Abstract
We report a novel method of increasing both the reaction rate and the sensitivity of low-abundance enzyme assay using a micro/nanofluidic preconcentration chip. The disposable preconcentration device made out of PDMS with a surface-patterned ion-selective membrane increases local enzyme/substrate concentrations for rapid monitoring of enzyme activity. As a model system, we used trypsin as the enzyme and BODIPY FL casein as the fluorogenic substrate. We demonstrated that the reaction rate of trypsin-BODIPY FL was significantly enhanced by increasing the local concentrations of both trypsin and BODIPY FL casein in the preconcentration chip. The reaction time required to turn over substrates at 1 ng/mL was only ~10 min compared to ~1 h without preconcentration, which demonstrates significantly higher reaction rate through the increase of the concentrations of both the enzyme and substrate. Furthermore, trypsin activity can be measured down to a concentration level of 10 pg/mL, which is ~100 fold enhancement in sensitivity compared to the result without the preconcentration step. This micro/nanofluidic preconcentrator chip could be used as a generic micro reaction platform to study any enzyme-substrate systems, or other biochemical reaction systems in low concentration ranges.
INTRODUCTION
In cellular systems, biological signals are transmitted via specific receptor-ligand binding and subsequent modification of the ligand. The most notable examples are various kinases in the cell signaling pathway. These enzymes are highly specific in their binding partners, and transmit the signal by phosphorylating/dephosphorylating their ligands (which may be another kinase, which in turn transmits the signal in the same manner). Therefore, experimentally monitoring the state of each and (preferably) every kinase (or other enzymes) along the signaling pathway would be critical in understanding complicated interconnections between different pathways. While efforts toward systems-level computational modeling have resulted in much deeper understanding regarding how cells work and transduce information, better measurement tools for cellular enzyme activities are still critically needed. Also, from the diagnostics point of view, a tool that measures enzyme activities from serum or saliva, especially for low-abundance ones, would have a tremendous value. 1–3 Many of the disease biomarkers are indeed enzymes, often associated with apoptosis/degradation processes relevant to disease progress. 4, 5
While biochemical analysis (either by mass spectrometry or immunoassays) would give chemical information such as the concentrations of the molecules of interest, often it is much more valuable to measure the activity of the enzyme directly, just so that one can obtain a much more accurate picture of the biological/pathological processes ongoing in the human body. Often, biochemical analysis techniques cannot discern subtle post-translational modifications on proteins (such as glycosylation/phosphorylation) and other subtle changes that could have a profound impact on the enzyme activity. Enzyme activities are currently measured by monitoring the turnover of enzyme substrates. For example, various kinase activities are measured by kinase assays,6–8 simply by isolating kinases by specific antibodies on the surface and then letting the enzyme turn over excess amounts of substrates. The amount of reaction product is then measured either by fluorescence or precipitation assays. Major issues of this kinase assay include the fact that the reaction occurs in sub-physiological ATP concentrations, and the requirement of radiolabeling, which makes the assay an inherently slow and low-throughput technique. Moreover, immobilization/capturing of target enzymes on a surface is often required,9, 10 which has to be carefully characterized for each enzyme carefully for an accurate quantification. Often, the activity of immobilized enzymes could be significantly different from the free solution activities because of both diffusional restrictions or interactions with the support 11, 12 and altered protein conformation, steric hindrance of the catalytic site,13 although a good correlation between the two are expected. More recently, a target-specific peptide chemosensor approach14 has been developed, which is a solution-based, flexible and compatible sensing directly from crude cell lysates. However, development of individual chemosensor peptides, specific to each target as required, limits generic application of the tool to other areas (outside of kinase activity monitoring for signaling pathways).
While these techniques have been successfully used for monitoring activities of a number of kinases in systems-biology studies, they share several common drawbacks. The most important one is that the target enzyme concentrations are typically low in the cell extract, and therefore reaction times required to turn over enough substrates for detection can be long, typically a few hours. The application of microchip (Lab-on-a-chip) for enzyme (kinase) studies has received great attention because it offers advantages in analytical speeds, performance, reduction of sample/reagent consumption, automation and integration. The use of enzyme (kinase) in microfluidic chip enables a variety of application for on-chip mixing as well as analyzing kinetics of the enzyme reactions for performing enzyme or inhibition assays.12, 15–19
Our group has developed novel nanofluidic biomolecule concentration devices which can be used to collect and trap proteins contained in a given sample into a much smaller volume, thereby increasing the local concentration significantly.20, 21 This device can take a sample volume of 1–10 μL and collect/trap any charged biomolecules such as proteins continuously, until a sufficient amount of sample is collected in a very small volume (1 pL-1 nL). While this device has a significant potential in immunoassays and biosensing applications, it is also clear that any kind of biochemical reactions (not just the binding reaction between antigen and antibody) could be enhanced by concentrating the sample inside this device.
In this paper, we demonstrate a simple method to increase the reaction rate and the sensitivity of low-abundance enzyme assay by concentrating the enzyme and the substrate in a disposable poly(dimethylsiloxane) (PDMS) preconcentration chip. For the proof of concept, we used trypsin as the model enzyme and BODIPY FL casein as the fluorogenic substrate. We observed significantly increased reaction rate of trypsin and BODIPY FL casein by increasing the local concentrations of both the enzyme and the substrate in the concentrated reaction zone. Furthermore, the preconcentration chip enabled a significantly higher sensitivity through an increased concentration of the fluorescent reaction products in the reaction zone. This PDMS device with its simplicity and efficient preconcentration capability could be used as a generic and powerful tool for improving enzyme assays for diagnostics and systems biology applications.
