Abstract
The goal of many plant scientists' research is to explain natural phenotypic variation in terms of simple changes in DNA sequence. Traditionally, linkage mapping has been the most commonly employed method to reach this goal: experimental crosses are made to generate a family with known relatedness, and attempts are made to identify cosegregation of genetic markers and phenotypes within this family. In vertebrate systems, association mapping (also known as linkage disequilibrium mapping) is increasingly being adopted as the mapping method of choice. Association mapping involves searching for genotype-phenotype correlations in unrelated individuals and often is more rapid and cost-effective than traditional linkage mapping. We emphasize here that linkage and association mapping are complementary approaches and are more similar than is often assumed. Unlike in vertebrates, where controlled crosses can be expensive or impossible (e.g., in humans), the plant scientific community can exploit the advantages of both controlled crosses and association mapping to increase statistical power and mapping resolution. While the time and money required for the collection of genotype data were critical considerations in the past, the increasing availability of inexpensive DNA sequencing and genotyping methods should prompt researchers to shift their attention to experimental design. This review provides thoughts on finding the optimal experimental mix of association mapping using unrelated individuals and controlled crosses to identify the genes underlying phenotypic variation.
GENETIC MAPPING: IT'S ALL ABOUT RECOMBINATION
The aim of many genetic mapping studies is to identify quantitative trait loci (QTL) that are responsible for phenotypic variation. Although often viewed as fundamentally different, linkage and association mapping share a common strategy that exploits recombination's ability to break up the genome into fragments that can be correlated with phenotypic variation. The key difference between the two methods is the control the experimenter has over recombination. On the one hand, linkage mapping is a highly controlled experiment: individuals are crossed to generate a mapping population in which relatedness is known. In plants, these are generally biparental crosses, while in humans these populations may be extended pedigrees. The experimenter thereby creates a closed system and uses a small number of genetic markers to infer the locations of the relatively few recombination breakpoints. With genotype data from across the genome, the experimenter can then determine if a chromosomal fragment between two specific breakpoints is associated with a phenotype. Association mapping, on the other hand, is not a controlled experiment, but rather a natural experiment. Genotype and phenotype data are collected from a population in which relatedness is not controlled by the experimenter, and correlations between genetic markers and phenotypes are sought within this population. This open system design provides higher mapping resolution compared with the closed system of controlled crosses, but it is difficult to infer when and where recombination has occurred. Moreover, the uncontrolled relatedness among individuals can result in spurious signals of association in downstream analyses.
For the rest of this review, we will avoid using the terms “linkage mapping” versus “association” or “linkage disequilibrum” mapping, as both of these approaches identify genotype-phenotype associations by identifying polymorphisms that are linked to functional alleles. As we move into a world of complete genome sequencing, the distinction between the two methods will disappear, but questions about the optimal experimental design and analysis will remain. We will refer to “family mapping” when mapping is conducted in progeny of a biparental cross and to “population mapping” when mapping is conducted in populations in which relatedness is unknown.
Using family mapping, an experimenter can only exploit the recombination events that have occurred during the establishment of the mapping population. In this case, recombination has not had enough time to shuffle the genome into small fragments, and QTL are generally localized to large chromosomal regions (10 to 20 centimorgans). In addition, family mapping can only identify QTL from the phenotypic diversity generated from the controlled cross, which may often represent only a small fraction of the phenotypically relevant variation in a species. Indeed, because different QTL segregate in different family mapping populations, QTL often are not consistent across mapping populations (Holland, 2007).
It has long been recognized that population mapping offers advantages over family mapping for the identification of QTL (Spielman et al., 1993; Risch and Merikangas, 1996; Long et al., 1997), and it has recently seen enormous success in human disease research (Donnelly, 2008). Although it is currently the method of choice for mapping human phenotypes, population mapping is just beginning to be widely applied to plant populations (Thornsberry et al., 2001; Breseghello and Sorrells, 2006; Zhao et al., 2007; Gonzalez-Martinez et al., 2008; Harjes et al., 2008). The main advantage of population mapping is that it exploits all of the recombination events that have occurred in the evolutionary history of a sample, which almost invariably results in a much higher mapping resolution compared with family mapping. In addition, the number of QTL one can map for a given phenotype is not limited to what segregates between parents of a cross, but rather by the number of real QTL underlying the trait and the degree to which the mapping population captures the total genetic diversity available in nature (Zhu et al., 2008). This approach is particularly useful in plant breeding where alleles associated with desirable phenotypes can be introduced efficiently into selected lines. Here, we focus on identifying QTL, but it is worth noting that genomic selection, which aims to estimate breeding values without identifying QTL, uses similar data and populations and is showing great promise for breeding programs (Meuwissen et al., 2001; Goddard and Hayes, 2009; Heffner et al., 2009). Perhaps the most attractive aspect of association mapping is its ease and cost-effectiveness compared with the laborious and often expensive process of establishing mapping families. This is especially the case for researchers who work on organisms that cannot be crossed, cloned, or have long generation times (Nordborg and Weigel, 2008). In some instances, however, population mapping can involve a significant phenotyping burden because of the large sample sizes required. Also, obtaining reliable phenotypic measurements from a population of plants that are adapted to different growing conditions may present limitations in the use of certain germplasm. Experimenters must consider these points carefully when trying to exploit the advantages of population mapping.
