Abstract
Background
The genome of Arthrobacter sp. strain FB24 contains a chromate resistance determinant (CRD), consisting of a cluster of 8 genes located on a 10.6 kb fragment of a 96 kb plasmid. The CRD includes chrA, which encodes a putative chromate efflux protein, and three genes with amino acid similarities to the amino and carboxy termini of ChrB, a putative regulatory protein. There are also three novel genes that have not been previously associated with chromate resistance in other bacteria; they encode an oxidoreductase (most similar to malate:quinone oxidoreductase), a functionally unknown protein with a WD40 repeat domain and a lipoprotein. To delineate the contribution of the CRD genes to the FB24 chromate [Cr(VI)] response, we evaluated the growth of mutant strains bearing regions of the CRD and transcript expression levels in response to Cr(VI) challenge.
Results
A chromate-sensitive mutant (strain D11) was generated by curing FB24 of its 96-kb plasmid. Elemental analysis indicated that chromate-exposed cells of strain D11 accumulated three times more chromium than strain FB24. Introduction of the CRD into strain D11 conferred chromate resistance comparable to wild-type levels, whereas deletion of specific regions of the CRD led to decreased resistance. Using real-time reverse transcriptase PCR, we show that expression of each gene within the CRD is specifically induced in response to chromate but not by lead, hydrogen peroxide or arsenate. Higher levels of chrA expression were achieved when the chrB orthologs and the WD40 repeat domain genes were present, suggesting their possible regulatory roles.
Conclusion
Our findings indicate that chromate resistance in Arthrobacter sp. strain FB24 is due to chromate efflux through the ChrA transport protein. More importantly, new genes have been identified as having significant roles in chromate resistance. Collectively, the functional predictions of these additional genes suggest the involvement of a signal transduction system in the regulation of chromate efflux and warrants further study.
Background
Arthrobacter species are high G+C Gram positive bacteria that are prevalent in both pristine and polluted soils [1-3]. Although Arthrobacter spp. have been noted for their high levels of resistance to a variety of toxic metals [4,5], very little is known about the genetic basis or regulatory mechanisms underlying metal resistance in this genus. Arthrobacter sp. FB24 was isolated from soils contaminated with lead-chromate salts and was selected for detailed study based on its high tolerance to a wide assortment of toxic heavy metals [6-8]. Most notably, this strain can survive in the presence of 200 mM potassium chromate in dilute nutrient broth [6]. Reported resistance levels for other Arthrobacter species range from 2 to 48 mM chromate [9,10].
The mechanism of chromium resistance in Arthrobacter strains remains enigmatic. Although some strains can reduce toxic Cr(VI) to less toxic Cr(III) [11,12], chromate reduction is not typically considered a resistance mechanism [13]. However, chromate efflux has only been biochemically identified as a resistance mechanism in Proteobacteria [14-17]. The earliest analyses of efflux-mediated chromate resistance have been performed in Cupravidus metallidurans and Pseudomonas aeruginosa, and until recently, these two organisms have served as the model organisms for chromate efflux. As a structural analog of sulfate (SO42-), chromate enters cells through sulfate uptake systems [18]. Chromate efflux occurs via the ChrA protein in P. aeruginosa and C. metallidurans and resulted in resistance levels of 4 and 0.3 mM, respectively [19-21]. It is important to note that the number and arrangement of chromate resistance genes differs between these two strains [13,15,20,21]. In addition, in 2007 at least 135 ChrA orthologs were noted in other bacteria as members of the CHR superfamily of chromate transporters [22,23]. There is considerable variation in the genomic context surrounding ChrA orthologs [22], which raises the question as to whether functional or regulatory differences in chromate efflux among organisms bearing ChrA orthologs also exist. Although the CHR superfamily includes representatives from all domains of life, at the time of its construction, the phylogeny was largely dominated by Proteobacteria (35 out of 72 organisms). Moreover, given the high levels of chromate resistance among Actinomycetales such as Arthrobacter [2-5], the 135 ChrA orthologs (which includes only three representatives within the order Actinomycetales, Corynebacterium glutamicum, C. efficiens and Kineococcus radiotolerans) reported by Ramirez-Diaz et al [22] is very likely an underestimate of the range of this protein family and warrants further investigation.
Chromate resistance levels reported for bacterial strains with ChrA orthologs are also highly variable, ranging from 0.3 to 200 mM Cr(VI). It is apparent that the mere presence of a chrA gene cannot explain this vast difference in resistance levels. Thus, further study of ChrA orthologs and their genomic neighborhoods in a greater diversity of chromate-resistant organisms will undoubtedly yield additional functional and regulatory elements that are relevant to different levels of chromium resistance found in diverse taxa. In this work, we examine such a chromate resistance determinant found in Arthrobacter sp. FB24.
Results
Identification of a chromate resistance determinant (CRD) in Arthrobacter sp. strain FB24
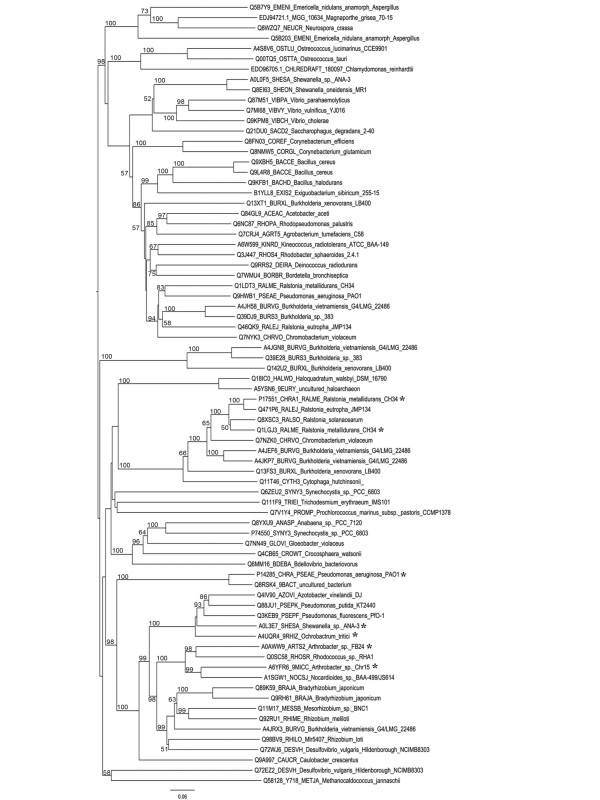
Arthrobacter sp. strain FB24 genome analysis deduced a 450 amino acid (aa) sequence Arth_4248 with similarity to chromate ion transporters. Phylogenetic analysis of the sequence with 512 other characterized and putative ChrA sequences (see Figure 1 and Additional files 1 and 2) suggests that it forms a new branch in the CHR superfamily [22] that is composed of Actinobacteria. This group likely has unique evolutionary features since the majority (70%) of ChrA ortholog sequences used in the comparison is from Proteobacteria yet it formed its own branch. In fact, most of the clades are composed of specific phyla/classes of biota (Additional file 1).
Figure 1.
Phylogenetic Tree of ChrA Orthologs. Phylogenetic tree of LCHR proteins generated from a subset of the alignment of 513 putative chromate ion transport sequences using ClustalX and default setting for Gonnet series for protein weight matrix (34). Neighbor Joining tree graphically viewed using the FigTree program http://tree.bio.ed.ac.uk/software/figtree/. Branched tips labeled with Uniprot protein accession number, sequence name and species name. Sequences with function supported with experimental data marked with asterisk. Scale bar indicates 0.06 amino acid substitutions per site. Branch ends labeled with bootstrap values >50%. Full tree available in the figure in Additional file 1 and all sequences used are listed in the table provided in Additional file 2.
The genome neighborhood of Arth_4248 consists of a 10.6-kb region of five putative chromate resistance genes and three proximal genes of unknown function located on a 96-kb plasmid (Figure 2). Of five genes similar to ones associated with Cr(VI) resistance in other organisms, two encode ChrA efflux protein orthologs (Arth_4248 and 4251) and three are similar to different regions of a putative regulatory protein, ChrB (Arth_4249, 4253 and 4254). The remaining three genes (Arth_4247, 4252 and 4255) have not been previously shown to be associated with chromate resistance. The region between Arth_4251 and Arth_4249 is an approximate 1.3 kb region of low complexity. Currently, there is no strong indication of functional genes within this region.
Figure 2.
