Abstract
Angiogenesis is crucial for development and metastasis of tumors and VEGF is a key mediator of this process. The importance of VEGF in tumorigenesis and tumor progression makes it an attractive target for the development of anticancer therapies. Inhibition of angiogenesis has shown promising clinical efficacy; however, not all patients treated with anti-angiogenic agents get benefited. Some patients are predisposed to refractory disease while others develop resistance after initial response. Patients may also have different severity of drug-related adverse events. Optimization of drug administration based on disease status and individual responsiveness is important in limiting the treatment failure and minimization of side-effects. Single nucleotide polymorphisms (SNPs) in VEGF may alter VEGF protein concentrations, influence the process of angiogenesis and may relate to inter-individual variation in the risk and progression of selected tumors, and their resistance to treatments. This review examines the role of SNPs in VEGF gene as predictive and prognostic markers for major solid tumors, including the breast, non-small cell lung (NSCLC), colorectal (CRC) and prostate cancers. Selected VEGF SNPs appear to be associated with risk of these cancers; however, there is lack of unanimity in findings, in part influenced by differences in study design and analysis.
Keywords: VEGF, gene, SNP, cancer, breast, lung, colon, prostate
Introduction
Antiangiogenic therapies have improved the clinical outcome of selected tumors by inhibition of tumor growth via restriction of new blood vessel formation. As tumor grows, the lack of sufficient blood supply creates an hypoxic environment, stimulating the release of factors like hypoxia inducible factor (HIF)−1α, which induces angiogenesis by activating the transcription of VEGF (1). VEGF and its receptors are frequently overexpressed in human tumors, including the breast, non-small cell lung, colorectal and prostate cancers (2–5). VEGF and associated signaling pathways have been the target for many newly developed anticancer drugs. These agents have shown promising efficacy; however, they are often very expensive and some patients experience drug resistance, limited activity, and severe toxicities. There is a strong need for identification of markers enabling a priori selection of patients who are likely to benefit from these agents. These markers might also carry prognostic value and may help in decisions about the overall treatment approach to manage more or less aggressive tumors with varying degrees of angiogenic involvement. This review evaluates the role of VEGF SNPs in predicting the cancer susceptibility, progression, and anti-VEGF therapeutic response in subjects with major solid tumors including the breast, non-small cell lung, colorectal and prostate cancers. For these tumors anti-VEGF or anti-VEGFR treatments are either approved or in advance stages of clinical development. We also performed meta-analysis, where methods of DerSimonian and Laird (6) were used to test the homogeneity of odds ratio across studies and for calculation of pooled OR and their confidence intervals. Standard error estimates for individual OR were calculated using exact methods from published genotype frequencies or were derived from published confidence intervals.
VEGF gene
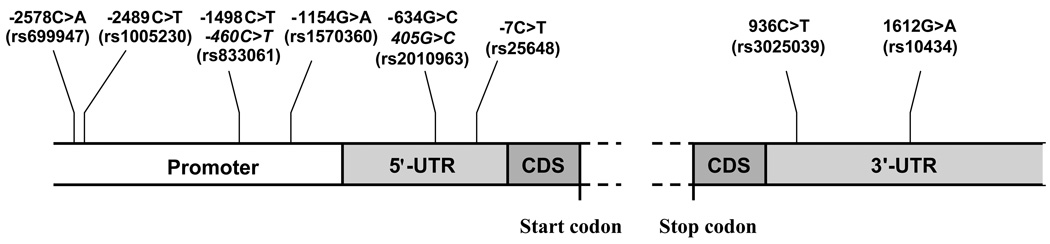
The VEGF gene is located on chromosome 6 at location 6p21.1 (7). Its coding region spans approximately 14 kilobases and consists of 8 exons (8, 9). Numerous SNPs in the promoter, 5'-, and 3'- untranslated regions (UTR), are present in VEGF. Some of the more frequent SNPs are shown in Figure 1 and their frequencies in control population are summarized in Table 1. The 5'- and 3'- UTR contain key regulatory elements which are sensitive to hypoxia (10), and contributes to high variability in VEGF production among tissues (11). For example −634G>C SNP in the 5'-UTR of VEGF affects the protein translation efficiency (12), and 936C>T SNP in the 3'-UTR influences the circulating plasma concentrations (13) and tumor tissue expression of VEGF (14). However, it is likely that only a small number of these polymorphisms and haplotypes (linearly linked SNPs) actually have a functional effect on VEGF translation, whereas others act as proxies (15).
Fig. 1.
Structure of VEGF gene and position of VEGF SNPs relative to translation start site. Italic positions indicate alternate names. Dashed lines indicate the region consisting of coding sequence (CDS) and seven introns. (adapted from reference (29)). UTR: Untranslated region.
Table 1.
Frequencies of selected VEGF polymorphisms in control subjects
| Polymorphism | Ethnicity | Controls | Variants | Allele Frequency | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| N (% Genotype Frequency) | p | q | ||||||
| 936 C>T | CC | CT | TT | |||||
| (rs3025039) | Austrian Caucasian | 500 | 353 (70.6) | 137 (27.4) | 10 (2.0) | (22) | ||
| Polish | 422 | 297 (70.4) | 114 (27.0) | 11 (2.6) | (24) | |||
| German | 163 | 128 (78.5) | 31 (19.0) | 4 (2.5) | (24) | |||
| Swedish | 934 | 720 (77.1) | 203 (21.7) | 11 (1.2) | (24) | |||
| Chinese | 1195 | 793 (66.4) | 351 (29.4) | 51 (4.3) | (24) | |||
| UK Caucasian | 708 | 531 (75.0) | 165 (23.3) | 12 (1.7) | (28) | |||
| 98% Caucasian | 479 | 353 (73.7) | 118 (24.6) | 8 (1.7) | (27) | |||
| NR | 60 | 57 (95.0) | 3 (5.0) | 0 (0.0) | (30) | |||
| Polish Caucasian | 290 | 202 (69.7) | 81 (27.9) | 7 (2.4) | (32) | |||
| Caucasian | 1458 | 1125 (77.2) | 308 (21.1) | 25 (1.7) | (39) | |||
| Korean | 229 | 169 (73.8) | 57 (24.9) | 3 (1.3) | (43) | |||
| Korean | 413 | 252 (61.0) | 149 (36.1) | 12 (2.9) | (45) | |||
| Austrian | 427 | 308 (72.1) | 108 (25.3) | 11 (2.6) | (48) | |||
| Tunisian | 100 | 72 (72.0) | 27 (27.0) | 1 (1.0) | (55) | |||
| Austrian | 804 | 0.85 | 0.15 | (29) | ||||
| −1154G>A | GG | GA | AA | |||||
| (rs1570360) | NR | 263 | 120 (45.6) | 109 (41.4) | 34 (12.9) | (23) | ||
| Polish | 423 | 199 (47.0) | 178 (42.1) | 46 (10.9) | (24) | |||
| German | 163 | 76 (46.6) | 69 (42.3) | 18 (11.0) | (24) | |||
| 98% Caucasian | 492 | 208 (42.3) | 227 (46.1) | 57 (11.6) | (27) | |||
| UK Caucasian | 263 | 120 (45.6) | 109 (41.4) | 34 (12.9) | (53) | |||
| Tunisian | 100 | 36 (36.0) | 50 (50.0) | 14 (14.0) | (55) | |||
| −634 G>C | GG | GC | CC | |||||
| (rs2010963) | Swedish | 941 | 492 (52.3) | 367 (39.0) | 82 (8.7) | (24) | ||
| 98% Caucasian | 500 | 232 (46.4) | 221 (44.2) | 47 (9.4) | (27) | |||
| Korean | 413 | 106 (25.7) | 223 (54.0) | 84 (20.3) | (45) | |||
| Austrian | 430 | 192 (44.7) | 195 (45.4) | 43 (10.0) | (48) | |||
| Tunisian | 100 | 44 (44.0) | 46 (46.0) | 10 (10.0) | (55) | |||
| Chinese | 1198 | 418 (34.9) | 598 (49.9) | 182 (15.2) | (26) | |||
| UK Caucasian | 498 | 209 (42) | 225 (45.2) | 64 (12.9) | (28) | |||
| Caucasian | 1458 | 650 (44.6) | 644 (44.2) | 164 (11.3) | (39) | |||
| Austrian | 804 | 0.66 | 0.34 | (29) | ||||
| −2578 C>A | CC | CA | AA | |||||
| (rs699947) | Polish | 423 | 106 (25.1) | 207 (48.9) | 110 (26.0) | (24) | ||
| German | 162 | 50 (30.9) | 72 (44.4) | 40 (24.7) | (24) | |||
| Swedish | 940 | 257 (27.3) | 451 (48.0) | 232 (24.7) | (24) | |||
| 98% Caucasian | 495 | 129 (26.1) | 236 (47.7) | 130 (26.3) | (27) | |||
| Korean | 203 | 106 (52.2) | 82 (40.1) | 15 (7.4) | (4) | |||
| Austrian | 427 | 85 (19.9) | 238 (55.7) | 104 (24.4) | (48) | |||
| Austrian | 804 | 0.53 | 0.47 | (29) | ||||
| −1498 C>T | CC | CT | TT | |||||
| (rs833061) | Chinese | 1222 | 78 (6.4) | 479 (39.2) | 665 (54.4) | (26) | ||
| UK Caucasian | 498 | 106 (21.3) | 251 (50.4) | 141 (28.3) | (28) | |||
| Caucasian | 1458 | 342 (23.5) | 694 (47.6) | 422 (28.9) | (39) | |||
| Taiwanese | 119 | 4 (3.4) | 72 (60.5) | 43 (36.1) | (54) | |||
| Turkish | 157 | 50 (31.9) | 94 (59.9) | 13 (8.3) | (56) | |||
| Japanese | 252 | 23 (9.1) | 97 (38.5) | 132 (52.4) | (51) | |||
| Austrian | 804 | 0.54 | 0.46 | (29) | ||||
| −7 C>T | CC | CT | TT | |||||
| (rs25648) | UK Caucasian | 493 | 332 (67.3) | 151 (30.7) | 10 (2.0) | (28) | ||
| Austrian | 804 | 0.84 | 0.16 | (29) | ||||
| −2489C>T | CC | CT | TT | |||||
| (rs1005230) | Austrian | 804 | 0.53 | 0.47 | (29) | |||
| 1612G>A | GG | GA | AA | |||||
| (rs10434) | Austrian | 804 | 0.54 | 0.46 | (29) | |||
NR: not reported
VEGF SNPs in breast cancer
VEGF has shown to be important for development, invasiveness and metastasis of breast cancer in both preclinical and clinical settings (16). Higher levels of VEGF have shown to be related with adverse prognosis, decreased overall survival and resistance to hormonal therapy (17–19). High microvessel density (MVD), a marker of VEGF expression and local activity, increases the likelihood of metastatic disease (20) and acts as a prognostic indicator for relapse-free and overall survival in node-negative breast cancer patients (21). These data suggest that SNPs in VEGF may play a substantial role in development and progression of breast cancer; studies evaluating this hypothesis are summarized in Table 2.