WHY LINKAGE DISEQUILIBRIUM MATTERS
The ultimate aim of most mapping studies is to identify the functional genetic variants, or the quantitative trait nucleotides, that are responsible for phenotypic variation. Current data sets are commonly obtained using genotyping microarrays and often consist of hundreds of thousands of genotypes from hundreds or even thousands of individuals. Even with such large numbers of markers, however, it is unlikely that sought after functional variant(s) will be among the markers genotyped. (Though in the near future, data sets will likely contain [nearly] all variants!) The experimenter often can only hope that genetic markers that are in strong linkage disequilibrium (LD) with the functional variant(s) have been genotyped. LD refers to the correlation between polymorphisms in a population. Thus, the genotyped markers become proxies, or sentinels, for the functional variant because their genotypes are highly correlated with the genotypes of the functional variant. The power of an association study depends on the strength of this correlation (i.e., on the degree of LD between the genotyped marker and the functional variant). Figure 1 depicts a scenario in which two markers have been genotyped at a locus, one of which is associated with the phenotype and is in LD with the functional variant and one of which is not in LD with the functional variant and is therefore not associated with the phenotype.
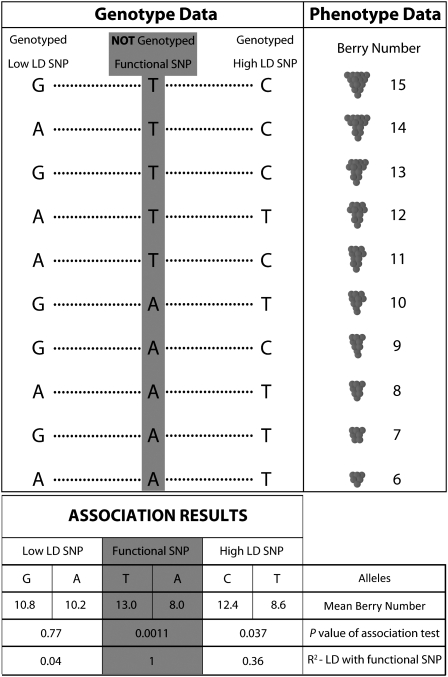
Figure 1.
A Fictional Depiction of a Simple Genotype-Phenotype Association Test.
The functional SNP responsible for variation in berry number in grapevine is in gray and is not genotyped. The genotyped SNPs lie on either side of the functional SNP. The genotyped SNP to the right is in high LD with the functional SNP, while the genotyped SNP to the left is not in LD with the functional SNP. The results of a simple association test (Pearson correlation) are shown in the bottom box. The C allele of the high LD SNP is significantly associated with berry number (P = 0.037), while there is no significant association for the low LD SNP (P = 0.77).
In general, the strength of the correlation between two markers is a function of the distance between them: the closer two markers are, the stronger the LD. The resolution with which a QTL can be mapped is a function of how quickly LD decays over distance. Therefore, the first step in the design of an association study is an analysis of the structure of LD in the population under study. The decay of LD has been shown to differ dramatically between species (Flint-Garcia et al., 2003), often due to differences in breeding systems. Selfing reduces opportunities for recombination (Nordborg, 2000), so inbreeders such as rice (Oryza sativa), for example, can have LD that extends to 100 kb or more (Garris et al., 2005). Conversely, in outcrossers, LD generally breaks down more rapidly. For example, LD decays within 300 bp in the grapevine (Vitis vinifera; Lijavetzky et al., 2007) and within 100 bp in Norway spruce (Picea abies; Heuertz et al., 2006). Even within a species, LD decay can vary significantly. In maize (Zea mays), for example, LD decays within 1 kb in land races (Tenaillon et al., 2001), within 2 kb in diverse inbred lines (Remington et al., 2001), and can extend up to 500 kb in commercial elite inbred lines (Rafalski, 2002; Jung et al., 2004). Finally, LD decay also varies among loci within a population, sometimes due to positive selection, which can generate LD that extends much farther than the genome-wide average (e.g., Whitt et al., 2002). Since the resolution with which QTL can be mapped is a function of LD decay, population mapping may offer little or no advantage over family mapping in cases where LD is extensive. It is therefore crucial that the experimenter choose a diverse set of germplasm that exploits the recombination events that have occurred in the history of the species of interest.
FROM CANDIDATE GENES TO GENOME-WIDE STUDIES
The first population mapping attempts within a species usually involve candidate gene studies: genetic markers are genotyped at a locus thought to be involved in some phenotype, and one tests for an association between these genetic markers and the phenotype. The candidate gene approach was widely used in the search for disease-gene associations in humans but has recently been declared woefully inadequate as most confirmed disease genes went undetected using this approach (Altshuler et al., 2008). In plants, population mapping has been successful for candidate genes in relatively simple pathways (Harjes et al., 2008; Zheng et al., 2008) and for candidate genes with extensive prior evidence of a role in the phenotype of interest (Werner et al., 2005). However, the choice of candidate genes and the markers within them often involves some guesswork, so the insights one can gain into the genetic control of the phenotype of interest will necessarily be limited.