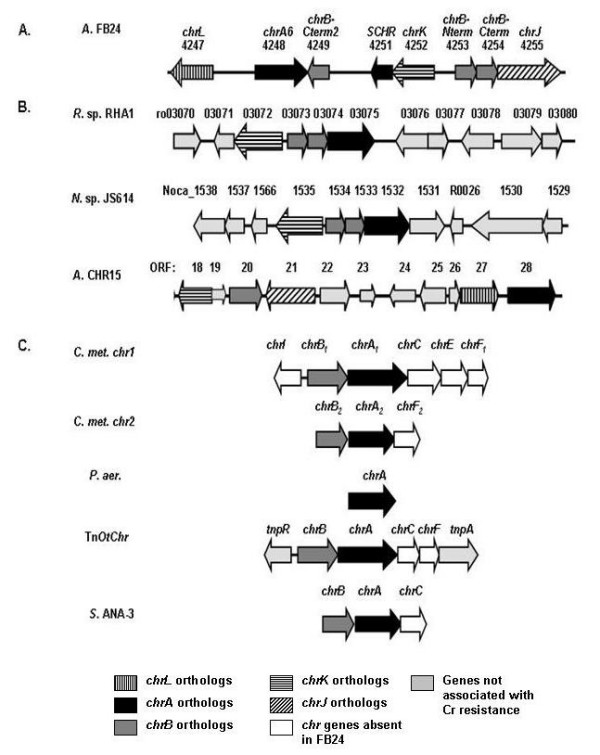
Comparison of genetic determinants of chromate resistance as studied in other bacterial strains versus Arthrobacter sp. strain FB24. R. sp. RHA1, Rhodococcus sp. RHA1 [GenBank: NC_008268]; N. sp. JS614, Nocardiodes sp. JS614 [GenBank: NC_008699]; A. CHR15, Arthrobacter sp. CHR15 plasmid pCHR15 [6,35]; C. met. chr1 and chr2, C. metallidurans chromate resistance determinants 1 (plasmid pMOL28) and 2 (chromosomal) [21]; P. aer., Pseudomonas aeruginosa plasmid pUM505 [20]; TnOtChr, transposable element from Ochrobactrum tritici 5bv11 [58]; S. ANA-3, Shewanella sp. strain chrBAC operon, plasmid 1 [GenBank: CP000470]. Drawing not to scale.
The chromate resistance determinant in Arthrobacter sp. strain FB24 has a similar genetic arrangement to that found in chromate-resistant Arthrobacter sp. CHR15, but is markedly different than in the two well-studied Proteobacteria, P. aeruginosa and C. metallidurans (Figure 2). More recently, a transposable element conferring chromate resistance in Ochrobactrum tritic was found to have a similar genetic makeup to the chr1 determinant in C. metallidurans [17], while a chromate resistance operon containing chrA, chrB and chrC was found in Shewanella sp. strain ANA-3 [16]. Additional genes involved in chromate resistance in C. metallidurans, such as the superoxide dismutase gene chrC, chrI and rpoH [21] are not present within the CRD of strain FB24. This could point to functional and regulatory differences in chromate resistance between these distantly related taxa. Thus, we were led to investigate Arth_4247, 4252 and 4255, as well as previously characterized chrA and chrB sequences. Due to the potential involvement of Arth_4247, 4252 and 4255 in chromate resistance, we have named these genes chrL, chrK and chrJ, respectively (Figures 2 and 3).
Figure 3.
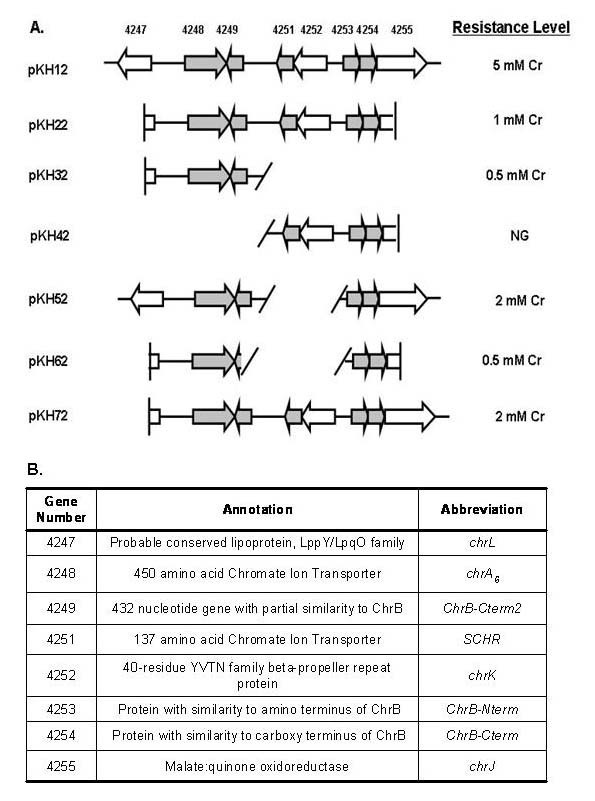
Schematic of constructs used in complementation experiments with strain D11. Panel A: 10.6 kb region of FB24 plasmid 3. Numbers correspond to the following genes: Arth_4255 (chrJ), Arth_4254 (ChrB-Cterm), Arth_4253 (ChrB-Nterm), Arth_4252 (chrK), Arth_4251 (SCHR), Arth_4249 (ChrB-Cterm2), Arth_4248 (ChrA6), Arth_4247 (chrL). Genes present in each of the constructs and chromate resistance levels on 0.1X NA plates. NG = No growth. D11 transformed with vector pART2 only did not grow on Cr. Panel B: Designated gene names and corresponding gene numbers used within text.
Sequence analysis of the CRD
Arth_4248, the putative 450 amino acid (aa) chromate ion transporter, is most similar to ChrA from Rhodococcus sp. RHA1 (79%). The protein is predicted to have 12 transmembrane helices and two CHR domains defined by a conserved GGX12VX4WX16PGPX10GX7G motif, placing it within the LCHR family of chromate ion transporters [22,24]. However, there is little sequence similarity (35% similarity across 106 of 225 amino acids) between the amino and carboxy halves of Arth_4248; hence, it does not appear to have arisen by direct tandem duplication of the same CHR domain-containing open reading frame. Because of the topological diversity of the CHR superfamily proteins [22,25] and the observed preponderance of conserved residues in the N-terminal half of the P. aeruginosa ChrA protein [26], it is expected that the amino and carboxy termini may carry out different functional roles in chromate efflux. Alignment of the amino acid sequence of Arth_4248 with that of P. aeruginosa ChrA indicated that residues which resulted in Cr(VI) sensitivity following mutation in P. aeruginosa [26] are also conserved in Arth_4248.
The other chrA ortholog Arth_4251 is predicted to be 137-aa protein with sequence similarity to known ChrA transporters. The protein sequence aligns to the N-terminus of C. metallidurans ChrA1 with 71% similarity across 49 amino acids. In comparison, Arth_4251 is only 52% similar to the N-terminus of Arth_4248 across 44 amino acids. The CHR domain in Arth_4251 contains the GGX12VX4W motif, but lacks the PGPX10GX7G motif. In addition, no definitive transmembrane helices were predicted for Arth_4251. Small (<200 aa) proteins containing a single CHR domain have been recognized as a separate group of proteins within the CHR family. Recently, Bacillus subtilis SCHR orthologs ywrA and ywrB were shown to confer chromate resistance in E. coli; however, both genes were required for the resistant phenotype [27]. Genes encoding SCHR proteins are usually present as pairs within a genome [22]. In FB24, though, there does not appear to be a partner SCHR gene for Arth_4251 and the aa sequence is more closely related to CHR domains from LCHR proteins than to those of the SCHR family [23].
Three open reading frames (ORFs) designated Arth_4253, Arth_4254 and Arth_4249 in the putative CRD region share sequence similarity to the C. metallidurans ChrB proteins. Arth_4253, which encodes a 171 aa protein, aligns with the N-terminal portion of the C. metallidurans ChrB1 protein [GenPept: YP_582012] (42% similarity across 133 aa). Arth_4254 is a predicted 143 aa protein that exhibits 53% similarity across 132 aa of the C-terminal portion of the C. metallidurans ChrB1 protein. Together, Arth_4253 and Arth_4254 appear to encode the complete sequence for a full-length ChrB gene, but the gene sequences overlap by 4 nucleotides and a potential Shine-Dalgarno sequence is present upstream of the predicted start codon of Arth_4254. Repeated sequencing of this region did not reveal any potential sequencing errors that could explain this observation. RT-PCR analysis revealed that Arth_4253 and Arth_4254 can form a dicistronic mRNA (operon structure analysis provided in Additional file 3). Arth_4249 contains 430 nucleotides, but does not yield any hits to known genes at the nucleotide level. A BLASTx search of the translated nucleotide sequence versus the protein database shows that the predicted amino acid sequence is 76% similar to Arth_4254 across 77 aa.