Table 2.
Summary of case-control studies assessing the VEGF polymorphisms in relation to risk of breast cancer
| Polymorphism | Ethnicity | Case/Control | Genotype | Association of Cancer Risk with respect to VEGF Genotype |
OR | OR Adjusted (Yes/No) |
L95%CI | U95%CI | Ref |
|---|---|---|---|---|---|---|---|---|---|
| 936 C>T1 | Austrian Caucasian | 500/500 | CC vs. (CT or TT)* | Yes (decreases risk) | 0.51 | No | 0.38 | 0.7 | (22) |
| TT vs. CC* | No | 0.77 | No | 0.29 | 2.03 | ||||
| Polish | 412/422 | CT vs. CC | No | 0.87 | No | 0.63 | 1.21 | (24) | |
| TT vs. CC | No | 1.27 | No | 0.53 | 3.05 | ||||
| CC vs. (CT or TT)* | No | 0.91 | No | 0.67 | 1.23 | ||||
| German | 153/163 | CT vs. CC | No | 1.07 | No | 0.59 | 1.93 | (24) | |
| TT vs. CC | No | 0.53 | No | 0.07 | 3.46 | ||||
| CC vs. (CT or TT)* | No | 1.01 | No | 0.58 | 1.75 | ||||
| Swedish | 924/934 | CT vs. CC | No | 1.02 | No | 0.81 | 1.28 | (24) | |
| TT vs. CC | No | 1.11 | No | 0.45 | 2.71 | ||||
| CC vs. (CT or TT)* | No | 1.03 | No | 0.82 | 1.28 | ||||
| Chinese | 1109/1195 | CT vs. CC | No | 1.01 | No | 0.85 | 1.21 | (26) | |
| TT vs. CC | Yes (decreases risk) | 0.65 | No | 0.41 | 1.02 | ||||
| CC vs. (CT or TT)* | No | 0.97 | No | 0.81 | 1.15 | ||||
| Caucasian (98%) | 488/479 | CC vs. (CT or TT) | No | 1.02 | Yes | 0.76 | 1.36 | (27) | |
| UK Caucasian | 848/708 | TT vs. CC* | No | 1.42 | No | 0.67 | 3.07 | (28) | |
| CC vs. (CT or TT)* | No | 1.08 | No | 0.85 | 1.36 | ||||
| Polish Caucasian | 319/290 | TT vs. CC* | No | 0.82 | No | 0.26 | 2.6 | (32) | |
| CC vs. (CT or TT) | Yes (decreases risk) | 0.69 | No | 0.48 | 0.99 | ||||
| NR | 60/60 | CC vs. (CT or TT)* | Yes (increases risk) | 6.91 | Yes | 1.84 | 32 | (30) | |
| −2578 C>A2 | Polish | 411/423 | CA vs. CC | No | 0.96 | No | 0.68 | 1.36 | (24) |
| AA vs. CC | No | 1.04 | No | 0.7 | 1.54 | ||||
| AA vs. (CA or CC)* | No | 1.07 | No | 0.78 | 1.46 | ||||
| German | 153/162 | CA vs. CC | No | 1.18 | No | 0.68 | 2.06 | (24) | |
| AA vs. CC | No | 0.97 | No | 0.5 | 1.86 | ||||
| AA vs. (CA or CC)* | No | 0.87 | No | 0.51 | 1.49 | ||||
| Swedish | 939/940 | CA vs. CC | No | 0.99 | No | 0.79 | 1.24 | (24) | |
| AA vs. CC | No | 1 | No | 0.77 | 1.29 | ||||
| AA vs. (CA or CC)* | No | 1 | No | 0.81 | 1.24 | ||||
| 98% Caucasian | 498/495 | CA vs. AA | No | 1.18 | Yes | 0.86 | 1.61 | (27) | |
| AA vs. CC† | No | 0.8 | Yes | 0.56 | 1.14 | ||||
| 89% Cau + 7% AA | NR (total 656) | AA vs. (CA or CC) | Yes (increases risk) | 1.99 | Yes | 1.06 | 3.74 | (31) | |
| −1154 G>A3 | NR | 134/263 | AA vs. GG* | No | 0.61 | No | 0.27 | 1.3 | (23) |
| Polish | 411/423 | GA vs. GG | No | 1.22 | No | 0.91 | 1.65 | (24) | |
| AA vs. GG | No | 1.23 | No | 0.76 | 1.97 | ||||
| German | 159/163 | GA vs. GG | No | 0.76 | No | 0.46 | 1.25 | (24) | |
| AA vs. GG | No | 0.85 | No | 0.39 | 1.89 | ||||
| 98% Caucasian | 495/492 | GA vs. AA | No | 1.01 | Yes | 0.66 | 1.55 | (27) | |
| AA vs. GG† | No | 0.78 | Yes | 0.51 | 1.19 | ||||
| −634 G>C4 | Chinese | 1095/1198 | CG vs. CC | No | 0.81 | No | 0.64 | 1.02 | (26) |
| CC vs. GG† | No | 1.12 | No | 0.88 | 1.44 | ||||
| UK Caucasian | 490/498 | CG vs. CC* | No | 1.03 | No | 0.68 | 1.56 | (28) | |
| CC vs. GG* | No | 0.82 | No | 0.54 | 1.26 | ||||
| GC vs. GG* | No | 0.85 | No | 0.65 | 1.12 | ||||
| Swedish | 936/941 | GC vs. GG | No | 1 | No | 0.82 | 1.21 | (24) | |
| CC vs. GG | No | 1.05 | No | 0.74 | 1.47 | ||||
| CG vs. CC* | No | 0.95 | No | 0.68 | 1.34 | ||||
| 98% Caucasian | 495/500 | GC vs. GG | No | 1.05 | Yes | 0.8 | 1.37 | (27) | |
| CC vs. GG | No | 1.18 | Yes | 0.76 | 1.83 | ||||
| −1498 C>T5 | Chinese | 1123/1222 | CT vs. TT | No | 0.94 | No | 0.79 | 1.12 | (26) |
| CC vs. TT | No | 1.23 | No | 0.89 | 1.7 | ||||
| CC vs. (CT or TT)* | No | 1.26 | No | 0.92 | 1.73 | ||||
| UK Caucasian | 493/498 | CT vs. TT* | No | 1.04 | No | 0.77 | 1.4 | (28) | |
| CC vs. TT* | No | 1.1 | No | 0.77 | 1.59 | ||||
| CC vs. (CT or TT)* | No | 1.07 | No | 0.8 | 1.45 | ||||
| 89% Cau + 7% AA | NR (total 656) | CC vs. (CT or TT) | Yes (increases risk) | 2.01 | Yes | 1.08 | 3.76 | (31) | |
| −1498T/−634G/936T | Chinese | Ref: −1498T/−634C/936C | Yes (decreases risk) | 0.67 | No | 0.43 | 1.04 | (26) | |
| −1498T/−634G/936C | No | 0.97 | No | 0.84 | 1.11 | ||||
| −1498C/−634G/936C | No | 1.02 | No | 0.86 | 1.21 | ||||
| −1498C/−634G/936T | No | 0.96 | No | 0.78 | 1.18 | ||||
| −1498T/−634C/936T | No | 0.98 | No | 0.76 | 1.25 | ||||
| −2578A/−1154A/−634G | 98% Caucasian | 1 HC vs. 0 | Yes (decreases risk) | 0.69 | Yes | 0.51 | 0.92 | (27) | |
| 2 HC vs. 0 | Yes (decreases risk) | 0.62 | Yes | 0.38 | 1 | ||||
| −2578A/−1154G/−634G | 1 HC vs. 0 | No | 0.97 | Yes | 0.71 | 1.33 | |||
| 2 HC vs. 0 | No | 1.71 | Yes | 0.7 | 4.15 | ||||
| −2578C/−1154G/−634G | 1 HC vs. 0 | No | 1.27 | Yes | 0.94 | 1.72 | |||
| 2 HC vs. 0 | No | 1.68 | Yes | 0.78 | 3.61 | ||||
| −2578C/−1154G/−634C | 1 HC vs. 0 | No | 1.07 | Yes | 0.8 | 1.43 | |||
Calculated odds ratio
OR inverted from original publication. HC-Haplotype copy numbers
Test for homogeneity of odds ratio: Pooled OR (95%CI); CT vs. CC, p=0.86, 0.99 (0.88–1.13), TT vs. CC, p=0.61, 0.86 (0.64–1.16), CC vs. (CT or TT), p=0.0010, 0.91 (0.76–1.10)
Test for homogeneity of odds ratio: Pooled OR (95%CI); CA vs. CC, p=0.82, 1.00 (0.84–1.20), AA vs. CC, p=0.73, 0.95 (0.80–1.13), AA vs. (CA or CC), p=0.20, 1.08 (0.86–1.35)
Test for homogeneity of odds ratio: Pooled OR (95%CI); AA vs. GG, p=0.36, 0.89 (0.67–1.18), GA vs. GG, p=0.11, 1.01 (0.64–1.59)
Test for homogeneity of odds ratio: Pooled OR (95%CI); GC vs. GG, p=0.52, 0.97 (0.85–1.11), GC vs. CC, p=0.52, 0.88 (0.74–1.05), CC vs. GG, p=0.62, 1.06 (0.89–1.25)
Test for homogeneity of odds ratio: Pooled OR (95%CI); CT vs. TT, p=0.57, 0.96 (0.83–1.12), CC vs. TT, p=0.66, 1.17 (0.92–1.49), CC vs. (CT or TT), p=0.21, 1.23 (1.00–1.51)