So why not just cover the entire genome with genetic markers? The strategy of a genome-wide association (GWA) study is to genotype enough markers across the genome so that functional alleles will likely be in LD with at least one of the genotyped markers. GWA has revolutionized genetic mapping in humans (Altshuler et al., 2008; Donnelly, 2008) and is increasingly being adopted in plants (Nordborg and Weigel, 2008). Of course, the first step in this process is the discovery of a large number of genetic markers, typically single nucleotide polymorphisms (SNPs), as a reference resource. In humans, the International HapMap Project currently boasts over three million SNPs (http://www.hapmap.org/), and similar projects are underway for Arabidopsis thaliana (http://walnut.usc.edu/2010), rice (http://irfgc.irri.org), and maize (http://www.panzea.org/). The number of markers and their density are defined by genome size and LD decay and will therefore vary considerably among species. For example, while 140,000 markers provide reasonable coverage of the 125 Mb Arabidopsis genome (Kim et al., 2007), a rough estimate suggests that over two million markers will be required to cover the 475 Mb genome of the grapevine, and 10 to 15 million may be necessary for diverse maize varieties. While genotyping microarrays have been the technology of choice so far, the decreasing costs of next-generation sequencing (such as Illumina's Genome Analyzer, Applied Biosystems' SOLiD, and Roche's 454) will make it possible for future projects cost-effectively to obtain full sequence data from large population samples. The construction of genotyping microarrays necessarily involves an ascertainment bias: SNPs are discovered in a small set of samples and are then genotyped in a larger set of samples. Thus, polymorphisms not present in the initial SNP discovery panel (i.e., primarily low frequency SNPs) remain undetected in the larger sample. Whole-genome sequencing will provide a significant advantage over the use of microarrays as it avoids the erosion of power due to ascertainment bias by detecting all polymorphisms in the mapping population (Clark et al., 2005).
FINDING THE MISSING HERITABILITY
Most GWA studies proceed first by identifying a set of reference SNPs that segregate at intermediate frequency in a small panel of individuals. These SNPs are then genotyped in large samples for which phenotype data are available. The motivation for this strategy is the assumption that common phenotypic variation will be caused by common genetic variation. In humans, a version of this assumption is known as the common disease-common variant hypothesis (Lander, 1996), and it was the impetus for the International HapMap project. Although GWA studies in humans have uncovered thousands of significant associations, they often account for very little of the variation in a phenotype. For example, human height is known to be 80 to 90% heritable, but the 40 variants associated with height from GWA studies account for just over 5% of height's heritability (Maher, 2008). Quantitative geneticists are beginning to ask themselves: where is the missing heritability?
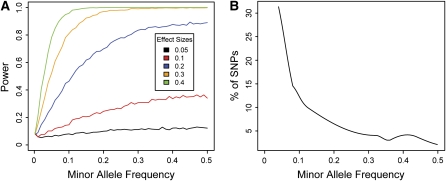
Low frequency functional alleles are among the likely culprits. The power to detect an association is a function of allele frequency: functional variants at low frequency have little influence on the population as a whole, and their signal is therefore difficult to detect (Figure 2A). Even if a low frequency allele has an enormous effect on the phenotype, population mapping normally will have little power to detect it. Unfortunately, it is a well-known result from population genetics theory that, in the majority of species, most alleles are rare (Figure 2B). For example, 30% of the polymorphisms in a diverse panel of 27 maize inbred lines are unique to a single line (www.panzea.org). This raises a fundamental biological question: Is the frequency distribution of functional alleles similar to that of random alleles in Figure 2B? If so, we will have difficulty accounting for most phenotypic variation using association mapping because most of it will be caused by rare alleles. Fortunately, family mapping can be used to identify such low frequency functional alleles. By creating crosses, the experimenter can artificially inflate the allele frequencies in the progeny to provide increased power for mapping. It is arguably only through the use of controlled crosses that we eventually will gain an understanding of the frequency distribution of functional alleles.
Figure 2.
Important factors affecting the power of population mapping studies.
(A) The power of an association test is a function of the allele frequency and the effect size.
(B) The allele frequency spectrum from 3641 SNPs genotyped in 25 diverse maize inbred lines (www.panzea.org) demonstrates that most alleles in a population are rare. Therefore, if the frequency spectrum of functional alleles is similar to the frequency spectrum of random SNPs, most functional alleles will remain undetected through population mapping because of low power. For (A), phenotype data were simulated for 1000 haploid samples as a normal distribution with mean = 0 and sd = 1 for one allele and mean 0 + effect size and sd = 1 for the other allele. Effect size is therefore defined as the difference between the mean phenotypic values of the two alleles. Power is defined as the proportion of association tests (Pearson correlation) significant at P < 0.05 out of 5000 simulated data sets.