Arth_4252 encodes a 344 aa protein containing a 40-residue YVTN family beta-propeller repeat and a WD40 repeat domain (with 81% sequence similarity to ORF18 in Arthrobacter sp. strain CHR15) with an N-terminal signal sequence. The function of Arth_4252 is presently unknown, but other proteins within the WD40 repeat domain family are associated with the regulation of signal transduction and sensing membrane stress [28,29]. Arth_4252 also shares 62% sequence similarity to Rmet_6194, which is located approximately 4 kb downstream of the C. metallidurans chrA1 gene, Rmet_6202. However, a functional role for Rmet_6194 in chromate resistance in this organism has not been established. Orthologs of Arth_4252 were also found in close proximity to chrA genes in Arthrobacter sp. strain CHR15 and several species of Burkholderia as revealed by a gene ortholog neighborhood search in the Integrated Microbial Genomes database http://img.jgi.doe.gov.
Arth_4247 has an expected protein sequence of 337 aa with a putative overlapping signal sequence and transmembrane helix at the N-terminus, which suggests that it is a membrane-anchored protein. The protein sequence shares 75% aa similarity with lipoproteins of the LppY/LpqO family, which were first described in Mycobacterium tuberculosis but have not been functionally characterized. Other mycobacterial lipoproteins have been shown to perform such diverse roles as binding solutes in ABC transporter complexes, sensing environmental stressors and participating in signal transduction mechanisms [30]. M. tuberculosis, like strain FB24, is a high GC% Gram positive bacterium of the order Actinomycetales. The role of lipoproteins in the response to Cr(VI) has not been established in other organisms. Other lipoproteins have been shown to participate in the response to divalent metals such as copper and lead [31,32]. In the case of copper, NlpE stimulated the CpxAR envelope stress response pathway in copper-exposed E. coli cells [32]; however, it is not known if an analogous function exists for LppY/LpqO proteins. As in the case for Arth_4252, orthologs of Arth_4247 are also present near chrA orthologs in Arthrobacter sp. strain CHR15 (81% similarity to ORF 27) and C. metallidurans (52% similarity to Rmet_6195).
Arth_4255 encodes a putative malate:quinone oxidoreductase of 517 aa with 77% similarity to Arthrobacter aurescens TC1 Mqo. This class of proteins generally functions in energy production, but the biochemical role of Arth_4255 in the context of Cr(VI) resistance is not known. In Agrobacterium tumefaciens, insertional inactivation of an operon specifying NADH:quinone oxidoreductases similar to malate:quinone oxidoreductases (MrpA, MrpC and MrpD) resulted in the loss of arsenite oxidation. The phenotype was recovered via complementation with the intact Mrp operon [33]. In other bacteria, NADH-dependent oxidoreductases have been shown to reduce Cr(VI) [34]; however, there is no conclusive evidence of Cr(VI) reduction in FB24, and it is unlikely that Arth_4255 is a Cr(VI) reductase.
Loss of plasmid DNA from strain FB24 results in metal sensitivity and increased intracellular chromium accumulation
A chromate-sensitive mutant (D11) was obtained after successive culturing of FB24 for 90 generations in the absence of chromate. Loss of plasmid DNA was assessed by Southern hybridization using a 10.6-kb probe for the CRD, and the results were validated by a PCR screen using gene-specific primers (data not shown). Strain D11 was hypersensitive to low levels (0.5 mM), whereas the wild type grew prolifically on 0.1X nutrient agar (NA) plates amended with 5 mM chromate. Strain D11 was also very sensitive to lead, zinc and cadmium. Jerke et al (2008) had shown that FB24 contained 3 plasmids, each with genes that confer resistance to lead, zinc and cadmium [35]. Whereas FB24 attained maximal cell densities in 200 μM lead, zinc and cadmium in mXBM, growth of strain D11 was strongly inhibited by 10 μM lead, 50 μM zinc and 1 μM cadmium (data not shown).
Total intracellular chromium content was measured in chromate-exposed cells of FB24 and D11 to determine if the loss of chromate resistance in strain D11 correlated with increased intracellular accumulation of chromium. There was a significant difference (p = 0.015) in chromium content between strain D11 (2.8 × 10-7 mol mg protein-1) and FB24 (9.2 × 10-8 mol mg protein-1). Chromium was undetectable in FB24 and D11 cells that were not exposed to chromate. Similar decreases in chromium accumulation were found between chromate-resistant and -sensitive strains of P. aeruginosa and C. metallidurans which contain ChrA efflux pumps [15,36]. The comparable change in chromium accumulation between resistant and sensitive strains of Arthrobacter sp. FB24 and the two organisms in which the operation of an efflux pump has been biochemically demonstrated supports the hypothesis that plasmid-encoded Cr(VI) resistance in strain FB24 results from the function of a ChrA-like efflux pump.
Complementation of strain D11 with CRD
To localize the essential determinants for chromate resistance within the CRD, a series of plasmids were designed and tested for their capacity to confer chromate resistance in the chromate-sensitive strain D11 (Figure 3). Only D11 transformed with pKH12 (the complete 10.6 kb region) was able to grow comparably to FB24 on 0.1X (NA) plates containing 5 mM chromate (Figure 3). The other transformants, in which regions of the CRD were deleted, were able to grow only at lower levels of chromate (0.5 to 2 mM). In particular, chrA produced a resistance level of 0.5 mM Cr(VI) regardless of the presence of chrB-Nterm and chrB-Cterm.
Expression of chromate resistance genes in strain FB24 under chromate stress
Quantitative RT-PCR was employed to determine if expression of the chromate resistance genes was inducible by and specific to Cr(VI). Transcription from each of the eight genes of the CRD was induced by increasing concentrations of chromate (Table 1). Five μM chromate was sufficient to detect enhanced expression of each gene. For most genes in the CRD, maximal expression was achieved at 0.1 mM Cr(VI). In the case of chrB-Nterm, Arth_4253, maximum transcript abundance occurred at 5 μM chromate and was maintained up to 20 mM Cr(VI). ChrB-Cterm2, Arth_4249, exhibited low (2-fold) induction at 5, 25 and 50 μM Cr, followed by a sharp increase in transcript levels at 0.1 mM Cr(VI). Specificity of induction of the CRD genes was assessed with lead, arsenate and hydrogen peroxide, all of which induced little or no expression (Table 2).
Table 1.
Expression of CRD genes under various levels of chromate stressa.
| CRD Gene | Basal Expression In 0 mM Cr(VI)b × 102 |
Relative Fold Differencec Cr(VI)/0 mM Cr(VI) |
||||||
|---|---|---|---|---|---|---|---|---|
| 0.005 | 0.025 | 0.05 | 0.1 | 5 | 20 | 100 | ||
| chrL | 4.20 (0.45) |
36.7* (9.3) |
95.2 (8.7) |
69.8 (12.1) |
95.1 (42.9) |
63.4 (29.7) |
45.1* (14.3) |
15.3* (3.5) |
| chrA6 | 2.25 (0.36) |
8.5* (1.3) |
16.2* (3.9) |
27.4* (2.5) |
42.1 (4.2) |
50.7 (14.5) |
37.6 (9.8) |
22.9 (8.2) |
| chrB-Cterm2 | 15.6 (4.95) |
2.0* (0.3) |
2.2* (0.5) |
2.5* (0.5) |
7.1 (2.6) |
6.3 (1.8) |
8.0 (3.2) |
2.0* (0.8) |
| SCHR | 8.50 (2.06) |
1.9* (0.5) |
4.7* (0.6) |
5.1* (0.7) |
7.8 (0.7) |
6.8 (1.9) |
5.1 (1.2) |
2.1* (0.9) |
| chrK | 21.9 (2.89) |
3.7* (0.5) |
6.1* (0.7) |
7.5 (1.9) |
10.1 (1.9) |
7.2 (1.6) |
6.9 (1.6) |
4.4 (1.4) |
| chrB-Nterm | 249 (86.4) |
8.0 (2.6) |
12.5 (4.0) |
13.8 (5.6) |
18.0 (8.0) |
16.9 (7.1) |
14.0 (6.5) |
4.2 (1.5) |
| chrB-Cterm | 0.51 (0.04) |
4.3* (0.7) |
8.4* (2.1) |
16.0* (1.5) |
21.3 (2.0) |
25.4 (4.4) |
30.9 (6.0) |
15.3 (5.5) |
| chrJ | 1.23 (0.40) |
7.2* (1.5) |
14.3* (2.8) |
19.0* (2.5) |
37.0 (15.0) |
92.4 (47.2) |
47.6 (13.2) |
19.2 (6.7) |
a The basal (0 mM Cr(VI)) transcript levels are given in copy number/ng total RNA. For the remaining concentrations of Cr(VI), the average fold difference relative to the 0 mM Cr(VI) basal state is given. Bold values indicate the concentration at which maximum expression was observed. *, significantly different than the maximum expression level.
b Mean transcript copy number/ng total RNA. SE is given in parentheses (n = 6).
c Mean Fold Difference calculated by dividing the average transcript copy number in each Cr(VI) condition by the average transcript copy number in 0 mM Cr(VI). SE is given in parentheses (n = 6).