A case-control study, with 500 subjects in each group, evaluating the role of the germline VEGF 936C>T SNP in Austrian subjects, found that subjects with variant 936T allele were at reduced risk of breast cancer (22). However, the 936C>T SNP was not related to prognostic factors such as tumor size, histological grades, lymph node status, estrogen and progesterone receptor status or age at diagnosis (22). In a small subset of 21 healthy, non-smoking postmenopausal women, the 936T allele was associated with significantly lower plasma concentrations of VEGF, which may partially explain, the reduced risk of cancer associated with this allele (22). A smaller study from the UK, including 144 cases and 263 controls, evaluated the effect of SNPs in the promoter regions of various cytokines, including the VEGF −1154G>A transition, on risk and prognosis of breast cancer. The −1154G>A allele was not associated with risk of breast cancer; however, carriers of heterozygous −1154AG genotype displayed a trend towards favorable prognosis (23). A larger case-control study including 571 patients with familial breast cancer from Germany and Poland, and 974 patients with sporadic breast cancer from Sweden failed to find a relation between VEGF SNPs (−2578C>A, −1154G>A, −634G>C, and 936C>T) or their haplotypes and risk of breast cancer (24). This finding is in contrast with the earlier report (22) suggesting protective role of 936T allele. However, patients with the −634CC genotype and −2578C/−634 C haplotype had larger, more histologically advanced tumors while −2578AA and −2578A/−634G had low histological grade tumors, suggesting that these SNPs might act as genetic marker for tumor aggressiveness (24).
Two different publications (25, 26) evaluated data from the Shanghai Breast Cancer Study, a large population based case-control study to evaluate the impact of germline VEGF −1498C>T, −634G>C, and 936C>T SNPs on survival and risk of breast cancer in Chinese women. Based on data from 1133 cases and 1233 age-matched controls, the −1498 CC allele was associated with shorter overall and disease-free survival (age adjusted hazard ratio of 1.5 for both) (25), but had no association with risk of breast cancer (26). The carriers of the −634GG allele had significantly shorter overall survival than non-carriers with an age adjusted hazard ratio of 1.6 but the disease-free survival and risk were not different between these groups (25, 26), while previously the complementary allele −634CC was shown to be related with advanced tumors (24). The 936C>T SNP was not related to overall or disease-free survival (25), but the 936TT genotype carriers were at reduced risk of cancer (OR, 0.65) (26), agreeing with Krippl et al. (22) despite of differences in ethnicity. The subset analysis based on menopausal status explained that the protective role of the 936TT genotype was restricted to pre-menopausal women (OR, 0.45). Haplotype analysis demonstrated that −1498T/−634C/936C was related with longer overall survival (hazard ratio (HR), 0.57), and pre-menopausal women with −1498T/−634G/936T were at reduced risk of breast cancer (OR, 0.47) compared to individuals with the reference −1498C/−634G/936T haplotype (25, 26).
A study evaluating the impact of four previously studied (24) VEGF SNPs (−2578C>A, −1154G>A, −634G>C and 936C>T) in Caucasian population from a different geographic location (i.e. USA), including 501 cases and 504 controls, also failed to find any association between these SNPs and the risk of breast cancer (27), disregarding the spread of disease. However, subgroup analysis in patients with invasive (n=380) and in-situ (n=107) breast cancer showed higher risk of invasive, but not of in-situ, cancer in carriers of −2578CC (OR, 1.46) and −1154GG (1.64) genotypes (27). Conversely, the 936C allele was not associated with invasive cancer, but was related to reduced risk of in-situ breast cancer (OR, 0.59) (27). Notably, this finding is in contrast with the majority of previous studies where patients with the 936T allele had lower risk of breast cancer (22, 26). The −634G>C SNP had no association with risk of either in-situ or invasive breast cancer. These results demonstrate the possible differences in genetic markers based on stage of disease (i.e., localized or metastatic). Haplotype analysis showed that one or two copies of −2578A/−1154A/−634G resulted in lower risk of invasive breast cancer compared to zero-copy carriers (OR, 0.69 and 0.62, respectively). Assessment of promoter diplotypes suggested no significant association with risk of invasive breast cancer (27). Another case-control study in 500 Caucasian patients and 500 matched controls analyzed the effect of germline VEGF −1498C>T, −634G>C, −7C>T and 936C>T SNPs on risk and severity of breast cancer (assessed by tumor size, tumor grade, nodal involvement, vascular invasion and oestrogen receptor status), survival of patients, circulating levels of VEGF and VEGF expression in tumor (28). Singularly, none of these SNPs were related to risk, but patients carrying the −1498T/−634C/−7C/936C haplotype were at lower risk of breast cancer than non-carriers. Except for −7C>T transition, which predicted longer overall survival, no other SNPs were related with survival (28). The serum and plasma VEGF levels and tumor VEGF expression were not related with any of these four SNPs (28). A large case-control study including 804 breast cancer patients and 804 age-matched healthy volunteers from Austria found that germline VEGF −2578C>A, −2489C>T, −1498C>T, −634G>C, −7C>T, 936C>T and 1612G>A SNPs or their haplotypes were not related with risk of breast cancer (29). However the −634C allele was related with small tumor size (29), contrasting an earlier report (24) showing −634CC allele’s relation with high tumor aggressiveness. A small case-control study with 60 patients and 60 age matched healthy volunteers found no carriers of the 936TT genotype in two groups, and the frequency of the 936CT genotype was significantly higher in patients compared to controls (30). The 936C>T SNP had no association with clinicopathological characteristics of the patients(30).
An epidemiological study with predominantly Caucasian subjects, including 715 controls and 520 breast cancer cases, evaluated the impact of alleles in seven genes, including the VEGF −2578C>A, −1498C>T, −634G>C, −1154G>A, and 936C>T SNPs on risk of breast cancer. The −2578AA and −1498CC genotypes had higher risk of breast cancer, independent of non-genetic risk factors estimated by the Gail model (respective adjusted OR, 1.99 and 2.01), while other VEGF SNPs had no association with disease risk (31). Finally, a case-control study in Polish Caucasian subjects, including 319 breast cancers, 146 ovarian cancers, and 290 unaffected controls, investigated the effect of the 936C>T SNP in modifying the hereditary risk of breast and ovarian cancer in women harboring a mutated BRCA1 gene (32). Carriers of the VEGF 936 T allele and BRCA1 mutation had reduced risk of breast cancer (OR, 0.63 for CT+TT genotype) but the risk of ovarian cancer was not affected (32). These findings suggest a protective role for the 936T allele in women at risk of breast cancer carrying BRCA1 mutations.