Perhaps the most important factor that accounts for the missing heritability, however, is the genetic architecture of the trait under study. The power to detect an association between a functional variant and a phenotype is also a function of the effect that the functional variant exerts on the phenotype (Figure 2A). Phenotypic variation controlled by numerous small effect QTL will be inherently more difficult to dissect to the genetic level than variation caused by a small number of large effect QTL. Determining the genetic architecture underlying phenotypic variation has generated enormous interest, but it is unclear whether there are any definitive trends. For example, a single QTL explains 86% of the variation in flowering time in an interspecific mapping family in sorghum (Sorghum bicolor; Lin et al., 1995), whereas 50 QTL are required to explain 50% of the variation in kernel oil concentration in maize (Laurie et al., 2004). To what extent the genetic architecture differs among species and among traits remains largely unknown (Flint and Mackay, 2009). Future studies with sufficient power to detect small effect QTL promise to elucidate the genetic architecture of common phenotypic variation.
RELATEDNESS: A COMMON CONFOUNDER
Except in population genetics theory, randomly mating populations probably do not exist. Nonrandom mating has generated complex patterns of population structure and relatedness in crops and wild plants (Flint-Garcia et al., 2005; Nordborg et al., 2005). In population mapping, complex patterns of genetic relatedness among individuals can be problematic when trying to map a phenotype whose variation is correlated with genetic relatedness. In such cases of genotype-phenotype covariance, many genetic markers across the genome will appear to be associated with the phenotype, when in fact these genetic markers simply capture the genetic relatedness among individuals. This problem is particularly apparent when trying to map traits that have been subject to local adaptation, like flowering time (Aranzana et al., 2005; Flint-Garcia et al., 2005) because variation in these phenotypes between populations is highly correlated with allele frequency differences between populations. Even for a set of common traits of agronomic interest in maize, such allele frequency differences account for an average of 9.3% of the phenotypic variation across all traits (Flint-Garcia et al., 2005). It has long been known that genotype-phenotype covariance can lead to spurious associations (Lander and Schork, 1994), and recent attempts to map such traits have resulted in extremely high false positive rates (Aranzana et al., 2005).
There has been intense interest in the development of methods to correct for genetic relatedness in population mapping studies. The methods used to correct for genetic relatedness involve using random markers throughout the genome to estimate relatedness among individuals within a mapping population. These estimates of relatedness provide baseline predictions of background QTL sharing among individuals and are used to determine whether a candidate marker actually explains more variation than a random marker does. These methods are most effective when the trait is complex and controlled by many QTL. The principle underlying these approaches is similar to the principle applied in family mapping, where background markers are used as covariates to control for QTL outside of the genomic region of interest.
The first generation of methods for correcting for relatedness focused on large-scale clinal or island-like population structure. Structured association (Pritchard et al., 2000b) involves using the program STRUCTURE (Pritchard et al., 2000a) to identify populations and then estimate the proportion of each individual's variation that came from a particular population. The matrix of these estimates is called Q, and the estimates are used as covariates to control for population structure in population mapping. Structured association was extended to quantitative traits and has been used in plants (Thornsberry et al., 2001). Alternatively, using principal components analysis (PCA) to reduce the high-dimensional genotype data to a small number of dimensions, one can then use the axes of variation from these dimensions to calculate ancestry-adjusted genotypes and phenotypes (Price et al., 2006). Estimation of the Q matrix using STRUCTURE is computationally intensive and is designed for unrelated individuals from populations in Hardy-Weinberg equilibrium, whereas PCA is fast, makes no assumptions about the structure of the populations, and performs similarly or better than STRUCTURE (Zhao et al., 2007).
The problem with both of these approaches is that individuals can only vary along a few axes of differentiation that may or may not be well captured by the STRUCTURE or PCA models. An extreme nonclinal type of relatedness is an extended pedigree, for example, where many individuals have close relationships that cannot be described by a single vector of relatedness. An alternative approach to capture this complex differentiation is to estimate the pairwise relatedness between all individuals in the sample. Like Q, one can use pairwise relatedness to control for the effects of relatedness in population mapping. The statistical approach used to relate the pairwise relatedness matrix to a phenotype is the mixed model, where the variance explained by pairwise relatedness is fit to the vector of phenotypes. This approach was originally developed for cattle breeding (Henderson, 1975). One can envision the mixed model as a statistical method to obtain a weighted average phenotypic prediction based on relatedness.
While the first generation of mixed models used pedigree information, random genetic markers are now most often used to generate a pairwise relatedness matrix called the kinship matrix, or simply K. This approach of using genetic markers in estimating relatedness has been used to predict breeding values in animals and plants (Meuwissen et al., 2001; Schaeffer, 2006; VanRaden, 2008; Heffner et al., 2009) and to correct for relatedness in population mapping studies in both human families and inbred maize lines (Yu et al., 2006). The application of mixed model methods using the K matrix in maize, human, mouse, Arabidopsis, and potato (Solanum tuberosum) demonstrates that the additional correction for pairwise relatedness significantly decreases false positives and false negatives over and above corrections involving only the Q matrix (Yu et al., 2006; Malosetti et al., 2007; Zhao et al., 2007; Kang et al., 2008). Intuitively, this makes good sense: While Q takes only a few axes of variation into account, the K matrix captures the relatedness between each possible pair of individuals in a sample. In general, the mixed model (K) is far superior to the clinal approaches (Q), but in many cases a combination (Q+K) of these approaches appears to be most powerful.