EXPERIMENTAL SECTION
Microchip fabrication
The microchip was fabricated using poly(dimethylsiloxane) (PDMS) (Sylgard 184, Dow Corning Inc., Midland, MI) bonded with a glass slide plate. Microchannels were molded in PDMS by the replica molding technique. The fabrication of the microchip includes the following fabrication steps: (1) master fabrication, (2) PDMS pouring/curing step, (3) patterning of Nafion resin on the slide glass and (4) irreversible bonding of the PDMS microchannel and the Nafion patterned glass slide via plasma bonding. One critical step for the nanochannel formation in PDMS chip is the generation of nanogaps, and several techniques have already been developed for this purpose.21, 22 In this work, we used Nafion film printing on the glass substrate to generate a submicron thick ion-selective membrane.23, 24
At first, SU8 photoresist (SU8-2025, MicroChem Inc., Newton, MA) was patterned on a silicon wafer to build a positive master. The positive master mold for the device contains channels that are 50 μm wide and 50 μm deep. The SU-8 master was further treated with a hexamethyldisilane (Sigma-Aldrich, St. Louis, MO) for 1 h to prevent adhesion with PDMS after molding. The hexamethyldisilane solution was evaporated and deposited on the master in a desiccator with a 5 psi vacuum.
In the second step, PDMS was poured on the master mold, which was degassed in a desiccator with a 5 psi vacuum for 1 h before pouring. After curing in an oven at 65 °C for 3 h, the PDMS layer was peeled off from the silicon master. To punch holes through the end of the channels, we used a metal syringe needle with an outer diameter of 1/16 in. (Hamilton Co., Reno, NV).
As for the printing of the ion-selective membrane, we used a Nafion ® perfluorinated ion-exchange resin 5 wt% solution in a lower alphatic alcohol/H2O mix (Sigma Aldrich, St. Louis, MO) and patterned a thin planar Nafion film on a standard glass slide via microcontact printing or alternatively, via microflow patterning inside the microchannel. In the first method, a PDMS microstamp is used to print the membrane. The major steps are: (1) ink the PDMS microstamp with Nafion resin, (2) transfer Nafion resin to a glass substrate, and (3) cure the patterned Nafion resin on a hotplate at 95 °C for 10 min. To achieve a good wetting of the Nafion resin on the PDMS stamp, we treated the surface of the PDMS stamp with oxygen plasma beforehand. We used standard pre-cleaned microscope glass slides (25 mm × 75 mm) and cleaned the surface with IPA and DI water thoroughly before printing. Preliminary tests showed that microcontact printing with a consistent thickness of the Nafion membrane between 200–400 nm was quite challenging. Depending on the pressure applied on the stamp and the pressure distribution when stamping on the glass surface, the thickness of the Nafion film varied between 0.5–1 μm. As an alternative patterning method, the microflow patterning technique was applied by using the microchannel to define the flow path of the resin. The channel used was 120 μm deep and 200 μm wide. By placing a drop of liquid Nafion resin at one open reservoir of the channel, the channel was immediately filled with it by the capillary force. Then, the liquid resin was completely flushed out of the microchannel by applying negative pressure through the syringe tubing on the other end of the channel. After this step, only a thin layer of Nafion remained on top of the hydrophilic glass slide. In this flow patterning process, the thickness of the membrane is controlled by the amount of the applied negative pressure. A repeatable thickness can be achieved as long as a constant pressure is applied through the tubing. To fully cure the patterned ion-selective film in a stripe shape, it was heated up at 95°C for 10 minutes. Finally, the poly(dimethylsiloxane) (PDMS) (Sylgard 184, Dow Corning Inc., Midland, MI) chip with microchannels was bonded on top of the patterned glass substrate by standard plasma bonding. Since an exact alignment was not required, the bonding could be performed without any help of microscope. After bonding, the PDMS device was heated up in an oven at 70 °C for 12 h to achieve higher bonding strength.
Surface treatment of microchannel
Since trypsin (Mw ~ 24 kDa, pI ~10.5) in 10 mM Tris-HCl buffer at pH 7.8 is positively charged, we applied bovine serum albumin (5% solution in 1× phosphate-buffered saline (PBS, pH=7.4), Sigma, St. Louis, MO) into the microchannel for 1 hour to prevent nonspecific binding of biomolecules on the PDMS surface. We then repeatedly washed the microchannel out with phosphate-buffered saline (PBS, pH 7.4). Due to this surface passivation, only a marginal non-specific binding (increase of the fluorescence intensity by only ~ 20 A.U.) occurred for the negatively charged BODIPY FL-labeled peptides on the channel wall. However, the surface adsorption posed a problem to the Nafion membrane for long operation times. We noticed a degradation of the Nafion membrane, especially caused by the surface absorption of the positively charged molecules on the membrane, which led to a degradation of the device performance.