Table 2.
Specificity of Induction of Chromate Resistance Genesa.
| Gene | Cr(VI) 5 mM |
Lead 5 μMb |
Arsenate 5 mMc |
H2O2 5 mMc |
|---|---|---|---|---|
| chrL |
63.4 (29.7) |
0.3 (0.02) |
0.6 (0.05) |
12.5 (3.50) |
| chrA6 |
50.7 (14.5) |
0.2 (0.02) |
0.8 (0.15) |
3.2 (0.87) |
| chrB-Cterm2 |
6.3 (1.9) |
0.1 (0.01) |
0.3 (0.03) |
0.1 (0.01) |
| SCHR |
6.8 (1.9) |
0.1 (0.01) |
0.3 (0.03) |
0.9 (0.12) |
| chrK |
7.2 (1.6) |
0.1 (0.01) |
0.2 (0.04) |
1.0 (0.21) |
| chrB-Nterm |
16.9 (7.1) |
0.1 (0.01) |
0.4 (0.08) |
0.5 (0.12) |
| chrB-Cterm |
25.4 (4.4) |
2.6 (0.12) |
5.3 (0.97) |
4.9 (0.70) |
| chrJ |
92.4 (47.2) |
0.7 (0.05) |
1.7 (0.10) |
6.6 (0.58) |
a Values shown for lead, arsenate and H2O2 represent the transcript copy number ng-1 total RNA in each experimental condition relative to transcript levels in 0.2X NB and the SE (parentheses, n = 6 qRT-PCR reactions per treatment). The relative expression of each gene in 5 mM Cr(VI) is shown for comparison.
b 0.5 and 50 μM lead also tested with similar results
c 0.5 and 50 mM Arsenate and H2O2 also tested with similar results
Potential regulatory element within the CRD
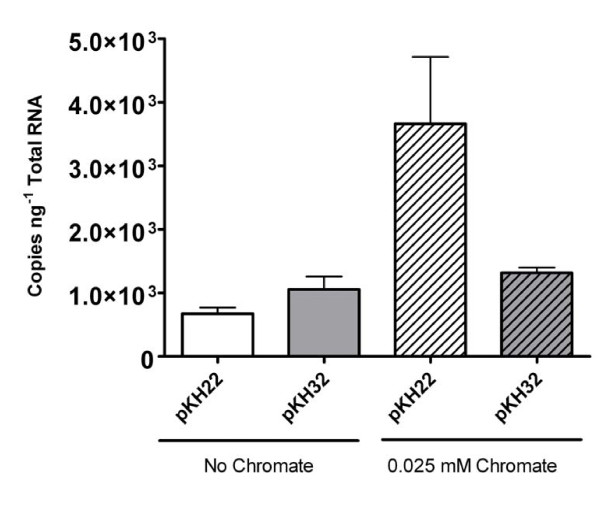
ChrB has been proposed to function as an activator of the chromate resistance determinants in C. metallidurans [21]. A bioinformatics analysis using protein function prediction software [37] suggested possible DNA-binding and kinase activities for ChrB-Cterm and ChrB-Nterm, respectively. In addition, proteins containing WD40 repeats, such as Arth_4252, have been associated with signal transduction and regulatory mechanisms [29,38]. To determine if chrK, chrB-Nterm and chrB-Cterm influence expression of chrA, strain D11 bearing plasmids pKH22 and pKH32 was grown in the presence and absence of chromate, and qRT-PCR was used to quantify chrA expression under these conditions. Expression of chrA was induced to higher levels by chromate in strain D11 bearing pKH22 than when the putative regulatory genes were absent (pKH32) (Figure 4). This difference is not likely to be attributable to differences in plasmid copy number provided that chrA expression in both strains without chromate was similar.
Figure 4.
Induction of chrA in D11 transformed with pKH22, pKH32. Error bars show the standard error (n = 6 qRT-PCR reactions per treatment)
Discussion
We have described a cluster of eight genes that confers chromate resistance in Arthrobacter sp. strain FB24 and appears to specifically respond to chromate. In other organisms, proteomic and genomic analyses revealed that chromate induces a variety of generalized stress-responsive systems, including those involved in the SOS response, DNA repair and protection against oxidative stress [39,40]. However, evidence suggests that induction of the FB24 CRD genes does not represent a general stress response. The concentrations of chromate (5 μM) that induced elevated levels of transcripts from CRD genes are 1000-fold lower than those that affect the growth rate of FB24, and the CRD genes were specifically induced in a dose-dependent manner in response to chromate exposure. Other classes of stressors (lead, arsenate or hydrogen peroxide) resulted in little or no induction of CRD genes. Furthermore, whereas other metal efflux systems, such as those in the cation diffusion facilitator (CDF) family, exhibit broad metal specificity [41,42], the lack of induction of the CRD genes by lead and arsenate supports the contention that this is a chromate-specific system.
Expression of the CRD in response to chromate was also verified at the proteomic level using tandem liquid chromatography-mass spectrometry [43]. In a global proteomic study, ORF-specific peptides were confirmed for all genes, with the exception of Arth_4249 and Arth_4250. Note that protein products were detected for the truncated genes of ChrA and ChrB (Arth_4253, 4254 and 4251). This is the first report that a SCHR gene product is synthesized in response to chromate. Although its exact function requires further experimentation, chromate-specific increases in transcript and protein abundance levels of Arth_4251 indicate that this gene, and perhaps its orthologs, plays a significant role in chromate resistance, as was seen recently with the ywrA and ywrB SCHR genes in B. subtilis [27]. It is important to note that SCHR in FB24 has greater sequence similarity to LCHR sequences than other SCHR sequences possibly explaining its maintenance of a chromate response. Arth_4251 may be an integral link to elucidate the evolution of chromate resistance mechanisms. It may represent a remnant precursor to the evolution of LCHR from gene duplication or the next step in evolution essential for the high chromate-resistance phenotype.
Our investigation of Arthrobacter sp. strain FB24 further suggests roles for three new genes (chrJ, chrK and chrL) in addition to catalytic and regulatory proteins found in those Proteobacteria and may help to explain the variability in chromate resistance levels across bacterial species. Whereas genetic studies in Proteobacteria [14,17,20,21] have pointed to the primacy of the chrA gene in conferring Cr(VI) resistance, the introduction of chrA alone into Cr(VI) sensitive strain D11 produced resistance levels that were only one-tenth of those found when the entire CRD was introduced. As of late, the chrA gene has only been intensively studied in two Proteobacteria, P. aeruginosa and C. metallidurans, and thus far, these systems have been the paradigm for understanding bacterial chromium resistance [13,23,44]. Recent studies with chrA orthologs from two additional Proteobacteria, Shewanella sp. strain ANA-3 [16] and Ochrobactrum tritici 5bvl1 [17], have also demonstrated that chrA and neighboring genes (Figure 2) confer resistance in Cr(VI)-sensitive strains. Aguilar-Barajas et al [16] were able to recover Cr(VI)-resistance in Cr(VI)-sensitive E. coli and P. aeruginosa strains by expressing the chr operon from Shewanella sp. strain ANA-3 on a low-copy plasmid. Similar to what was shown for Arthrobacter FB24, though, expression of chrA alone resulted in lower resistance levels in E. coli than strains bearing the entire ANA-3 chrBAC operon. The ANA-3 chrA gene conferred chromate resistance in P. aeruginosa, and this phenotype was enhanced by the presence of the host chrR regulatory gene [16], thus emphasizing the importance of accessory genes in achieving higher levels of chromate resistance.