Recently two studies (33, 34) evaluated the relationship of VEGF plasma levels or somatic VEGF and VEGFR-2 SNPs with treatment efficacy or toxicity. One of them compared the effect of paclitaxel with paclitaxel and bevacizumab combination in metastatic breast cancer by enrolling 183 patients in each arm (33). The VEGF −2578AA had longer median overall survival (HR 0.58) compared to the −2578CA+CC genotype in the combination arm (33). Similarly −1154A allele had longer overall survival (HR 0.62) in the combination arm, which increased additively with increase in number of A alleles (33). VEGF −634CC and −1498TT had significantly less likelihood of developing grade 3 or 4 hypertension in the combination arm compared to −634 GC+GG and −1498 CT+CC respectively (33). The VEGF 936C>T, VEGFR-2 889G>A and VEGFR-2 1416A>T had no significant relationship with either the efficacy or toxicity. Another study enrolled 56 breast cancer patients to examine the role of plasma VEGF levels as predictor of treatment outcome with bevacizumab and vinorelbine therapy (34). Lower levels of baseline VEGF predicted longer progression free survival (34).
In conclusion, VEGF −634G>C (24, 26–29, 31), −7C>T (26, 28, 29), and −2489C>A (26, 29) SNPs were not related with risk of breast cancer. VEGF −1154G>A had no relation with overall risk of breast cancer (23, 24, 27, 31), however the G allele was related with increased risk of invasive disease (23, 27, 31). VEGF −1498C>T was shown to have no relation (26, 28, 29) or predicted higher risk of breast cancer (31). Results for VEGF −2578C>A were inconsistent, i.e., no association with risk (23, 24, 27, 29) and higher risk of invasive disease with the C allele (23, 27, 31) or A allele (31). For the 936C>T SNP, three studies have shown a protective role of the 936T allele(22, 26, 32), four failed to find any correlation with risk (24, 28, 29, 31) and one found 936 C as a protective allele for in-situ breast cancer (27). Differences in geographic location of the studied population did not explain the differences in the role of the 936C>T SNP. Results of meta-analysis (Table 2) show that, except for 936 CC vs. (CT or TT), odds ratios for all other genotype comparisons were homogenous to be combined across studies. Despite a small number of studies that disagree with the others, the pooled analyses for these genotypes appear not to have found any significant associations with breast cancer risk.
VEGF SNPs in NSCLC
High VEGF mRNA or protein expression in tumor (35) or in serum have shown to be associated with advanced tumor stage (IIIB and IV) (36), shorter survival (37) and prognosis of NSCLC (38). VEGF mRNA and protein expression were found to be significantly higher in adenocarcinomas than in squamous-cell carcinomas (35). These observations suggest that SNPs in the VEGF gene may affect the prognosis or clinical outcome of NSCLC by altering the VEGF levels; studies evaluating this hypothesis are summarized in Table 3.
Table 3.
Summary of case-control studies assessing the VEGF polymorphisms in relation to risk of NSCLC
| Polymorphism | Ethnicity | Case/Control | Genotype | Association of Cancer Risk with respect to VEGF Genotype |
Gender | OR | L95%CI | U95%CI | Ref |
|---|---|---|---|---|---|---|---|---|---|
| 936 C>T | Caucasian | 1900/1458 | (CT or TT) vs. CC | No | All | 1.05 | 0.87 | 1.27 | (39) |
| No | Male | 1.13 | 0.87 | 1.5 | |||||
| No | Female | 0.99 | 0.27 | 1.29 | |||||
| −1498 C>T | Caucasian | 1900/1458 | (CT or CC) vs. TT | No | All | 1.16 | 0.96 | 1.4 | (39) |
| No | Male | 1.07 | 0.81 | 1.4 | |||||
| No | Female | 1.27 | 0.98 | 1.65 | |||||
| −634 G>C | Caucasian | 1900/1458 | (CG or CC) vs. GG | No | All | 1.14 | 0.96 | 1.36 | (39) |
| Yes (increases risk, lung adenocarcinoma) | Male | 1.27 | 0.99 | 1.63 | |||||
| No | Female | 1.04 | 0.82 | 1.32 | |||||
| −1498T/−634C/936C | Caucasian | 1900/1458 | Ref −1498C/−634G/936C | No | All | 1.02 | 0.88 | 1.17 | (39) |
| No | Male | 1.15 | 0.94 | 1.42 | |||||
| No | Female | 0.91 | 0.74 | 1.11 | |||||
| −1498T/−634G/936C | Caucasian | 1900/1458 | Ref −1498C/−634G/936C | No | All | 0.88 | 0.75 | 1.04 | (39) |
| Yes (decreases risk, lung adenocarcinoma) | Male | 0.81 | 0.63 | 1.02 | |||||
| No | Female | 0.93 | 0.74 | 1.17 | |||||
| −1498C/−634G/936T | Caucasian | 1900/1458 | Ref −1498C/−634G/936C | No | All | 0.98 | 0.74 | 1.31 | (39) |
| No | Male | 0.9 | 0.59 | 1.36 | |||||
| No | Female | 1.04 | 0.69 | 1.55 | |||||
| −1498T/−634C/936T | Caucasian | 1900/1458 | Ref −1498C/−634G/936C | No | All | 1.14 | 0.84 | 1.55 | (39) |
| No | Male | 1.15 | 0.72 | 1.85 | |||||
| No | Female | 1.04 | 0.69 | 1.57 | |||||
A small study on DNA samples extracted from tumor and normal lung tissues from 36 surgically resected patients, examined the effect of VEGF −2578C>A, −634G>C, −1154G>A, and 936C>T SNPs on tumor VEGF expression and vascular density (14). The −2578 CC, −634 GG and −1154 AA and GA genotypes were associated with low VEGF expression, while high VEGF levels were detected in samples carrying the −2578 CA, −634 GC and −1154 GG genotypes (14). The MVD in tumors bearing the −2578 CC and −634 GG genotypes was significantly lower compared to the −2578 CA and −634 CC+GC genotypes, respectively. VEGF −1154G>A and 936C>T were not related with tumor VEGF expression and MVD (14). A large case-control study on 1900 NSCLC Caucasian patients and 1458 healthy controls failed to find a correlation between germline VEGF −1498C>T, −634C>G and 936C>T SNPs taken singularly on risk of NSCLC after adjusting for age, gender, smoking status, pack-years of smoking, and years since smoking cessation (39). However, male patients bearing the −634CC+CG genotype and−1498T/−634G/936C haplotype respectively had higher and reduced risk of developing lung adenocarcinoma (39). The lack of association in female patients was unclear and was not explained by differences in smoking status between genders (39). Another study on 462 patients with early-stage NSCLC treated with surgical resection, examined the impact of germline 936C>T, −1498C>T, and −634G>C SNPs on survival (3). Carriers of −634C and 936T alleles had longer five-year overall survival than non-carriers, with respective survival percent of 61% vs. 51% and 67% vs. 54% (3). While considering combined effect of 936C>T and −634G>C SNPs, increased number of pooled variant alleles resulted in significantly longer survival, with adjusted HR for one, two and three variant allele(s) being 0.76, 0.61, and 0.61, respectively compared to the zero variant allele (3). The −1498C>T SNP had no association with overall survival in these patients (3).
In conclusion, male subjects carrying the −634C allele appear to be at increased risk of lung adenocarcinoma (39). Considering both males and females together, −634G>C, 936C>T and −1498C>T had no significant association with risk of disease (39) but variant allele for these SNPs predicted longer overall survival (3). The −1498T/−634G/936C haplotype may predict the risk of NSCLC. The −2578C>A, −1154G>A, and −634G>C SNPs were associated with VEGF expression and angiogenesis, but these observations were limited by small sample size (14). There were not enough studies (Table 3) to perform meta-analysis.
VEGF SNPs in colorectal cancer
Like other solid tumors, overexpression of VEGF mRNA and protein has shown to be associated with tumor progression and poor prognosis of colon carcinoma (40). VEGF has shown to be an important predictor of prognosis in cases of advanced CRC (41). High MVD and VEGF expression were shown to be related with poor relapse free and overall survival (42). Therefore, VEGF SNPs may play important role in determining the risk, prognosis and survival in colorectal cancer patients; studies evaluating this hypothesis are summarized in Table 4.
Table 4.