Materials
To investigate the ability of our microfluidic preconcentration chip as a reaction enhancing tool for the low-abundance enzymes, we used trypsin (from porcine pancreas, Aldrich, MO) as the enzyme and BODIPY FL casein as the fluorogenic substrate (EnzChek® Protease Assay Kits, Molecular Probes, Eugene, OR). It is well-known that protease-catalyzed hydrolysis releases highly fluorescent BODIPY FL-labeled peptide and the accompanying increase in fluorescence signal is proportional to protease activity.25 For all experiments, we used 10mM Tris-HCl with pH 7.8 as the buffer solution.
Measurement instrument and image analysis
An inverted epifluorescence microscope IX 71 (Olympus, Melville, NY) equipped with a thermoelectrically cooled CCD camera (Hamamatsu Co., Japan) was used for fluorescence imaging. We measured the average peak value over a square window (30 μm × 30 μm) in the preconcentration zone as a function of time. To reduce the photobleaching effect, we used a mechanical shutter (Olympus, Melville, NY) which automatically block UV light and opens only when images are taken. Also, we used a neutral density filter (Olympus, Melville, NY) to prevent the CCD array from saturation so that we can achieve an increased dynamic range and reduce the photobleaching effect. Fluorescence images were taken every 30 sec and were analyzed using IPLab software (Scanalytics, Fairfax, VA). Since we performed the assay with regard to the fluorescence signal detection of proteolytic activity in the low enzyme concentration regime, we took the peak height instead of the peak area to characterize the reaction rate. For the plotting of both hyperbolic fitting curves and slope curves, we used Sigmaplot® 10 software (Systat Software Inc. CA) and Mathematica V 5.2 (Wolfram Research, Inc., IL).
Dynamic range assessment
We used the commerically available EnzChek Protease assay kit for our experiment and its manufacturer provided the limit of detection (LOD) as the enzyme concentration required to cause a 10~20% signal intensity change above background signal in their product manual.25, 26 According to the sample standard curve obtained with the EnzChek protease assay, the detection limit of this assay was between ctrypsin = 1–5 ng/mL for trypsin in 10 mM Tris-HCl at pH 7.8 with a fluorescence microplate reader using 200 μL reaction mixtures. We set the concentration range for our experiment between ctrypsin = 1 ng/mL and ctrypsin = 1 μg/mL to include the above-mentioned detection limit. For the fluorescence signal detection in our experiment, we defined the LOQ (limit of quantitation) with more standard threshold, namely 10σ above the background RMS noise.
As a preliminary test of enzyme assay in microchannel, we used csubstrate = 10 μg/mL BODIPY FL casein, as suggested by the supplier,25, 26 for four different trypsin concentrations from ctrypsin = 1 ng/mL to 1 ctrypsin = 1 μg/mL in the microchannel. However, the BODIPY FL casein was not sufficiently hydrolyzed to produce a detectable signal for this trypsin concentration range in the microchannel. This result seems to be related to the low sample volume we used in the microchannel. The standard assays provided by the vendor use 200 μL reaction mixtures, while the sample volume in our experiment was only tens of nanoliters. To compensate for the low sample volume, we increased the concentration of BODIPY FL casein from csubstrate = 10 μg/mL to csubstrate = 50 μg/mL in 10 mM Tris-HCl buffer at pH 7.8 and were able to acquire a signal difference with different enzyme concentrations in the microchannel.
In order to measure the increased reaction rate with preconcentration accurately, the enzyme and the substrate had to be injected into the preconcentration device using two separate channels. Due to the laminar flow in microchannel, the mixing between them will be only minimal and the reaction will take place mainly in the concentrated reaction zone. However, since our device had only one input channel, we mixed the enzyme and substrate off-chip in a vial instead and injected the premixed solution into the preconcentration device. This approach can be justified by the low reaction rate in the enzyme concentration range we investigated. The preliminary experiment showed that the reaction time required for turning over enough substrates at ctrypsin = 10 ng/mL was up to ~1 hour.