In the case of Ochrobactrum, Cr(VI)-sensitive strains transformed with a plasmid carrying the chrA and chrB genes from TnOtChr showed similar growth in chromate as the wild-type O. tritici strain. However, no additional growth advantage was provided by the presence of chrC and chrF [17]. In C. metallidurans, deletion of chrC resulted in a slight decrease in chromate resistance compared to the wild-type strain (0.3 mM chromate minimal inhibitory concentration versus 0.35 mM, respectively). In the same study, deletion of chrF2 did not affect chromate resistance levels [21]. In these organisms, it appears that chrB makes a significant contribution to chromate resistance, but the exact contributions made by chrC and chrF are not so apparent and may vary depending on the host strain. This is in stark contrast to the chrJ, chrK and chrL accessory genes in strain FB24, whose deletion results in a noticeable decrease in chromate resistance. A conclusion that can be drawn from these observations is that, although chromate efflux appears to be the overarching mode for resistance, the intricacies of the exact biochemical and regulatory mechanisms controlling efflux differ among bacterial strains, and these differences await full characterization.
Since most work regarding chromate efflux has been done in Proteobacteria, we were interested in whether CRD orthologs were present in strains more closely related to Arthrobacter sp. strain FB24. In searching for organisms with gene neighborhoods similar to the Arthrobacter FB24 CRD, it was discovered that other actinomycetes share a similar genetic makeup (Figure 2). Rhodococcus sp. RHA1 and Nocardiodes sp. JS614 both contain chrK, chrB-Nterm and chrB-Cterm orthologs in the near vicinity of chrA, while the chromate-resistant Arthrobacter sp. CHR15 harbors chrJ, chrK and chrL orthologs near chrA and chrB. The chromate resistance status of Nocardiodes sp. JS614 and Rhodococcus sp. RHA1 is not known; however, both species are known PCB degraders and are considered important environmental Actinobacteria [45-47]. The distinct genomic context between Proteobacteria and Actinobacteria suggests that functional and regulatory differences in efflux-mediated chromate resistance likely exist in distantly related taxa. This demands genetic and biochemical studies in a greater diversity of organisms in order to fully understand the breadth of physiological strategies that have evolved to confer chromium resistance.
Conclusion
This study increases our knowledge of the genetics of chromate resistance by identifying three novel genes that play a significant role in chromate resistance in Arthrobacter sp. strain FB24: chrJ, chrK, and chrL. Future work should focus on elucidating the exact physiological function of these genes. However, our research is an important first step in characterizing potential regulatory networks controlling efflux-mediated chromate resistance. We further illustrate the value of examining the genomic context of already characterized metal resistance genes in identifying new players in metal resistance mechanisms.
Methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 3. Arthrobacter strains were cultured in 0.1X or 0.2X nutrient broth (NB) [Difco, Sparks, MD], Luria-Bertani (LB) medium pH 7.0, or modified Xenobiotic Basal Medium (mXBM). Modified XBM contained 10 mM glycerol phosphate, 10 mM KNO3, 6.0 mM NH4NO3, 0.01 mM CaCl2, 2 ml L-1 of EDTA Fe Citrate Solution [7.4 mM FeCl3, 11.4 mM Na2EDTA, 12.8 mM sodium citrate (C6H5O7Na3), 100 mM MgSO4, 5% NH4Cl2, 0.05 M CaCl2, 1.0 M NaCl, 1 M NaHCO3], 10 ml L-1 of vitamin solution (see Jerke [48] and Additional file 4 for components), 1 ml L-1 SL-7 trace elements [49], with glucose (1.7 mM) as a carbon and energy source.
Table 3.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Reference |
|---|---|---|
| Arthrobacter | ||
| FB24 | CrR | [6] |
| D11 | CrS derivative of FB24 | This work |
| E. coli | ||
| JM110 | dam-dcm- | Stratagene |
| Plasmids | ||
| pAOWA10128 | 7.3 kb insert in pMCL200 obtained from DOE-JGI. Contains Arth_4248-Arth_4254. | DOE-JGI |
| pBluescript II SK+ | 3.0 kb, ApR, lacZ, used for sublconing inserts prior to ligation into pART2. | Promega |
| pART2 | 4.6 kb, KmR, pCG100 ori, ColE1 ori, vector for expression in Arthrobacter | [55] |
| pKH11 | 10.6 kb PCR product from FB24 plasmid 3 (CP000457) containing Arth_4247-4255 in pBluescript II SK+ | This worka |
| pKH12 | Insert from pKH11 cloned into pART2 | This work |
| pKH21 | 7.3 kb insert from pAOWA10128 in pBluescript II SK+ | This work |
| pKH22 | Insert from pKH21 cloned into pART2 | This work |
| pKH32 | 3.7 kb EcoRI-KpnI fragment from pKH21 cloned into pART2. Contains Arth_4248-4249. | This work |
| pKH42 | 3.8 kb XhoI-BglII fragment from pKH21 cloned into pART2. Contains Arth_4251-Arth_4254. | This work |
| pKH52 | 8.3 kb insert from MluI-BglII digest of pKH11 to delete Arth_4252 and Arth_4252 cloned into pART2 | This work |
| pKH62 | pKH22 digested with SfiI to delete Arth_4249-Arth_4252. | This work |
| pKH72 | pKH12 digested with ScaI and XbaI to delete Arth_4247. | This work |
aA schematic of each construct is presented in Figure 3.
Induction of Cr(VI) resistance genes was assessed in Arthrobacter sp. strain FB24 cells by culturing in 150 ml NB to early mid-log phase (OD600, 0.3) at 30°C with shaking at 200 rpm. Cells were harvested by centrifugation, washed once with 0.2X NB and suspended in 15 ml 0.2X NB. Chromate (K2CrO4) was added to final concentrations of 0, 0.005, 0.025, 0.05, 0.1, 5, 20 or 100 mM. To test for specificity of induction, additional cultures were incubated in the presence of 0, 0.5, 5 and 50 μM PbNO3 in mXBM; 0, 0.5, 5 and 50 mM Na2HAsO4·7 H2O in 0.2X NB; and 0, 0.5, 5, 50 mM hydrogen peroxide (H2O2) in 0.2X NB. Cells were incubated for 2.5 hours at 30°C with agitation. Induction experiments with Cr(VI)-sensitive strain D11 transformed with pKH22, pKH23 and pKH24 were carried out in the same manner with the following exceptions: kanamycin was added to a concentration of 30 μg ml-1 and chromate was added to one culture at a concentration of 0.025 mM.
Generation of chromate-sensitive FB24 derivative
The lead- and chromate-sensitive mutant, D11, was generated from the resistant wild-type strain FB24 by growing cells in LB without chromate. Cultures were transferred daily by diluting cells 1:1000 into fresh media. Transfers were maintained for approximately 90 generations at 30°C with shaking at 200 rpm and then screened for cells sensitive to 75 μM lead on mXBM agar plates. Lead-sensitive colonies were then tested for Cr(VI) sensitivity on 0.1X nutrient agar (NA) plates supplemented with 0.5, 1, 2 and 5 mM K2CrO4. Loss of plasmid DNA in strain D11 was assessed by Southern hybridization and rep-PCR. Loss of the CRD genes was confirmed by PCR using gene-specific primers.
Total genomic DNA was extracted from cultures grown overnight in NB with appropriate selection. Cells were harvested by centrifugation, suspended in TE buffer, and treated with lysozyme (1 mg ml-1) for one hour followed by treatment with proteinase K (10 mg ml-1). Cells were lysed using a FastPrep instrument (Qbiogene, Carlsbad, CA) at a setting of 4 for 30 s with 0.64 cm ceramic beads. Genomic DNA was purified by phenol: chloroform: isoamyl alcohol extraction and precipitated with isopropanol [50]. DNA was digested with restriction enzymes (SacI and XcmI) and separated on a 0.7% agarose gel and transferred to Hybond-N+ membrane (Amersham Pharmacia, Pisscataway, NJ) using a Trans-blot semi dry transfer cell (Bio-Rad, Hercules, CA) following the manufacturer's recommendations for voltage and transfer time. A digoxigenin-labeled probe targeting the 10.6-kb CRD on Arthrobacter sp. strain FB24 pFB24-104 [GenBank: NC_008539] was generated by PCR with primers C42/F and C42/R (Table 4) using the TripleMaster PCR system (Eppendorf North America, Inc., Westbury, NY) according to the manufacturer's reaction mixture and cycling specifications for long-range PCR. Hybridization and chromogenic detection was carried out under high stringency conditions as described in the DIG Application Manual for Filter Hybridization (Roche Applied Science, Indianapolis, IN).
Table 4.