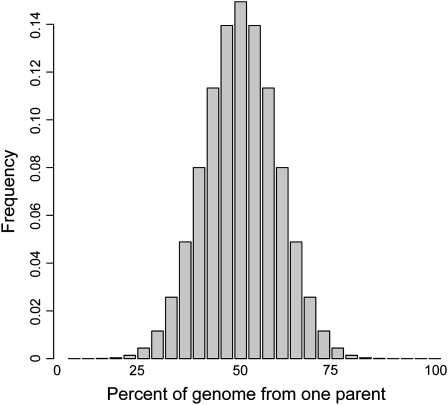
It is not trivial, however, to define the K matrix. One may expect, for example, that the pedigree data available for many mapping populations could provide accurate estimates of the K matrix. However, marker-based kinship coefficients are more accurate than pedigree-based estimates because they account for deviations from expected parental contributions due to independent assortment (Mendelian sampling) or segregation distortion (selection) (Bernardo, 1993; Bernardo et al., 1996). Figure 3 demonstrates that the expected contribution from one parent to the progeny of a recombinant line (RIL) population varies widely from 20 to 80% (Figure 3). The distribution in Figure 3 only takes the effects of independent assortment into account. Selection applied during breeding can widen the tails of this distribution. This demonstrates that marker-based estimates of kinship are highly preferred over pedigree-based estimates.
Figure 3.
The Expected Genetic Contribution from Each Parent to the Progeny of a Biparental RIL Family with a Genetic Map Size of Maize.
Progeny can be much more closely related to one parent than another. In fact, ∼9% of progeny are ≥2 times more closely related to one parent than the other.
Currently, there are two main difficulties with the application of the mixed model that are being addressed by statistical geneticists. First, more sophisticated methods are being developed to refine marker-based estimates of relatedness in the generation of the K matrix, as previously used estimates of relatedness were rather simplistic (Yu et al., 2006; Zhao et al., 2007). The difficulty lies in determining whether alleles that are identical by state (i.e., the same genotype) are also identical by descent (IBD; i.e., inherited from a common ancestor). A recently proposed restricted maximum likelihood estimate of the probability of two alleles at the same locus being identical by state but not IBD improves the power of the mixed model (Stich et al., 2008). The second difficulty is computational speed. The original mixed model mapping method presented by Yu et al. (2006) is computationally intensive and is prohibitively time-consuming in analyses of most large, genome-wide data sets. A more recent method, efficient mixed model association, substantially increases computation speed (Kang et al., 2008), and recently developed methods in our laboratory promise to make the mixed model method accessible to most genome-wide data sets (Z. Zhang, E. Ersoz, C.-Q. Lai, R.J. Todhunter, H.K. Tiwari, M.A. Gore, P.J. Bradbury, J. Yu, D.K. Arnett, J.M. Ordovas, and E.S. Buckler, unpublished data).
Correcting for various types of relatedness is beginning to allow successful weeding out of false from true positives in several studies in plants. For example, previously reported associations in maize and Arabidopsis were subsequently found to be likely due to the confounding effects of relatedness (Wilson et al., 2004; Zhao et al., 2007). Thornsberry et al. (2001) reduced the number of false-positive associations with flowering time traits in maize by almost fivefold using the Q matrix to correct for population structure. However, the associations between variants in dwarf8 and flowering time in maize have never been formally proven or disproven, as all the analyses are confounded with relatedness despite every attempt to control for it statistically (Thornsberry et al., 2001; Andersen et al., 2005; Camus-Kulandaivelu et al., 2006). Through a combination of family mapping and mixed model estimation, it can be shown that effects at dwarf8 were overestimated by at least 10-fold using a simple Q model and may not be significant (S. Larsson and E.S. Buckler, unpublished data). The evidence for effects on flowering time from variation at Vgt1 in maize is unequivocal, but effect estimates for this locus using Q alone are overestimates, while a correction using Q+K generates relatively accurate effect estimates (Buckler et al., 2009). It is becoming increasingly clear that corrections using only Q often are inadequate, especially in species with complex patterns of relatedness. Currently, mixed model approaches should be the method of choice among plant scientists for population mapping studies. Software for mixed model analyses is freely available at http://www.maizegenetics.net/tassel and http://mouse.cs.ucla.edu/emma.