RESULTS AND DISCUSSION
Device scheme and operation
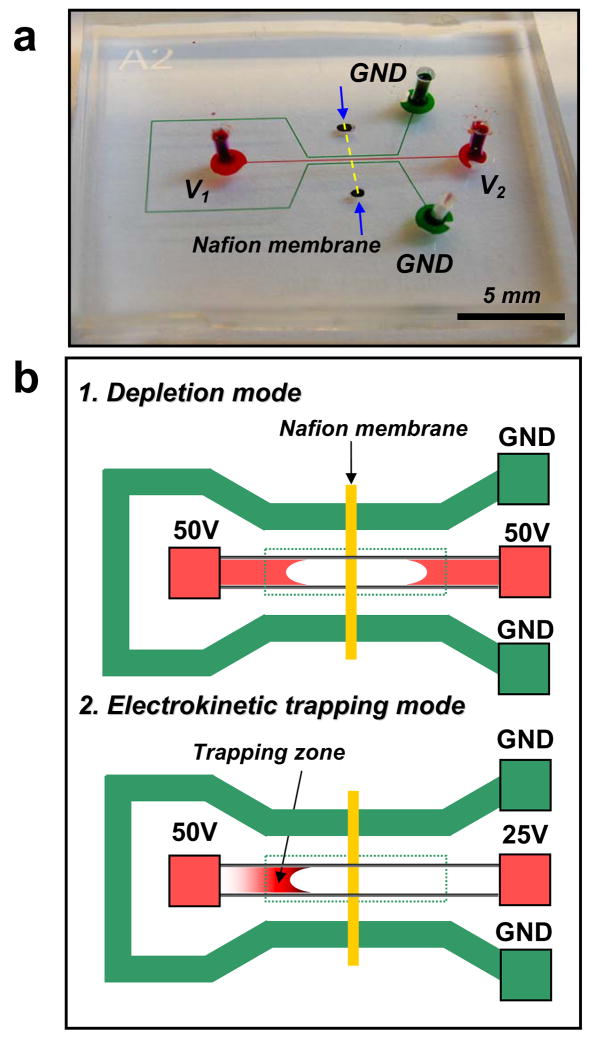
The PDMS preconcentrator device with its operation scheme for the enhancement of low-abundance enzyme reaction is shown in Figure 1. The micro/nanofluidic preconcentration chip was composed of two microchannels with a planar ion-selective membrane (e.g., Nafion) across the microchannels, as shown in Figure 1a (dashed line). The scanning electron microscope (SEM) images showed a well-defined Nafion membrane on the glass substrate (see S1 in supporting material). The thickness of the Nafion membrane on the glass substrate was only 191 nm. The middle channel, shown as red in Figure 1a, was loaded with a mixture of trypsin (enzyme) and BODIPY FL casein (substrate), while the side channel, shown as green in Figure 1a, was filled with 10 mM Tris-HCl buffer solution. For preconcentration, we applied a potential difference across the middle and the side channel in combination with an electrokinetically-driven tangential flow inside the middle channel to trap the molecules in front of the depletion region.20, 21 First, we applied V1 = V2 = 50 V to two anodic reservoirs, while the others were grounded, as shown in Figure 1b. In this way, we induced a depletion region near the ion-selective Nafion membrane by using the concentration polarization effect. In the second step, we applied V1 = 50 V and V2 = 25 V. With this potential difference, we initiated the tangential electroosmotic flow to trap both the trypsin and BODIPY FL casein in front of the depletion region (see the bottom image in Figure 1b). During this operation, we kept a balance between anion repulsion from the space charge layer and electroosmotic flow from the reservoir to preconcentrate the molecules, as described in previous reports.20, 21
Figure 1.
(a) Optical image and (b) operation of PDMS preconcentration chip as a reaction enhancing tool for low-abundance enzyme assay. The microchannel in the center (red) is loaded with enzyme/substrate mixture and the side channel (green) is filled with a buffer solution. To concentrate the premixed solution of enzyme/substrate, a potential difference is applied across the middle and the side channel in combination with an electrokinetic flow. In the depletion mode, the depletion region is formed due to the concentration polarization and then in the trapping mode, the molecules driven with an electroosmotic flow are electrokinetically trapped in front of the depletion region, resulting in increased concentration.
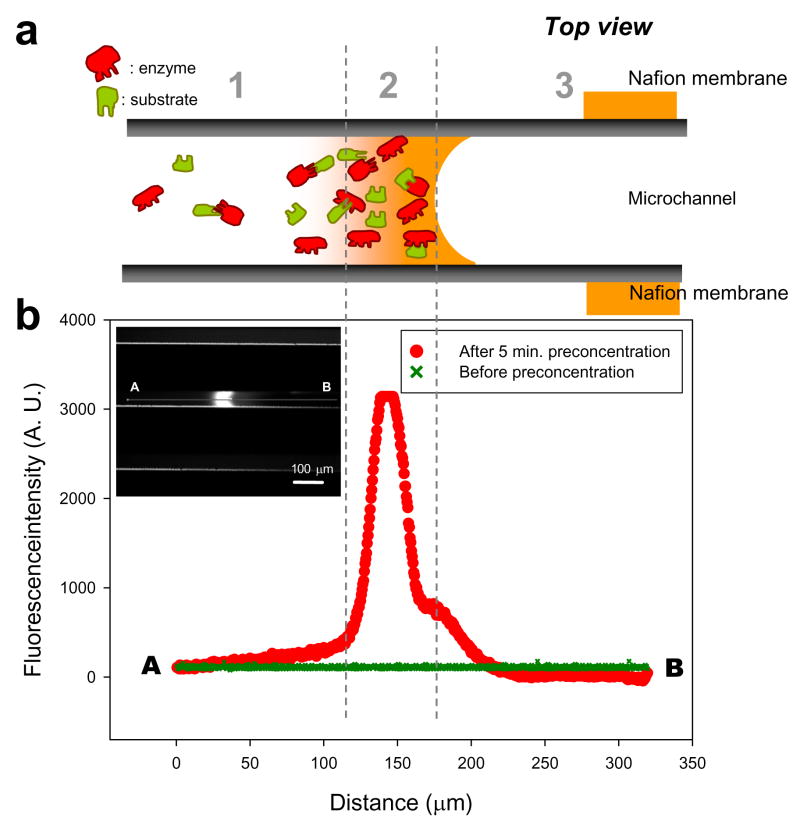
Figure 2a shows schematically the electrokinetic trapping of the enzyme and the substrate molecules in the concentrated reaction zone near the planar Nafion membrane (zone 2 in Figure 2). Both molecules are electroosmotically flown from zone 1, in which the reaction takes place without preconcentration. Zone 3 represents the ion depletion region which repels all charged ions. The location of the concentrated plug where the reaction between the enzyme and the substrate takes place can be easily controlled by the potential difference across the Nafion membrane. We measured the fluorescence intensity profile along the microchannel to obtain the concentration distribution of the reaction products in three different zones after 5 min. of preconcentration. As shown in Figure 2b, the enzyme-substrate reaction rate was drastically increased in the concentrated reaction zone, indicated by a high intensity plug in the fluorescence image and the corresponding intensity peak. For comparison, the intensity profile taken before preconcentration is also shown as a green reference line in Figure 2b.