PCR and qRT-PCR primers used in this study.
| Primer | Sequence (5'→3') | Description |
|---|---|---|
| C42/F | CCCAAAGCTTGGGTCCTGCTCATCACCAGAAACTCCA | HindIII site, used to construct pKH11 |
| CF2/R | GCTCTAGAGCAACCGCTTTCAGGCACTGTTGTTC | XbaI site, used to construct pKH11 |
| Primers for RT-PCR--asterisk denotes primer for cDNA synthesis | ||
| Primer | Sequence (5'→3') | Gene |
| MQO RT/A* | AGGCCTGCCCGTAGACTTTC | Arth_4255 |
| MQO RT/B | TCTTCACCGCCGGTATGAG | Arth_4255 |
| ChrB RT/A* | CGAGGATGAGGGATCGTTTG | Arth_4254 |
| ChrB RT/B | TGATCCGCAGGAACATCG | Arth_4254 |
| SP RT/F | CCCGGGAGCACTTCGACTGGA | Arth_4253 |
| SP RT/R* | CCTGGCGCGTTCGGTTGCAT | Arth_4253 |
| Cog4RT/F | AAGGCCTACGTCTCCAACGAACA | Arth_4252 |
| Cog4RT/R* | ATTCTGTCGGTGACCGTGTCAGT | Arth_4252 |
| ChrAP RT/A | GCTTCATCCTGTGCTTCTTG | Arth_4251 |
| ChrAP RT/B* | TGTTCATGATGCCGGTACTG | Arth_4251 |
| BP RT/F | CCTGCGCCGCTACGAACTCACCGAT | Arth_4249 |
| BP RT/R* | GCGCTGGTGTTCGTACAGCCCGTC' | Arth_4249 |
| ChrAC RT/A* | AAGTACAGGGCCAGGTTC | Arth_4248 |
| ChrAC RT/B | TGCGGTCCTGTCCTATATC' | Arth_4248 |
| Lppy RT/F | AGTGACCACGGCCATCAATTTCCA | Arth_4247 |
| Lppy RT/R* | TCAGGGAATGATTGTGCACGGAGA | Arth_4247 |
Chromate resistance determination
Starter cultures were grown overnight at 30°C in 0.2X NB with appropriate selection. Cultures for minimal inhibitory concentration (MIC) determination were diluted 1:1000 in 3 ml of 0.1X NB for chromate cultures or mXBM plus glucose for divalent cationic metals in borosilicate glass tubes and maintained at 30°C with shaking at 200 rpm. The OD600 was measured daily for a period of 3 days until growth stabilized. Divalent cationic metals used in MIC experiments were added as lead nitrate (Pb(NO3)2, zinc chloride (ZnCl2), or cadmium sulfate (CdSO4) at concentrations ranging from 0 to 200 μM. Cultures were prepared in triplicate for each growth or MIC experiment. D11 transformants were screened for chromate resistance by streaking single colonies onto 0.1X nutrient agar plates containing 0, 0.5, 1, 2, or 5 mM chromate.
Sequence analysis of putative chromate efflux gene
The genome sequence is available in the GenBank database under accession numbers NC_008537 to NC_008539 and NC_008541. The genome was sequenced by the Department of Energy Joint Genome Initiative (DOE-JGI) and can be accessed at http://genome.jgi-psf.org/finished_microbes/art_f/art_f.home.html. The annotated sequence at this site was used for locating the CRD and construction of primer sequences. Multiple sequence alignment of Arth_4248 (ChrA) with other described and putative members of the CHR family of chromate efflux proteins [24] was performed using the ClustalX program and default setting for Gonnet series for protein weight matrix [51] and bootstrap Neighbor Joining tree options with 1000 resamplings. Output trees were visualized in Fig Tree http://tree.bio.ed.ac.uk/software/figtree/. Sequences were retrieved from the UniProt database [52] by conducting a search for ChrA sequences according to Diaz-Perez et al [22]. Some additional eukaryotic sequences were found by conducting a similar search of the GenBank database [53]. All short ChrA (SCHR) sequences (<350 amino acids) were excluded from the alignment. A total of 513 sequences (Additional files 1 and 2) were retrieved and aligned. Transmembrane helices were predicted using the TMHMM 2.0 server [54].
Cloning of chromate resistance determinant
A series of constructs were created to test expression of chromate resistance in strain D11. All constructs, except for pKH62 and pKH72, were prepared by subcloning into pBluescript SK+ (Stragene, La Jolla, CA) prior to cloning into pART2 [55]. Recombinant plasmid DNA was transformed into strain D11 by electroporation as described elsewhere [56]. Ampicillin was used for selection at a concentration of 100 μg ml-1 for pBluescript-derived transformants, and kanamycin was used at a concentration of 40 μg ml-1 for pART2-derived transformants. Plasmids were submitted to the Purdue University Core Genomics Center for validation of insert sequences.
Plasmid pKH11 was generated by amplifying a 10.6 kb fragment bearing bases 72880 to 83464 of pFB24-104 using the TripleMaster PCR system (Eppendorf North America, Inc., Westbury, NY) according to the manufacturer's specifications and primers C42/F and C42/R. The PCR product was digested with HindIII and XbaI and ligated into pBluescript SK+ to give pKH11. Plasmid pKH21 contains a 7.3 kb insert bearing bases 74642 to 81771 from FB24-104; the insert was isolated by digesting pAOWA10128 (obtained from DOE-JGI) with XbaI and HindIII. The remaining constructs (Table 3) were generated by restriction digestion of either pKH11 or pKH21 using standard cloning procedures [50].
Expression analysis by quantitative reverse transcriptase PCR (qRT-PCR)
Primer sequences for qRT-PCR are listed in Table 4. Total RNA was extracted from Arthrobacter cell pellets using the FastRNA PRO Blue Kit (MP Biomedical, Solon, OH) and treated with Turbo DNA-Free DNAse (Ambion, Austin, TX) to remove contaminating DNA. RNA concentrations were quantified by measuring the A260 on a Smart Spec 3000 spectrophotometer (Bio-Rad, Hercules, CA). cDNA was synthesized from 100 ng total RNA using ImProm II reverse transcriptase (RT) (Promega, Madison, WI) following the manufacturer's reaction conditions. PCR was performed using the following conditions: 98°C for 5 min, followed by 30 cycles of 94°C for 30 s, 56-58°C (depending on the primer pair) for 30 s, 72°C for 1 min, with a final extension step at 72°C for 10 min.
For real-time PCR, 1 μl of the reverse transcription reaction mixtures prepared as described above was used as the template. The PCR mixture contained 1 U of HotMaster Taq (Eppendorf North America, Inc., Westbury, NY), 1× HotMaster Taq PCR buffer with 25 mM MgCl2, 1% bovine serum albumin, 0.2 mM each of dNTPs, 0.25 mM each of a forward and reverse primer, SYBR Green (1:30,000; Molecular Probes, Eugene, OR) and 10 nM FITC (Sigma, St. Louis, MO) in a final volume of 25 μl. Reactions were carried out using a Bio-Rad MyIQ single-color real time PCR detection system, and data were analyzed using the MyIQ Optical System software version 2.0. Transcript copy numbers were calculated from a standard curve using known concentrations of pKH11. Plots of transcript copy number ng-1 total RNA versus chromate concentration were prepared in GraphPad Prism 4 (GraphPad Software, San Diego, CA). The mean and standard errors were determined from 6 qRT-PCR reactions per chromate treatment (3 independent cultures × 2 reactions per culture). Significant differences among chromate treatments for each gene were determined by generating least square means in PROC GLIMMIX with the LS MEANS option in SAS version 9.1. Multiple comparisons were adjusted using Tukey's test. To normalize the variance of the model residuals, a negative binomial distribution was used for each set of gene expression data.
Chromium content in chromate-exposed cells
Arthrobacter strains FB24 and D11 were grown to mid-log phase (OD600, ~0.2) in 50 ml 0.2X NB at which time four replicate cultures were amended with 2 mM chromate (final concentration). One culture per strain was incubated without chromate. All cultures were incubated for an additional 2 h. Aliquots of 40 ml of cells were harvested by centrifugation and washed 4 times with ddH2O. Cell pellets were solubilized in concentrated nitric acid (cHNO3) and heated at 95°C for 2 h. Samples were adjusted to a final concentration of 2% HNO3 with double distilled water and analyzed for total chromium content at the Purdue University Mass Spectrometry Center. The 52Cr inductively coupled argon plasma mass spectrometry (ICPMS) results were obtained using an ELEMENT-2 (ThermoFinnigan, Bremen, Germany) mass spectrometer in the medium resolution mode. The samples were introduced into the plasma using an Aridus desolvating system with a T1H nebulizer (Cetac Technologies, Omaha NE), which is used to enhance sensitivity and reduce oxide and hydride interferences. The argon sweep gas and nitrogen of the Aridus is adjusted for maximum peak height and stability using 7Li, 115In and 238Upeaks obtained from a multi-element standard (1 ng/ml, Merck & Co.). Chromium concentration was normalized per mg protein. Total soluble cell protein concentration was determined using the Lowry method [57] after collecting cells by centrifugation and extracting protein with 1N NaOH at 100°C. Student's t-test was used to determine statistically significant differences in the average chromium content between strains D11 and FB24 at the 95% confidence level.