Summary of case-control studies assessing the VEGF polymorphisms in relation to risk of Colon cancer
| Polymorphism | Ethnicity | Case/Control | Genotype | Association of Cancer Risk with Respect to VEGF Genotype |
OR | L95%CI | U95%CI | Ref |
|---|---|---|---|---|---|---|---|---|
| 936C>T1 | Korean | 262/229 | CT vs. CC | No | 1.45 | 0.97 | 2.16 | (43) |
| TT vs. CC | No | 2.98 | 0.79 | 11.21 | ||||
| (CT or TT) vs. CC | Yes (increases risk) | 1.52 | 1.03 | 2.25 | ||||
| NR | 465/413 | CT vs. CC | No | 0.89 | 0.67 | 1.18 | (45) | |
| TT vs. CC | No | 1.19 | 0.5 | 2.35 | ||||
| (CT or TT) vs. CC | No | 0.92 | 0.7 | 1.21 | ||||
| T vs. C | No | 0.96 | 0.76 | 1.21 | ||||
| Austrian | 427/427 | CT vs. CC* | No | 0.76 | 0.55 | 1.05 | (48) | |
| TT vs. CC* | No | 0.68 | 0.27 | 1.7 | ||||
| (CT or TT) vs. CC* | No | 0.75 | 0.55 | 1.02 | ||||
| −2578C>A2 | Korean | 246/203 | CA vs. CC | No | 0.72 | 0.49 | 1.07 | (44) |
| AA vs. CC | No | 0.66 | 0.31 | 1.43 | ||||
| (CA or AA) vs. CC | No | 0.71 | 0.49 | 1.04 | ||||
| Austrian | 433/427 | CA vs. CC* | No | 1 | 0.7 | 1.43 | (48) | |
| AA vs. CC* | No | 1.31 | 0.88 | 1.95 | ||||
| (CA or AA) vs. CC* | No | 1.1 | 0.78 | 1.54 | ||||
| −634G>C3 | NR | 465/413 | GC vs. GG | Yes (decreases risk) | 0.55 | 0.41 | 0.75 | (45) |
| CC vs. GG | No | 0.81 | 0.55 | 1.18 | ||||
| (GC or CC) vs. GG | Yes (decreases risk) | 0.62 | 0.47 | 0.83 | ||||
| C vs. G | No | 0.86 | 0.71 | 1.04 | ||||
| Austrian | 432/430 | GC vs. GG* | No | 0.98 | 0.74 | 1.3 | (48) | |
| CC vs. GG* | No | 1.09 | 0.69 | 1.72 | ||||
| (GC or CC) vs. GG* | No | 1 | 0.76 | 1.31 | ||||
| −634C/936C | NR | Ref: −634G/936C | Yes (decreases risk) | 0.53 | 0.43 | 0.66 | (45) | |
| −634G/936T | NR | Ref: −634G/936C | Yes (decreases risk) | 0.56 | 0.42 | 0.74 | ||
Calculated odds ratio. NR-Not Reported
Test for homogeneity of odds ratio: Pooled OR (95%CI); CT vs. CC, p=0.042, 0.98 (0.70–1.37), TT vs. CC, p=0.20, 1.17 (0.57–2.39), (CT or TT) vs. CC, p=0.019, 1.00 (0.69–1.44)
Test for homogeneity of odds ratio: Pooled OR (95%CI); CA vs. CC, p=0.22, 0.86 (0.62–1.19), AA vs. CC, p=0.12, 1.01 (0.53–1.93), (CA or AA) vs. CC, p=0.095, 0.89 (0.58–1.36)
Test for homogeneity of odds ratio: Pooled OR (95%CI); GC vs. GG, p=0.0070, 0.74 (0.42–1.30), CC vs. GG, p=0.33, 0.91 (0.68–1.22), (GC or CC) vs. GG, p=0.018, 0.79 (0.49–1.26)
A study that analyzed VEGF SNPs in peripheral blood and their impact on prognosis in 125 patients with locally advanced colon cancer found that the VEGF 936CC genotype had a significantly shorter median time to relapse compared to CT or TT genotypes (2.6 vs. 11.1 years) (40). A case-control study on the VEGF 936C>T germline SNP in Korean colon cancer patients (n=262) and healthy controls (n=229) demonstrated that the frequency of T allele-bearing genotypes (CT+TT) was higher in cases than in controls. When stratified by gender and age, the frequency of CT+TT was significantly associated with increased risk for colon cancer in women and patients younger than 55 years, thus suggesting that the VEGF 936C>T SNP might be a genetic determinant for colon cancer in Korean patients (43). Another case-control study on the VEGF −2578C>A germline SNP was carried out in a cohort of 246 patients with colon cancer and 203 healthy controls and found that VEGF −2578C>A SNP had no association with susceptibility to colon cancer, although gender stratification indicated that the −2578CA and AA genotypes had a marginal protective effect for colon cancer in women. Therefore, the VEGF −2578C>A SNP, at least in Koreans, appears to have a borderline role in colon cancer risk (44). The analysis of two VEGF SNPs (−634G>C and 936C>T) in colorectal tissue from 465 patients and peripheral blood lymphocytes from 413 healthy controls demonstrated that the −634 GC and CC genotypes were associated with a decreased risk of cancer, indicating a dominant role of the C allele, whereas the 936TT genotype correlated with advanced stage, high serum levels of CA19-9, and higher histologic grades (45). Haplotype analysis showed that −634C/936C and −634G/936T were associated with decreased susceptibility of colorectal cancer (45). The VEGF −1498C>T, −634G>C and −7C>T SNPs, were determined in 36 Japanese patients and demonstrated that the −1498CC allele, but not the −634G>C and −7C>T SNPs, was predictive of poorly-differentiated tumors and thereby poor prognosis (46). A larger study on 445 patients with colorectal cancer examined the VEGF −2578C>A, −634G>C, and 936C>T SNPs and showed that the survival for patients with the −634GC or CC genotypes was better than for those with the −634GG genotype (47). Furthermore, the 936CT or TT genotypes were associated with a worse survival compared with the 936CC genotype and −2578C>A was not related with survival (47). Finally, in haplotype analysis, the −2578A/−634G/936T exhibited a significantly less favorable survival when compared with the wild type −2578C/−634G/936C haplotype (47). A case-control study examined VEGF 936C>T, −2578C>A, and −634G>C SNPs in 427 patients and 427 age-and sex-matched healthy control subjects and found no correlation between all these variants and tumor size, histological grading, positive regional lymph node metastases or tumor stage (48). Patients treated with cetuximab were genotyped to assess the possible predictive role of SNPs of cyclin D1, cyclooxygenase 2, EGF, EGFR, IL-8 and VEGF genes in blood samples. While a significant association was found between cyclin D1 870A>G and the EGF 61A>G SNPs and survival, VEGF 936C>T showed no association (49). Finally, a study on RNA from 25 colorectal cancer tissues demonstrated the presence of VEGF121, VEGF145, VEGF165 and VEGF189 splice variants and an array of novel mutations within the conserved expression site of the gene. Five samples exhibited single nucleotide changes and in one sample a 2-nucleotide deletion was detected. In addition to this, a 1- and 2-base deletion with frameshift and protein truncation in exon 3 at positions +172 and +171/172 were shown, respectively, as well as a transition mutation in exon 3 at position +248 and two transition mutations in exon 4 at positions +398 and +403. The biologic and clinical impact of these variants were not demonstrated, however (50).
In conclusion, 936 T allele has shown to be associated with increased risk (43), advanced stage of disease (45), worse survival (47) and longer time to relapse (40), while other studies have shown no correlation with tumor size, grade and stage (48) and no relation with survival (49) in patients with colorectal cancer. The −634 C allele was predictive of decreased risk (45) and better survival (47), and was not related with tumor size, grade and stage (48). The VEGF −2578C>A SNP was not related with risk (44), survival (47), tumor size and stage (48), but −2578 CA and AA had borderline protective role in Korean women (44). The results of meta-analysis shown in Table 4 suggest that ORs for several genotypic comparisons were not homogeneous to be pooled across studies (p-value for test of homogeneity, <0.05). The 95% CI for all the genotypic comparison for which ORs were homogeneous and could be pooled together included 1, suggesting no significant correlation of selected VEGF SNPs with risk of colorectal cancer.
VEGF SNPs in prostate cancer
Like some solid tumors, progressive growth and metastasis of prostate cancer is also dependent on angiogenesis. VEGF expression and vascular density were significantly higher in prostate cancer tissue compared to benign prostate hypertrophy (BPH) or normal prostate tissue (5, 51). Metastatic prostate cells had enhanced VEGF production and tumor vascularity than cells with low metastatic potential (52), suggesting the importance of VEGF in prostate cancer metastasis. Studies evaluating the role of VEGF SNPs in prostate cancer susceptibility, progression and metastasis are summarized in Table 5.
Table 5.