Figure 2.
(a) Schematic of electrokinetic trapping of enzyme and substrate and (b) intensity profile along the microchannel as a measure of the enzyme-substrate reaction product after 5 minutes of preconcentration. It shows an increased concentration of the enzyme-substrate reaction product in the concentrated reaction zone generated by electrokinetic trapping (zone 2). Zone 1 contains the mixture of trypsin and BODIPY FL casein outside of the concentrated reaction zone. The difference of the fluorescence signals between zone 1 and 2 indicates an enhanced enzyme-substrate reaction through the preconcentration. The increase of the intensity was partly due to the stacking of the reaction products flowing from zone 1. The Zone 3 illustrates the depletion zone, allowing a control experiment to estimate the background noise that can be generated from the adsorption of the reaction products on the side wall of the microchannel.
We can express the reaction rate of the assay by the Michaelis-Menten equation as below,27
| (1) |
| (2) |
Here, [P] denotes the product concentration, [E0] and [S] are the starting amounts of enzyme and substrate concentration, respectively, while k1, k2, k3 are the rate constants for the individual steps. Km is the Michaelis constant which is defined as the concentration where the rate of the enzyme reaction is half Vmax. Vmax is the maximum velocity in reaction rate versus substrate concentration curve. According to eq. (2), we increased the reaction rate in the reaction zone (zone 2 in Figure 2) by increasing the concentrations of enzyme ([E0]) as well as of substrate ([S]). Furthermore, we increased the amount of the fluorescent reaction products through the preconcentration step which led to the intensity peak in the concentrated reaction zone. This enabled higher sensitivity of the assay. In the depleted zone 3, however, the fluorescence intensity dropped down below the reference level.
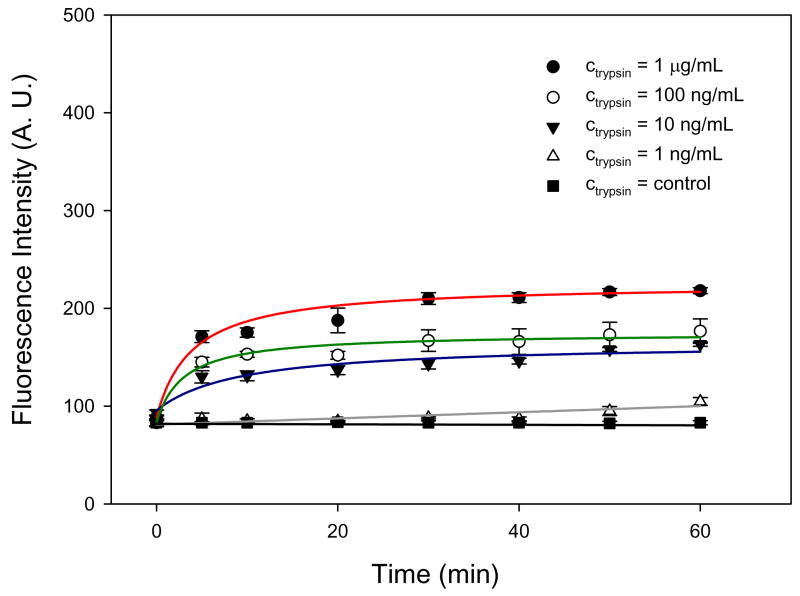
Figure 3 shows the fluorescence intensity of the reaction product measured as a function of reaction time for four different trypsin concentrations (ctrypsin = 1 ng/mL to ctrypsin = 1 μg/mL) without preconcentration. For this experiment, we filled the preconcentrator chip with different premixed enzyme/substrate solutions and measured the fluorescence signal near the Nafion membrane without applying any electrical potential. The progress curves shown in Figure 3 correspond to the typical enzyme-substrate reaction curve suggested,27 which demonstrated that the reaction rate decreases with time after reaching the initial maximum value. Obviously, the intensity values after 60 min. increased with higher initial enzyme concentrations.
Figure 3.
Fluorescence signal intensity of products as a function of enzyme-substrate reaction time for different trypsin concentrations ranging from ctrypsin = 1 ng/mL and ctrypsin = 1 μg/mL without preconcentration. BODIPY FL casein with csubstrate = 50 μg/mL was used as a fluorogenic substrate to monitor the turnover rate. It shows that the limit of detection was between ctrypsin = 1 ng/mL and ctrypsin = 10 ng/mL (> 10% change of the signal intensity above control) and the reaction time required to turn over enough substrates was ~ 1 hours.