Authors' contributions
KH conceived and carried out the molecular genetic, gene expression and growth studies and performed the majority of manuscript writing. CN participated in study design and coordination, performed sequence analysis of the chromate efflux gene, alignment of chromate efflux amino acid sequences and generated the phylogenetic trees. DT participated in study design and coordination. AK participated in study design and coordination. All authors participated in drafting the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplemental Figure S1. Radial phylogenetic tree of LCHR proteins generated from an alignment of 513 putative ChrA, chromate ion transport sequences (see Supplemental Table S1) using ClustalX. Neighbor Joining tree graphically viewed using the FigTree program http://tree.bio.ed.ac.uk/software/figtree/. Branched tips labeled with protein accession number followed by species name. Scale bar indicates 0.06 amino acid substitutions per site. Branches colors are fungi-brown, algae-green, Archaea-red, Proteobacteria (alpha-pink, beta-magenta, delta-blue, gamma-purple), Cyanobacteria-torquoise, Firmicutes-yellow, Actinobacteria-red and all other Bacteria-black.
Supplemental Table S1. Sequence accession numbers, taxa name and sequence length of putative ChrA sequences used in phylogenetic analysis.
Supplemental Figure S2. Operon structure analysis of the Arthrobacter sp. strain FB24 CRD. RT-PCR was used to determine co-transcription of the genes within the chromate resistance determinant. A: Location of primer pairs. Primer sequences are listed in table 4. Primer numbers correspond to the following primers: 1-MQO RT/A, 2-BC RT/A, 3-SP RT/F, 4-SP RT/R, 5-COG4RT/F, 6-COG4RT/R, 7-ChrAP RT/A, 8-ChrAP RT/B, 9-BP RT/R. B: RT-PCR results with listed primer pairs. C: RT-PCR products of reactions performed with primer pair 2 + 4 (lanes 2 and 3) and primer pair 5 + 8 (lanes 8 and 9). Lanes 1 and 7-100 bp PCR ruler, dark band is 1 kb; Lanes 4 and 10-no template controls; Lanes 5 and 11-No RT controls; Lanes 6 and 12 positive PCR control using pKH12 as template.
Supplemental Table S2. Recipe for vitamin solution added to mXBM.
Contributor Information
Kristene L Henne, Email: khenne@purdue.edu.
Cindy H Nakatsu, Email: cnakatsu@purdue.edu.
Dorothea K Thompson, Email: dthomps4@utk.edu.
Allan E Konopka, Email: allan.konopka@pnl.gov.
Acknowledgements
This work was supported by a grant from the Department of Energy's Environmental Remediation Science Program (grant DE-FG02-98ER62681). K.H. received support from the Purdue Research Foundation and the Purdue Graduate School Bilsland Doctoral Fellowship. We would like to thank Karl Wood and Arlene Rothwell of the Purdue Mass Spectrometry Center for performing the ICP-MS analysis, Jillian Detweiler for assistance with statistical analyses and Gene Wickham, Kurt Jerke for phylogenetic and technical assistance and Militza Carrero-Colon for thoughtful discussion. Vector pART2 was a kind gift from Cristinel Sandu.
References
- Jones D, Keddie RM. In: The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community, release 3.0 edn. 3. Dworkin, et al, editor. New York: Springer-Verlag; 1999. The Genus Arthrobacter. [Google Scholar]
- Crocker FH, Fredrickson JK, White DC, Ringelberg DB, Balkwill DL. Phylogenetic and physiological diversity of Arthrobacter strains isolated from unconsolidated subsurface sediments. Microbiology. 2000;146(Pt 6):1295–1310. doi: 10.1099/00221287-146-6-1295. [DOI] [PubMed] [Google Scholar]
- van Waasbergen LG, Balkwill DL, Crocker FH, Bjornstad BN, Miller RV. Genetic diversity among Arthrobacter species collected across a heterogeneous series of terrestrial deep-subsurface sediments as determined on the basis of 16S rRNA and recA gene sequences. Appl Environ Microbiol. 2000;66(8):3454–3463. doi: 10.1128/AEM.66.8.3454-3463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyehuda G, Coombs J, Ward PL, Balkwill D, Barkay T. Metal resistance among aerobic chemoheterotrophic bacteria from the deep terrestrial subsurface. Canadian Journal of Microbiology. 2003;49(2):151–156. doi: 10.1139/w03-012. [DOI] [PubMed] [Google Scholar]
- Margesin R, Schinner F. Heavy metal resistant Arthrobacter sp.--a tool for studying conjugational plasmid transfer between gram-negative and gram-positive bacteria. J Basic Microbiol. 1997;37(3):217–227. doi: 10.1002/jobm.3620370312. [DOI] [PubMed] [Google Scholar]
- Beasley FC. Masters. West Lafayette: Purdue University; 2004. Characterization of Diversity, Chromate Resistance and Aromatic Hydrocarbon Degradation Among Arthrobacter Isolates from Mixed Waste Soil. [Google Scholar]
- Joynt JL. Bacterial community in a metal and organic contaminated soil. West Lafayette: Purdue University; 2000. [Google Scholar]
- Nakatsu CH, Carmosini N, Baldwin B, Beasley F, Kourtev P, Konopka A. Soil microbial community responses to additions of organic carbon substrates and heavy metals (Pb and Cr) Appl Environ Microbiol. 2005;71(12):7679–7689. doi: 10.1128/AEM.71.12.7679-7689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FA, Bento FM, Okeke BC, Frankenberger WT. Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J Environ Qual. 2003;32(4):1228–1233. doi: 10.2134/jeq2003.1228. [DOI] [PubMed] [Google Scholar]
- Megharaj M, Avudainayagam S, Naidu R. Toxicity of hexavalent chromium and its reduction by bacteria isolated from soil contaminated with tannery waste. Curr Microbiol. 2003;47(1):51–54. doi: 10.1007/s00284-002-3889-0. [DOI] [PubMed] [Google Scholar]
- Camargo FA, Bento FM, Okeke BC, Frankenberger WT. Hexavalent chromium reduction by an actinomycete, Arthrobacter crystallopoietes ES 32. Biol Trace Elem Res. 2004;97(2):183–194. doi: 10.1385/BTER:97:2:183. [DOI] [PubMed] [Google Scholar]
- Horton RN, Apel WA, Thompson VS, Sheridan PP. Low temperature reduction of hexavalent chromium by a microbial enrichment consortium and a novel strain of Arthrobacter aurescens. BMC Microbiol. 2006;6(1):5. doi: 10.1186/1471-2180-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes C, Silver S. Plasmid chromate resistance and chromate reduction. Plasmid. 1992;27(1):65–71. doi: 10.1016/0147-619X(92)90007-W. [DOI] [PubMed] [Google Scholar]
- Nies A, Nies DH, Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990;265(10):5648–5653. [PubMed] [Google Scholar]
- Pimentel BE, Moreno-Sanchez R, Cervantes C. Efflux of chromate by Pseudomonas aeruginosa cells expressing the ChrA protein. FEMS Microbiol Lett. 2002;212(2):249–254. doi: 10.1111/j.1574-6968.2002.tb11274.x. [DOI] [PubMed] [Google Scholar]
- Aguilar-Barajas E, Paluscio E, Cervantes C, Rensing C. Expression of chromate resistance genes from Shewanella sp. strain ANA-3 in Escherichia coli. FEMS Microbiol Lett. 2008;285(1):97–100. doi: 10.1111/j.1574-6968.2008.01220.x. [DOI] [PubMed] [Google Scholar]
- Branco R, Chung AP, Johnston T, Gurel V, Morais P, Zhitkovich A. The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium(VI) and superoxide. J Bacteriol. 2008;190(21):6996–7003. doi: 10.1128/JB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake H, Cervantes C, Silver S. Decreased chromate uptake in Pseudomonas fluorescens carrying a chromate resistance plasmid. J Bacteriol. 1987;169(8):3853–3856. doi: 10.1128/jb.169.8.3853-3856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes C, Ohtake H. Plasmid-determined resistance to chromate in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1988;56:173–176. doi: 10.1111/j.1574-6968.1988.tb03172.x. [DOI] [Google Scholar]
- Cervantes C, Ohtake H, Chu L, Misra TK, Silver S. Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J Bacteriol. 1990;172(1):287–291. doi: 10.1128/jb.172.1.287-291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhnke S, Peitzsch N, Hubener N, Grosse C, Nies DH. New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch Microbiol. 2002;179(1):15–25. doi: 10.1007/s00203-002-0492-5. [DOI] [PubMed] [Google Scholar]
- Diaz-Perez C, Cervantes C, Campos-Garcia J, Julian-Sanchez A, Riveros-Rosas H. Phylogenetic analysis of the chromate ion transporter (CHR) superfamily. Febs J. 2007;274(23):6215–6227. doi: 10.1111/j.1742-4658.2007.06141.x. [DOI] [PubMed] [Google Scholar]