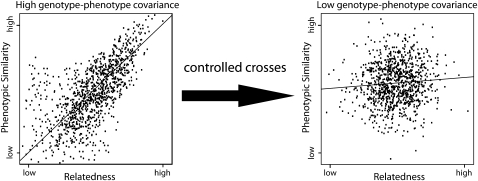
Although the mixed model provides a robust method to correct for relatedness in population mapping studies, attempts to map phenotypes that are strongly correlated with relatedness will remain problematic. Using population mapping, there is simply no way statistically to determine whether a genetic variant is a true QTL if the phenotype is so strongly correlated with relatedness that random genetic variants throughout the genome associate equally well with the trait. As is the case for detecting low frequency functional variants, family mapping can come to the rescue when encountering the confounding effects of relatedness (e.g., Balasubramanian et al., 2006; Manenti et al., 2009). The generation of controlled crosses can break up the covariance between genotypes and phenotypes and enhance power to detect QTL. Thus, in cases where Q+K explain most of the phenotypic variance, population mapping will be severely underpowered, and the experimenter will need to consider family mapping to detect the underlying QTL. In fact, many QTL will remain practically undetectable without the help of controlled crosses. Figure 4 provides an illustration of how controlled crosses can be used to break up the genotype-phenotype covariance to enhance power to detect QTL.
Figure 4.
Genotype-Phenotype Covariance Can Be Broken up by Generating Controlled Crosses.
The left panel is a scenario of an extreme correlation between relatedness and phenotypic similarity. Individuals, represented by dots, who are closely related have similar phenotypes, and distantly related individuals are more phenotypically dissimilar. In these cases, random genetic markers throughout the genome will be strongly associated with the phenotype, and population mapping will therefore lack power to detect real QTL. By generating controlled crosses, this genotype-phenotype covariance can be broken, and a population of individuals can be generated in which this correlation is weakened. In the right panel, phenotypic differences between individuals are no longer strongly associated with relatedness, and the power to detect QTL is significantly enhanced.
JOINT FAMILY POPULATION MAPPING
As we have previously discussed, there are scenarios in which population mapping will have little power to detect an association (e.g., low frequency alleles and QTL with small effect) or will generate an excess of false positives (e.g., genotype-phenotype covariance). In these scenarios, we suggested that the experimenter manipulate allele frequencies and population structure to their advantage by generating controlled crosses and using family mapping to enhance power. In the past few years, several elegant statistical approaches have been developed to combine family and population mapping, often called joint linkage-association mapping (e.g., Wu and Zeng, 2001; Meuwissen et al., 2002; Wu et al., 2002; Blott et al., 2003). The primary statistical challenge encountered with these methods involves the estimation of probabilities of IBD across the genome. However, with near full genome sequence data, these challenges can readily be overcome. Computationally intensive estimation of IBD probabilities will no longer be necessary when near complete genome sequence data are available. Experimenters must now consider that, by the time the phenotypic data are collected in 3 to 4 years, full sequence data will in many cases be available for an experiment planned today. Thus, the relevant question is not what the nature of the genotype data will be, but rather how to select germplasm that will maximize allelic diversity and the power to dissect complex traits.
With this in mind, the maize community has developed the largest publicly available platform for complex trait dissection of any species. In the maize Nested Association Mapping population (NAM), 25 diverse maize lines were crossed to the reference maize inbred B73 (Yu et al., 2008). The reference design was chosen to reduce the confounding physiology effects of having too much flowering time variation. From each cross, 200 RILs were generated, for a total of 5000 lines. The controlled crosses in NAM reduce the confounding effects of population structure, while the large numbers of progeny derived from the crosses allow for family mapping with substantial statistical power. Moreover, with full sequence information from the 26 founder lines and low-density genotyping in the progeny, the genotype information from the founders can be projected to the progeny, optimizing genotyping costs. This design provides significant power to identify QTL underlying complex traits and estimate additive effects and some epistatic interactions among QTL.
A publicly available platform for joint linkage association also exists in mouse, where 1000 RILs are derived from eight founder lines (Churchill et al., 2004). With only eight founder lines, however, functional alleles segregating in mouse, but not found in the founders, will remain undetected. In addition, the number of ancestral recombination events captured in such a design is a function of the number of founder lines used to generate the population. Thus, although joint family population mapping approaches show great promise for the dissection of complex traits, questions about optimal experimental design remain. For example, the founder lines in NAM were chosen largely to maximize genetic variation. Would NAM be more powerful if the founder lines had been chosen to maximize physiological variation? Should more than one reference founder be used in the NAM design? Also, how many founder lines are required to capture the desired amount of genetic variation and number of ancestral recombination events in the species of interest? How should experimental design change according to the population structure of the species under study and the genetic architecture of the phenotypes of interest? Simulation studies help address these issues by investigating the effects of experimental design (e.g., number of crosses and sample size) on QTL detection power under varying genetic architectures (Wu et al., 2002; Verhoeven et al., 2005). These simulations have limitations, however, in that they are forced to make assumptions about the sharing of QTL among individuals and their frequency distributions. In the future, the results from current large-scale QTL mapping projects, such as those in maize and mouse, will provide valuable guidelines for the design of future joint family population experiments.