To estimate the reaction rates, we calculated the slopes of the fitting curves shown in Figure 3. First, all data points in Figure 3 were fitted with a hyperbolic function. Then, we calculated the slopes of the fitting curves and compared their initial slope values as a measure of reaction rate. We provided the equation of each fitting and slope curve as well as the graph of the corresponding slope curves in supporting material S2. The initial slopes were 155 A.U./min., 121 A.U./min., 153 A.U./min., and 4.2 A.U./min. for four different trypsin concentrations of ctrypsin = 1 μg/mL, ctrypsin = 100 ng/mL, ctrypsin = 10 ng/mL, ctrypsin = 1 ng/mL, respectively. A significant difference in the initial slopes was noticeable only between ctrypsin = 10 ng/mL and ctrypsin = 1 ng/mL. Above ctrypsin = 10 ng/mL, the difference of the initial slope values was rather small due to the generally low intensity values without preconcentration. At ctrypsin = 1 ng/mL, the signal intensity was 99 A.U. after 60 min., which was higher than the 10σ of the background RMS (= 95.5 A.U.). This limit of quantitation was in good agreement with the value provided by the assay vendor, which was approximately between 1–5 ng/mL.
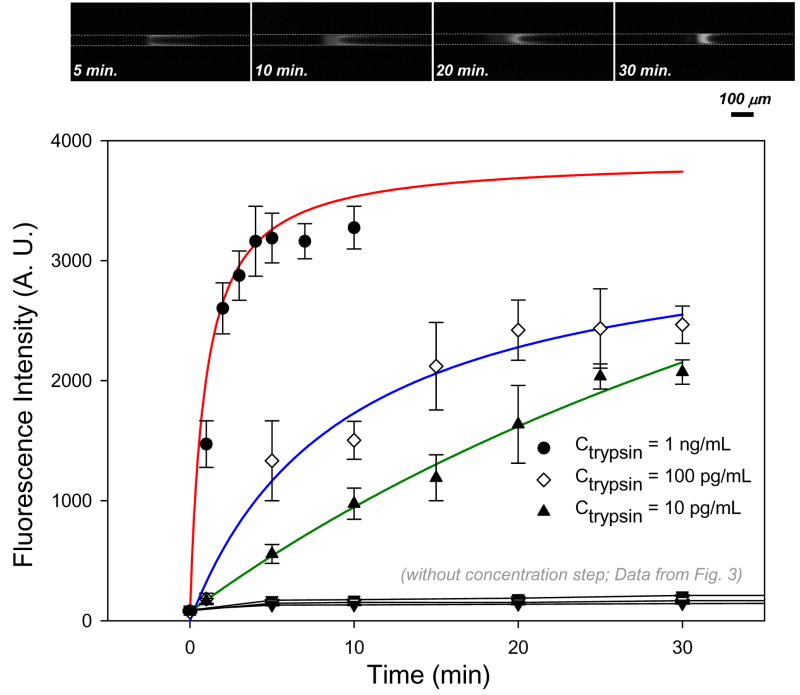
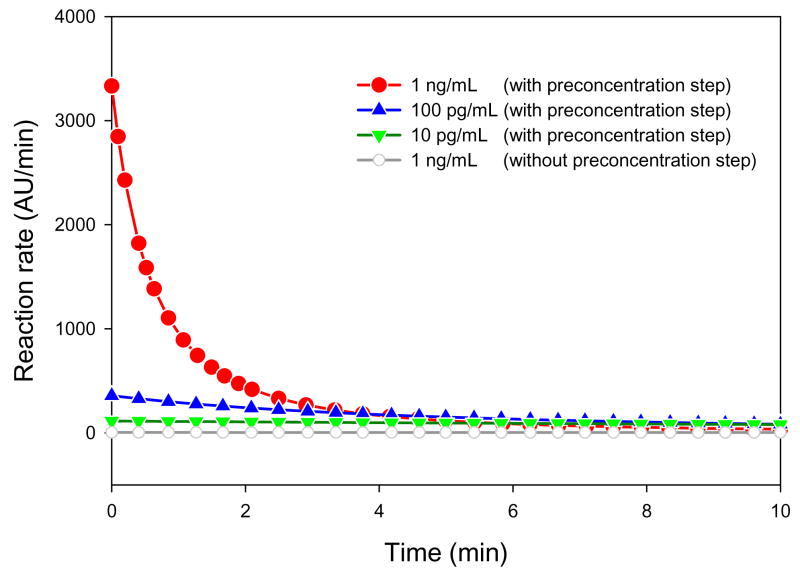
To investigate the influence of the preconcentration on the reaction rate, we repeated the same experiment with preconcentration. The highest initial concentration at which we were able to measure the signal intensity as a function of time was ctrypsin = 1 ng/mL. Above ctrypsin = 1 ng/mL, the fluorescence signal intensity immediately reached the saturation value of the CCD camera (= 4096 A.U.) as soon as the preconcentration was started. Therefore, we reduced the trypsin concentrations down to ctrypsin = 10 pg/mL, ctrypsin = 100 pg/mL, and ctrypsin = 1 ng/mL, with the lowest concentration being two orders of magnitude lower than that used in Figure 3. Because of this concentration shift, a direct comparison between the measurements without and with preconcentration was only possible at ctrypsin = 1 ng/mL. The enhancing effect of preconcentration on the trypsin-catalyzed reaction is shown in Figure 4. Again, all data points in Figure 4 were fitted with a hyperbolic function (see supporting material S3). The reaction time required to turn over the substrate at 1 ng/ml was reduced from 1 hour to ~ 10 min, which means a 6-time increase in the reaction rate compared to that of non-preconcentrated sample. To elucidate the effect of preconcentration on the reaction rate, the slope curves are plotted in Figure 5. The slope of ctrypsin = 1 ng/mL (red; circle), 100 pg/mL (blue; triangle up), and 10 pg/mL (green; triangle down) with preconcentration was calculated from the data of Figure 4, while ctrypsin = 1 ng/mL (grey; open circle) without preconcentration was calculated from Figure 3. The slope curve for ctrypsin = 1 ng/mL without preconcentration is given as a reference. The calculated initial slopes of the curves were 3333 A.U./min, 357 A.U./min, and 114 A.U./min for three different trypsin concentrations, ctrypsin = 1 ng/mL, ctrypsin = 100 pg/mL, ctrypsin = 10 pg/mL, respectively. In case of ctrypsin = 1 ng/mL with preconcentration, for instance, the initial slope was increased by ~794 folds compared to that without preconcentration. This increase in reaction rate was due to the increase of the enzyme concentration [E] as well as of the substrate [S] in the concentrated reaction zone. Since we used a premixed enzyme and substrate, the increase of the fluorescence signal can be partially attributed to the products flowing from the reservoir into the concentrated reaction (trapping) zone. However, this contribution was negligible compared to that of the products generated in the concentrated reaction zone since the reaction rate outside the concentrated reaction zone was significantly low, as shown for ctrypsin = 1 ng/mL in Figure 3. At ctrypsin = 10 pg/mL, the initial slope value was comparable to that of ctrypsin = 10 ng/mL without preconcentration (153 A.U./min). This result demonstrated that the preconcentrator chip can increase the reaction rate of the enzyme assay up to the level of three-order of magnitude higher enzyme concentrations.