- Ramirez-Diaz MI, Diaz-Perez C, Vargas E, Riveros-Rosas H, Campos-Garcia J, Cervantes C. Mechanisms of bacterial resistance to chromium compounds. Biometals. 2007;21(3):321–332. doi: 10.1007/s10534-007-9121-8. [DOI] [PubMed] [Google Scholar]
- Nies DH, Koch S, Wachi S, Peitzsch N, Saier MH Jr. CHR, a novel family of prokaryotic proton motive force-driven transporters probably containing chromate/sulfate antiporters. J Bacteriol. 1998;180(21):5799–5802. doi: 10.1128/jb.180.21.5799-5802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mejia R, Campos-Garcia J, Cervantes C. Membrane topology of the chromate transporter ChrA of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2006;262(2):178–184. doi: 10.1111/j.1574-6968.2006.00386.x. [DOI] [PubMed] [Google Scholar]
- Aguilera S, Aguilar ME, Chavez MP, Lopez-Meza JE, Pedraza-Reyes M, Campos-Garcia J, Cervantes C. Essential residues in the chromate transporter ChrA of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2004;232(1):107–112. doi: 10.1016/S0378-1097(04)00068-0. [DOI] [PubMed] [Google Scholar]
- Diaz-Magana A, Aguilar-Barajas E, Moreno-Sanchez R, Ramirez-Diaz MI, Riveros-Rosas H, Vargas E, Cervantes C. Short-chain CHR (SCHR) Proteins from Bacillus subtilis Confer Chromate Resistance in Escherichia coli. J Bacteriol. 2009;191(171):5441–5445. doi: 10.1128/JB.00625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24(5):181–185. doi: 10.1016/S0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Gonzalez L, Phalip V. Survey, analysis and genetic organization of genes encoding eukaryotic-like signaling proteins on a cyanobacterial genome. Nucleic Acids Res. 1998;26(16):3619–3625. doi: 10.1093/nar/26.16.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28(5):645–659. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Borremans B, Hobman JL, Provoost A, Brown NL, Lelie D van Der. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol. 2001;183(19):5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005;56(1):215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- Kashyap DR, Botero LM, Lehr C, Hassett DJ, McDermott TR. A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J Bacteriol. 2006;188(4):1577–1584. doi: 10.1128/JB.188.4.1577-1584.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerley DF, Gonzalez CF, Park CH, Blake R 2nd, Keyhan M, Matin A. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl Environ Microbiol. 2004;70(2):873–882. doi: 10.1128/AEM.70.2.873-882.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerke K, Nakatsu CH, Beasley F, Konopka A. Comparative analysis of eight Arthrobacter plasmids. Plasmid. 2008;59(2):73–85. doi: 10.1016/j.plasmid.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Nies A, Nies DH, Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171(9):5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T, Luban S, Kihara D. Enhanced automated function prediction using distantly related sequences and contextual association by PFP. Protein Sci. 2006;15(6):1550–1556. doi: 10.1110/ps.062153506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Janda L, Stoytcheva Z, Tichy P. PkwA, a WD-repeat protein, is expressed in spore-derived mycelium of Thermomonospora curvata and phosphorylation of its WD domain could act as a molecular switch. Microbiology. 2000;146(Pt 12):3259–3267. doi: 10.1099/00221287-146-12-3259. [DOI] [PubMed] [Google Scholar]
- Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A. Effect of chromate stress on Escherichia coli K-12. J Bacteriol. 2006;188(9):3371–3381. doi: 10.1128/JB.188.9.3371-3381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol. 2005;187(24):8437–8449. doi: 10.1128/JB.187.24.8437-8449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- Munkelt D, Grass G, Nies DH. The chromosomally encoded cation diffusion facilitator proteins DmeF and FieF from Wautersia metallidurans CH34 are transporters of broad metal specificity. J Bacteriol. 2004;186(23):8036–8043. doi: 10.1128/JB.186.23.8036-8043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne KL, Turse JE, Nicora CD, Lipton MS, Tollaksen S, Lindberg C, Babnigg G, Giometti CS, Nakatsu CH, Thompson DK. Global Proteomic Analysis of the Chromate Response in Arthrobacter sp. Strain FB24. J Proteome Res. 2009;8(4):1704–1716. doi: 10.1021/pr800705f. [DOI] [PubMed] [Google Scholar]
- Cervantes C. Bacterial interactions with chromate. Antonie Van Leeuwenhoek. 1991;59(4):229–233. doi: 10.1007/BF00583675. [DOI] [PubMed] [Google Scholar]
- Coleman NV, Mattes TE, Gossett JM, Spain JC. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microbiol. 2002;68(12):6162–6171. doi: 10.1128/AEM.68.12.6162-6171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes TE, Coleman NV, Spain JC, Gossett JM. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch Microbiol. 2005;183(2):95–106. doi: 10.1007/s00203-004-0749-2. [DOI] [PubMed] [Google Scholar]
- McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103(42):15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerke KH Physiological and Genetic Analysis of Plasmid-Mediated Metal Resistance in Arthrobacter sp strain AK-1 2006West Lafayette: Purdue University; 17087467 [Google Scholar]
- Biebl H, Pfenning N. In: The Prokaryotes. Starr MP, Stolp H, Truber HG, Balows A, Schlegel HG, editor. Berlin: Springer-Verlag KG; 1981. Isolation of members of the family Rhodospirillaceae. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. 2. Cold Spring Laboratory, Cold Spring Harbor, NY; 1989. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009. pp. D169–174. [DOI] [PMC free article] [PubMed]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009. pp. D26–31. [DOI] [PMC free article] [PubMed]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Sandu C, Chiribau CB, Sachelaru P, Brandsch R. Plasmids for nicotine-dependent and -independent gene expression in Arthrobacter nicotinovorans and other Arthrobacter species. Appl Environ Microbiol. 2005;71(12):8920–8924. doi: 10.1128/AEM.71.12.8920-8924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartemann KH, Eichenlaub R. Isolation and characterization of IS an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis. J Bacteriol. 1409;183(12):3729–3736. doi: 10.1128/JB.183.12.3729-3736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- Branco R, Chung AP, Morais PV. Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiol. 2008;8:95. doi: 10.1186/1471-2180-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Radial phylogenetic tree of LCHR proteins generated from an alignment of 513 putative ChrA, chromate ion transport sequences (see Supplemental Table S1) using ClustalX. Neighbor Joining tree graphically viewed using the FigTree program http://tree.bio.ed.ac.uk/software/figtree/. Branched tips labeled with protein accession number followed by species name. Scale bar indicates 0.06 amino acid substitutions per site. Branches colors are fungi-brown, algae-green, Archaea-red, Proteobacteria (alpha-pink, beta-magenta, delta-blue, gamma-purple), Cyanobacteria-torquoise, Firmicutes-yellow, Actinobacteria-red and all other Bacteria-black.
Supplemental Table S1. Sequence accession numbers, taxa name and sequence length of putative ChrA sequences used in phylogenetic analysis.
Supplemental Figure S2. Operon structure analysis of the Arthrobacter sp. strain FB24 CRD. RT-PCR was used to determine co-transcription of the genes within the chromate resistance determinant. A: Location of primer pairs. Primer sequences are listed in table 4. Primer numbers correspond to the following primers: 1-MQO RT/A, 2-BC RT/A, 3-SP RT/F, 4-SP RT/R, 5-COG4RT/F, 6-COG4RT/R, 7-ChrAP RT/A, 8-ChrAP RT/B, 9-BP RT/R. B: RT-PCR results with listed primer pairs. C: RT-PCR products of reactions performed with primer pair 2 + 4 (lanes 2 and 3) and primer pair 5 + 8 (lanes 8 and 9). Lanes 1 and 7-100 bp PCR ruler, dark band is 1 kb; Lanes 4 and 10-no template controls; Lanes 5 and 11-No RT controls; Lanes 6 and 12 positive PCR control using pKH12 as template.
Supplemental Table S2. Recipe for vitamin solution added to mXBM.