of Campylobacter laridis and allied thermophilic campylobacters from the natural environment. J Appl Bacteriol. 1988;65:69–78. doi: 10.1111/j.1365-2672.1988.tb04319.x. [DOI] [PubMed] [Google Scholar]
- Bezian MC, Ribou G, Barberis-Giletti C, Megraud F. Isolation of a urease positive thermophilic variant of Campylobacter lari from a patient with urinary tract infection. Eur J Clin Microbiol Infect Dis. 1990;9:895–897. doi: 10.1007/BF01967506. [DOI] [PubMed] [Google Scholar]
- Kaneko A, Matsuda M, Miyajima M, Moore JE, Murphy PG. Urease-positive thermophilic strains of Campylobacter isolated from seagulls (Larus spp.) Lett Appl Microbiol. 1999;29:7–9. doi: 10.1046/j.1365-2672.1999.00565.x. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Kaneko A, Stanley T, Miller BC, Miyajima M, Murphy PG, Moore JE. Characterization of urease-positive thermophilic Campylobacter subspecies by multilocus enzyme electrophoresis typing. Appl Environ Microbiol. 2003;69:3308–3310. doi: 10.1128/AEM.69.6.3308-3310.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IG, Moore JE. Presence of Salmonella spp. and Campylobacter spp. in shelfish. Epidemiol Infect. 1996;116:147–153. doi: 10.1017/S0950268800052377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endtz HP, Vliegenthart JS, Vandamme P, Waverink HW, Braak NP Van den, Verbrug HA, Belkum AV. Genotypic diversity of Campylobacter lari isolated from mussels and oysters in The Netherlands. Int J Food Microbiol. 1997;34:79–88. doi: 10.1016/S0168-1605(96)01174-9. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Kaneko A, Fukuyama M, Itoh T, Shingaki M, Inoue M, Moore JE, Murphy PG, Ishida Y. First finding of urease-positive thermophilic strains of Campylobacter in river water in the Far East, namely, in Japan, and their phenotypic and genotypic characterization. J Appl Bacteriol. 1996;81:608–612. [Google Scholar]
- Matsuda M, Shibuya T, Itoh Y, Takiguchi M, Furuhata K, Moore JE, Murayama O, Fukuyama M. First isolation of urease-positive thermophilic Campylobacter (UPTC) from crows (Coruvs levaillantii) in Japan. Int J Hyg Environ Health. 2002;205:321–324. doi: 10.1078/1438-4639-00157. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Moore JE. Urease-positive thermophilic Campylobacter species. Appl Environ Microbiol. 2004;70:4415–4418. doi: 10.1128/AEM.70.8.4415-4418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröman G, Switalski LM, Faris A, Wadstom T, Hook M. Binding of Escherichia coli to fibronectin. J Biol Chem. 1984;259:14899–14905. [PubMed] [Google Scholar]
- Konkel ME, Garvis SG, Tipton SL, Anderson DE Jr, Cieplak W Jr. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- Myhre EB, Kuusela P. Binding of human fibronectin to group A, C, and G streptococci. Infect Immun. 1983;40:29–34. doi: 10.1128/iai.40.1.29-34.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Putten JP, Duensing TD, Cole RL. Entry of OpaA+ gonococci into HEp-2 cells requires concerted action of glycosaminoglycans, fibronectin and integrin receptors. Mol Microbiol. 1998;29:369–379. doi: 10.1046/j.1365-2958.1998.00951.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol. 1999;37:510–517. doi: 10.1128/jcm.37.3.510-517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, Deboy RT. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin L. Genes VII. Oxford; Oxford University Press; 2000. [Google Scholar]
- Konkel ME, Christensen JE, Keech AM, Monteville MR, Klena JD, Garvis SG. Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Mol Microbiol. 2005;57:1022–1035. doi: 10.1111/j.1365-2958.2005.04744.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- Krause-Gruszczynska M, van Alphen LB, Oyarzabal OA, Alter L, Hanel I, schliephake A, Konig W, van Putten JM, Konkel ME. Expression patterns and role of the CadF protein in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett. 2007;274:9–16. doi: 10.1111/j.1574-6968.2007.00802.x. [DOI] [PubMed] [Google Scholar]
- Yu F, Lyer D, Anaya C, Lewis JP. Identification and characterization of a cell surface protein of Prevotella intermedia 17 with broad spectrum binding activity for extra cellular matrix proteins. Proteomics. 2006;6:6023–6032. doi: 10.1002/pmic.200600177. [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;47:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning; a laboratory manual. 3. Cold Spring Harbor Laboratory Press., Cold Spring harbor, N. Y; 2001. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]