Figure 4.
Fluorescence signal intensity of products versus preconcentration time shows an enhanced trypsin-catalyzed reaction with preconcentration. We used three different trypsin concentrations ranging from ctrypsin = 10 pg/ml to ctrypsin = 1 ng/ml (lower concentrations than those used in Figure 3) for enzymes while maintaining the same substrate concentration at csubstrate = 50 μg/mL. The reaction time required to turn over the substrate at a concentration of 1 ng/ml was only ~ 10 min, which was 6 times faster than that without preconcentration. It also showed that the limit of detection was lowered down to ctrypsin = 10 pg/mL, which means a ~1000 folds sensitivity enhancement compared with the results without preconcentration. The fluorescence images were taken at ctrypsin = 10 pg/mL after different concentration times.
Figure 5.
Reaction rate versus preconcentration time, calculated from the fitting curve (Figure 4). In case of ctrypsin = 1 ng/mL, the initial slope was drastically increased through preconcentration (compare with the grey line (open circle) which shows the slope curve of the same concentration without preconcentration). At ctrypsin = 100 pg/mL, the initial slope was still higher than any of those measured without preconcentration. In case of the lowest concentration tested, ctrypsin = 10 pg/mL, the initial reaction rate was comparable to the initial slope of ctrypsin = 10 ng/mL without preconcentration.
Furthermore, the fluorescence intensity was increased by 20 folds through the preconcentration. This increase in fluorescence intensity enabled even detection at an enzyme concentration of ctrypsin = 10 pg/mL possible. As a result of preconcentration, the sensitivity of the enzyme assay was improved at least by two orders of magnitude (the LOQ was at ctrypsin = 1 ng/mL without preconcentration).
The experimental data in Figure 3 and 4 show the average run-to-run values of three devices and relatively large errors in Figure 4 than in Figure 3. These relatively higher standard deviations in case of preconcentration are due to the fact that the electrokinetic trapping process with its “dynamic” character (e.g., force balance between the depletion region and trapping region) is more susceptible to the variations of the device such as the thickness of the Nafion membrane, bonding strength and surface conditions of the microchannel than the electroosmotic flow without preconcentration operation. We characterized the chip-to-chip reproducibility with the coefficient of variance (CV) value. The coefficient of variance (CV) for the measurement with preconcentration was 11.5 %, showing a good reproducibility of our PDMS devices even with the “dynamic” preconcentration process.
CONCLUSION
In this paper, we reported a simple, disposable PDMS microfluidic platform to assay the enzyme-substrate reaction. This device contains a planar surface-patterned cation-selective membrane across the microchannels and enables an electrokinetic trapping of the enzyme and the substrate, near the ion-selective membrane, thereby increasing the concentrations in the trapping zone. Since the molecules can be electrokinetically trapped, there is no need to immobilize the enzymes inside the channel.