CONCLUSIONS
The days when the design and implementation of genotyping assays were both time-consuming and expensive will soon be behind us. Full resequencing of mapping populations is likely to be within reach in the next decade. Now is the time to concentrate on experimental design, so that the deluge of genotype data can be fully exploited when it arrives in the future. While it is often optimal or even necessary to generate specific mapping populations, experimenters should also consider mining useful variation from the massive germplasm collections we already have. For example, the National Plant Germplasm System of the USDA currently holds >500,000 accessions (http://www.ars-grin.gov/npgs). Paying careful attention to the selection of germplasm to maximize genetic diversity will pay dividends in the end. Finally, as the proportion of an experiment's costs dedicated to genotyping approaches the negligible, it is clear that the collection of high-quality phenotypes will often be the main bottleneck to many mapping studies. Experimenters should now be concerned with determining the appropriate experimental design that maximizes their phenotyping efforts. It is imperative that experimenters begin to select germplasm of appropriate levels of relatedness and to generate high-quality phenotype data, as these factors will be major determinants of the power to identify QTL in the future world of inexpensive, large-scale genotyping.
Acknowledgments
We thank Jean-Luc Jannink and Keyan Zhao for helpful discussions.
References
- Altshuler, D., Daly, M.J., and Lander, E.S. (2008). Genetic mapping in human disease. Science 322: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, J.R., Schrag, T., Melchinger, A.E., Zein, I., and Lübberstedt, T. (2005). Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). Theor. Appl. Genet. 111: 206–217. [DOI] [PubMed] [Google Scholar]
- Aranzana, M.J., et al. (2005). Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, S., Sureshkumar, S., Agrawal, M., Michael, T.P., Wessinger, C., Maloof, J.N., Clark, R., Warthmann, N., Chory, J., and Weigel, D. (2006). The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo, R. (1993). Estimation of coefficient of coancestry using molecular markers in maize. Theor. Appl. Genet. 85: 1055–1062. [DOI] [PubMed] [Google Scholar]
- Bernardo, R., Murigneux, A., and Karaman, Z. (1996). Marker-based estimates of identity by descent and alikeness in state among maize inbreds. Theor. Appl. Genet. 93: 262–267. [DOI] [PubMed] [Google Scholar]
- Blott, S., et al. (2003). Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics 163: 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello, F., and Sorrells, M.E. (2006). Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E.S., et al. (2009). The genetic architecture of maize flowering time. Science, in press. [DOI] [PubMed]
- Camus-Kulandaivelu, L., Veyrieras, J.-B., Madur, D., Combes, V., Fourmann, M., Barraud, S., Dubreuil, P., Gouesnard, B., Manicacci, D., and Charcosset, A. (2006). Maize adaptation to temperate climate: Relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172: 2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G.A., et al. (2004). The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Clark, A.G., Hubisz, M.J., Bustamante, C.D., Williamson, S.H., and Nielsen, R. (2005). Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15: 1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly, P. (2008). Progress and challenges in genome-wide association studies in humans. Nature 456: 728–731. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia, S.A., Thornsberry, J.M., and Buckler, E.S. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54: 357–374. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia, S.A., Thuillet, A.-C., Yu, J., Pressoir, G., Romero, S.M., Mitchell, S.E., Doebley, J., Kresovich, S., Goodman, M.M., and Buckler, E.S. (2005). Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 44: 1054–1064. [DOI] [PubMed] [Google Scholar]
- Flint, J., and Mackay, T.F. (2009). Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, A.J., Tai, T.H., Coburn, J., Kresovich, S., and McCouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, M.E., and Hayes, B.J. (2009). Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10: 381. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez, S.C., Huber, D., Ersoz, E., Davis, J.M., and Neale, D.B. (2008). Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 101: 19–26. [DOI] [PubMed] [Google Scholar]
- Harjes, C.E., Rocheford, T.R., Bai, L., Brutnell, T.P., Kandianis, C.B., Sowinski, S.G., Stapleton, A.E., Vallabhaneni, R., Williams, M., Wurtzel, E.T., Yan, J., and Buckler, E.S. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319: 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner, E.L., Sorrells, M.E., and Jannink, J.-L. (2009). Genomic selection for crop improvement. Crop Sci. 49: 1–12. [Google Scholar]
- Henderson, C. (1975). Best linear unbiased estimation and prediction under a selection model. Biometrics 31: 423–447. [PubMed] [Google Scholar]
- Heuertz, M., De Paoli, E., Kallman, T., Larsson, H., Jurman, I., Morgante, M., Lascoux, M., and Gyllenstrand, N. (2006). Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce Picea abies (L.) Karst. Genetics 174: 2095–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J.B. (2007). Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 10: 156–161. [DOI] [PubMed] [Google Scholar]
- Jung, M., Ching, A., Bhattramakki, D., Dolan, M., Tingey, S., Morgante, M., and Rafalski, A. (2004). Linkage disequilibrium and sequence diversity in a 500-kbp region around the adh1 locus in elite maize germplasm. Theor. Appl. Genet. 109: 681–689. [DOI] [PubMed] [Google Scholar]