Summary of case-control studies assessing the VEGF polymorphisms in relation to risk of prostate cancer
| Polymorphism | Ethnicity | Case/ Control |
Genotype | Association of Cancer Risk with Respect to VEGF Genotype |
OR | L95%CI | U95%CI | OR Adjusted | L95%CI | U95%CI | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1498C>T1 | Taiwanese | 96/119 | CT vs. CC | No | 0.4 | 0.1 | 1.9 | 0.3 | 0.1 | 1.5 | (54) |
| TT vs. CC | No | 1.4 | 0.3 | 5.9 | 1.1 | 0.2 | 4.9 | ||||
| T vs. C | Yes (increases risk) | 2.3 | 1.4 | 3.8 | 2.2 | 1.3 | 3.8 | ||||
| Turkish | 133/157 | CT vs. CC | No | 1.44 | 0.84 | 2.42 | (56) | ||||
| TT vs. CC | No | 1.28 | 0.51 | 3.2 | |||||||
| (CT or TT) vs. CC | No | 1.42 | 0.84 | 2.37 | |||||||
| Japanese | 270/252 | CT vs. TT | No | 0.98 | 0.68 | 1.4 | (51) | ||||
| TT vs. CC† | No | 1.02 | 0.97 | 1.39 | |||||||
| (CT or TT) vs. CC* | No | 1.03 | 0.57 | 1.88 | |||||||
| −1154G>A2 | Tunisian | 101/100 | GA vs. GG | Yes (decreases risk) | 0.46 | 0.24 | 0.87 | (55) | |||
| AA vs. GG | Yes (decreases risk) | 0.27 | 0.08 | 0.83 | |||||||
| (GA or AA) vs. GG | Yes (decreases risk) | 0.42 | 0.23 | 0.76 | |||||||
| (GA or GG) vs. AA* | No | 0.39 | 0.12 | 1.14 | |||||||
| UK Caucasian | 238/263 | GA vs. GG* | No | 1.05 | 0.72 | 1.55 | (53) | ||||
| (GA or GG) vs. AA | Yes (decreases risk) | 0.45 | 0.24 | 0.86 | |||||||
| AA vs. GG* | Yes (decreases risk) | 0.46 | 0.22 | 0.93 | |||||||
| (GA or AA) vs. GG* | No | 0.91 | 0.63 | 1.32 | |||||||
| −634G>C | Tunisian | 101/100 | GC vs. GG | No | 1.88 | 0.98 | 3.62 | (55) | |||
| CC vs. GG | No | 2.28 | 0.82 | 6.38 | |||||||
| (GC or CC) vs. GG | Yes (increases risk) | 1.95 | 1.04 | 3.65 | |||||||
| Austrian | 702/702 | CC vs. GG* | No | 1.08 | 0.86 | 1.35 | (58) | ||||
| 936C>T | Tunisian | 101/100 | CT vs. CC | No | 0.68 | 0.33 | 1.37 | (55) | |||
| TT vs. CC | No | 1.82 | 0.13 | 51.93 | |||||||
| (CT or TT) vs. CC | No | 0.72 | 0.36 | 1.43 | |||||||
| Austrian | 702/702 | TT vs. CC* | No | 1.16 | 0.91 | 1.48 | (58) | ||||
| −2578C>A | Austrian | 702/702 | CC vs. AA* | No | 0.96 | 0.75 | 1.24 | (58) | |||
| −2489C>T | Austrian | 702/702 | CC vs. TT* | No | 0.97 | 0.75 | 1.25 | (58) | |||
| −1498C>T | Austrian | 702/702 | CC vs. TT* | No | 1 | 0.77 | 1.28 | (58) | |||
| −7C>T | Austrian | 702/702 | CC vs. TT* | No | 1.09 | 0.86 | 1.39 | (58) | |||
| 1612G>A | Austrian | 702/702 | GG vs. AA* | No | 0.95 | 0.74 | 1.23 | (58) | |||
| −1154G/−634C | Tunisian | 101/100 | Ref: −1154G/−634G | No | 1.12 | 0.67 | 1.88 | (55) | |||
| −1154A/−634G | Tunisian | Yes (decreases risk) | 0.48 | 0.28 | 0.84 | ||||||
| −1154A/−634C | Tunisian | No | 0.91 | 0.28 | 2.99 | ||||||
Calculated odds ratio
OR inverted from original publication
Test for homogeneity of odds ratio: Pooled OR (95%CI); CT vs. CC, p=0.069, 0.80 (0.18–3.54), TT vs. CC, p=0.90, 1.05 (0.78–1.40), (CT or TT) vs. CC, p=0.42, 1.24 (0.84–1.83)
Test for homogeneity of odds ratio: Pooled OR (95%CI); GA vs. GG, p=0.029, 0.72 (0.32–1.63), AA vs. GG, p=0.41, 0.40 (0.22–0.72), (GA or AA) vs. GG, p=0.030, 0.64 (0.30–1.38)
A case-control study on 247 cancer patients and 263 controls from the UK examined SNPs in five cytokines including VEGF −1154 G>A and demonstrated that −1154 AA genotype had reduced risk of prostate cancer (OR, 0.45) (53). Another small case-control study on the VEGF −1498C>T germline SNP in 96 prostate cancer patients and 119 controls of Taiwanese descent found that carriers of −1498T allele were at 2.3 times higher risk of prostate cancer than non-carriers (54). However, this SNP was not related with prognosis (i.e., clinical stage and pathological tumor grade) and response to hormonal therapy (54). A case-control study in Tunisian subjects evaluated the influence of germline −1154G>A, −634G>C, and 936C>T VEGF SNPs on risk and prognosis of prostate cancer in 101 patients and 100 age-matched healthy controls (55). Carriers of at least one −1154 A allele were at reduced risk (OR, 0.42) and lower susceptibility to high-grade (OR, 0.25) and advanced prostate cancer (OR, 0.37). The odds ratio for −1154 GA and AA genotypes were 0.46 and 0.27, respectively (55), agreeing with McCarron et al. (53) despite of differences in ethnicity. The VEGF −634 GC+CC genotypes were at increased risk of prostate cancer (OR, 1.95), and the −634GC and CC genotypes were related with high-grade tumors (OR, 3.83 and 4.89, respectively) (55). Haplotype analysis showed that −1154A/−634G had reduced disease risk (OR, 0.48), high-grade tumor (OR, 0.22) and advanced disease (OR, 0.45) compared to −1154G/−634G (55). No correlation was observed between 936C>T SNP and risk or aggressiveness of tumor (55). These results suggest that both VEGF −1154G>A and −634G>C SNPs may act as important genetic marker of risk and aggressiveness of prostate cancer in Tunisian subjects. A case-control study in 133 sporadic prostate cancer patients and 157 healthy controls of Turkish descent evaluated the germline −1498C>T SNP and found no relation with susceptibility to disease (56), which was contrary to the findings for Taiwanese subjects (54). Further, the −1498C>T SNP was not related with the TNM staging, Gleason scores and serum prostate specific antigen (PSA) levels of patients (56). The impact of the same germline VEGF −1498C>T SNP in Japanese population was examined in a case-control study enrolling 270 patients and 252 controls. Similar to the Turkish population (56), the −1498C>T SNP was not related with disease risk, tumor grade and stage, Gleason scores, VEGF expression, MVD and age at diagnosis (51). The rate of PSA relapse after radical prostatectomy was higher in patients with the 1498CC or CT genotype (51), but the TT genotype had significantly worse cancer-specific and overall survival (51).
Another study in Tunisian subjects determined the combined effect of four germline SNPs which are involved in the process of angiogenesis, i.e., VEGF −1154G>A, VEGF −634G>C, MMP9 −1562C>T and TSP1 −8831A>G, in predicting the risk of prostate cancer by enrolling 101 patients and 74 controls (57). The VEGF −1154AA, VEGF −634GG, MMP9 −1562CC and TSP1 −8831AA genotypes were considered as low-risk genotypes and were used as reference, and the remaining genotypes for each SNP were considered as high-risk genotypes (57). Patients with 1, 2, or 3 high-risk genotypes in VEGF −1154/VEGF −634/MMP9 group had increasingly high risk of prostate cancer (OR, 2.79, 4.57 and 7.11 respectively), compared to zero high-risk genotype group (57). In the VEGF-1154/634/TSP1 group, patients carrying three high-risk genotypes had 6 times higher risk of prostate cancer (OR, 6.0) and were 20 times more prone to have high-grade tumors when compared with reference groups (OR, 20.75) (57). Overall, their findings show that the gene-gene interactions between high-risk angiogenic polymorphisms may increase the risk of prostate cancer and tumor aggressiveness (57). Finally, a case-control study in 702 patients and 702 age matched healthy controls from Austria analyzed 7 VEGF SNPs (-2578C>A, −2489C>T, −1498C>T, −634G>C, −7C>T, 936C>T and 1612G>A) and their haplotypes, and failed to find any relation between these SNPs and the risk of prostate cancer or VEGF plasma levels (58).
To conclude, studies from Austria, Japan, Taiwan, Tunisia, Turkey and the UK evaluated the importance of VEGF SNPs in prostate cancer. The VEGF −1154 A allele appears to predict the reduced risk (53, 55), which was consistent among studies as shown by meta-analysis (Table 5), where the pooled OR (95%CI) for −1154 AA genotype was 0.40 (0.22–0.72). The VEGF −1154A allele also imparted protection against high-grade tumor and tumor aggressiveness (55). Except for Taiwanese subjects, where −1498T allele predicted high risk of prostate cancer (54), the −1498C>T SNP was not related with risk (51, 56, 58) and disease prognosis (i.e., with tumor grade, stage, Gleason scores, VEGF expression and MVD) (51, 56, 58). The −634C allele was predictive of increased risk and high tumor grades (55) in Tunisian subjects but had no relation with risk in Austrian subjects (58). The results of meta-analysis showed no significant association between any other VEGF polymorphism and prostate cancer risk or clinical outcome.