Using the fluorescence intensity measurement, we could establish a 6-time faster reaction between the trypsin and BODIPY FL casein in the concentrated zone. In addition to the rapid enzymatic reaction, the preconcentration device lowers the limit of quantitation of the enzyme by two orders of magnitude from ctrypsin = 1 ng/mL down to ctrypsin = 10 pg/mL. These significant enhancements in terms of the reaction rates and sensitivity have an important implication for the enzyme kinetic studies as well as for enzyme-based diagnostics. In the future, this type of PDMS preconcentrator chip with its simple design and operation can find widespread applications as a general and efficient reaction enhancing platform for low-abundance kinase assays as well as for other enzyme assays involving low-abundance molecules or slow reactions. Since our PDMS preconcentrators can be also built in a massively parallel fashion, this array-type reaction platform will enable high-throughput analysis of enzymatic processes in molecular engineering. Immediate application could be found in the study of cell signaling pathways, where multiple, specific kinase/phosphatase activity monitoring from a small volume of sample would be highly desirable.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants R01-EB005743 and P30-ES002109-28 (MIT-CEHS). J. Han and Y. A. Song were partially supported by NSF CAREER (CTS- 0347348) programs. Microfabrication of the device was done in the Microsystems Technology Laboratories of MIT, with the help of its staff members.
References
- 1.Sobel BE, Shell WE. Circulation. 1972;45:471–482. doi: 10.1161/01.cir.45.2.471. [DOI] [PubMed] [Google Scholar]
- 2.Hortin GL, Jortani SA, Ritchie JC, Jr, Valdes R, Jr, Chan DW. Clinical Chemistry. 2006;52:1218–1222. doi: 10.1373/clinchem.2006.067280. [DOI] [PubMed] [Google Scholar]
- 3.Allard L, Burkhard PR, Lescuyer P, Burgess JA, Walter N, Hochstrasser DF. Clin Chem. 2005;51:2043–2051. doi: 10.1373/clinchem.2005.053942. [DOI] [PubMed] [Google Scholar]
- 4.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MSS, Flavell RA. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 5.Miura M, Zhu H, Rotello R, Hartwieg EA, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 6.Whitmarsh AJ, Davis RJ. Methods Enzymology. 2001;332:319–336. doi: 10.1016/s0076-6879(01)32212-7. [DOI] [PubMed] [Google Scholar]
- 7.Hall JP, Davis RJ. Methods Enzymology. 2002;345:413–425. doi: 10.1016/s0076-6879(02)45034-3. [DOI] [PubMed] [Google Scholar]
- 8.Janes KA, Albeck JG, Peng LX, Sorger PK, Lauffenburger DA, Yaffe MB. Mol Cell Proteomics. 2003;2:463–473. doi: 10.1074/mcp.M300045-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Logan TC, Clark DS, Stachowiak TB, Svec F, Frechet JMJ. Anal Chem. 2007;79:6592–6598. doi: 10.1021/ac070705k. [DOI] [PubMed] [Google Scholar]
- 10.Holden MA, Jung SY, Cremer PS. Anal Chem. 2004;76:1838–1843. doi: 10.1021/ac035234q. [DOI] [PubMed] [Google Scholar]
- 11.Goddard JM, Hotchkiss JH. Progress in Polymer Science. 2007;32:698–725. [Google Scholar]
- 12.Seong GH, Heo J, Crooks RM. Anal Chem. 2003;75:3161–3167. doi: 10.1021/ac034155b. [DOI] [PubMed] [Google Scholar]
- 13.DeLouise LA, Miller BL. Anal Chem. 2005;77:1950–1956. doi: 10.1021/ac0486185. [DOI] [PubMed] [Google Scholar]
- 14.Shults MD, Janes KA, Lauffenburger DA, Imperiali B. Nature Methods. 2005;2:277–283. doi: 10.1038/nmeth747. [DOI] [PubMed] [Google Scholar]
- 15.Wang J. Electrophoresis. 2002;23:713–718. doi: 10.1002/1522-2683(200203)23:5<713::AID-ELPS713>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Chatrathi MP, Tian B. Anal Chem. 2001:1296–1300. doi: 10.1021/ac001205t. [DOI] [PubMed] [Google Scholar]
- 17.Hadd AG, Jacobson SC, Ramsey JM. Anal Chem. 1999;71:5206–5212. [Google Scholar]
- 18.Hadd AG, Raymond DE, Halliwell JW, Jacobson SC, Ramsey JM. Anal Chem. 1997;69:3407–3412. doi: 10.1021/ac970192p. [DOI] [PubMed] [Google Scholar]
- 19.Kerby MB, Legge RS, Tripathi A. Anal Chem. 2006;78:8273–8280. doi: 10.1021/ac061189l. [DOI] [PubMed] [Google Scholar]
- 20.Wang YC, Stevens AL, Han J. Anal Chem. 2005;77:4293–4299. doi: 10.1021/ac050321z. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Chung S, Kim SJ, Han J. Anal Chem. 2007;79:6868–6873. doi: 10.1021/ac071162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SM, Burns MA, Hasselbrink F. Anal Chem. 2006;78:4779–4785. doi: 10.1021/ac060031y. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, Song YA, Kim SJ, Han J. MicroTAS 2007 Conference. 2007;2:1198–1200. [Google Scholar]
- 24.Lee JH, Song Y-A, Han J. Lab Chip. 2008 doi: 10.1039/B717900F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.http://probes.invitrogen.com/media/pis/mp06638.pdf.
- 26.Jones LJ, Upson RH, Haugland RP, Panchuk-Voloshina N, Zhou M, Haugland RP. Analytical Biochemistry. 1997;251:144–152. doi: 10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- 27.Pandey A, Webb C, Soccol CR, Larroche C. Enzyme Technology. Springer; New Delhi: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.