- Kang, H.M., Zaitlen, N.A., Wade, C.M., Kirby, A., Heckerman, D., Daly, M.J., and Eskin, E. (2008). Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Plagnol, V., Hu, T.T., Toomajian, C., Clark, R.M., Ossowski, S., Ecker, J.R., Weigel, D., and Nordborg, M. (2007). Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lander, E.S. (1996). The new genomics: Global views of biology. Science 274: 536–539. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., and Schork, N.J. (1994). Genetic dissection of complex traits. Science 265: 2037–2048. [DOI] [PubMed] [Google Scholar]
- Laurie, C.C., Chasalow, S.D., LeDeaux, J.R., McCarroll, R., Bush, D., Hauge, B., Lai, C., Clark, D., Rocheford, T.R., and Dudley, J.W. (2004). The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168: 2141–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky, D., Cabezas, J.A., Ibanez, A., Rodriguez, V., and Martinez-Zapater, J.M. (2007). High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics 8: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.R., Schertz, K.F., and Paterson, A.H. (1995). Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141: 391–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A.D., Grote, M.N., and Langley, C.H. (1997). Genetic analysis of complex diseases. Science 275: 1328. [PubMed] [Google Scholar]
- Maher, B. (2008). Personal genomes: The case of the missing heritability. Nature 456: 18–21. [DOI] [PubMed] [Google Scholar]
- Malosetti, M., van der Linden, C.G., Vosman, B., and van Eeuwijk, F.A. (2007). A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175: 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti, G., Galvan, A., Pettinicchio, A., Trincucci, G., Spada, E., Zolin, A., Milani, S., Gonzalez-Neira, A., and Dragani, T.A. (2009). Mouse genome-wide association mapping needs linkage analysis to avoid false-positive loci. PLoS Genet. 5: e1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen, T.H.E., Hayes, B.J., and Goddard, M.E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen, T.H.E., Karlsen, A., Lien, S., Olsaker, I., and Goddard, M.E. (2002). Fine mapping of a quantitative trait locus for twinning rate using combined linkage and linkage disequilibrium mapping. Genetics 161: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M. (2000). Linkage disequilibrium, gene trees and selfing: An ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., and Weigel, D. (2008). Next-generation genetics in plants. Nature 456: 720–723. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A.L., Patterson, N.J., Plenge, R.M., Weinblatt, M.E., Shadick, N.A., and Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K., Stephens, M., and Donnelly, P. (2000. a). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J.K., Stephens, M., Rosenberg, N.A., and Donnelly, P. (2000. b). Association mapping in structured populations. Am. J. Hum. Genet. 67: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A. (2002). Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5: 94–100. [DOI] [PubMed] [Google Scholar]
- Remington, D.L., Thornsberry, J.M., Matsuoka, Y., Wilson, L.M., Whitt, S.R., Doebley, J., Kresovich, S., Goodman, M.M., and Buckler IV, E.S. (2001). Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch, N., and Merikangas, K. (1996). The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- Schaeffer, L.R. (2006). Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 123: 218–223. [DOI] [PubMed] [Google Scholar]
- Spielman, R.S., McGinnis, R.E., and Ewens, W.J. (1993). Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 52: 506–516. [PMC free article] [PubMed] [Google Scholar]
- Stich, B., Mohring, J., Piepho, H.-P., Heckenberger, M., Buckler, E.S., and Melchinger, A.E. (2008). Comparison of mixed-model approaches for association mapping. Genetics 178: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M.I., Sawkins, M.C., Long, A.D., Gaut, R.L., Doebley, J.F., and Gaut, B.S. (2001). Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J.M., Goodman, M.M., Doebley, J., Kresovich, S., Nielsen, D., and Buckler, E.S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- VanRaden, P.M. (2008). Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. [DOI] [PubMed] [Google Scholar]
- Verhoeven, K.J.F., Jannink, J.L., and McIntyre, L.M. (2005). Using mating designs to uncover QTL and the genetic architecture of complex traits. Heredity 96: 139–149. [DOI] [PubMed] [Google Scholar]
- Werner, J.D., Borevitz, J.O., Uhlenhaut, N.H., Ecker, J.R., Chory, J., and Weigel, D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt, S.R., Wilson, L.M., Tenaillon, M.I., Gaut, B.S., and Buckler, E.S.I.V. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 99: 12959–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L.M., Whitt, S.R., Ibanez, A.M., Rocheford, T.R., Goodman, M.M., and Buckler, E.S.I.V. (2004). Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R., Ma, C.-X., and Casella, G. (2002). Joint linkage and linkage disequilibrium mapping of quantitative trait loci in natural populations. Genetics 160: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R., and Zeng, Z.-B. (2001). Joint linkage and linkage disequilibrium mapping in natural populations. Genetics 157: 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Holland, J.B., McMullen, M.D., and Buckler, E.S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Pressoir, G., Briggs, W.H., Vroh Bi, I., Yamasaki, M., Doebley, J.F., McMullen, M.D., Gaut, B.S., Nielsen, D.M., Holland, J.B., Kresovich, S., and Buckler, E.S. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zhao, K., Aranzana, M.J., Kim, S., Lister, C., Shindo, C., Tang, C., Toomajian, C., Zheng, H., Dean, C., Marjoram, P., and Nordborg, M. (2007). An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, P., et al. (2008). A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 40: 367–372. [DOI] [PubMed] [Google Scholar]
- Zhu, C., Gore, M., Buckler, E.S., and Yu, J. (2008). Status and prospects of association mapping in plants. Plant Genome 1: 5–20. [Google Scholar]