Conclusions
Many studies have investigated the role of VEGF polymorphisms as genetic determinant for susceptibility and outcome of breast, prostate, NSCL and colorectal cancer. Several polymorphisms have been reported within the promoter (-2578C>A, −2489C>T, −1498C>T and −1154G>A), 5’-UTR (-634G>C and −7C>T) and 3’-UTR (936C>T and 1612G>A) for the VEGF gene. The variant allele for −1154G>A and 936C>T results in lower VEGF expression, whereas the variant allele for −1498C>T and −7C>T results in increased concentrations of VEGF mRNA (15, 59). The functional role of −2578C>A and −634G>C is not in agreement among studies, which report low or high VEGF production for variant alleles (15). Results of association studies of these polymorphisms with risk and outcome in breast, prostate, NSCL and colorectal cancer are also variable (Table 2–Table 5, Supplementary Table 6). For example the −936T allele has shown to impart protection against breast cancer (22, 26, 32), but conversely increases the risk of colorectal cancer (43). Similarly, the −634C allele was predictive of higher risk of NSCLC (39) and prostate cancer (55), but predicts reduced risk for colon cancer (45) and has no relation with risk of breast cancer (24, 26–29, 31). The lack of consensus about the role of these SNPs could be because of their linkage with other unknown functional SNPs in the VEGF gene or unknown SNPs in other factors of the angiogenesis pathway. Perhaps, association with haplotypes will be more predictive than individual SNPs. More understanding about the functionality of these polymorphisms and their role in tumor biology is essential to discern the reasons for inconsistencies in the literature (60), which may also help in designing association studies in the future. Other possible reasons are differences in study population (i.e., ethnicity and tumor type), small sample size (insufficient power), and failure to correct for multiple testing when studying the correlation with more than one SNP, which is important to avoid chance correlations. However, likelihood of chance correlations should be assessed with consideration to the biological plausibility of any observed correlation (60).
Independently, several of these studies reported association between VEGF SNPs and risk of cancer (Table 2–Table 5); however, the findings from these studies appear to be inconsistent. We performed meta-analysis to look at the combined results from studies addressing the same hypothesis. Except VEGF −1154G>A, no other VEGF SNP was found to be related with risk of studied tumors. The VEGF −1154A allele was at significantly reduced risk of prostate cancer (OR, 0.40; 95%CI, 0.22–0.72). Haplotype −2578A/−1154A/−634G and −1154A/−634G predicted reduced risk of breast (26) and prostate cancer (55).
The VEGF-mediated angiogenesis pathway involves a complex signaling network regulated by multiple factors and it is likely that final outcome is governed by interaction and cross-talk between these signaling molecules/receptors rather than the VEGF gene alone. In that case, a pathway approach, which includes combined analysis of variations in different genes in the angiogenesis pathway, may provide better predictive markers. For example carriers of three high risk genotypes i.e., VEGF −1154AA/VEGF −634GG/MMP9CC, had 7.11 times high risk of prostate cancer than VEGF −1154GG/VEGF −634CC/MMP9TT group (57). Genome-wide association (GWA) studies, performed to identify the candidate genes and chromosomal sites linked to cancer, showed that locus including the VEGF gene was not associated with risk of breast, NSCLC, colon and prostate cancer (61–71). However, our meta-analysis suggests that VEGF SNPs predict the risk of prostate cancer, indicating their role in vascular growth, at the minimal in prostate cancer, although they may not be as strongly predictive (i.e. penetrant) as other alleles in the genome. It should also be noted that there are other limitations to GWA approach, such as multiple testing in small population and low-density genotyping (72, 73), which may limit the identification of true susceptibility genes.
Most of the published studies assessing the role of VEGF polymorphisms compare the cases with controls to identify the disease markers. There is critical need to conduct more studies evaluating the relation of genotype with drug concentrations and response. These studies are equally important and helpful for identification of genetic markers to guide the choice of treatment and supportive care specific to each individual.
Acknowledgments
Financial support: Supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health (NIH), U.S. Department of Health and Human Services
Abbreviations
- VEGF
vascular endothelial growth factor
- SNP
single nucleotide polymorphism
- NSCLC
non-small cell lung cancer
- CRC
colorectal cancer
- MVD
micro-vessel density
- OR
odds-ratio
Footnotes
Conflict of Interest: There are no potential conflicts of interests to be disclosed
REFERENCES
- 1.Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 2.Schneider BP, Sledge GW., Jr Drug insight: VEGF as a therapeutic target for breast cancer. Nat Clin Pract Oncol. 2007;4:181–189. doi: 10.1038/ncponc0740. [DOI] [PubMed] [Google Scholar]
- 3.Heist RS, Zhai R, Liu G, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:856–862. doi: 10.1200/JCO.2007.13.5947. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi T, Bando H, Mori T, et al. Overexpression of soluble vascular endothelial growth factor receptor 1 in colorectal cancer: Association with progression and prognosis. Cancer Sci. 2007;98:405–410. doi: 10.1111/j.1349-7006.2007.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrer FA, Miller LJ, Andrawis RI, et al. Angiogenesis and prostate cancer: in vivo and in vitro expression of angiogenesis factors by prostate cancer cells. Urology. 1998;51:161–167. doi: 10.1016/s0090-4295(97)00491-3. [DOI] [PubMed] [Google Scholar]
- 6.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 7.Pharm GKB. [Accessed on 03/20/2009]; http://www.pharmgkb.org/ [Google Scholar]
- 8.Brogan IJ, Khan N, Isaac K, Hutchinson JA, Pravica V, Hutchinson IV. Novel polymorphisms in the promoter and 5' UTR regions of the human vascular endothelial growth factor gene. Hum Immunol. 1999;60:1245–1249. doi: 10.1016/s0198-8859(99)00132-9. [DOI] [PubMed] [Google Scholar]
- 9.Tischer E, Mitchell R, Hartman T, et al. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J Biol Chem. 1991;266:11947–11954. [PubMed] [Google Scholar]
- 10.Minchenko A, Salceda S, Bauer T, Caro J. Hypoxia regulatory elements of the human vascular endothelial growth factor gene. Cell Mol Biol Res. 1994;40:35–39. [PubMed] [Google Scholar]
- 11.Schultz A, Lavie L, Hochberg I, et al. Interindividual heterogeneity in the hypoxic regulation of VEGF: significance for the development of the coronary artery collateral circulation. Circulation. 1999;100:547–552. doi: 10.1161/01.cir.100.5.547. [DOI] [PubMed] [Google Scholar]
- 12.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–1235. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 13.Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000;37:443–448. doi: 10.1159/000054076. [DOI] [PubMed] [Google Scholar]
- 14.Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46:293–298. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Pander J, Gelderblom H, Guchelaar HJ. Pharmacogenetics of EGFR and VEGF inhibition. Drug Discov Today. 2007;12:1054–1060. doi: 10.1016/j.drudis.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Schneider BP, Miller KD. Angiogenesis of breast cancer. J Clin Oncol. 2005;23:1782–1790. doi: 10.1200/JCO.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Linderholm B, Tavelin B, Grankvist K, Henriksson R. Vascular endothelial growth factor is of high prognostic value in node-negative breast carcinoma. J Clin Oncol. 1998;16:3121–3128. doi: 10.1200/JCO.1998.16.9.3121. [DOI] [PubMed] [Google Scholar]
- 18.Foekens JA, Peters HA, Grebenchtchikov N, et al. High tumor levels of vascular endothelial growth factor predict poor response to systemic therapy in advanced breast cancer. Cancer Res. 2001;61:5407–5414. [PubMed] [Google Scholar]
- 19.Gasparini G, Toi M, Gion M, et al. Prognostic significance of vascular endothelial growth factor protein in node-negative breast carcinoma. J Natl Cancer Inst. 1997;89:139–147. doi: 10.1093/jnci/89.2.139. [DOI] [PubMed] [Google Scholar]
- 20.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis--correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 21.Weidner N, Folkman J, Pozza F, et al. Tumor angiogenesis: a new significant and independent prognostic indicator in early-stage breast carcinoma. J Natl Cancer Inst. 1992;84:1875–1887. doi: 10.1093/jnci/84.24.1875. [DOI] [PubMed] [Google Scholar]
- 22.Krippl P, Langsenlehner U, Renner W, et al. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer. 2003;106:468–471. doi: 10.1002/ijc.11238. [DOI] [PubMed] [Google Scholar]
- 23.Smith KC, Bateman AC, Fussell HM, Howell WM. Cytokine gene polymorphisms and breast cancer susceptibility and prognosis. Eur J Immunogenet. 2004;31:167–173. doi: 10.1111/j.1365-2370.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 24.Jin Q, Hemminki K, Enquist K, et al. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res. 2005;11:3647–3653. doi: 10.1158/1078-0432.CCR-04-1803. [DOI] [PubMed] [Google Scholar]
- 25.Lu H, Shu XO, Cui Y, et al. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65:5015–5019. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka N, Cai Q, Wen W, et al. Population-based case-control study of VEGF gene polymorphisms and breast cancer risk among Chinese women. Cancer Epidemiol Biomarkers Prev. 2006;15:1148–1152. doi: 10.1158/1055-9965.EPI-05-0871. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs EJ, Feigelson HS, Bain EB, et al. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res. 2006;8:R22. doi: 10.1186/bcr1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balasubramanian SP, Cox A, Cross SS, Higham SE, Brown NJ, Reed MW. Influence of VEGF-A gene variation and protein levels in breast cancer susceptibility and severity. Int J Cancer. 2007;121:1009–1016. doi: 10.1002/ijc.22772. [DOI] [PubMed] [Google Scholar]
- 29.Langsenlehner U, Wolf G, Langsenlehner T, et al. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res Treat. 2008;109:297–304. doi: 10.1007/s10549-007-9655-z. [DOI] [PubMed] [Google Scholar]
- 30.Eroglu A, Ozturk A, Cam R, Akar N. Vascular endothelial growth factor gene 936 C/T polymorphism in breast cancer patients. Med Oncol. 2008;25:54–55. doi: 10.1007/s12032-007-0046-4. [DOI] [PubMed] [Google Scholar]
- 31.Schneider BP, Radovich M, Sledge GW, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat. 2008;111:157–163. doi: 10.1007/s10549-007-9755-9. [DOI] [PubMed] [Google Scholar]
- 32.Jakubowska A, Gronwald J, Menkiszak J, et al. The VEGF_936_C>T 3'UTR polymorphism reduces BRCA1-associated breast cancer risk in Polish women. Cancer Lett. 2007 doi: 10.1016/j.canlet.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 33.Schneider BP, Wang M, Radovich M, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26:4672–4678. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burstein HJ, Chen YH, Parker LM, et al. VEGF as a marker for outcome among advanced breast cancer patients receiving anti-VEGF therapy with bevacizumab and vinorelbine chemotherapy. Clin Cancer Res. 2008;14:7871–7877. doi: 10.1158/1078-0432.CCR-08-0593. [DOI] [PubMed] [Google Scholar]
- 35.Yuan A, Yu CJ, Chen WJ, et al. Correlation of total VEGF mRNA and protein expression with histologic type, tumor angiogenesis, patient survival and timing of relapse in non-small-cell lung cancer. Int J Cancer. 2000;89:475–483. doi: 10.1002/1097-0215(20001120)89:6<475::aid-ijc2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 36.Kaya A, Ciledag A, Gulbay BE, et al. The prognostic significance of vascular endothelial growth factor levels in sera of non-small cell lung cancer patients. Respir Med. 2004;98:632–636. doi: 10.1016/j.rmed.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Dudek AZ, Mahaseth H. Circulating angiogenic cytokines in patients with advanced non-small cell lung cancer: correlation with treatment response and survival. Cancer Invest. 2005;23:193–200. doi: 10.1081/cnv-200055949. [DOI] [PubMed] [Google Scholar]
- 38.Kaya A, Oner F, Fitoz S, Erden I, Numanoglu N. Metastatic lung cancer: presenting with ocular symptoms. Tuberk Toraks. 2005;53:386–389. [PubMed] [Google Scholar]
- 39.Zhai R, Liu G, Zhou W, et al. Vascular endothelial growth factor genotypes, haplotypes, gender, and the risk of non-small cell lung cancer. Clin Cancer Res. 2008;14:612–617. doi: 10.1158/1078-0432.CCR-07-1655. [DOI] [PubMed] [Google Scholar]
- 40.Lurje G, Zhang W, Schultheis AM, et al. Polymorphisms in VEGF and IL-8 predict tumor recurrence in stage III colon cancer. Ann Oncol. 2008 doi: 10.1093/annonc/mdn368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaio E, Tanaka S, Kitadai Y, et al. Clinical significance of angiogenic factor expression at the deepest invasive site of advanced colorectal carcinoma. Oncology. 2003;64:61–73. doi: 10.1159/000066511. [DOI] [PubMed] [Google Scholar]
- 42.Des Guetz G, Uzzan B, Nicolas P, et al. Microvessel density and VEGF expression are prognostic factors in colorectal cancer. Meta-analysis of the literature. Br J Cancer. 2006;94:1823–1832. doi: 10.1038/sj.bjc.6603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae SJ, Kim JW, Kang H, Hwang SG, Oh D, Kim NK. Gender-specific association between polymorphism of vascular endothelial growth factor (VEGF 936 C>T) gene and colon cancer in Korea. Anticancer Res. 2008;28:1271–1276. [PubMed] [Google Scholar]
- 44.Park HM, Hong SH, Kim JW, et al. Gender-specific association of the VEGF −2578C > A polymorphism in Korean patients with colon cancer. Anticancer Res. 2007;27:2535–2539. [PubMed] [Google Scholar]
- 45.Chae YS, Kim JG, Sohn SK, et al. Association of vascular endothelial growth factor gene polymorphisms with susceptibility and clinicopathologic characteristics of colorectal cancer. J Korean Med Sci. 2008;23:421–427. doi: 10.3346/jkms.2008.23.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamori M, Taniguchi M, Maeda S, et al. VEGF T-1498C polymorphism, a predictive marker of differentiation of colorectal adenocarcinomas in Japanese. Int J Med Sci. 2008;5:80–86. doi: 10.7150/ijms.5.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JG, Chae YS, Sohn SK, et al. Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with colorectal cancer. Clin Cancer Res. 2008;14:62–66. doi: 10.1158/1078-0432.CCR-07-1537. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann G, Langsenlehner U, Renner W, et al. Common single nucleotide polymorphisms in the vascular endothelial growth factor gene and colorectal cancer risk. J Cancer Res Clin Oncol. 2008;134:591–595. doi: 10.1007/s00432-007-0322-x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Gordon M, Press OA, et al. Cyclin D1 and epidermal growth factor polymorphisms associated with survival in patients with advanced colorectal cancer treated with Cetuximab. Pharmacogenet Genomics. 2006;16:475–483. doi: 10.1097/01.fpc.0000220562.67595.a5. [DOI] [PubMed] [Google Scholar]
- 50.Uthoff SM, Duchrow M, Schmidt MH, et al. VEGF isoforms and mutations in human colorectal cancer. Int J Cancer. 2002;101:32–36. doi: 10.1002/ijc.10552. [DOI] [PubMed] [Google Scholar]
- 51.Fukuda H, Tsuchiya N, Narita S, et al. Clinical implication of vascular endothelial growth factor T-460C polymorphism in the risk and progression of prostate cancer. Oncol Rep. 2007;18:1155–1163. [PubMed] [Google Scholar]
- 52.Balbay MD, Pettaway CA, Kuniyasu H, et al. Highly metastatic human prostate cancer growing within the prostate of athymic mice overexpresses vascular endothelial growth factor. Clin Cancer Res. 1999;5:783–789. [PubMed] [Google Scholar]
- 53.McCarron SL, Edwards S, Evans PR, et al. Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res. 2002;62:3369–3372. [PubMed] [Google Scholar]
- 54.Lin CC, Wu HC, Tsai FJ, Chen HY, Chen WC. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for prostate cancer. Urology. 2003;62:374–377. doi: 10.1016/s0090-4295(03)00268-1. [DOI] [PubMed] [Google Scholar]
- 55.Sfar S, Hassen E, Saad H, Mosbah F, Chouchane L. Association of VEGF genetic polymorphisms with prostate carcinoma risk and clinical outcome. Cytokine. 2006;35:21–28. doi: 10.1016/j.cyto.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Onen IH, Konac E, Eroglu M, Guneri C, Biri H, Ekmekci A. No association between polymorphism in the vascular endothelial growth factor gene at position −460 and sporadic prostate cancer in the Turkish population. Mol Biol Rep. 2008;35:17–22. doi: 10.1007/s11033-006-9046-2. [DOI] [PubMed] [Google Scholar]
- 57.Sfar S, Saad H, Mosbah F, Chouchane L. Combined effects of the angiogenic genes polymorphisms on prostate cancer susceptibility and aggressiveness. Mol Biol Rep. 2007 doi: 10.1007/s11033-007-9149-4. [DOI] [PubMed] [Google Scholar]
- 58.Langsenlehner T, Langsenlehner U, Renner W, et al. Single nucleotide polymorphisms and haplotypes in the gene for vascular endothelial growth factor and risk of prostate cancer. Eur J Cancer. 2008;44:1572–1576. doi: 10.1016/j.ejca.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 59.Yamamori M, Sakaeda T, Nakamura T, et al. Association of VEGF genotype with mRNA level in colorectal adenocarcinomas. Biochem Biophys Res Commun. 2004;325:144–150. doi: 10.1016/j.bbrc.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 60.Daly AK, Day CP. Candidate gene case-control association studies: advantages and potential pitfalls. Br J Clin Pharmacol. 2001;52:489–499. doi: 10.1046/j.0306-5251.2001.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 63.Huang YT, Heist RS, Chirieac LR, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim TM, Yim SH, Lee JS, et al. Genome-wide screening of genomic alterations and their clinicopathologic implications in non-small cell lung cancers. Clin Cancer Res. 2005;11:8235–8242. doi: 10.1158/1078-0432.CCR-05-1157. [DOI] [PubMed] [Google Scholar]
- 65.Kronenberg F. Genome-wide association studies in aging-related processes such as diabetes mellitus, atherosclerosis and cancer. Exp Gerontol. 2008;43:39–43. doi: 10.1016/j.exger.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 66.Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Hum Genet. 2009;17:722–731. doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roukos DH. Genome-wide association studies: how predictable is a person's cancer risk? Expert Rev Anticancer Ther. 2009;9:389–392. doi: 10.1586/era.09.12. [DOI] [PubMed] [Google Scholar]
- 68.Seng KC, Seng CK. The success of the genome-wide association approach: a brief story of a long struggle. Eur J Hum Genet. 2008;16:554–564. doi: 10.1038/ejhg.2008.12. [DOI] [PubMed] [Google Scholar]
- 69.Spinola M, Leoni VP, Galvan A, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251:311–316. doi: 10.1016/j.canlet.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 70.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009 doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ambrosone CB. The promise and limitations of genome-wide association studies to elucidate the causes of breast cancer. Breast Cancer Res. 2007;9:114. doi: 10.1186/bcr1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360 doi: 10.1056/NEJMra0808700. 1759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]